Background: mTORC1 recruits its substrate 4E-BP1 via Raptor/4E-BP1 interaction. Chemical cross-linking/mass spectrometry permits characterization of protein-protein interactions.
Results: Cross-linked peptides between Raptor and 4E-BP1 were identified. Raptor intramolecular cross-links were also identified.
Conclusion: Raptor N-terminal region containing RNC1 is implicated in the interaction with the central region of 4E-BP1.
Significance: Our study provides novel insight into how mTORC1 recognizes 4E-BP1.
Keywords: mTOR, mTOR Complex (mTORC), Protein Cross-linking, Protein Kinases, Signaling, 4E-BP1, TOS Motif, Cross-linking Mass Spectrometry, Raptor
Abstract
mTORC1 plays critical roles in the regulation of protein synthesis, growth, and proliferation in response to nutrients, growth factors, and energy conditions. One of the substrates of mTORC1 is 4E-BP1, whose phosphorylation by mTORC1 reverses its inhibitory action on eIF4E, resulting in the promotion of protein synthesis. Raptor in mTOR complex 1 is believed to recruit 4E-BP1, facilitating phosphorylation of 4E-BP1 by the kinase mTOR. We applied chemical cross-linking coupled with mass spectrometry analysis to gain insight into interactions between mTORC1 and 4E-BP1. Using the cross-linking reagent bis[sulfosuccinimidyl] suberate, we showed that Raptor can be cross-linked with 4E-BP1. Mass spectrometric analysis of cross-linked Raptor-4E-BP1 led to the identification of several cross-linked peptide pairs. Compilation of these peptides revealed that the most N-terminal Raptor N-terminal conserved domain (in particular residues from 89 to 180) of Raptor is the major site of interaction with 4E-BP1. On 4E-BP1, we found that cross-links with Raptor were clustered in the central region (amino acid residues 56–72) we call RCR (Raptor cross-linking region). Intramolecular cross-links of Raptor suggest the presence of two structured regions of Raptor: one in the N-terminal region and the other in the C-terminal region. In support of the idea that the Raptor N-terminal conserved domain and the 4E-BP1 central region are closely located, we found that peptides that encompass the RCR of 4E-BP1 inhibit cross-linking and interaction of 4E-BP1 with Raptor. Furthermore, mutations of residues in the RCR decrease the ability of 4E-BP1 to serve as a substrate for mTORC1 in vitro and in vivo.
Introduction
The mammalian target of rapamycin (mTOR)3 signaling pathway has attracted attention because of its involvement in a variety of cellular processes in response to stimuli such as growth factors, nutrients, and energy conditions (1–4). Up-regulation of this signaling pathway is frequently found in a variety of human cancers (5–7). Single amino acid changes can confer constitutive activation of mTOR, and we have identified activating mutations R2505P and S2215Y by mining the human cancer genome database (8). mTOR is a protein kinase belonging to the PI3K-related kinase family that includes PI3K-, ATM-, ATR-(ATM- and Rad3-related), and DNA-dependent protein kinase (9). Protein kinases in this group share similar structural features including the presence of the HEAT repeat, FAT, FATC, and the kinase domains. mTOR forms two different multiprotein complexes, mTORC1 and mTORC2 (10, 11); mTORC1 consists mainly of mTOR, Raptor, and mLST8 (GβL), whereas mTORC2 consists of mTOR, Rictor, mLST8, and Sin1. Substrates of mTORC1 include 4E-BP1 and S6K, whose phosphorylation causes promotion of protein synthesis. In the case of 4E-BP1, its phosphorylation results in relieving inhibitory action of this protein on eIF4E, a cap-binding protein (1–4, 12). S6K stimulates protein synthesis by phosphorylating ribosomal protein S6. In cancer cells, it is believed that 4E-BP1 has an important role in growth and proliferation (13, 14).
Phosphorylation of 4E-BP1 by mTOR is assisted by Raptor, which functions as a scaffold protein. Raptor recruits substrate proteins to mTORC1 so that they can be phosphorylated by mTOR (15, 16). Raptor is a protein of 150 kDa that contains the Raptor N-terminal conserved (RNC) motif, the HEAT repeat, and the WD40 motif (15, 16). Mutations in the RNC motif decrease interaction with 4E-BP1 (15, 16). We wanted to further investigate how mTORC1 interacts with 4E-BP1. Specifically, we sought to gain insight into the region of proximity between Raptor and 4E-BP1.
The recognition of substrate proteins by Raptor is believed to involve a five-amino acid sequence called the TOR signaling (TOS) motif, because mutations of this motif result in decreased interaction with Raptor (17–19). Another motif called “RAIP” is also proposed to be involved (17–19). Although these motifs are important, other regions of 4E-BP1 are also implicated in the interaction with Raptor (19).
Chemical cross-linking coupled with mass spectrometry analysis (CXMS) has emerged as a powerful method to characterize protein-protein interactions (20–26). A basic principle of the method is to use chemical cross-linkers to link reactive amino acid residues that are closely located. After protease digestion, cross-linked peptide pairs are separated by liquid chromatography and can be identified using mass spectrometry. Because only the residues located in close proximity can be cross-linked, intramolecular cross-links can provide insights into the folding of a protein, and intermolecular cross-links can reveal the site(s) of interaction between two proteins. As such, CXMS has been applied to gain information about subunit interactions within multiprotein complexes, which is particularly advantageous for protein complexes that are difficult to purify and crystallize. One of the challenges concerning CXMS is to develop a software tool for identification of cross-linked peptides from their fragmentation spectra (21, 25). The pLink software (21) has been optimized using a large data set and shown to be effective on a variety of samples including protein complexes.
In this paper, we report analysis of Raptor-4E-BP1 interaction using CXMS. We show that Raptor cross-links with 4E-BP1 and that several cross-linked peptides can be identified. Our results show that the cross-links mainly involve the RNC1 domain of Raptor and the central region of 4E-BP1, suggesting their close proximity. Identification of intramolecular cross-links raises the possibility that Raptor contains two structured domains. Finally, we have been able to block the formation of Raptor-4E-BP1 cross-links by synthetic peptides corresponding to the sequence within the RCR of 4E-BP1. We also show that mutations of 4E-BP1 residues within the RCR decrease the ability of 4E-BP1 to serve as a substrate of mTORC1 in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Reagents
Cross-linkers, bis[sulfosuccinimidyl] suberate (BS3) and disuccinimidyl suberate were obtained from Pierce. Anti-FLAG M2 affinity gel and 3× FLAG peptide were obtained from Sigma. Full-length recombinant 4E-BP1 protein was obtained from Santa Cruz Biotechnology (Dallas, TX). Phosphorylated 4E-BP1 purified from Sf9 cells was obtained from CalBiochem (EMD Millipore, Billerica, MA). Anti-mTOR, anti-mLST8, and anti-4E-BP1 antibodies were obtained from Cell Signaling (Danvers, MA). Anti-Raptor antibody was obtained from BETHYL Lab (Montgomery, TX). Peptides were synthesized by Peptibody Inc. (Charlotte, NC).
