Background: Box jellyfish produce a unique family of toxic venom proteins.
Results: The toxins are structurally similar, yet two subgroups confer different cytolytic activities in red blood cells and cardiovascular effects in rats.
Conclusion: Diversification within the toxin family may influence toxin function/specificity.
Significance: Characterization of the toxins provides new insight into their potential roles in human envenoming.
Keywords: Bioinformatics, Cardiovascular, Endotoxin, Protein Purification, Toxins, Cubozoa, Cytolytic, Jellyfish Toxin, Nematocyst, Venom
Abstract
The box jellyfish Chironex fleckeri produces extremely potent and rapid-acting venom that is harmful to humans and lethal to prey. Here, we describe the characterization of two C. fleckeri venom proteins, CfTX-A (∼40 kDa) and CfTX-B (∼42 kDa), which were isolated from C. fleckeri venom using size exclusion chromatography and cation exchange chromatography. Full-length cDNA sequences encoding CfTX-A and -B and a third putative toxin, CfTX-Bt, were subsequently retrieved from a C. fleckeri tentacle cDNA library. Bioinformatic analyses revealed that the new toxins belong to a small family of potent cnidarian pore-forming toxins that includes two other C. fleckeri toxins, CfTX-1 and CfTX-2. Phylogenetic inferences from amino acid sequences of the toxin family grouped CfTX-A, -B, and -Bt in a separate clade from CfTX-1 and -2, suggesting that the C. fleckeri toxins have diversified structurally and functionally during evolution. Comparative bioactivity assays revealed that CfTX-1/2 (25 μg kg−1) caused profound effects on the cardiovascular system of anesthetized rats, whereas CfTX-A/B elicited only minor effects at the same dose. Conversely, the hemolytic activity of CfTX-A/B (HU50 = 5 ng ml−1) was at least 30 times greater than that of CfTX-1/2. Structural homology between the cubozoan toxins and insecticidal three-domain Cry toxins (δ-endotoxins) suggests that the toxins have a similar pore-forming mechanism of action involving α-helices of the N-terminal domain, whereas structural diversification among toxin members may modulate target specificity. Expansion of the cnidarian toxin family therefore provides new insights into the evolutionary diversification of box jellyfish toxins from a structural and functional perspective.
Introduction
Chironex fleckeri (Cnidaria: Cubozoa) is a large, venomous, Australasian box jellyfish that preys on fish and crustaceans but also inflicts painful and potentially fatal stings to humans. Contact with the jellyfish tentacles triggers the explosive discharge of nematocysts (i.e. stinging capsules) that inject extremely potent and rapidly acting venom into the victim or prey. The effects of C. fleckeri envenoming can involve severe localized and systemic effects, including cutaneous pain, inflammation and necrosis, hypertension followed by hypotension, cardiovascular collapse, and cardiac arrest (1, 2).
A number of bioactive fractions have been isolated from C. fleckeri venom (reviewed in Ref. 3); however, few individual toxins have been unequivocally identified. The first toxins in C. fleckeri venom to be sequenced were CfTX-1 and -2 (4). These highly abundant venom proteins belong to a family of taxonomically restricted cnidarian toxins (42–46 kDa) that includes CqTX-A, CrTX-A, and CaTX-A from box jellyfish species Chironex yamaguchii (5) (as Chiropsalmus quadrigatus (6)), Carybdea rastonii (7), and Alatina moseri (8) (as Carybdea alata (9)), respectively, as well as other representatives from Cubozoa, Scyphozoa, and Hydrozoa. In cubozoans, the toxin family is associated with potent hemolytic activity and pore formation in mammalian erythrocytes as well as nociception, inflammation, dermonecrosis, cardiovascular collapse, and lethality in rats (5–7, 10, 11). Although hemolysis has not been reported in human envenoming, the in vivo effects in rats suggest that these toxins may be the primary cause of similar effects in humans.
A recent proteomic study confirmed the presence of CfTX-1 and -2 in C. fleckeri venom and also identified a large number of potential homologues of CqTX-A, CrTX-A, and CaTX-A using tandem mass spectrometry and de novo sequencing (12). Although clearly related to CfTX-1 and -2, these new homologues do not cross-react with CfTX-1 and -2 antibodies and are thus likely to be structurally and functionally different from the characterized toxins. In this study, we describe the purification and molecular characterization of two CfTX-like toxins from C. fleckeri venom that are closely related in sequence to CaTX-A as well as a third, putative toxin that is also homologous to CaTX-A. Through computational analyses and bioactivity assays, we examine the structural and functional characteristics of the new toxins, explore the molecular diversity of the expanded toxin family, and discuss the implications for the biological role of these toxins in box jellyfish stings.
EXPERIMENTAL PROCEDURES
Sample Collection and Venom Preparation
Jellyfish were collected from coastal waters near Weipa (Queensland, Australia). Nematocysts were isolated from excised jellyfish tentacles (13) and purified from tentacle debris in a discontinuous gradient of Percoll (10). C. fleckeri venom was extracted from purified nematocysts into ice-cold nematocyst extraction buffer (NEB3; 20 mm PO43−, 0.15 m NaCl, pH 6.7) using bead mill homogenization (4). The extracted venom was centrifuged (18,000 × g, 30 min, 4 °C), filtered (0.22-μm Millex-GP filters, Millipore), and kept on ice.
Size Exclusion Chromatography (SEC)
Crude venom (1.5 mg of total protein) was applied to a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) that was pre-equilibrated and eluted with NEB using a Shimadzu HPLC system (0.5 ml min−1, 4 °C). The column was calibrated using molecular mass standards (GE Healthcare/Sigma), including blue dextran (>2000 kDa), apoferritin (440 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumen (66 kDa), albumin (45 kDa), carbonic anhydrase (29 kDa), cytochrome c (12 kDa), and vitamin B12 (1.4 kDa). Protein elution was monitored by UV detection (280 nm), and fractions (1 ml) were collected and retained on ice. Following SDS-PAGE analysis on individual chromatography fractions, fractions corresponding to Peaks 1–6 were pooled and retained on ice.
Cation Exchange Chromatography (CEX)
Pooled fractions corresponding to SEC Peak 3 were applied to a 1-ml Uno-S1 column (GE Healthcare), pre-equilibrated with NEB and connected to a Shimadzu HPLC system (0.5 ml min−1, 4 °C). The column was washed with NEB (10 column volumes), and retained proteins were eluted with stepwise increases in NaCl concentration in the phosphate buffer (10–15 column volumes each of 0.25, 0.5, and 1 m NaCl). Protein elution was monitored by UV detection (280 nm), and fractions (1 ml) were collected and stored on ice.
SDS-PAGE and Western Blot Analysis
Reducing SDS-PAGE (14) was conducted on the crude venom and chromatography fractions, and proteins were visualized using Coomassie Brilliant Blue R-250 (Bio-Rad). For Western blot analysis, proteins in the crude venom and chromatography fractions were separated using reducing SDS-PAGE and transferred to Immobilon-P membranes (Millipore). Membranes were blocked (5% skim milk powder in TBST, 0.5 h) and incubated overnight with toxin-specific rabbit antibodies diluted in blocking solution (1:2000). Membranes were washed (3 × 5 min in TBST) and incubated (2 h) with goat anti-rabbit alkaline phosphatase-conjugated antibodies (Sigma) diluted in TBST (1:5000). Following membrane washing, antibody-bound proteins were visualized using nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Promega).
Bradford Assay
The protein concentration of crude venom and purified toxins was determined using Coomassie Plus assay reagent (Pierce, Thermo Scientific), as per the manufacturer's standard microplate protocol. BSA was used as the calibration standard.
MTS Cell Proliferation Assay
The cytotoxicity of crude venom and partially purified venom proteins was determined on cultured rat aorta smooth muscle cells (A7r5 cell line) using the CellTiter 96® AQueous One solution cell proliferation assay (MTS) (Promega). A7r5 cells (ATTC, Manassas, VA) were cultured in DMEM supplemented with 5% (v/v) fetal bovine serum and 1% (w/v) penicillin/streptomycin (culture medium) as described previously (15). Cultured cells were seeded into wells of a flat-bottomed 96-well cell culture plate (50,000 cells/well to a final volume of 100 μl). The plates were incubated (48 h, 37 °C) in a humidifying 5% (v/v) CO2 atmosphere. Culture media were removed from the wells, and the cells were washed once with prewarmed PBS (37 °C). Crude C. fleckeri venom and chromatography peak fractions were diluted in culture medium to final protein concentrations of 2, 0.2, 0.02, and 0.002 μg ml−1. Diluted samples (100 μl) were added in quadruplicate to wells of the cell culture plate. Control cells, in which medium was added instead of C. fleckeri samples, were prepared in quadruplicate and used as a reference for 100% cell viability. Medium blanks (no cells, no venom) were also prepared in quadruplicate. The plates were incubated (37 °C, 24 h) in 5% CO2 and washed three times with prewarmed PBS (37 °C). Fresh culture medium (50 μl) and MTS solution (10 μl) were added to each well, and the plates were incubated (37 °C, 3 h) in 5% CO2. The absorbance of formazan product formed in the cell cultures was measured at A492. The percentage of viable cells present following exposure to C. fleckeri proteins was calculated as the absorbance of the samples relative to the control cells.
