Background: Eukaryotic translation elongation factor 3 (eEF3) is a ribosome binding ATPase essential for fungal protein synthesis.
Results: Mutations in the chromodomain-like insertion in an ATPase domain of eEF3 reduce ATPase activity but not ribosome binding.
Conclusion: The chromodomain-like insertion affects the ATPase activity of eEF3 and translation.
Significance: The chromodomain-like insertion of eEF3 may be targeted to develop new antifungal compounds.
Keywords: ATPases, Ribosomes, Translation, Translation Elongation Factors, Yeast, Chromodomain
Abstract
Translation elongation is mediated by ribosomes and multiple soluble factors, many of which are conserved across bacteria and eukaryotes. During elongation, eukaryotic elongation factor 1A (eEF1A; EF-Tu in bacteria) delivers aminoacylated-tRNA to the A-site of the ribosome, whereas eEF2 (EF-G in bacteria) translocates the ribosome along the mRNA. Fungal translation elongation is striking in its absolute requirement for a third factor, the ATPase eEF3. eEF3 binds close to the E-site of the ribosome and has been proposed to facilitate the removal of deacylated tRNA from the E-site. eEF3 has two ATP binding cassette (ABC) domains, the second of which carries a unique chromodomain-like insertion hypothesized to play a significant role in its binding to the ribosome. This model was tested in the current study using a mutational analysis of the Sac7d region of the chromodomain-like insertion. Specific mutations in this domain result in reduced growth rate as well as slower translation elongation. In vitro analysis demonstrates that these mutations do not affect the ability of eEF3 to interact with the ribosome. Kinetic analysis revealed a larger turnover number for ribosomes in comparison to eEF3, indicating that the partial reactions involving the ribosome are significantly faster than that of eEF3. Mutations in the chromodomain-like insertion severely compromise the ribosome stimulated ATPase of eEF3, strongly suggesting that it exerts an allosteric effect on the hydrolytic activity of eEF3. The chromodomain-like insertion is, therefore, vital to eEF3 function and may be targeted for developing novel antifungal drugs.
Introduction
Translation is a multistep process during which the information in the mRNA is decoded into polypeptides by the ribosome with the help of numerous soluble factors. During translation initiation, in a series of highly regulated steps catalyzed by the eukaryotic initiation factors (eIFs), a fully functional 80 S ribosome is assembled at the start codon of the mRNA open reading frame (1). This is followed by translation elongation, a cyclical process that involves aminoacyl-tRNA (aa-tRNA)2 delivery, peptide bond formation, and translocation, facilitated by the eukaryotic elongation factors (eEFs) (2). The canonical GTPase elongation factors that are also highly conserved in bacteria function at the ribosome and play an important role in the accuracy of gene expression by maintaining the correct reading frame of the mRNA and assuring the access of cognate aa-tRNA to the ribosomal A-site (3). eEF1A is responsible for the delivery of aa-tRNA to the ribosomal A-site (4), whereas eEF2 acts as the ribosomal translocase (5). Fungal translation elongation is unique in its absolute requirement for an ATPase factor called eEF3. In the yeast Saccharomyces cerevisiae, deletion of the single YEF3 gene encoding eEF3 is lethal, and strains harboring temperature sensitive forms of eEF3 display defects in protein synthesis and translation elongation (6–8).
eEF3 is a member of the ATP binding cassette (ABC) family of proteins, the majority of which are integral membrane transporters involved in the import or export of diverse substrates across lipid bilayers (9). They are characterized by their ability to utilize ATP binding or hydrolysis to do mechanical work and represent important pharmacological targets because they have been implicated in multidrug resistance of cancer (10) and are key players in several genetic diseases such as cystic fibrosis, adrenoleukodystrophy, and diabetes mellitus (11). eEF3 is a soluble factor lacking a transmembrane domain and has two ABC domains arranged in tandem. The intrinsic ATPase activity of eEF3 is low but strongly stimulated by the ribosome (12, 13). Mutations of highly conserved residues in either of the two ABC domains have been reported to be non-functional in vivo and to compromise ribosome-dependent ATPase activity in vitro, indicating that both domains are critical for its function (14). eEF3 has been shown to promote the release of deacylated tRNA from the post-translocation ribosomal E-site in an ATP-dependent manner and consequently stimulate A-site aa-tRNA binding mediated by eEF1A, supporting the allosteric three-site model of translating ribosomes (15). eEF3 has also been shown to bind directly to eEF1A in the presence of ADP, suggesting that upon ATP hydrolysis, eEF3 interacts with eEF1A to promote aa-tRNA delivery to the ribosome A-site (8).