Purification of mTORC1 and Raptor
Cells stably expressing FLAG-Raptor protein were established using the pLJM1 FLAG-Raptor plasmid (Addgene no. 26633). The lentivirus containing this plasmid was generated by the UCLA Vector Core Facility. HEK293T cells were infected with the lentivirus, and cells stably expressing FLAG-Raptor were obtained by puromycin selection. The resulting cell line, FLAG-Raptor 2, was cultured using Dulbecco's modified Eagle's medium enriched with 10% fetal bovine serum and 1× penicillin/streptomycin at 37 °C and 5% CO2. Cells expressing FLAG-Raptor were grown, washed with PBS, and stored at −80 °C for subsequent experiments. Cells (3.3 × 107) were lysed in 1 ml of 50 mm HEPES, pH 7.4, 150 mm NaCl, 0.4% CHAPS, 1× Complete, EDTA-free protease inhibitor mixture (Roche Applied Science), and 1 mm Na3VO4. In experiments where Raptor was isolated without the other components of mTORC1, 1% Triton X-100 was substituted for 0.4% CHAPS in the lysis buffer. The cell suspension was cleared of insoluble debris by centrifugation at 16,000 × g for 10 min before being mixed with anti-FLAG M2 affinity gel (Sigma) for affinity purification. The affinity gel was collected, washed twice with wash buffer including ATP (50 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm DTT, 2 mm ATP, 0.1% CHAPS), and washed three times with wash buffer without ATP (50 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm DTT, 0.1% CHAPS). Bound proteins were eluted using 62 μg/ml 3× FLAG peptide (Sigma) in 50 mm HEPES, pH 7.4, 150 mm NaCl, 0.4% CHAPS. Eluted proteins were concentrated using Amicon Ultra 0.5-ml centrifugal filters NMWL 100K (EMD Millipore, Billerica, MA). If necessary, we subjected the preparation to another round of affinity purification using anti AU1 beads. In this case, we transfected HEK293T cells stably expressing FLAG-Raptor with AU1-mTOR DNA (8) before collecting cells. The presence of mTOR, Raptor, and mLST8 in the final preparation was evaluated by staining and Western blotting.
mTORC1 Kinase Assay
Protein kinase activity of mTORC1 was examined by carrying out kinase reaction using unphosphorylated 4E-BP1 (Santa Cruz Biotechnology, Dallas, TX) as described previously (27). Briefly, mTORC1 purified above was incubated with 0.25 μg of 4E-BP1 protein in kinase buffer (20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 0.2 mm ATP) for 20 min at 37 °C. The product was analyzed by Western blot using antibodies against phospho-4E-BP1 (Thr37/46), phospho-4E-BP1 (Ser65), or total 4E-BP1.
4E-BP1 Binding Assay
Binding of 4E-BP1 to mTORC1 was examined by the pulldown of 4E-BP1 with mTORC1 bound to beads. mTORC1 was purified from HEK293T cells expressing FLAG-Raptor using anti-FLAG beads. The beads were mixed with 4E-BP1 in a buffer containing 20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 100 mm NaCl. After the incubation, the beads were collected, washed, and then suspended in Laemmli buffer. The sample was run on a SDS-polyacrylamide gel and probed with an antibody against 4E-BP1 as described (27).
Cross-linking Procedure
Purified proteins were combined with 4E-BP1 (Santa Cruz Biotechnology, Dallas, TX) and cross-linked using BS3 or disuccinimidyl suberate. The reaction condition was the same as that used for the kinase reaction (27), except that ATP and MgCl2 were omitted and Tris was replaced with HEPES. The water-soluble characteristics of BS3 made this cross-linker more compatible with the experimental work flow than its water-insoluble counterpart disuccinimidyl suberate. The cross-linking reagent was reacted with the proteins at room temperature for 20 min. Then the solution was adjusted to 20 mm NH4HCO3 and kept at room temperature for 20 min to inactivate the remaining free cross-linker reagent. The cross-linked proteins were stored at −80 °C in Laemmli buffer. Cross-linked proteins were separated by SDS-PAGE and stained with GelCode Blue (Pierce) according to the manufacturer's instructions. Protein bands corresponding to areas of interest identified by Western blot were excised and subjected to mass spectrometry analyses. For peptide inhibition experiments, peptides with the amount indicated in the legend to Fig. 6 were added to cross-linking reactions.
FIGURE 6.
Inhibition of cross-linking between Raptor and 4E-BP1 using peptides. A, inhibition of Raptor-4E-BP1 cross-linking by peptide 1. Raptor and 4E-BP1 were incubated in the absence (lane 1) and in the presence of BS3 (lanes 2–6). 24 μm (lane 3) and 48 μm (lane 4) of peptide 1 was added, whereas the same amount of control scrambled peptide (control 1) was added to lanes 5 and 6). No peptide was added to lane 1. After the incubation, cross-linked band was detected by Western blot probed with anti-4E-BP1. B, experiments similar to those described in A were carried out using peptide 2 and its scrambled version (control 2). The concentrations used are 15 and 30 μm. C, varying concentrations of peptide 2 and its scrambled version were added to the cross-linking reaction, and the Raptor-4E-BP1 cross-link band was quantitated by the analysis with ImageJ. Cross-linking Raptor and 4E-BP1 without peptide was set as 100%, whereas a reaction without BS3 was set as 0%. D, binding of 4E-BP1 to mTORC1 was examined by using mTORC1 attached to the beads. The beads were collected and washed, and the amount of 4E-BP1 bound to the beads was examined by Western against 4E-BP1. The amount of bound 4E-BP1 is compared when mTORC1 and 4E-BP1 were incubated in the presence of 30 μm of peptide 2 (lane 2) or control peptide (lane 3). No peptides were added in lane 1. This result is representative of two independent experiments.
Mass Spectrometry Analysis and Database Search
In-gel digestions were carried out as described before (28) with slight modifications. Briefly, gel slices of cross-linked protein bands were destained and reduced with 200 μl of 10 mm DTT at 56 °C for 40 min. After the removal of the DTT solution, the gel slices were incubated with 200 μl of 55 mm iodoacetimide in the dark for 1 h. Washed and dehydrated gel pieces were rehydrated with trypsin alone or with trypsin and ASP-N solution (10 ng/μl for each protease).
Extracted peptides were loaded onto a precolumn (75-μm inner diameter, 6 cm long, packed with 10 μm of C18 resin) before it was connected with an analytic column (75-μm inner diameter, 10 cm long, packed with 1.8 μm of C18 resin). The HPLC gradient was generated by Easy-nLC 1000 (Thermo Fisher Scientific): 300 nl/min flow rate, a 45-min linear gradient from 100% buffer A (0.1% formic acid in water) to 30% buffer B (100% acetonitrile, 0.1% formic acid), followed by a 5-min gradient from 30% to 100% buffer B, then from 100% buffer B to 0% buffer B over 5 min, and lastly a 10-min wash with buffer A. Mass spectrometry data were collected on Q-Exactive (Thermo Fisher Scientific) as follows: full scans in the orbitrap (r = 70,000) followed by 10 higher energy collisional dissociation scans at r = 17,500 and normalized collisional energy = 27; precursors of charge state +1, +2, or unassigned were excluded from MS2 scans; monoisotopic screening was enabled; and dynamic exclusion was set to repeat count, 1; exclusion list size, 200; and exclusion duration, 60 s.
Using the pLink search engine (21), we searched the mass spectrometry data against a protein sequence database containing human Raptor, human mTOR, and rat 4E-BP1. The following pLink parameters were used: precursor mass tolerance, 50 ppm; fragment mass tolerance, 20 ppm; cross-linker BS3 (cross-linking sites K and protein N terminus; xlink mass shift, 138.0680796; monolink mass shift, 156.0786442); fixed modification C, 57.02146; peptide length minimum, 4 amino acids per chain; peptide length maximum, 100 amino acids per chain; peptide mass minimum, 400 Da per chain; and peptide mass maximum, 10,000 Da per chain). No restriction on peptide terminal residues was applied in pLink search (i.e., treat the peptides as protein digestion products using a nonspecific protease). The search results were filtered with a false discovery rate of less than 5% and E values of less than 0.01.