In Vivo Cardiovascular Effects of Fractionated Venom in Rats
Male Sprague-Dawley rats (250–300 g) were anesthetized with pentobarbitone sodium (60–100 mg kg−1, intraperitoneal, supplemented as required) (Jurox Pty Ltd.). Cannulae were inserted into the trachea, jugular vein, and carotid artery for artificial respiration (if required), administration of sample, and measurement of blood pressure, respectively. Samples were administered via the jugular vein at a protein concentration of 25 μg kg−1 rat. NEB was used as a negative control. Arterial blood pressure was recorded using a Gould Statham P23 pressure transducer connected to a Power Lab system. At the conclusion of the experiment, animals were euthanized by an overdose of pentobarbitone. Pulse pressure was defined as the difference between systolic and diastolic blood pressures. Mean arterial pressure (MAP), defined as diastolic blood pressure plus one-third of pulse pressure, and average heart rate (HR) were calculated using LabChart (ADInstruments). Ethical approval for all animal experiments was obtained from the Monash University Animal Ethics Committee.
Hemolytic Activity
Hemolytic activity of crude nematocyst venom and purified toxins was determined using a sheep erythrocyte microplate assay. Heparinized sheep blood was centrifuged (3000× g, 4 °C, 10 min) and the sedimented erythrocytes were repeatedly washed in sterile PBS and recovered by centrifugation (3000 × g, 4 °C, 10 min). Aliquots (300 μl) of washed, diluted erythrocytes (1% in PBS) were added to PBS-diluted samples (10 μl) in quadruplicate in a 96-well plate on ice and incubated (30 min, 37 °C) with gentle agitation. Samples were chilled on ice (5 min) and centrifuged (3000 × g, room temperature, 5 min), and supernatants (200 μl) were transferred to another 96-well plate. The absorbance of released hemoglobin was measured at A540. Quadruplicates of diluted Triton X-100 (1% in PBS) and PBS alone were used as references for 100 and 0% lysis, respectively. Hemolysis results were calculated as a percentage relative to complete lysis. HU50 values, defined as the concentration of protein that causes 50% lysis, were determined for the crude venom and pooled chromatography fractions from plots of percentage hemolysis as a function of the final concentration of assayed protein. Hemolysis dose-response plots were each fitted with a four-parameter logistic curve (SigmaPlot version 12.5, Systat Software Inc.).
Edman Sequencing
N-terminal and internal peptide sequences were determined for two unidentified venom proteins (hereafter referred to as CfTX-A and -B) that were present in hemolytic SEC Peak 3 fractions. SDS-polyacrylamide gel fragments of CfTX-A and -B (40 and 42 kDa, respectively) were destained (50% (v/v) acetonitrile, 50 mm ammonium bicarbonate), reduced with DTT (25 mm, 56 °C, 0.5 h), and alkylated with acrylamide (100 mm, room temperature, 1 h). For N-terminal sequencing, the proteins (100 pmol each) were passively eluted from the gel using an SDS elution buffer (100 mm sodium acetate, 0.1% SDS, 10 mm DTT; 37 °C, 16 h), loaded onto Prosorb filter cartridges (Applied Biosystems), and washed twice with 0.1% TFA (100 μl). The proteins, bound to PVDF membrane, were subjected to automated Edman sequencing using an Applied Biosystems 494 Procise Protein Sequencing System. For internal sequencing, destained, reduced, and alkylated gel bands (100 pmol each) were washed with ammonium bicarbonate (50 mm) and incubated with trypsin (37 °C, 16 h). Peptides were extracted from the gel pieces in 0.1% TFA in 10% acetonitrile and separated using reversed phase HPLC. Selected peptides were loaded onto precycled Biobrene-treated discs and subjected to automated Edman sequencing.
Full-length cDNA Cloning of CfTX-A and -B
Partial cDNA fragments encoding CfTX-A and -B were amplified from a C. fleckeri tentacle cDNA library (4) using degenerate PCR. A series of forward and reverse degenerate oligonucleotide primers were designed according to the CfTX-A and -B N termini sequences, DVDEVTSDINQLVNQLNNVQ and SSSEINAEIDGLIQQ, respectively, and their internal peptides FFGLPDPPR and SIVDEAFKR, respectively. Degenerate PCR was conducted for each combination of primers using hot start PCR (94 °C for 4 min, 30 cycles of 94 °C for 0.5 min, 50 °C for 0.5 min, and 72 °C for 1.5–2 min and then 72 °C for 10 min), MyTaq DNA polymerase (Bioline) and 1 μl of cDNA library. Two cDNA fragments (890 and 363 bp) were amplified using primer pairs A-F1/A-R1 and B-F1/B-R1, respectively (Table 1). The 5′- and 3′-ends of CfTX-A cDNA were amplified from the library using primers T3/A-R2 and A-F2/T7, respectively. The 5′-end of CfTX-B cDNA was amplified in two PCRs using primers T3/B-R2 and M13R/B-R3; the 3′-end was amplified using B-F2/T7. To verify the nucleotide sequences encoding mature CfTX-A and -B, cDNA was amplified from the library using gene-specific primer pairs, A-F3/A-R3 and B-F3/B-R4, and high fidelity BIO-X-ACT Short DNA polymerase (Bioline) under hot start PCR conditions (94 °C for 4 min, 35 cycles of 94 °C for 0.5 min, 55 °C for 0.5 min, and 70 °C for 1 min 10 s and then 70 °C for 10 min). All PCR products were gel-purified (QIAgen) and TOPO TA-cloned into pCR2.1-TOPO (Invitrogen), and their nucleotide sequences were determined in both directions (Macrogen Inc.). Nucleotide sequences were trimmed and aligned using Sequencher (Gene Codes).
TABLE 1.
Oligonucleotide primer sequences for CfTX-A and CfTX-B
| Toxin | Primer ID | Sequence (5′–3′) |
|---|---|---|
| CfTX-A | A-F1 | GAYGTHGAYGARGTHACWTCTG |
| A-F2 | CATCTGGTGACGCATTCAAC | |
| A-F3 | AACAGACGACGAGCCAAAAG | |
| A-R1 | CKWGGWGGRTCWGGHAGTCC | |
| CfTX-B | A-R2 | GCTCCTGCTCGGTTACATTC |
| A-R3 | CTGCTTCTCTTATTACAACAGACG | |
| B-F1 | GARATHAAYGCHGAAATHGATGG | |
| B-F2 | CTGTCGCTTCATTGTTATCG | |
| B-F3 | ATTCCGAATCGAGCAAAAAGG | |
| B-R1 | YTTRAAIGCYTCRTCDACDATGG | |
| B-R2 | GTTGCAATCGATGAAAGG | |
| B-R3 | TGAAGAAATTCTTGGATCC | |
| B-R4 | CGAAATTTTAAGGCTATGGC |
Polyclonal Antibodies against C. fleckeri Toxins
Polyclonal antibodies against purified toxins CfTX-1 and -2 were available from a previous study (10). In addition, a rabbit was immunized with four doses of combined and emulsified gel fragments corresponding to purified toxins CfTX-A and CfTX-B (50, 85, and 2 × 100 μg of protein) at 3-week intervals (IMVS Veterinary Services Division, Australia) under the aegis of the IMVS Animal Ethics Committee (License 155). Toxin-specific antibodies were purified from rabbit serum using protein A-Sepharose chromatography according to the manufacturer's instructions (GE Healthcare).
Analysis of CfTX-A and -B Using Liquid Chromatography and Tandem Mass Spectrometry (LC-MS/MS)
Tryptic fragments from in-gel digestion of SDS-polyacrylamide gel bands containing CfTX-A and -B were prepared and subjected to MS/MS as described previously (12, 16). Briefly, tryptic fragments from in-gel digests were chromatographically separated on a Dionex Ultimate 3000 HPLC system using an Agilent Zorbax 300SB-C18 (3.5 μm, 150 mm × 75 μm) column and a linear gradient of 0–80% solvent B over 60 min and directly introduced into the source of a QSTAR Elite Hybrid MS/MS system (AB Sciex) operated in positive ion electrospray mode. Protein constituents of gel slices were identified using Mascot (Matrix Science). Search parameters allowed for trypsin as the enzyme, methionine oxidation and carbamidomethylation as variable and fixed modifications, respectively, mass tolerances of +0.1 Da on peptide and fragment ions, two missed cleavages, and charge states +2 and +3. For spectral searches, the deduced amino acid sequences of the new toxins were added to a custom-made database of all cnidarian sequences in the GenBankTM non-redundant protein database. The criteria for accepting peptide identifications were subjected to an e-value threshold of less than 0.05.