Both an x-ray crystal structure of apolipoprotein S. cerevisiae eEF3 and a cryo-EM structure of the eEF3-ATP-80 S ribosome complex are available (16). The cryo-EM structure of eEF3 bound to post-translocation ribosome indicates that eEF3 binds the ribosome close to the E-site. The N-terminal HEAT (Huntingtin, Elongation factor 3, subunit A of protein phosphatase 2A, and TOR1) domain (17) makes extensive contacts with the 40 S subunit via helix 39 of the 18 S rRNA, rpS18, and rpS19. Although the ABC1 domain does not contact the ribosome, the ABC2 domain binds to the central protuberance of the 60 S subunit. Within the ABC2 domain a unique chromodomain-like insertion consisting of a five-stranded β-sheet traversed by an α-helix is positioned proximal to the E-site potentially influencing L1 stalk movement and thereby E-site tRNA affinity. Proteins containing the chromodomain fold have been reported to have molecular interactions with diverse targets including histones, DNA, and RNA (18). The eEF3 chromodomain-like insertion is predicted to interact with the 40 S subunit through rpS18 and rpS25 and the 60 S subunit via rpL11 and 5 S rRNA (16).
As an integral component of the 60 S subunit the 5 S rRNA plays a role in the maintenance of translational fidelity despite its distance from the functional centers of the ribosome (19). S. cerevisiae eEF3 residues 803–808 correspond to the DNA binding region of a homologous chromodomain protein Sac7d and were proposed to contact the 5 S rRNA based on its close proximity to the latter on the cryo-EM structure (Fig. 1A). We hypothesized that previously identified translation defects of 5 S rRNA mutants may be mediated through its interaction with the eEF3 chromodomain-like insertion. In the current study we show that this domain is important for the translation function of eEF3 and its stable expression. Although mutations in this domain do not affect the ability of eEF3 to interact with the ribosomes, they result in compromised ATPase activity, indicative of an allosteric effect on the ATP hydrolysis and exchange activity of eEF3.
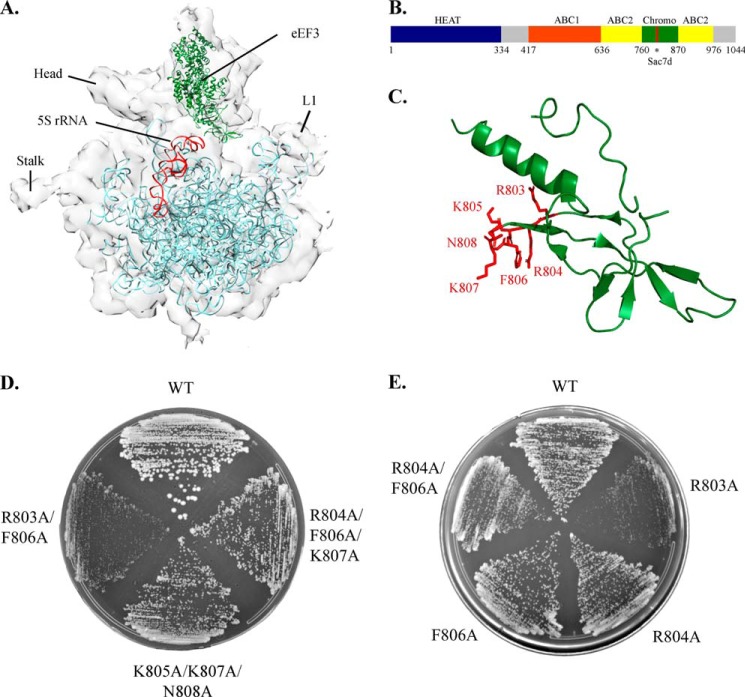
FIGURE 1.
Mutations in the eEF3 Sac7d homology region perturb growth. A, cryo-EM map of eEF3 bound post-translocation ribosome (EMD 1233) with 60 S rRNA (PDB ID 3IZF) in blue and eEF3 (PDB ID 2IX8) in green. The 5 S rRNA is highlighted in red. The image was rendered using Chimera. B, schematic diagram of eEF3 showing the domains. The Sac7d homology region (residues 803–808, marked by) lies within the chromodomain-like insertion in the ABC2 domain. C, ribbon diagram of the eEF3 chromodomain spanning residues 764–863 (green) showing the Sac7d homology residues (red). The image was rendered using PyMOL. D and E, yeast strains expressing alanine substitution mutants at indicated positions as the only form of eEF3 were streaked on YEPD plates and incubated at 30 °C. Growth was monitored after 50 h (D) for slow growing mutants and after 30 h (E) for the others.
EXPERIMENTAL PROCEDURES
Strains, Growth, and Drug Sensitivity
S. cerevisiae strains and their genotypes used in this study are listed in Table 1. Escherichia coli DH5α cells were used for plasmid preparation. Standard protocols were followed for yeast and bacterial cell growth and genetic manipulations (20). Yeast cells were grown in either YEPD (1% Bacto-yeast extract, 2% peptone, 2% dextrose) or defined synthetic complete media (C or C−) supplemented with 2% dextrose. Yeast strains were transformed by the lithium acetate method (21). Growth was assayed by streaking on YEPD plates. The doubling times in liquid culture (mean ± S.E.) was determined by monitoring the A600 of at least two independent colonies of each strain cultured at 30 °C in YEPD to mid-log phase, diluted to A600 of 0.05, and grown in triplicate in a 96-well microtiter assay plates incubated with shaking at 30 °C. Sensitivity to paromomycin was assayed by measuring the zone of inhibition of growth as previously described (22).