Structure Modeling
The tertiary structures for Raptor RNC1 domain and 4E-BP1 fragment were modeled using the I-TASSER server (29). The online program implements a homolog threading protocol followed by fragment assembly and refinement. Five three-dimensional models for each protein were generated based on C-score, a quality measurement for I-TASSER prediction, and the one with the highest C-score was used for subsequent modeling. The structure of the RNC1·4E-BP1 complex was modeled with XPLOR-NIH (30), by refining against knowledge-based torsional angle potential (31) and distance restraints derived from both intra- and intermolecular cross-linking. For each cross-linking, the distances between lysine Cα atoms were constrained <22 Å, and the distances between NZ atoms were constrained <10 Å. The secondary structures, mainly helices, from I-TASSER prediction were kept fixed, and the complex was heated up to 3000 K before gradual cooling to 25 K in a simulated annealing protocol (32). A set of structure models was generated, all of which satisfy experimental data. The most compact structure that appears most energetically reasonable was selected using program ZRANK (33), which utilizes a scoring function consisting of van der Waals, electrostatics, and desolvation. The structure model was illustrated with the program PyMOL (34).
Construction of Mutant 4E-BP1 Plasmid
pET14b PHAS-I plasmid (Addgene 15679) was used as template for construction of mutant 4E-BP1. A QuikChange Lightning multisite-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) was used to construct double-site mutations in the region of 4E-BP1 that cross-links with Raptor. PCR primers were designed as 5′-gtcctttgggggtgttaaggccacaggcgagttc-3′ and 5′-ccgacactccatcaggaataaccggtcatagatgattctg-3′ (obtained from Invitrogen) to double mutate lysine 56 and lysine 68 to leucine. Original plasmids were digested with DpnI restriction enzyme. Mutations were confirmed by sequencing analysis (GENEWIZ, Inc.) (supplemental Fig. S2). Mutant plasmids were transformed into BL21 Star DE3 One Shot® chemically competent cells (Invitrogen) for protein expression.
Expression and Purification of Mutant 4E-BP1
Cultures were grown to an A600 of 0.5, induced with 1 mm isopropyl β-d-thiogalactopyranoside, harvested by centrifugation, and frozen at −20 °C overnight. The pellet was then thawed on ice and resuspended in lysis buffer (4 ml/g). 1 mg/ml lysozyme was added, and the suspension was incubated on ice for 30 min followed by three periods of sonication. Soluble and insoluble fractions were separated by centrifugation at 100,000 × g for 30 min. The supernatant was retained for purification of soluble proteins. Soluble proteins were purified with nickel-nitrilotriacetic acid-agarose slurry according to the manufacturer's protocol (Qiagen). Purity was confirmed by SDS-PAGE followed by GelCode Blue staining.
Raptor/4E-BP1 Far Western Blot
Far Western blot (overlay) was carried out following published protocol (19). Briefly, 0.2–2 μg of bacterially expressed His-tagged 4E-BP1 was separated on a 15% SDS-PAGE gel and transferred onto a PVDF membrane by electroblotting. The membrane was blocked for 1 h with 5% (w/v) fat-free milk in PBS and 0.02% (v/v) Tween 20. The membrane was then incubated overnight at 4 °C with FLAG-Raptor lysate diluted in blocking buffer (1:10). The FLAG-Raptor lysate was obtained by lysing FLAG-Raptor expressing HEK293T cells in a 15-cm dish with 1 ml of lysis buffer (50 mm HEPES, pH 7.4, 150 mm NaCl, 0.1% CHAPS) and centrifuged for 30 min to remove insoluble fraction. The PVDF membrane was washed in blocking buffer and probed with anti-Raptor antibody (1:500) for 1 h, washed again, probed with secondary HRP-conjugated anti-rabbit antibody (1:10,000) (GE Healthcare) for 1 h, incubated with enhanced chemiluminescence solution (ECL) (Pierce) for 60s, and exposed to autoradiography film.
Construction of FLAG Epitope-tagged 4E-BP1 Wild Type and Mutant Plasmids
N-terminal FLAG epitope-tagged forms of 4E-BP1 were created by PCR with the following primers: 5′-gagaggtaccatgggcagcagcgattacaaggatgacgacgataagagcagcggcctggtgccgcgcg-3′ (forward) and 5′-tggctgggcccttaaatgtccatctc-3′ (reverse) with the above mentioned pET14b PHAS-I wild type and mutant plasmids as templates. The PCR product was digested with KpnI and ApaI, purified, and ligated into the corresponding restriction sites in pcDNA3 so that the 4E-BP1 produced contained an N-terminal FLAG tag. FLAG and 4E-BP1 insert (wild type and mutant) were confirmed by sequencing analysis (GENEWIZ, Inc.). These plasmids were transformed into maximum efficiency DH5α competent cells, and plasmids were purified with Purelink HiPure plasmid maxi prep kit (Life Technologies).
Transfection and Immunoprecipitation
FLAG-4E-BP1 (wild type and mutant) was transfected with Lipofectamine 2000 into HEK293T cells (4 μg of plasmid DNA/6-cm plate). 24 h after transfection, cells were harvested in ice-cold PBS and lysed in lysis buffer (40 mm HEPES, pH 7.4, 150 mm NaCl, 0.4% CHAPS), supplemented with complete protease inhibitors (Roche Applied Science) without EDTA, 1 mm Na3VO4. The lysate were then sonicated and centrifuged. The supernatants were incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) for 2 h at 4 °C. The beads were extensively washed and then boiled in SDS sample buffer (50 mm Tris, pH 6.8, 10% SDS, 10% glycerol, 0.1% bromphenol blue) for 5 min to elute binding proteins. The eluted proteins were analyzed by Western blot.
RESULTS
Cross-linking of Raptor with 4E-BP1 and Identification of Cross-linked Peptides by Mass Spectrometry
mTORC1 consists of three major proteins: mTOR, Raptor, and mLST8(GβL). This multiprotein complex functions to phosphorylate substrate proteins including 4E-BP1. This occurs when Raptor recognizes 4E-BP1, which results in the recruitment of 4E-BP1 to the mTOR kinase. To investigate the interaction between mTORC1 and 4E-BP1, we first purified mTORC1 from HEK293T cells stably expressing FLAG-tagged Raptor (established by infecting cells with lentivirus carrying FLAG-Raptor). Cells were lysed in a buffer containing 0.4% CHAPS, and mTORC1 was affinity-purified by using anti-FLAG beads and eluted by using FLAG peptides as described under “Experimental Procedures.” The preparation contained mTOR, Raptor, and mLST8, as detected by staining and Western analysis (Fig. 1, A and B). The purified mTORC1 was active in phosphorylating 4E-BP1, as evidenced by the appearance of phospho-4E-BP1 after incubating mTORC1 with unphosphorylated 4E-BP1 (Fig. 1C). The kinase activity was robust in the presence of Mg2+ as well as in the presence of Mn2+.
FIGURE 1.