Bioinformatics
Nucleotide and deduced amino acid sequences were subjected to BLAST analysis (17). Global and local pairwise sequence alignments were performed using EMBOSS Needle and Water, respectively (18). Multiple sequence alignment, phylogenetic analysis, and tree construction (MUSCLE, PhyML, TreeDyn, respectively) were performed via the Phylogeny.fr platform (19); optional alignment curation was performed using Gblocks, and branch support for phylogenetic trees was evaluated using the approximate likelihood ratio test, Shimodaira-Hasegawa-like. Conserved motifs in amino acid sequences were detected using the MEME Suite Web server (20). Secondary structure predictions, structural homology detection, and three-dimensional modeling were performed using the I-TASSER server (21). Membrane helix predictions were performed using MEMSAT3 and MEMSAT-SVM (22) via the PSIPRED server.
RESULTS
SEC and CEX
Crude C. fleckeri venom proteins eluted from the Superdex 200pg SEC column in six major peaks with native molecular masses ranging from 30 to >600 kDa (Fig. 1a). SDS-PAGE analysis of SEC fractions revealed that the most abundant toxins in C. fleckeri venom (CfTX-1 and -2) were co-purified in Peak 2 (360 kDa) (Fig. 1b), consistent with previous experiments (10). Western blot analysis of the SEC peaks using α-CfTX-1/2 antibodies confirmed the presence of CfTX-1/2 predominantly in Peak 2 but also in the other peak fractions in smaller amounts (Peak 3 > 4 > 1 ≈ 5 > 6) (Fig. 1c). CfTX-A and -B (40 and 42 kDa, respectively) were partially co-purified in Peak 3 (130 kDa), but similar amounts of CfTX-1/2 were also present. Significant improvement in CfTX-A and -B purity was achieved by fractionating pooled Peak 3 fractions using a Uno S1 CEX column. The majority of CfTX-1 and -2 eluted as column flow-through (Peak 1; 150 mm NaCl), whereas CfTX-A and -B were selectively eluted at higher salt concentration (Peak 2; 250 mm NaCl) (Fig. 1b). Western blot analysis using toxin-specific antibodies showed negligible contamination of CfTX-A and -B with CfTX-1 and -2 after CEX purification (Fig. 2). Conversely, CfTX-A and -B were not detected in SEC-purified fractions containing CfTX-1 and -2 (SEC Peak 2).
FIGURE 1.
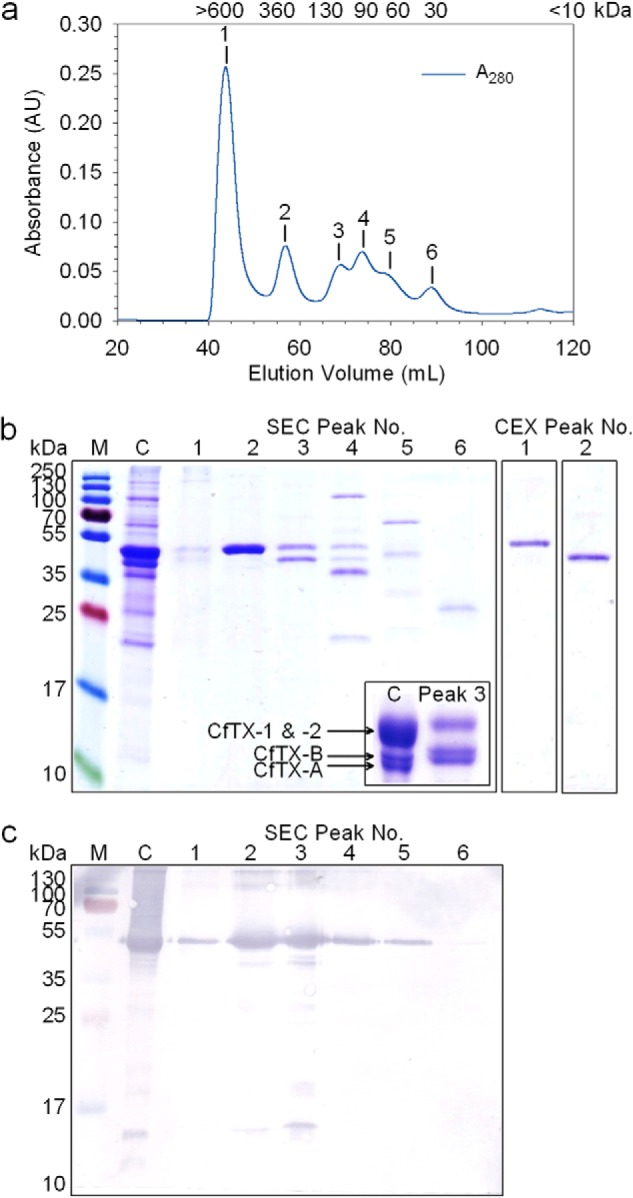
Purification of C. fleckeri toxins using size exclusion and cation exchange chromatography. a, a typical chromatogram of crude venom fractionated on a Superdex 200pg column; protein elution was monitored at 280 nm. The estimated native molecular masses of SEC Peaks 1–6 are indicated above each peak in kDa. b, 15% denaturing SDS-PAGE protein profiles of crude venom (C) and chromatography fractions corresponding to SEC Peaks 1–6 and CEX Peaks 1 and 2. M, protein ladder; the molecular masses of the protein standards are shown alongside. Inset, 12.5% SDS-PAGE profile of crude venom and SEC Peak 3 showing increased resolution of CfTX-A and -B. c, Western blot of crude venom and SEC Peaks 1–6 using CfTX-1/2-specific antibodies.
FIGURE 2.

Comparison of 12.5% SDS-PAGE and Western blot profiles during purification of CfTX-A/B. a, Coomassie-stained proteins; b, proteins bound to CfTX-1/2-specific antibodies; c, proteins bound to CfTX-A/B-specific antibodies. Lane 1, protein standards with molecular masses (kDa) indicated to the left; lane 2, crude C. fleckeri venom; lane 3, SEC Peak 2; lane 4, SEC Peak 3; lane 5, CEX Peak 2.
MTS Cell Proliferation Assay
The cytotoxicity of crude venom and SEC Peaks 1–6 (Fig. 1) on cultured A7r5 cells was determined using the MTS cell proliferation assay. Crude C. fleckeri venom elicited potent cytotoxic effects, reducing cell viability to less than 10% at the lowest concentration of protein tested (2 ng ml−1; Fig. 3). In contrast, venom proteins from Peak 1 produced relatively minor cytotoxic effects, with 82 ± 8% cells remaining viable after exposure to a protein concentration of 2 μg ml−1. Peaks 2 and 3, which contained the majority of CfTX-1/2 and CfTX-A/B (Fig. 1b), were the most cytotoxic of the SEC fractions, reducing cell viability to 57 ± 6 and 39 ± 12%, respectively, at 2 ng ml−1 protein concentration. Average cell viability following exposure to Peak 4, 5, and 6 proteins was greater than 80% at concentrations of 0.02, 0.2, and 2 μg ml−1, respectively, indicating a progressive decrease in cytotoxicity concomitant with a decrease in CfTX-1/2 concentration (Fig. 1c) as lower molecular mass proteins eluted from the column.
FIGURE 3.
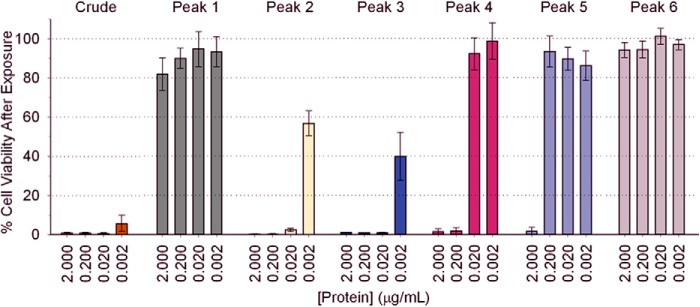
The in vitro cytotoxic effects of crude C. fleckeri venom and SEC-fractionated venom (Peaks 1–6) on cultured A7r5 cells, as determined by the MTS assay. The percentage of viable cells remaining in culture following exposure to the venom proteins compared with the negative control (100% cell viability) correlates inversely to cytotoxicity. Quadruplicate assays were performed at four different protein concentrations per sample. Error bars, S.E.