mTORC1 purification and characterization. A, silver staining of purified mTORC1 reveals the presence of mTOR and Raptor bands. B, Western blot of mTORC1 probed with mTOR antibody, Raptor antibody, and mLST8 antibody. C, kinase activity of mTORC1 as examined by the phosphorylation of 4E-BP1. Lanes 2 and 3, two different amounts of mTORC1 preparation were mixed with unphosphorylated 4E-BP1 in the presence of ATP, and the reaction mixture was analyzed on a SDS/PAGE using antibody specific for phosphorylated 4E-BP1 (Thr37/46) as described (27). Lane 1, no mTORC1. Lane 4, the kinase reaction was carried out using the same reaction condition as that in lane 3 except that Mn2+ was added. Lane 5, phosphorylated 4E-BP1 purified from Sf9 cells is loaded as a phosphorylated 4E-BP1 control. Note that this protein is fully phosphorylated.
To examine the interaction of 4E-BP1 with mTORC1, we mixed mTORC1 with 4E-BP1 in the presence of increasing concentration of a cross-linker BS3 that can covalently connect two closely spaced lysine residues (Cα-Cα distance, <24 Å). Western blotting was carried out to identify 4E-BP1 bands on a SDS-polyacrylamide gel. As can be seen in Fig. 2A, incubation with BS3 led to the appearance of new bands that had molecular weights much higher than that of 4E-BP1. A prominent band was detected just above the molecular weight of Raptor. Western analyses using anti-Raptor antibody confirmed the appearance of BS3-dependent Raptor bands that coincide with the bands detected by the anti-4E-BP1 Western (data not shown).
FIGURE 2.

Identification of Raptor·4E-BP1 cross-linked bands. A, cross-linking of mTORC1 and 4E-BP1. Purified mTORC1 was mixed with 4E-BP1 in the presence of 0 (lane 1), 0.25 (lane 2), 0.5 (lane 3), and 1 mm (lane 4) BS3, and the reaction mixture was analyzed by Western blot. Anti-4E-BP1 antibody identified high molecular weight bands when BS3 was added. B, isolated Raptor cross-links with 4E-BP1. Purified Raptor was mixed with 0 (lane 1), 0.05 (lane 2), and 0.25 mm (lane 3) BS3, and reaction mixtures were analyzed by Western blot as described in A.
To evaluate whether free Raptor can be cross-linked to 4E-BP1, we prepared Raptor protein separated from the complex. Because Triton X-100 is known to disrupt mTORC1 complex (36), we lysed HEK293T cells stably expressing FLAG-Raptor in a buffer containing 1% Triton X-100 and purified Raptor by the use of anti-FLAG beads and then eluted Raptor using FLAG peptides. Analysis of this preparation by SDS-polyacrylamide gel electrophoresis showed only a single band corresponding to the size of Raptor (data not shown). Raptor was then incubated with 4E-BP1 in the presence of BS3. As shown in Fig. 2B, a pattern of cross-linked bands obtained was similar to that seen when mTORC1 was used, indicating that it is Raptor that cross-links with 4E-BP1.
The major cross-linked band was cut out, digested with proteases, and subjected to mass spectrometry. Database search was carried out using the pLink program (21). Fig. 3 shows four representative, high resolution fragmentation spectra of cross-linked peptides between Raptor and 4E-BP1. For example, the cross-linked peptide spectrum shown in Fig. 3A depicts a cross-link between Raptor (KLCTSLR) and 4E-BP1 (NSPVAKTPPK). Other spectra are shown in supplemental Fig. S1.
FIGURE 3.
Representative high resolution fragmentation spectra of cross-linked peptides between Raptor and 4E-BP1. A, 4E-BP1[NSPVAKTPPK(6)]-Raptor[KLCTSLR(1)]. B, 4E-BP1[KFLMECR(1)]-Raptor[NAKEER(3)]. C, 4E-BP1[NSPVAKTPPK(6)]-Raptor[NAKEER(3). D, 4E-BP1[NSPVAKTPPK(6)]-Raptor[YQKSL(3)]. The numbers in parentheses indicate the positions of the cross-linked lysine residues in the peptide sequences. Observed C-terminal fragments (y ions, typically the dominant species) resulting from a peptide bond cleavage of the α peptide (the one with a higher mass) are colored purple, and those of the β peptide are dark yellow. The corresponding N-terminal fragments are called b ions, from which a further loss of carbonyl results in a ions. The b ions are colored green if the cleavage occurs on the α peptide or light blue if the cleavage occurs on the β peptide. The a ions are highlighted dark purple if generated from the breakage of the α peptide or dark blue if from the β peptide.
The Raptor/4E-BP1 Cross-links Involve the N-terminal Conserved Region 1 of Raptor and a Central Region of 4E-BP1
Table 1 summarizes all the cross-linked peptides between Raptor and 4E-BP1 that we have identified. A total of seven intermolecular cross-links were identified, and six of the spectra show excellent spectral quality. The table also shows how many times these peptide pairs were identified (spectral counts) and the best E values associated with their detection. Fig. 4A shows these cross-links. It is striking that six of the seven cross-links are clustered in a small region on each protein, namely, the RNC1 and the central region of 4E-BP1.
TABLE 1.
Cross-linked peptide pairs between Raptor and 4E-BP1
Filtering cutoff: FDR < 0.05; E value < 0.01; spectral count ≥ 2. Manual validation criteria: High, for each peptide, more than three continuous fragment peaks can be assigned to it; none or few of the high intensity peaks in the spectrum are left unassigned; and matched fragment peaks are significantly higher than noise peaks. Middle, one peptide has more than three whereas the other has only three continuous fragment peaks; several high intensity peaks in the spectrum cannot be assigned. Low, one or both peptides have less than three continuous fragment ions.
| Raptor |
4EBP1 |
Spectral counts | Best E value | Spectral quality (manual evaluation) | ||
|---|---|---|---|---|---|---|
| Sequence | Cross-linked lysine | Sequence | Cross-linked lysine | |||
| KLCTSLR | 132 | NSPVAKTPPK | 68 | 4 | 7.09 10−11 | High (Fig. 3A) |
| NAKEER | 142 | KFLMECR | 56 | 4 | 1 × 10−8 | High (Fig. 3B) |
| DPLSMGPQKAL | 97 | NSPVAKTPPK | 68 | 4 | 2.36 × 10−7 | High (Fig. S1A) |
| NAKEER | 142 | NSPVAKTPPK | 68 | 5 | 5.56 × 10−6 | High (Fig. 3C) |
| DLIEKIPGR | 297 | NSPVAKTPPK | 68 | 2 | 3.23 × 10−5 | High (Fig. S1B) |
| YKQSL | 120 | NSPVAKTPPK | 68 | 6 | 3.75 × 10−4 | High (Fig. 3D) |
| PTVNGEVWVFNKNYTQYIPLSIY | 169 | KTPPK | 68 | 2 | 2.37 × 10−4 | Low (Fig. S1C) |
FIGURE 4.
A, an overview of Raptor-4E-BP1 cross-linked peptides detected in this study. Displayed are the cross-linked peptides between Raptor and 4E-BP1 that we have identified from the cross-linking reactions containing 4E-BP1 and mTORC1 or isolated Raptor. Continuous underlines represent the sequence and location of the cross-linked peptides in the protein primary sequence. Cross-linked lysine pairs are connected with dotted lines. Lys297 of Raptor (shown by a star) is the only site outside RNC1 that cross-links to 4E-BP1 (15). RNC1 is between residues 48 and 205. RNC2 is between residues 239 and 390. RNC3 is between residues 408 and 511. B, structural model representing the interaction between Raptor RNC1 and 4E-BP1 fragment. RNC1 is colored in cyan, 4E-BP1 is in purple; lysine side chains are shown as sticks. The distances between lysine Cα atoms in the model are marked.