In Vivo Cardiovascular Effects of C. fleckeri Toxins in Anesthetized Rats
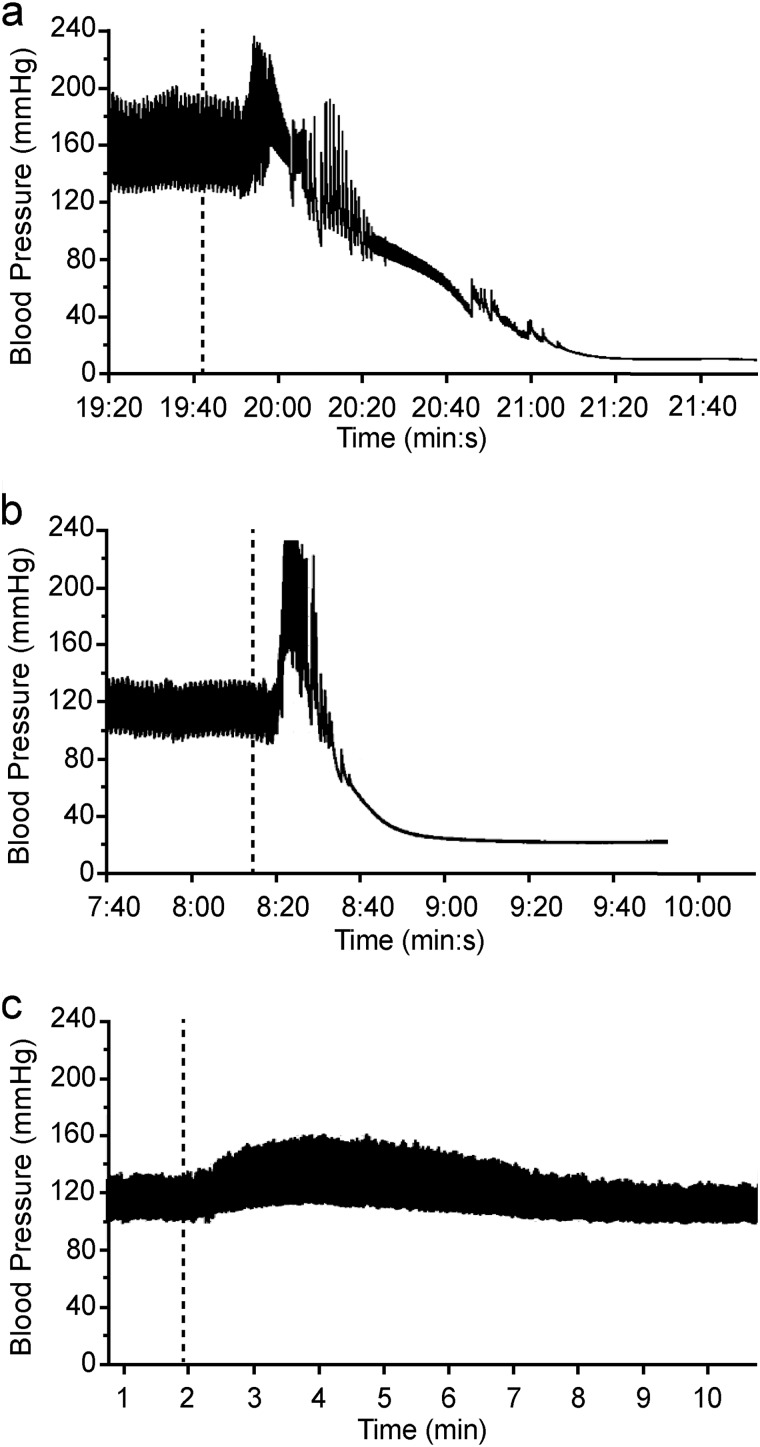
The cardiovascular effects of C. fleckeri venom and partially purified toxins (25 μg kg−1 intravenously) were tested in anesthetized rats. SEC Peak 3 was not tested because the majority of the sample was required for further purification of CfTX-A/B. Venom caused a transient increase in MAP (44 ± 11 mm Hg; n = 3) 8–15 s after injection, followed by cardiovascular collapse in all animals within 2–3 min (Fig. 4a). At maximum MAP, the HR varied by less than 1% compared with basal levels in two of the animals, whereas the HR of the third animal decreased by 6%. SEC Peak 2, containing CfTX-1/2, produced an initial increase in MAP of variable magnitude (12–70 mm Hg) 8–11 s after injection, followed by cardiovascular collapse in all animals within 1 min of injection (Fig. 4b). No significant change in HR from basal levels was observed at maximum MAP in all animals. SEC Peak 4 elicited an increase in MAP of 51 ± 6 mm Hg (n = 4) 10–27 s after injection, followed by cardiovascular collapse in 3 of 4 animals. In the fourth animal, a hypotensive effect was observed (i.e. MAP decreased to 34 mm Hg, and HR decreased to 262 b.p.m.), followed by partial recovery in MAP (77 mm Hg) and HR (325 b.p.m.) compared with basal levels (128 mm Hg and 381 b.p.m., respectively). Initial increases in MAP above basal levels were observed in all animals injected with SEC Peak 1 (i.e. an increase of 31 ± 5 mm Hg; n = 3), SEC Peak 5 (i.e. an increase of 41 ± 2 mm Hg; n = 3), and SEC Peak 6 (i.e. an increase of 30 ± 3 mm Hg; n = 3) within 23–54 s after injection. At maximum MAP, HR decreased by 1–8%. In 2 of 3 animals injected with SEC Peak 1, MAP and HR subsequently returned to (or near to) basal levels. In the third animal, HR returned to basal levels (410 b.p.m.), but MAP transiently decreased to 111 mm Hg before returning to basal levels (122 mm Hg). In all animals injected with Peak 5, no further decrease in HR was observed, but MAP decreased by 18 ± 5 mm Hg (n = 3) below basal levels before both HR and MAP returned to normal. In 2 of 3 animals injected with SEC Peak 6, no hypotensive effect was observed, and MAP and HR subsequently returned to basal levels. In the third animal, MAP and HR decreased to 99 mm Hg and 338 b.p.m., respectively, before recovering to basal levels (i.e. 132 mm Hg and 384 b.p.m., respectively). CEX-purified CfTX-A and -B (CEX Peak 2; Fig. 4c) had no significant effects on HR but produced relatively small increases in MAP in 2 of 3 animals (9 and 17 mm Hg) that subsequently returned to basal levels. No significant effects on MAP or HR were observed in the third animal. Similarly, no significant effects on MAP or HR were observed in animals injected with buffer only (i.e. 126 ± 10 mm Hg and 400 ± 36 b.p.m. preinjection; 127 ± 10 mm Hg and 410 ± 35 b.p.m. postinjection; n = 4).
FIGURE 4.
Representative arterial blood pressure traces from anesthetized rats injected with 25 μg kg−1 (intravenously) of crude C. fleckeri venom (a), SEC-purified CfTX-1/2 (Peak 2) (b), and CEX-purified CfTX-A/B (Peak 2) (c). A dotted line indicates the time of injection.
In Vitro Hemolytic Activity
Microplate hemolysis assays were performed to compare the hemolytic activity of crude venom with the CfTX toxins that were isolated using SEC and CEX. Concentration-response curves (Fig. 5) showed similar hemolytic activity for crude venom and co-purified CfTX-1/2 (SEC Peak 2) (HU50 = 148 and 161 ng ml−1, respectively). SEC Peak 3, which contained a mixture of CfTX-1/2 and CfTX-A/B, was at least 6 times more hemolytic (HU50 = 22 ng ml−1) than purified CfTX-1/2 or crude venom. Purified CfTX-A and -B (CEX Peak 3) exhibited the greatest potency, with hemolytic activity 30 times greater than the original crude venom (HU50 = 5 ng ml−1).
FIGURE 5.
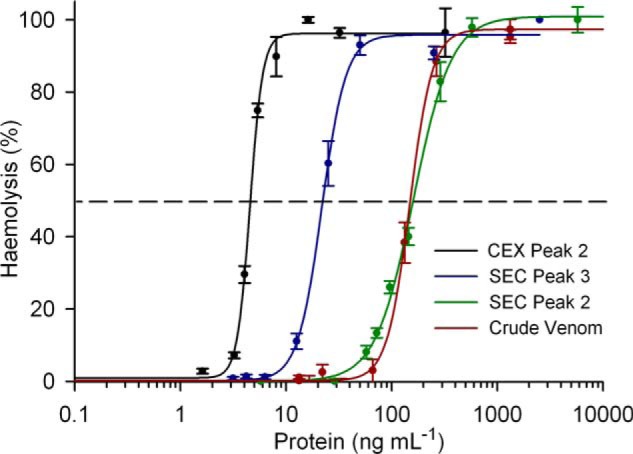
Hemolytic activity concentration-response curves for crude C. fleckeri venom and isolated CfTX toxins. The curve for crude venom is indicated in red. SEC Peak 2, containing purified CfTX-1/2, is indicated in green. SEC Peak 3, containing similar amounts of CfTX-1/2 and CfTX-A/B, is indicated in blue. CEX Peak 2, containing CfTX-A/B purified from SEC Peak 3, is indicated in black. Error bars, S.E. from four independent assays at each protein concentration.
Edman Sequencing
The N-terminal sequences obtained for CfTX-A and CfTX-B were DVDEVTSDINQLVNQLNNVQ and SSSEINAEIDGLIQQ, respectively. Two internal peptides, (S)ALE(E)L(G)TEVSA and FFGLPDPPR, were obtained for CfTX-A, and a single peptide, SIVDEAFK(R), was obtained for CfTX-B; tentative residue identifications are shown in parentheses.