Raptor, a protein of 1335 amino acids, has 52 lysine residues that are located throughout the protein. Only six of these were cross-linked to 4E-BP1. The protein has multiple structural features, the RNC regions 1–3, the HEAT repeats in the middle, and the WD40 repeats that are located in the C terminus. The sites of cross-linking with 4E-BP1 are all located in the RNC1 region except for one that is located in the RNC2 region. For 4E-BP1, we find that all the cross-links with Raptor took place at the central region of this protein. 4E-BP1 is a small protein of 117 amino acid residues, including four lysine residues that are located at the N-terminal and the C-terminal regions as well as two that are located in the internal regions. One of them, Lys68, was the major cross-linking residue, whereas another, Lys56, also cross-linked to Raptor. These results suggest that the central region of 4E-BP1 defined as the RCR is located closely to Raptor.
In Fig. 4B, we present a model of Raptor/4E-BP1 interaction based on the above cross-linking study as well as by using structure prediction described under “Experimental Procedures.” RNC1 was predicted to have two helices, encompassing residues 95–112 and 127–140, arranged in a parallel fashion. For the 4E-BP1 fragment, residues 57–62 likely form a short helix, followed by a loop structure. When interacting with RNC1, the short helix of 4E-BP1 mainly interacts with the second helix of RNC1 with a large angle between the two helices, whereas the helix C terminus and the following loop swing toward the first helix of RNC1. The structure of a slightly shorter fragment of 4E-BP1 in complex with eukaryotic translation initiation factor 4E has been previously determined using x-ray crystallography in several independent studies (37–42). The structural model of Raptor·4E-BP1 complex bears high similarity to these high resolution structures, which further corroborates our findings.
Raptor Intramolecular Cross-links Define Two Regions of Structured Domains in Raptor
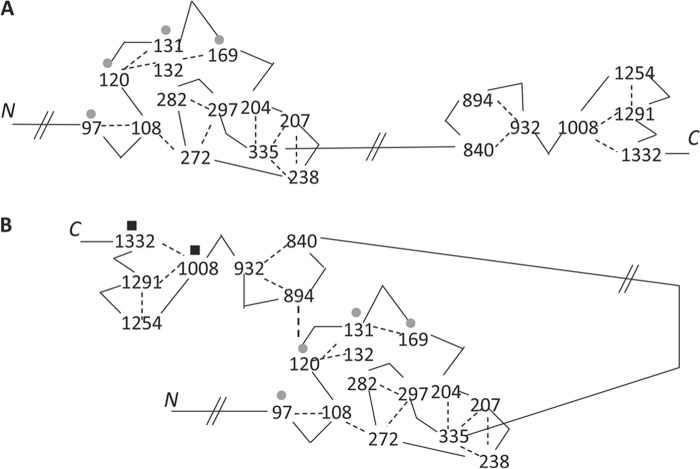
In addition to the cross-linked peptides between Raptor and 4E-BP1, our analyses revealed the presence of Raptor-Raptor cross-links. A total of 17 different cross-links were identified, and they are listed in Table 2. Because we cut out the bands that were located just above the Raptor band of 150 kDa but well below 250 kDa, these cross-links most likely represent those that are formed within the same polypeptide, not from a Raptor dimer. Some of the cross-links were repeatedly identified. For example, the cross-link between Lys207 and Lys335 was observed 132 times. Most cross-links were formed between lysine residues that are both in the N-terminal half of the protein or the C-terminal half except for the cross-link between Lys120 and Lys894. Fig. 5 depicts these intramolecular cross-links within a Raptor molecule, based on the idea that two lysine residues will cross-link only if they are located nearby. Fig. 5A depicts an extended Raptor structure to indicate the N-terminal and C-terminal regions. Two structured domains, one located in the N-terminal region and the other located in the C-terminal region, are seen. The central region of Raptor (residues 336–689) that includes the HEAT domain had no cross-linked residues, although this area contains six lysines. Interestingly, the sites where the interaction with 4E-BP1 takes place (Lys120, Lys131, and Lys169) appear to be clustered in one area within the N-terminal portion. Because we have identified a cross-link between residues 120 and 894, it is possible that the N-terminal and C-terminal regions are located near each other. This is depicted in Fig. 5B, and we mention its possible significance under “Discussion.”
TABLE 2.
Intramolecular lysine-lysine cross-links identified in the Raptor protein
Filtering cutoff: FDR < 0.05; E value < 0.01; spectral count ≥ 2. Manual validation criteria: High, for each peptide, more than three continuous fragment peaks can be assigned to it; none or few of the high intensity peaks in the spectrum are left unassigned; and matched fragment peaks are significantly higher than noise peaks. Middle, one peptide has more than three while the other has only three continuous fragment peaks; several high intensity peaks in the spectrum cannot be assigned. Low, one or both peptides have less than three continuous fragment ions.
| Position of one lysine | Position of the other lysine | Spectral counts | Best E value | Spectral quality (manual evaluation) |
|---|---|---|---|---|
| 207 | 335 | 132 | 5.4 × 10−14 | High |
| 1332 | 1008 | 43 | 9.3 × 10−17 | High |
| 272 | 297 | 36 | 3.3 × 10−18 | High |
| 132 | 120 | 26 | 3.2 × 10−6 | Low |
| 1291 | 1254 | 12 | 1.5 × 10−5 | High |
| 131 | 120 | 11 | 9.69 × 10−5 | Middle |
| 1008 | 1291 | 7 | 2.4 × 10−6 | High |
| 238 | 335 | 7 | 9.75 × 10−6 | Middle |
| 272 | 108 | 5 | 5.5 × 10−7 | High |
| 108 | 97 | 4 | 9.65 × 10−7 | High |
| 282 | 297 | 3 | 1.62 × 10−5 | Low |
| 894 | 932 | 3 | 4.59 × 10−6 | Low |
| 132 | 169 | 2 | 3.34 × 10−6 | High |
| 335 | 204 | 2 | 2.9 × 10−4 | Middle |
| 894 | 120 | 2 | 8.7 × 10−5 | High |
| 238 | 207 | 2 | 2.3 × 10−4 | Low |
| 840 | 932 | 2 | 4.5 × 10−7 | Low |
FIGURE 5.
Two views of Raptor based on the identification of Raptor-Raptor cross-linked peptides. Because the residues that are cross-linked suggest that they are in close proximity, it is possible to place cross-lined residues closeby. A, a scheme showing a stretched out structure to demonstrate two structured domains. To emphasize the presence of two structured domains, the cross-link between residues 120 and 894 is not shown on this figure. B, a scheme showing a folded structure based on the proximity of residues 120 and 894. Gray circles show the lysine residues that are identified to be cross-linked with 4E-BP1. Black squares show the lysine residues in Raptor that cross-linked with mTOR.