Isolation of cDNA Encoding Full-length CfTX-A and CfTX-B Isoforms
CfTX-A and -B full-length clones were amplified from a C. fleckeri cDNA library using PCR and a combination of degenerate, gene-specific, and universal primers. The CfTX-A cDNA was 1506 bp in length and was composed of a 24-bp stretch of 5′-untranslated region (UTR), a 1362-bp open reading frame (ORF), a 96-bp 3′-UTR, and a poly(A) tail (Fig. 6). The ORF translated into a 454-residue precursor protein featuring an 18-residue putative signal peptide (SignalP 4.1 server (23)), an N-terminal heptamer (ending in KR), and a 429-residue mature protein. In comparison, the CfTX-B cDNA was 1574 bp in length and contained an 81-bp 5′-UTR, a 1383-bp ORF, a 93-bp 3′-UTR, and a poly(A) tail (Fig. 7). The ORF translated into a 461-residue precursor protein containing a 24-residue putative signal peptide, an N-terminal heptamer (ending in KR), and a 430-residue mature protein. The theoretical molecular masses of mature CfTX-A and CfTX-B were calculated to be 47,577 and 47,655 Da, respectively, and their isoelectric points (pI) were calculated as 6.3 and 7.1, respectively (ProtParam (24)).
FIGURE 6.
Nucleotide and deduced amino acid sequences of CfTX-A (GenBankTM accession number JN695597). An 18-residue signal peptide is indicated with a dashed line; the start codon (ATG) is highlighted in boldface type. A 7-residue propeptide located between the signal peptide and the N terminus is shown in italic type. The N-terminal sequence (N) and two internal peptides (1 and 2) obtained by Edman sequencing are underlined. Peptide matches retrieved through LC-MS/MS analysis and Mascot searches are indicated in boldface type. An asterisk indicates the first stop codon in-frame with the start codon.
FIGURE 7.
Nucleotide and deduced amino acid sequences of CfTX-B (GenBankTM accession number JN695598). A 24-residue signal peptide is indicated with a dashed line; the start codon (ATG) is highlighted in boldface type. A 7-residue propeptide located between the signal peptide, and the N terminus is shown in italic type. The N-terminal sequence (N) and an internal peptide (1) obtained by Edman sequencing are underlined. Peptide matches retrieved through LC-MS/MS analysis and Mascot searches are indicated in boldface type. An asterisk indicates the first stop codon in-frame with the start codon. Blue nucleotides correspond to a stretch of nucleotides that are absent in CfTX-Bt (Fig. 8); arrows indicate the deletion boundaries. Nucleotide variations between CfTX-B and CfTX-Bt are highlighted in red; for non-synonymous substitutions, the affected residue is also highlighted in red.
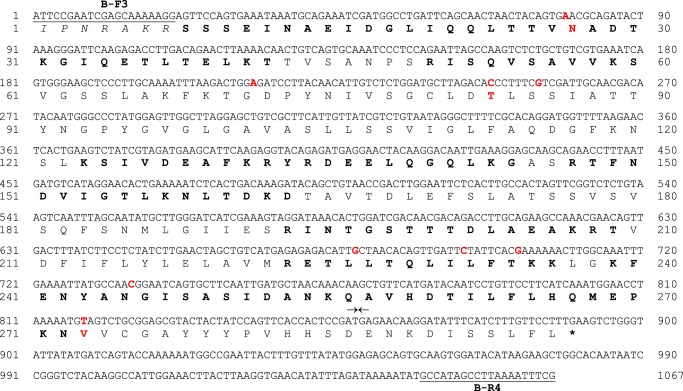
Two distinct cDNA populations were obtained following amplification of cDNA encoding mature CfTX-B with gene-specific primers that flanked the N terminus coding region and the stop codon (B-F3/B-R4). The two cDNA products were separately cloned and sequenced. The most abundant PCR product was 1359 bp, consistent with the expected length of CfTX-B cDNA (Fig. 7), whereas the second less abundant product was significantly shorter (1067 bp). The latter encoded a protein homologous to a truncated form of CfTX-B (296 residues), referred to hereafter as CfTX-Bt (Fig. 8). The theoretical molecular mass and pI of the mature protein were calculated as 31,293 Da and 5.2, respectively. The cDNA encoding CfTX-Bt is differentiated from CfTX-B cDNA by nine single nucleotide variations (all transitions, three non-synonymous) and a 292-bp deletion after nucleotide 1011 in CfTX-B (Fig. 7).
FIGURE 8.
Nucleotide and deduced amino acid sequences of CfTX-Bt (GenBankTM accession number KF583451). Gene-specific primers for CfTX-B and CfTX-Bt (B-F3/B-R4) are underlined. An asterisk indicates the stop codon. Arrows correspond to nucleotide deletion boundaries in CfTX-B (Fig. 7). Nucleotide variations between CfTX-B and CfTX-Bt are indicated in red; for non-synonymous substitutions, the affected residue is also indicated in red.
LC-MS/MS and Peptide Mapping
To confirm the identity of CfTX-A and -B in C. fleckeri venom, SDS-polyacrylamide gel bands corresponding to CfTX-A and -B were excised and analyzed after in-gel tryptic digest using tandem MS. Mascot searches against a custom database that included the sequences of CfTX-A and -B identified 22 (20 unique) and 21 (19 unique) significant (p < 0.05) tryptic peptides from CfTX-A and -B, respectively. Sequence coverage for CfTX-A and -B was 56 and 67%, respectively.
Bioinformatics and Phylogenetic Analysis
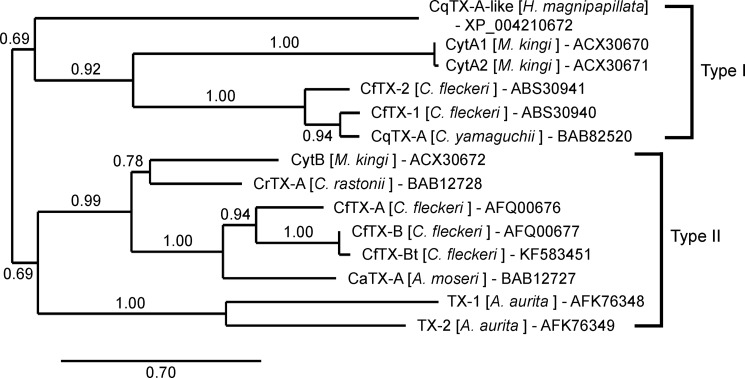
Global pairwise sequence alignment of the CfTX-A and -B precursor proteins revealed that the toxins share 77% sequence similarity (61% identity). Although mature CfTX-B and -Bt vary in length by 134 residues, local alignment of the overlapping sequences indicated 99% similarity and 98% identity between the two proteins. BLASTP analysis against public protein databases revealed that CfTX-A, CfTX-B, and CfTX-Bt are related to an expanding family of lethal, pore-forming cnidarian toxins, which includes CfTX-1 and -2, representatives from other Cubozoa, and putative toxins from Scyphozoa (Aurelia aurita) and Hydrozoa (Hydra magnipapillata). The newly sequenced toxins all share the highest sequence similarity with CaTX-A from A. moseri (71–72% similarity, 52–55% identity). In comparison, sequence similarities to CfTX-1 and -2 are substantially lower (45–48% similarity, 23–25% identity). A phylogenetic tree constructed using 11 available protein sequences from five cubozoan species, two sequences from A. aurita (TX-1 and TX-2), and an example from H. magnipapillata (CqTX-A-like) infers that the toxin family has diversified into two broad groupings (Type I and II) within the phylum Cnidaria (Fig. 9). Within each group, the cubozoan toxins form smaller clades, signifying additional toxin diversification. Notably, CfTX-1 and -2 (Type I) are grouped separately from CfTX-A and -B (Type II), suggesting that each pair of toxins varies in structure and possibly function. Likewise, the Cytotoxin A isoforms are grouped separately from Cytotoxin B, implying that similar patterns of toxin diversity occur in other cubozoan species.
FIGURE 9.
Phylogenetic relationships of CfTX-like cnidarian toxins. The tree was constructed using the Phylogeny.fr pipeline (MUSCLE, PhyML, TreeDyn); branch support was evaluated using the approximate likelihood ratio test, Shimodaira-Hasegawa-like, statistical test. Sequence accession numbers are included after the species names.
A multiple-sequence alignment of the cubozoan amino acid sequences (Fig. 10) revealed that only 17 aligned residues are conserved across all of the toxins. With the exception of Cys108, no cysteine residues are highly conserved, suggesting that disulfide bridges are not a conserved structural feature of the toxin family. However, several proline residues (e.g. Pro310, Pro431, and Pro469) are highly conserved and may influence structural conformation/stability or modulate protein aggregation (25). Using the MEME algorithm, three of the most highly conserved regions (sequence motifs) in the cubozoan sequences corresponded to alignment ranges 80–110, 138–187, and 427–460 (e-values ≥ 1.1E−051) (Fig. 10).
FIGURE 10.
Multiple sequence alignment of cubozoan toxin amino acid sequences. The sequences were aligned using MUSCLE, and the alignment was visualized using Jalview. Residue shading is based on the default Clustal protein color scheme. Identical residues are indicated with an asterisk, and dashes represent gaps introduced for better alignment. Signal peptides and proparts are underlined in black and red, respectively. Conserved sequence motifs are indicated below the alignment.
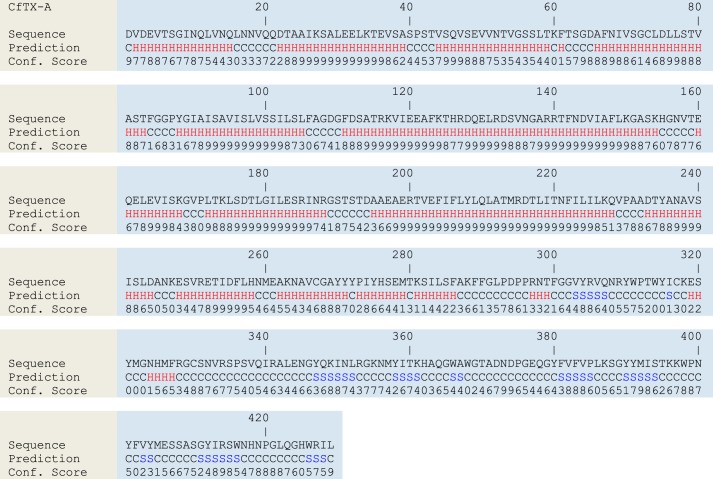
Structural Organization and Molecular Modeling
Secondary and tertiary structural analyses using the I-TASSER Server predicted that CfTX-A and -B are similar to all previously characterized box jellyfish toxins (3). The mature toxins form two putative domains. The N-terminal domain (290–300 residues) is dominated by α-helices (81–82%) and loop structures (17–19%), with β-strands constituting 0–1% (Figs. 11 and 12). The smaller C-terminal domain is composed mostly of loop structures (60–63%) and β-strands (29–34%); α-helices constitute 6–8%. CfTX-Bt is predicted to form a single domain structurally similar to the N-terminal domains of the other CfTX-like toxins (77% α-helices, 18% loop structures, 5% β-strands) (Fig. 13). Due to the truncated nature of CfTX-Bt, a C-terminal domain analogous to those in the dual-domain toxins is absent. Consequently, the C terminus of CfTX-Bt may provide a guide to domain demarcation in the dual-domain toxins. MEMSAT predictions suggested that the N-terminal domains of mature CfTX-A, -B, and -Bt each contain one or two consecutive transmembrane helices between residues 61 and 105, which is in agreement with predictions for other CfTX-like toxins (4). An additional transmembrane helix spanning residues 205–224 was less confidently predicted in each protein; nevertheless, the region consistently exhibited above average hydrophobicity. In the precursor molecules, the corresponding residue ranges are 86–130 and 230–249 (CfTX-A) and 92–136 and 237–255 (CfTX-B and -Bt).
FIGURE 11.
Secondary structure predictions for CfTX-A. H, S, and C, α-helical, β-strand, and coiled (loop) structures, respectively. Confidence scores range from 0 (low) to 9 (high).
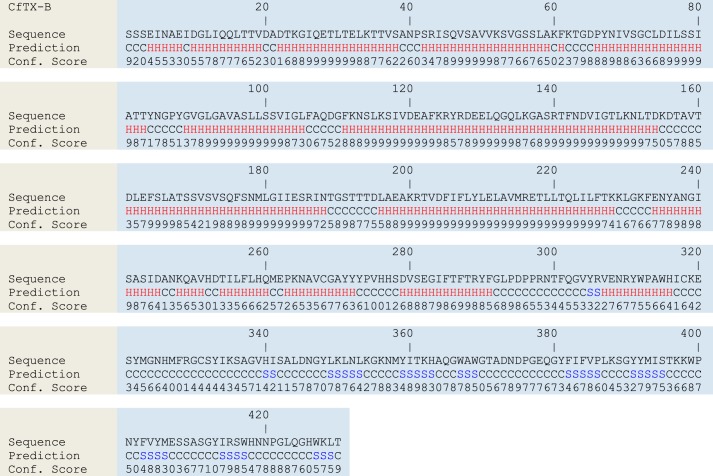
FIGURE 12.
Secondary structure predictions for CfTX-B. H, S, and C represent α-helical, β-strand, and coiled (loop) structures, respectively. Confidence scores range from 0 (low) to 9 (high).
FIGURE 13.
Secondary structure predictions for CfTX-Bt. H and C represent α-helical and coiled (loop) structures, respectively. Confidence scores range from 0 (low) to 9 (high).
The top three-dimensional models generated by I-TASSER for mature CfTX-A, -B, and -Bt, superimposed with a close structural analog in the RSCB Protein Data Bank, are presented in Fig. 14. Confidence scores for the predicted models were −1.2, −3.2, and −3.1, respectively. Confidence scores typically range between −5 and +2, where a higher value reflects a model of better quality. When confidence scores are ≥−1.5, both false-positive and false-negative rates are <0.1, indicating that >90% of the quality predictions are correct (21). The highest ranking structural analogues of CfTX-A in the Protein Data Bank were Bacillus thuringiensis insecticidal three-domain Cry (3d-Cry) toxins Cry8Ea (3EB7), Cry3Aa (1DLC), Cry3Bb (1JI6), Cry1Aa (1CIY), Cry2Aa (1I5P), and Cry4Aa (2C9K), with TM scores all ≥0.7 (structures with TM scores >0.5 share the same fold). The same Cry toxins were also the highest ranked structural analogues of CfTX-B and CfTX-Bt (TM scores 0.6–0.7). As indicated in Fig. 14, structural similarity is highest between the N-terminal domains of the CfTX models and the N-terminal domain (Domain I) of the Cry toxin, suggesting that these domains could share a similar functional role. Greater structural variability is observed between the C-terminal domains of the CfTXs (not present in CfTX-Bt) and the central domain (Domain II) of Cry toxins, whereas domains analogous to the C-terminal domain of 3d-Cry toxins (Domain III) are absent in all CfTX models.
FIGURE 14.
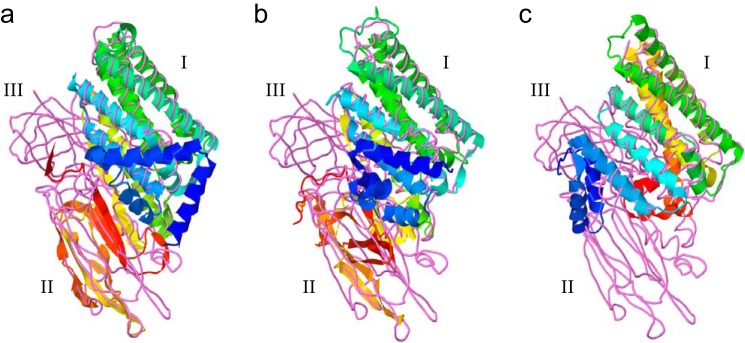
Predicted three-dimensional models of mature CfTX-A (a), CfTX-B (b), and CfTX-Bt (c), respectively, superimposed with the B. thuringiensis insecticidal 3d-Cry toxin, Cry8Ea1 (Protein Data Bank code 3EB7). Models were generated using the I-TASSER protocol and visualized using Jmol. The CfTXs are depicted in schematic form; the colors of secondary structures transition from blue (N terminus) to red (C terminus). Cry8Ea1 is depicted as a backbone trace (violet). The N-terminal, central, and C-terminal domains characteristic of 3d-Cry toxins are indicated as I–III, respectively.
DISCUSSION
Expansion of the cnidarian toxin family to include CfTX-A, CfTX-B, and CfTX-Bt has provided further insight into the molecular, structural, and functional diversity of the major toxins produced by box jellyfish. Phylogeny-based predictions suggest that the cubozoan toxins have diversified into at least two toxin groups (Type I and II), with the newly described CfTX-A, -B, and Bt grouped among the Type II toxins (Fig. 9). The Type I and Type II toxins are all predicted to contain signal peptides and, with the exception of CfTX-Bt, form dual-domain mature proteins. However, unlike their Type I counterparts, Type II toxins contain a short propart (5–7 residues) ending with the classical dibasic proteolytic cleavage site (RR/KR) between the signal peptide and mature protein. N-terminal proparts are a common feature in precursor proteins and fulfill a variety of important roles during protein biosynthesis and activation. The propart can facilitate controlled and efficient transport within the secretory pathway, promote correct folding, direct posttranslational modifications, and, in protoxins, prevent unwanted toxicity to the host cell prior to propart cleavage and toxin activation (26–30). Well known toxins that contain N-terminal proparts include ricin (28) and 3d-Cry insecticidal toxins (29), and they are also common in the precursor toxins of cnidarians (e.g. see Refs. 31–33), spiders (30), and cone snails (34). The purpose of the propart found exclusively in the Type II toxins remains unclear, but its presence may modulate toxin folding, posttranslational modifications, or intracellular trafficking that in turn leads to diversified function/specificity or variations in toxin localization between the two toxin groups. Alternatively, the Type II toxins may be more toxic to cnidarians (invertebrates) than Type I toxins, and thus the propart may provide additional protection to the host organism during toxin production.