Peptides Harboring the RCR of 4E-BP1 Inhibit Raptor-4EBP1 Cross-link
To gain further insight into the site of interaction between Raptor and 4E-BP1, we decided to use synthetic peptides and ask whether they inhibit cross-linking between the two proteins. To do this, we first synthesized a peptide corresponding to 4E-BP1 (amino acids 56–72), which has the sequence KFLMECRNSPVAKTPPK (peptide 1), and a scrambled version of this sequence as a control (control 1). Two different concentrations of these peptides were added separately to the cross-linking reactions, and the amount of cross-linked Raptor-4E-BP1 was examined on a SDS-polyacrylamide gel. As can be seen in Fig. 6A, 48 μm of peptide 1 indeed inhibited the cross-link between Raptor and 4E-BP1. On the other hand, the same concentration of the control peptide (control 1) did not inhibit the cross-link. We then synthesized another peptide that is located slightly closer to the N terminus. This peptide “GTRIILDRKFLMECRNS” (peptide 2) corresponds to the 4E-BP1 sequence between residues 48 and 64. A scrambled version of this peptide (control 2) was also synthesized. As shown in Fig. 6B, this peptide exhibited strong inhibition of the Raptor-4E-BP1 cross-link. Almost complete inhibition was seen in these experiments. In contrast, little inhibition was observed with the scrambled control peptide (control 2). To obtain IC50 values of the inhibition, we varied the concentration of peptides. As shown in Fig. 6C, peptide 2 inhibited the formation of Raptor-4E-BP1 cross-link at an IC50 value of 15 μm. To examine whether peptide 2 inhibits binding of 4E-BP1 to mTORC1, we purified mTORC1 from HEK293T cells expressing FLAG-Raptor by using anti-FLAG beads. After mixing 4E-BP1 with the beads in the presence of peptide 2 or control peptide, the amount of 4E-BP1 pulled down was examined by Western analysis. As shown in Fig. 6D, the amount of 4E-BP1 bound to mTORC1 was significantly decreased by peptide 2, whereas this effect was less with the control peptide. These results support the results obtained from the CXMS analyses.
Mutations of Residues within the RCR of 4E-BP1 Decrease Its Ability to Serve as a Substrate for mTORC1 in Vitro
We mutated the residues within the RCR of 4E-BP1 to assess the significance of this region for its interaction with Raptor. 4E-BP1 is a small protein that functions without folded structure (43). By PCR-mediated in vitro mutagenesis, we changed two lysine residues within the RCR of 4E-BP1. The mutant protein was purified with His6 nickel-nitrilotriacetic acid matrix (supplemental Fig. S2). Both the wild type and mutant 4E-BP1 were used for kinase assay to detect phosphorylation of 4E-BP1. As shown in Fig. 7A, although the wild type 4E-BP1 showed strong phosphorylation of 4E-BP1 as detected by the use of anti-phospho (Thr37/46) antibody as well as by anti-phospho (Ser65) antibody, the phosphorylation was dramatically reduced with the mutant 4E-BP1 protein. We also examined the binding of the 4E-BP1 mutant to mTORC1. The results shown in Fig. 7B demonstrate that the mutant protein binds much less efficiently than the wild type protein. Furthermore, Far Western blot study shows that the mutant 4E-BP1 interacts with FLAG-Raptor significantly less than the wild type (Fig. 7C). The Western blot in the upper panel shows the amount of total 4E-BP1 protein loaded onto the gel, whereas the lower panel shows the amount of FLAG-Raptor binding to the wild type or the mutant 4E-BP1.
FIGURE 7.

Mutations of 4E-BP1 residues within the RCR (Raptor cross-linking region) decrease the ability of 4E-BP1 to serve as a substrate of mTORC1. A, mutant 4E-BP1 (Mut) and wild type 4E-BP1 (WT) were used as a substrate for kinase assay using mTORC1 purified as described under “Experimental Procedures.” Phosphorylation of the wild type (lanes 1–3) and the mutant 4E-BP1 (lanes 4–6) was examined using anti-phospho (Thr37/46) as well as anti-phospho (Ser65) antibody as described on the right hand side of the figure. Because these antibodies were raised against the human protein, Rattus 4E-BP1 residue numbering should be shifted by one residue (−1 residue). The amount of mTORC1 used was 0 μl (lanes 1 and 4), 2 μl (lanes 2 and 5), and 4 μl (lanes 3 and 6) of purified protein. Anti-4E-BP1 antibody was used to estimate the amount of total 4E-BP1. B, binding of the mutant and the wild type 4E-BP1 with mTORC1. The ability of 4E-BP1 to bind mTORC1 was examined by using mTORC1 attached on beads as described under “Experimental Procedures.” The same amount of the mutant and the wild type protein was used for this assay. C, binding of the mutant and wild type 4E-BP1 protein with Raptor. The wild type and mutant 4E-BP1 were run on a 15% SDS-PAGE gel and probed for total 4E-BP1 with anti-4E-BP1 antibody (upper panel). Protein was loaded in incrementally increasing concentrations. The lower panel shows the result of Far Western blot (overlay) in which the mutant and the wild type 4E-BP1 were run on a 15% SDS-PAGE gel, incubated with the lysate of FLAG-Raptor expressing HEK293T cells, and probed with anti-Raptor antibody to detect binding interactions taking place between Raptor and 4E-BP1.
Mutations of Residues within the RCR of 4E-BP1 Decrease Its Ability to Serve as a Substrate for mTORC1 in Vivo
Next, we expressed the mutant 4E-BP1 in HEK293T cells. For this purpose, we inserted FLAG epitope-tagged 4E-BP1 (wild type and mutant) into pcDNA3 plasmid by PCR as described under “Experimental Procedures.” FLAG-4E-BP1 plasmids (wild type and mutant) were then transfected with Lipofectamine 2000 into HEK293T cells. 24 h after transfection, cells were collected and lysed, and FLAG-4E-BP1 was immunoprecipitated with anti-FLAG M2 affinity gel. The eluted proteins were analyzed by Western blot for phosphorylation of 4E-BP1 as well as for interaction with Raptor. As shown in Fig. 8A, phosphorylation of 4E-BP1 was dramatically reduced with the mutant 4E-BP1 protein compared with the wild type. We also examined their co-immunoprecipitation with Raptor. Significant reduction of Raptor co-immunoprecipitation with the mutant 4E-BP1 compared with the wild type was observed (Fig. 8B).
FIGURE 8.

In vivo effects of mutating 4E-BP1 residues within the RCR. A, FLAG-tagged mutant and wild type 4E-BP1 were expressed in HEK293T cells, and the FLAG-tagged proteins were immunoprecipitated with anti-FLAG M2 affinity gel. Phosphorylation of 4E-BP1 was examined using anti-phospho (Thr37/46) antibody (WT, wild type; Mut, mutant) as described on the right hand side of the figure. Because the antibody was raised against the human protein, Rattus 4E-BP1 residue numbering should be shifted by one residue (−1 residue). B, in vivo interaction of Raptor with 4E-BP1 was examined. FLAG tag immunoprecipitated proteins were analyzed with anti-Raptor antibody to detect binding of Raptor with 4E-BP1.
DISCUSSION
In this paper, we characterized the interaction of mTORC1 and its substrate 4E-BP1 using the CXMS technology. First, we found that 4E-BP1 could be cross-linked with Raptor using purified mTORC1 as well as free Raptor. We then characterized the cross-linked band by mass spectrometry and identified cross-linked peptides that contain both Raptor and 4E-BP1 sequences. Multiple cross-linked peptides were identified that were clustered primarily to a single region for each protein. These results represent the first observation of Raptor and 4E-BP1 cross-links, and the study revealed that the RNC of Raptor and the RCR of 4E-BP1 are in close proximity. Computational structural prediction (Fig. 4B) can be made regarding the residues in the two proteins.
Our cross-link results were further substantiated by first carrying out peptide inhibition studies. Two peptides that encompass the Raptor interaction site of 4E-BP1 inhibited Raptor-4E-BP1 cross-links, whereas the control peptides each having a scrambled sequence did not. Peptide 2 gave strong inhibition at 15 μm concentration. Second, we have mutated residues within the 4E-BP1 region we defined by our cross-linking study and showed that that the mutations dramatically decrease binding of 4E-BP1 to mTORC1 and reduce phosphorylation by mTORC1. In cells, phosphorylation of the mutant 4E-BP1 was significantly decreased compared with that of the wild type protein. In addition, Raptor binding was significantly decreased with the mutant 4E-BP1.