Another difference between the toxin groups is that mature Type II toxins are typically shorter in sequence length and consequently lower in theoretical molecular mass than Type I toxins (Fig. 15). This variation in molecular mass is consistent with experimental observations for CfTX-1 and -2 (Type I) and CfTX-A and -B (Type II) separated by reducing SDS-PAGE (Fig. 1b). It is also noted that the theoretical molecular masses of all mature CfTX-like proteins, including those of other cubozoans, are consistently higher than the apparent molecular mass estimated by SDS-PAGE (3). Notwithstanding the inherent inaccuracies of SDS-PAGE-based estimations, the discrepancies in molecular mass may result from additional posttranslational proteolytic processing during toxin maturation/activation.
FIGURE 15.
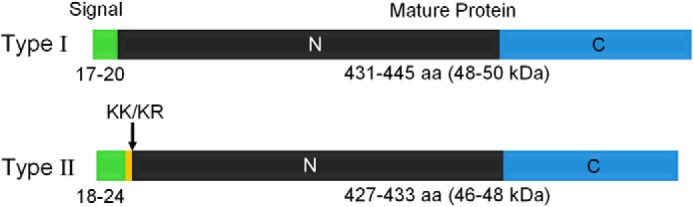
Structural organization of Type I and Type II CfTX-like proteins. Signal peptides are indicated in green; the number of residues is indicated below. Putative N-terminal (N) and C-terminal domains (C) of the mature proteins are indicated in black and blue, respectively. The residue and theoretical molecular mass ranges of the mature toxins are indicated below each toxin type. A short propart (5–7 residues) present only in the Type II toxins is indicated in yellow. An arrow indicates the dibasic proteolytic cleavage site (KK/KR) at the C-terminal end of the propart.
Despite only moderate amino acid sequence similarities between the Type I and II toxins, both toxin types are predicted to form similar secondary and tertiary structures. In this study, high structural homology was predicted between the N-terminal domains of the CfTX-A, -B, and -Bt and 3d-Cry toxins, which suggests that their domains share a similar functional role. B. thuringiensis 3d-Cry toxins are a family of proteins that exhibit specific biocidal activities against insect larvae from the orders Lepidoptera (butterflies and moths), Diptera (flies and mosquitoes), and Coleoptera (beetles) (35). The structures of 3d-Cry toxins are highly similar and contain three distinct domains (e.g. see Refs. 36–39). The N-terminal domain (Domain I) forms a seven- or eight-helix bundle in which a central hydrophobic helix is surrounded by outer helices. The central and C-terminal domains (Domains II and III) contain mostly β-sheets. The molecular mechanism underlying 3d-Cry toxin activity is complex and is proposed to involve several steps, including protoxin proteolytic processing, multireceptor toxin binding, oligomerization, and pore formation, which subsequently leads to midgut cell lysis and larval death (35). Numerous studies have implicated Domain I in toxin oligomerization, membrane insertion, and pore formation, whereas Domains II and III are implicated in receptor recognition, binding, and toxin specificity (reviewed in Ref. 35). Given that the N-terminal domains of CfTX-like and 3d-Cry toxins are structurally analogous, it is therefore plausible that the CfTX-like toxins are involved in oligomerization and pore formation in cardiocytes, erythrocytes, and other susceptible cells. The formation of relatively large ring-shaped pores (12-nm inner diameter/25-nm outer diameter) in human erythrocyte cell membranes following exposure to purified CfTX isoforms (11) provides evidence that toxin oligomerization at the cell surface is integral to pore formation. Furthermore, as demonstrated here and in a previous study (3), the CfTX proteins have a propensity to oligomerize into high molecular mass quaternary structures, implying that although each toxin is individually secreted, they assemble as larger heterogeneous Type I (CfTX-1/2) or Type II (CfTX-A/B) holotoxins.
The functional role(s) of the C-terminal domain in CfTX-like toxins is less clear, but due to its structural similarity (albeit weaker) to Domain II of 3d-Cry toxins, it may be involved in receptor binding and/or toxin specificity. Earlier experimental studies found that purified CaTX-A bound to specific carbohydrates (40), which implicates these carbohydrates as potential sugar moieties in toxin-binding receptors. However, further functional studies are still necessary to establish which domain(s) are involved in receptor binding and whether the carbohydrate-binding affinity of CfTX-like toxins influences target specificity. The discovery of the truncated isoform CfTX-Bt, in which the C-terminal domain is missing, also raises questions about the functional relevance of the C-terminal domain.
Although the cubozoan toxins share a conserved structural scaffold, evolutionary diversification of toxin family members into two broad groups infers that the toxins vary in function and/or specificity. This hypothesis is supported by our data indicating that Type I and II toxin-specific antibodies are not cross-reactive (Fig. 2), presumably due to the absence of mutual epitopes, and that the cardiovascular and cytolytic activities associated with Type I and II toxins are different. Purified CfTX-A and -B (CEX Peak 2) caused relatively minor in vivo cardiovascular effects in anesthetized rats (25 μg kg−1, intravenously), whereas CfTX-1 and -2 (SEC Peak 2) caused cardiovascular collapse within 1 min at the same dose. Fractions from the other SEC peaks caused less potent cardiovascular effects than SEC Peak 2, which could be attributed to CfTX-1 and -2 contamination, as detected by Western blot analysis, and/or the presence of other unidentified cardioactive toxins. Together, these findings suggest that the Type I toxins have a higher specificity for vertebrate cardiac cells than Type II toxins and therefore are more likely to be the primary toxins involved in human envenoming. The variability of the in vivo cardiovascular effects is also consistent with previous studies on the in vitro effects of C. fleckeri fractionated venom on human cardiac myocytes (41, 42), where only SEC Peak 2 fractions (purportedly containing CfTX-1 and -2) caused rapid cell detachment and death. To date, only Type II toxins have been identified in the venoms of A. moseri, C. rastonii, and C. yamaguchii, which may also explain why C. fleckeri is exceptionally more dangerous to humans than other box jellyfish species. Nonetheless, it is also feasible that Type I toxins are expressed in the other species but at much lower levels.
In contrast to the rat studies, in vitro hemolysis assays demonstrated that Type II toxins CfTX-A and -B elicit more potent hemolytic activity (HU50 = 5 ng ml−1) than Type I toxins CfTX-1 and -2 (HU50 = 161 ng ml−1). Variability in hemolytic activity is also apparent when comparing toxin family members of other box jellyfish. In studies of A. moseri, C. rastonii, and C. yamaguchii, HU50 values for purified CaTX-A and CrTX-A (Type II toxins) were at least 10-fold lower than purified CqTX-A (Type I toxin) (5, 7, 8). However, as mentioned previously, hemolysis has never been reported as a clinical feature of human envenoming, which suggests that the cubozoan toxins preferentially target other cell types in vivo. Although the rat studies suggest that Type I toxins elicit more potent cardiovascular effects in vertebrates than Type II toxins, some experimental evidence suggests that the Type II toxins elicit more potent effects in invertebrates. For example, in earlier studies on crustaceans, researchers found that the LD50 values of Type II toxins CaTX-A and CrTX-A (5–25 μg kg−1) were lower than the Type I toxin CqTX-A (80 μg kg−1) (43). Comparative studies on the invertebrate toxicity of purified CfTX-1/2 and CfTX-A/B have yet to be published, but they would undoubtedly provide important information on the target specificity of the two toxin types. Similarly, as more toxin sequences and structural/functional data are acquired for other venomous cnidarians, our understanding of the molecular diversity and actions of this unique toxin family can be further refined.
Acknowledgments
We thank Avril Underwood (James Cook University), Andrew Hart (Monash University), Georgina Giannikopoulos (Australian Proteome Analysis Facility), and Annabel Good (IMVS) for assistance with jellyfish collection, rat bioassays, Edman sequencing, and antibody production, respectively.
This research project was funded by a grant from the Herman Slade Foundation and facilitated by access to the Australian Proteome Analysis Facility supported under the Australian Government's National Collaborative Research Infrastructure Strategy (NCRIS).
- NEB
- nematocyst extraction buffer
- b.p.m.
- beats/min
- MTS
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt
- TBST
- Tris-buffered saline plus Tween 20
- SEC
- size exclusion chromatography
- CEX
- cation exchange chromatography
- MAP
- mean arterial pressure
- HR
- heart rate
- 3d-Cry
- three-domain Cry.
REFERENCES
- 1. Currie B. J., Jacups S. P. (2005) Prospective study of Chironex fleckeri and other box jellyfish stings in the “Top End” of Australia's Northern Territory. Med. J. Aust. 183, 631–636 [DOI] [PubMed] [Google Scholar]
- 2. Lumley J., Williamson J. A., Fenner P. J., Burnett J. W., Colquhoun D. M. (1988) Fatal envenomation by Chironex fleckeri, the north Australian box jellyfish. The continuing search for lethal mechanisms. Med. J. Aust. 148, 527–534 [DOI] [PubMed] [Google Scholar]
- 3. Brinkman D. L., Burnell J. N. (2009) Biochemical and molecular characterisation of cubozoan protein toxins. Toxicon 54, 1162–1173 [DOI] [PubMed] [Google Scholar]
- 4. Brinkman D., Burnell J. (2007) Identification, cloning and sequencing of two major venom proteins from the box jellyfish, Chironex fleckeri. Toxicon 50, 850–860 [DOI] [PubMed] [Google Scholar]
- 5. Nagai H., Takuwa-Kuroda K., Nakao M., Oshiro N., Iwanaga S., Nakajima T. (2002) A novel protein toxin from the deadly box jellyfish (sea wasp, Habu-kurage) Chiropsalmus quadrigatus. Biosci. Biotechnol. Biochem. 66, 97–102 [DOI] [PubMed] [Google Scholar]
- 6. Lewis C., Bentlage B. (2009) Clarifying the identity of the Japanese Habu-kurage, Chironex yamaguchii, sp. nov. (Cnidaria: Cubozoa: Chirodropida). Zootaxa 2030, 59–65 [Google Scholar]
- 7. Nagai H., Takuwa K., Nakao M., Ito E., Miyake M., Noda M., Nakajima T. (2000) Novel proteinaceous toxins from the box jellyfish (sea wasp) Carybdea rastoni. Biochem. Biophys. Res. Commun. 275, 582–588 [DOI] [PubMed] [Google Scholar]
- 8. Nagai H., Takuwa K., Nakao M., Sakamoto B., Crow G. L., Nakajima T. (2000) Isolation and characterization of a novel protein toxin from the Hawaiian box jellyfish (sea wasp) Carybdea alata. Biochem. Biophys. Res. Commun. 275, 589–594 [DOI] [PubMed] [Google Scholar]
- 9. Gershwin L. (2005) Carybdea alata auct. and Manokia stiasnyi, reclassification to a new family with description of a new genus and two new species. Mem. Qld. Mus. 51, 501–523 [Google Scholar]
- 10. Brinkman D., Burnell J. (2008) Partial purification of cytolytic venom proteins from the box jellyfish, Chironex fleckeri. Toxicon 51, 853–863 [DOI] [PubMed] [Google Scholar]
- 11. Yanagihara A. A., Shohet R. V. (2012) Cubozoan venom-induced cardiovascular collapse is caused by hyperkalemia and prevented by zinc gluconate in mice. PLoS One 7, e51368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brinkman D. L., Aziz A., Loukas A., Potriquet J., Seymour J., Mulvenna J. (2012) Venom proteome of the box jellyfish Chironex fleckeri. PLoS One 7, e47866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bloom D. A., Burnett J. W., Alderslade P. (1998) Partial purification of box jellyfish (Chironex fleckeri) nematocyst venom isolated at the beachside. Toxicon 36, 1075–1085 [DOI] [PubMed] [Google Scholar]
- 14. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 15. Konstantakopoulos N., Isbister G. K., Seymour J. E., Hodgson W. C. (2009) A cell-based assay for screening of antidotes to, and antivenom against Chironex fleckeri (box jellyfish) venom. J. Pharmacol. Toxicol. Methods 59, 166–170 [DOI] [PubMed] [Google Scholar]
- 16. Mulvenna J., Hamilton B., Nagaraj S. H., Smyth D., Loukas A., Gorman J. J. (2009) Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol. Cell Proteomics 8, 109–121 [DOI] [PubMed] [Google Scholar]
- 17. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST. A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rice P., Longden I., Bleasby A. (2000) EMBOSS. The European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 19. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., Claverie J.-M., Gascuel O. (2008) Phylogeny.fr. Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S. (2009) MEME SUITE. Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER. A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nugent T., Ward S., Jones D. T. (2011) The MEMPACK α-helical transmembrane protein structure prediction server. Bioinformatics 27, 1438–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0. Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 24. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M., Appel R., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M., ed) pp. 571–607, Humana Press, Totowa, NJ [Google Scholar]
- 25. Steward A., Adhya S., Clarke J. (2002) Sequence conservation in Ig-like domains. The role of highly conserved proline residues in the fibronectin type III superfamily. J. Mol. Biol. 318, 935–940 [DOI] [PubMed] [Google Scholar]
- 26. Chen Y.-J., Inouye M. (2008) The intramolecular chaperone-mediated protein folding. Curr. Opin. Struct. Biol. 18, 765–770 [DOI] [PubMed] [Google Scholar]
- 27. Tang B., Nirasawa S., Kitaoka M., Marie-Claire C., Hayashi K. (2003) General function of N-terminal propeptide on assisting protein folding and inhibiting catalytic activity based on observations with a chimeric thermolysin-like protease. Biochem. Biophys. Res. Commun. 301, 1093–1098 [DOI] [PubMed] [Google Scholar]
- 28. Jolliffe N. A., Di Cola A., Marsden C. J., Lord J. M., Ceriotti A., Frigerio L., Roberts L. M. (2006) The N-terminal ricin propeptide influences the fate of ricin A-chain in tobacco protoplasts. J. Biol. Chem. 281, 23377–23385 [DOI] [PubMed] [Google Scholar]
- 29. Bravo A., Sanchez J., Kouskoura T., Crickmore N. (2002) N-terminal activation is an essential early step in the mechanism of action of the Bacillus thuringiensis Cry1Ac insecticidal toxin. J. Biol. Chem. 277, 23985–23987 [DOI] [PubMed] [Google Scholar]
- 30. Wong E. S., Hardy M. C., Wood D., Bailey T., King G. F. (2013) SVM-based prediction of propeptide cleavage sites in spider toxins identifies toxin innovation in an Australian tarantula. PLoS One 8, e66279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anderluh G., Podlesek Z., Macek P. (2000) A common motif in proparts of Cnidarian toxins and nematocyst collagens and its putative role. Biochim. Biophys. Acta 1476, 372–376 [DOI] [PubMed] [Google Scholar]
- 32. Uechi G., Toma H., Arakawa T., Sato Y. (2010) Molecular characterization on the genome structure of hemolysin toxin isoforms isolated from sea anemone Actineria villosa and Phyllodiscus semoni. Toxicon 56, 1470–1476 [DOI] [PubMed] [Google Scholar]
- 33. Frazão B., Vasconcelos V., Antunes A. (2012) Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins. An overview. Mar. Drugs 10, 1812–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buczek O., Bulaj G., Olivera B. M. (2005) Conotoxins and the posttranslational modification of secreted gene products. Cell Mol. Life Sci. 62, 3067–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pardo-López L., Soberón M., Bravo A. (2013) Bacillus thuringiensis insecticidal three-domain Cry toxins. Mode of action, insect resistance, and consequences for crop protection. FEMS Microbiol. Rev. 37, 3–22 [DOI] [PubMed] [Google Scholar]
- 36. Li J. D., Carroll J., Ellar D. J. (1991) Crystal structure of insecticidal δ-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature 353, 815–821 [DOI] [PubMed] [Google Scholar]
- 37. Grochulski P., Masson L., Borisova S., Pusztai-Carey M., Schwartz J. L., Brousseau R., Cygler M. (1995) Bacillus thuringiensis CryIA(a) insecticidal toxin. Crystal structure and channel formation. J. Mol. Biol. 254, 447–464 [DOI] [PubMed] [Google Scholar]
- 38. Galitsky N., Cody V., Wojtczak A., Ghosh D., Luft J. R., Pangborn W., English L. (2001) Structure of the insecticidal bacterial δ-endotoxin Cry3Bb1 of Bacillus thuringiensis. Acta Crystallogr. D 57, 1101–1109 [DOI] [PubMed] [Google Scholar]
- 39. Guo S., Ye S., Liu Y., Wei L., Xue J., Wu H., Song F., Zhang J., Wu X., Huang D., Rao Z. (2009) Crystal structure of Bacillus thuringiensis Cry8Ea1. An insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela. J. Struct. Biol. 168, 259–266 [DOI] [PubMed] [Google Scholar]
- 40. Chung J. J., Ratnapala L. A., Cooke I. M., Yanagihara A. A. (2001) Partial purification and characterization of a hemolysin (CAH1) from Hawaiian box jellyfish (Carybdea alata) venom. Toxicon 39, 981–990 [DOI] [PubMed] [Google Scholar]
- 41. Saggiomo S. L., Seymour J. E. (2012) Cardiotoxic effects of venom fractions from the Australian box jellyfish Chironex fleckeri on human myocardiocytes. Toxicon 60, 391–395 [DOI] [PubMed] [Google Scholar]
- 42. McClounan S., Seymour J. (2012) Venom and cnidome ontogeny of the cubomedusae Chironex fleckeri. Toxicon 60, 1335–1341 [DOI] [PubMed] [Google Scholar]
- 43. Nagai H. (2003) Recent progress in jellyfish toxin study. J. Health Sci. 49, 337–340 [Google Scholar]