Our results on 4E-BP1 suggest that a region encompassing residues 56 and 72 is in close proximity with Raptor. This region is highly conserved among 4E-BP family members (44). The sequence KFLMECRNSPVAKTPP in 4E-BP1 corresponds to the sequence KFLLDRRNSPMAQTPP in 4E-BP2 and KFLLECKNSPIARTPP in 4E-BP3. Two mTORC1-mediated phosphorylation sites (45) are present in this region. A sequence that is known to interact with eIF4E, as defined by the co-crystallization of eIF4E and a 4E-BP1 peptide (46), is located on the N-terminal side of the region. The TOS and RAIP motifs have been proposed to play important roles for the interaction between Raptor and mTORC1 substrates. However, the location of the TOS motif in 4E-BP1 is distant from the Raptor cross-linking site that we have identified. It should be noted that our cross-linking method provides information on close proximity of residues, and our results do not exclude the possibility that the TOS and RAIP motifs are playing important roles in the interaction with Raptor. In fact, in earlier studies with 4E-BP1, it has been suggested that regions other than the TOS and RAIP are also involved in Raptor interaction (19). It may be the case that multiple sites of 4E-BP1 are involved in the interaction with Raptor. Further analysis is needed to fully understand how Raptor interacts with 4E-BP1.
On the Raptor side, our results point to the importance of the RNC region. We found that the 4E-BP1 cross-links are clustered within the first RNC domain (RNC1) of Raptor close to the N terminus. The significance of the RNC1 region for interaction with substrate proteins was further supported by our recent preliminary experiments that sought to identify cross-links between Raptor and S6K. In this experiment, unphosphorylated S6K was purified from HEK293T cells transfected with FLAG-S6K and treated with mTOR inhibitor pp242. Incubation of this protein with mTORC1 in the presence of a cross-linker BS3 identified cross-linked peptides that were between Lys120 of Raptor and Lys450 of S6K. Therefore, the same RNC1 of Raptor was involved in the interaction with S6K and 4E-BP1. This site on S6K that interacts with Raptor is distant from the TOS motif that is located close to the N terminus.
Our study also revealed intramolecular cross-links that occur within the Raptor protein. The locations of these intramolecular cross-links suggest that the Raptor protein has two separate structured domains: one in the N-terminal half and the other in the C-terminal half of the protein. The N-terminal structured domain encompasses RNC1 and RNC2 domains. Interestingly, the lysine residues found to be cross-linked with 4E-BP1 are all located in a small region within this structured domain (Fig. 5). On the other hand, our recent preliminary results raise the possibility that the interaction with mTOR takes place mainly at the C-terminal region. We have recently identified Raptor-mTOR cross-linked peptides. Three cross-linked peptides were identified, and all cross-linked lysines in Raptor were located within the C-terminal structured domain (Lys1332 and Lys1008). Two cross-linked peptides had Raptor cross-linked to the mTOR residue 2507, a residue located within the kinase domain of mTOR. These results suggest that 4E-BP1 and mTOR interact with different regions of Raptor: 4E-BP1 at the N-terminal structured region and mTOR at the C-terminal structured region. Based on our identification of Raptor120-Raptor894 cross-links, Raptor is likely to be folded as depicted in Fig. 5B. If this were the case, the N-terminal region and the C-terminal region of Raptor are located in close proximity, and this will result in bringing the mTOR kinase domain and 4E-BP1 together.
4E-BP1 in its nonphosphorylated form functions to inhibit protein synthesis by binding to eIF4E, a cap-binding protein that facilitates initiation of protein synthesis. eIF4E is also an oncogene as demonstrated by its ability to confer transformed phenotypes and tumor formation in mice (47), and overexpression of eIF4E is observed in a variety of cancer cells, most notably in acute myelogenous leukemia (48). Ectopic expression of nonphosphorylated form of 4E-BP1 partially reverses transformed phenotypes (49). Thus, blocking the interaction between Raptor and 4E-BP1 is expected to increase the concentration of nonphosphorylated 4E-BP1, which in turn would inhibit eIF4E and tumor growth when the tumor is driven by the overexpression of eIF4E. Small molecule inhibitors have been identified to inhibit the action of eIF4E (35, 50). Further understanding of how Raptor interacts with 4E-BP1 may open up a novel approach to inhibit eIF4E-mediated tumor formation.
Supplementary Material
Acknowledgments
We thank David Lopez for discussion on structured domains of Raptor. We thank the UCLA Vector Core Facility for the preparation of lentivirus carrying FLAG-Raptor.
This work was supported, in whole or in part, by National Institutes of Health Grant CA41996 (to F. T.). This work was also supported by Ministry of Science and Technology of China Grants 2010CB835203 (to M.-Q. D.) and 2013CB910200 (to C. T.).

This article contains supplemental Figs. S1 and S2.
- mTOR
- mammalian target of rapamycin
- mTORC1
- mTOR complex 1
- CXMS
- chemical cross-linking coupled with mass spectrometry analysis
- BS3
- bis[sulfosuccinimidyl] suberate
- RNC
- Raptor N-terminal conserved
- RCR
- Raptor cross-linking region
- TOS
- TOR signaling.
REFERENCES
- 1. Laplante M., Sabatini D. M. (2009) mTOR signaling at a glance. J. Cell Sci. 122, 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Polak P., Hall M. N. (2009) mTOR and the control of whole body metabolism. Curr. Opin. Cell Biol. 21, 209–218 [DOI] [PubMed] [Google Scholar]
- 3. Wullschleger S., Loewith R., Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 4. Tamanoi F., Hall M. N. (2010) Structure, Function and Regulation of TOR Complexes from Yeasts to Mammals Part B: The Enzymes, Vol. 28, Academic Press/Elsevier [Google Scholar]
- 5. Karbowniczek M., Spittle C. S., Morrison T., Wu H., Henske E. P. (2008) mTOR is activated in the majority of malignant melanomas. J. Invest. Dermatol. 128, 980–987 [DOI] [PubMed] [Google Scholar]
- 6. Robb V. A., Karbowniczek M., Klein-Szanto A. J., Henske E. P. (2007) Activation of the mTOR signaling pathway in renal clear cell carcinoma. J. Urol. 177, 346–352 [DOI] [PubMed] [Google Scholar]
- 7. Molinolo A. A., Hewitt S. M., Amornphimoltham P., Keelawat S., Rangdaeng S., Meneses García A., Raimondi A. R., Jufe R., Itoiz M., Gao Y., Saranath D., Kaleebi G. S., Yoo G. H., Leak L., Myers E. M., Shintani S., Wong D., Massey H. D., Yeudall W. A., Lonardo F., Ensley J., Gutkind J. S. (2007) Dissecting the Akt/mammalian target of rapamycin signaling network. Emerging results from the head and neck cancer tissue array initiative. Clin. Cancer Res. 13, 4964–4973 [DOI] [PubMed] [Google Scholar]
- 8. Sato T., Nakashima A., Guo L., Coffman K., Tamanoi F. (2010) Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene 29, 2746–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abraham R. T. (2004) PI 3-kinase related kinases. “Big” players in stress-induced signaling pathways. DNA Repair 3, 883–887 [DOI] [PubMed] [Google Scholar]
- 10. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 11. Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and Raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 12. Gingras A. C., Gygi S. P., Raught B., Polakiewicz R. D., Abraham R. T., Hoekstra M. F., Aebersold R., Sonenberg N. (1999) Regulation of 4E-BP1 phosphorylation. A novel two-step mechanism. Genes Dev. 13, 1422–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. She Q. B., Halilovic E., Ye Q., Zhen W., Shirasawa S., Sasazuki T., Solit D. B., Rosen N. (2010) 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell 18, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsieh A. C., Costa M., Zollo O., Davis C., Feldman M. E., Testa J. R., Meyuhas O., Shokat K. M., Ruggero D. (2010) Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP1-eIF4E. Cancer Cell 17, 249–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) mTOR interacts with Raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 16. Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 17. Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J. (2003) TOS motif-mediated Raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13, 797–806 [DOI] [PubMed] [Google Scholar]
- 18. Eguchi S., Tokunaga C., Hidayat S., Oshiro N., Yoshino K., Kikkawa U., Yonezawa K. (2006) Different roles for the TOS and RAIP motifs of the translational regulator protein 4E-BP1 in the association with Raptor and phosphorylation by mTOR in the regulation of cell size. Genes Cells 11, 757–766 [DOI] [PubMed] [Google Scholar]
- 19. Lee V. H., Healy T., Fonseca B. D., Hayashi A., Proud C. G. (2008) Analysis of the regulatory motifs in eukaryotic initiation factor 4E binding protein 1. FEBS J. 275, 2185–2199 [DOI] [PubMed] [Google Scholar]
- 20. Tabb D. L. (2012) Evaluating protein interactions through cross-linking mass spectrometry. Nat. Methods 9, 879–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang B., Wu Y. J., Zhu M., Fan S. B., Lin J., Zhang K., Li S., Chi H., Li Y. X., Chen H. F., Luo S. K., Ding Y. H., Wang L. H., Hao Z., Xiu L. Y., Chen S., Ye K., He S. M., Dong M. Q. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 [DOI] [PubMed] [Google Scholar]
- 22. Sinz A. (2003) Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J. Mass Spectrom. 38, 1225–1237 [DOI] [PubMed] [Google Scholar]
- 23. Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. (2010) Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rinner O., Seebacher J., Walzthoeni T., Mueller L. N., Beck M., Schmidt A., Mueller M., Aebersold R. (2008) Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singh P., Panchaud A., Goodlett D. R. (2010) Chemical cross-linking and mass spectrometry as a low-resolution protein structure determination technique. Anal. Chem. 82, 2636–2642 [DOI] [PubMed] [Google Scholar]
- 27. Sato T., Nakashima A., Guo L., Tamanoi F. (2009) Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J. Biol. Chem. 284, 12783–12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pandey A., Andersen J. S., Mann M. (2000) Use of mass spectrometry to study signaling pathways. Sci. STKE 2000, PL1. [DOI] [PubMed] [Google Scholar]
- 29. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER. A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 31. Bermejo G. A., Clore G. M., Schwieters C. D. (2012) Smooth statistical torsion angle potential derived from a large conformational database via adaptive kernel density estimation improves the quality of NMR protein structures. Protein Sci. 21, 1824–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang C., Clore G. M. (2006) A simple and reliable approach to docking protein-protein complexes from very sparse NOE-derived intermolecular distance restraints. J. Biomol. NMR 36, 37–44 [DOI] [PubMed] [Google Scholar]
- 33. Pierce B., Weng Z. (2007) ZRANK. Reranking protein docking predictions with an optimized energy function. Proteins 67, 1078–1086 [DOI] [PubMed] [Google Scholar]
- 34. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
- 35. Moerke N. J., Aktas H., Chen H., Cantel S., Reibarkh M. Y., Fahmy A., Gross J. D., Degterev A., Yuan J., Chorev M., Halperin J. A., Wagner G. (2007) Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128, 257–267 [DOI] [PubMed] [Google Scholar]
- 36. Yip C. K., Murata K., Walz T., Sabatini D. M., Kang S. A. (2010) Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell 38, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tomoo K., Matsushita Y., Fujisaki H., Abiko F., Shen X., Taniguchi T., Miyagawa H., Kitamura K., Miura K., Ishida T. (2005) Structural basis for mRNA cap-binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, x-ray crystal structural, and molecular dynamics simulation methods. Biochim. Biophys. Acta 1753, 191–208 [DOI] [PubMed] [Google Scholar]
- 38. Rosettani P., Knapp S., Vismara M. G., Rusconi L., Cameron A. D. (2007) Structures of the human eIF4E homologous protein, h4EHP, in its m7GTP-bound and unliganded forms. J. Mol. Biol. 368, 691–705 [DOI] [PubMed] [Google Scholar]
- 39. Brown C. J., McNae I., Fischer P. M., Walkinshaw M. D. (2007) Crystallographic and mass spectrometric characterisation of eIF4E with N7-alkylated cap derivatives. J. Mol. Biol. 372, 7–15 [DOI] [PubMed] [Google Scholar]
- 40. Liu W., Zhao R., McFarland C., Kieft J., Niedzwiecka A., Jankowska-Anyszka M., Stepinski J., Darzynkiewicz E., Jones D. N., Davis R. E. (2009) Structural insights into parasite eIF4E binding specificity for m7G and m2,2,7G mRNA caps. J. Biol. Chem. 284, 31336–31349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu W., Jankowska-Anyszka M., Piecyk K., Dickson L., Wallace A., Niedzwiecka A., Stepinski J., Stolarski R., Darzynkiewicz E., Kieft J., Zhao R., Jones D. N., Davis R. E. (2011) Structural basis for nematode eIF4E binding an m(2,2,7)G-Cap and its implications for translation initiation. Nucleic Acids Res. 39, 8820–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Siddiqui N., Tempel W., Nedyalkova L., Volpon L., Wernimont A. K., Osborne M. J., Park H. W., Borden K. L. (2012) Structural insights into the allosteric effects of 4EBP1 on the eukaryotic translation initiation factor eIF4E. J. Mol. Biol. 415, 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fletcher C. M., McGuire A. M., Gingras A. C., Li H., Matsuo H., Sonenberg N., Wagner G. (1998) 4E binding proteins inhibit the translation factor eIF4E without folded structure. Biochemistry 37, 9–15 [DOI] [PubMed] [Google Scholar]
- 44. Poulin F., Gingras A. C., Olsen H., Chevalier S., Sonenberg N. (1998) 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 273, 14002–14007 [DOI] [PubMed] [Google Scholar]
- 45. Gingras A. C., Raught B., Gygi S. P., Niedzwiecka A., Miron M., Burley S. K., Polakiewicz R. D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. (2001) Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15, 2852–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsuo H., Li H., McGuire A. M., Fletcher C. M., Gingras A. C., Sonenberg N., Wagner G. (1997) Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4, 717–724 [DOI] [PubMed] [Google Scholar]
- 47. Lazaris-Karatzas A., Montine K. S., Sonenberg N. (1990) Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345, 544–547 [DOI] [PubMed] [Google Scholar]
- 48. Ruggero D., Pandolfi P. P. (2003) Does the ribosome translate cancer? Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 49. Rousseau D., Gingras A. C., Pause A., Sonenberg N. (1996) The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene 13, 2415–2420 [PubMed] [Google Scholar]
- 50. Assouline S., Culjkovic B., Cocolakis E., Rousseau C., Beslu N., Amri A., Caplan S., Leber B., Roy D. C., Miller W. H., Jr., Borden K. L. (2009) Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML). A proof-of-principle clinical trial with ribavirin. Blood 114, 257–260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.