TABLE 1.
List of yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| TKY1617 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yef3::HIS3 pYEF3 URA3 CEN | This work |
| TKY1653 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yef3::HIS3 pHis6YEF3 TRP1 CEN | This work |
| TKY1654 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yef3::HIS3 pHis6yef3 (R803A) TRP1 CEN | This work |
| TKY1655 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yef3::HIS3 pHis6yef3 (R803A/F806A) TRP1 CEN | This work |
| TKY1656 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yef3::HIS3 pHis6yef3 (K805A/K807A/N808A) TRP1 CEN | This work |
| TKY1660 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yef3::HIS3 pHis6yef3 (R804A) TRP1 CEN | This work |
| TKY1661 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yef3::HIS3 pHis6yef3 (F806A) TRP1 CEN | This work |
| TKY1662 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yef3::HIS3 pHis6yef3 (R804A/F806A) TRP1 CEN | This work |
| TKY1695 | MATα leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 yef3::HIS3 pHis6yef3 (R804A/F806A/K807A) TRP1 CEN | This work |
| JD1370 | MATa trp1 ura3 leu2 pep4::HIS3 nuc1::LEU2 | Ref. 31 |
Translation Assays
For in vivo [35S]methionine incorporation assays, yeast strains were grown in liquid cultures (100 ml) in C-Met at 30 °C to mid-log phase and assayed as described (22). All time points were analyzed in triplicate. Yeast polyribosome profile analysis was performed as previously described with a modified lysis buffer containing 20 mm Tris-Cl, pH 7.5), 50 mm KCl, 10 mm MgCl2, 1 mm DTT, and 1 mm PMSF (23).
Isolation of eEF3 Chromodomain Mutants
Plasmid pTKB1115 expressing an N terminally His6-tagged YEF3 on a CEN TRP1 plasmid served as a template to generate the various eEF3 chromodomain-like insertion mutants using QuikChange site-directed mutagenesis (Stratagene) as per the manufacturer's recommendations. All mutations were confirmed by sequencing. The mutant plasmids were transformed into TKY1617 and counter-selected on 5-fluoroorotic acid. The expression of the tagged eEF3 mutants was confirmed by Western blot using mouse anti-His6 antibody (BD Pharmingen) as per the manufacturer's recommendations.
Purification of His6-eEF3 and Reconstituted 80 S Ribosomes
His6-tagged eEF3, R803A, F806A, and R803A/F806A eEF3 were purified from the respective yeast strains on a HisTrap Column (GE Healthcare). Total yeast extracts were clarified and loaded on the column in buffer A (50 mm K3PO4, pH 8.0, 1 m KCl, 1% Tween 20, 1 mm DTT, and 0.2 mm PMSF) with 10 mm imidazole. The protein was eluted with buffer A plus 120 mm imidazole and dialyzed into buffer B (20 mm Tris, pH 7.5, 100 mm KCl, 0.1 mm EDTA, 1 mm DTT, and 0.2 mm PMSF) plus 10% glycerol followed by gel filtration using a Superdex 200 10/300 GL column (GE Healthcare) with buffer B plus 20% glycerol. Reconstituted 80 S ribosomes were purified from strain JD1370 as previously described with the following minor modification (24). A 5–25% sucrose gradient was used to separate the ribosomal subunits after puromycin treatment of the salt-washed ribosomes.
Ribosome Binding Assay
The ribosome binding assay was performed based on the method described by Moreno et al. (25) with minor modifications. 50-μl reactions containing specified amounts of protein and reconstituted 80 S ribosomes in binding buffer (20 mm Tris-HCl, pH 7.5, 50 mm ammonium acetate, 10 mm magnesium acetate, and 2 mm DTT) plus 1 mm ATP were incubated for 5 min at room temperature, chilled on ice for 5 min, layered on top of a 200-μl sucrose cushion (20% sucrose in binding buffer), and centrifuged at 80,000 rpm for 1 h at 4 °C in a S80-AT2 (Sorvall) rotor. The pellet (bound fraction) and supernatant (free fraction) were resuspended in Laemmli loading buffer and subjected to SDS-PAGE followed by staining with Brilliant Blue R (Sigma).
ATP Hydrolysis Assay and Data Fitting
ATP hydrolysis was performed using the PiColorLock Gold Phosphate Detection System (Innova Biosciences) as per the manufacturer's instruction with the following minor modifications. The reactions were carried out at room temperature for 30 min and contained 50 mm Tris, pH 7.5, 5 mm MgCl2, and 1 mm ATP unless otherwise specified. Kinetic constants were determined by plotting the curves using Sigmaplot and are expressed as the mean ± S.E. of at least three independent experiments. Kinetic schemes for ribosome-stimulated ATPase reactions were analyzed by simultaneously fitting the data from multiple experiments to different mechanistic models using KinTek Global Kinetic Explorer (26, 27). The average of at least three independent traces per experiment was used for the global-fitting analysis.
RESULTS
The Chromodomain-like Insertion in the ABC2 Domain Is Essential for eEF3 Function
To understand the functional role of the chromodomain-like insertion in the ABC2 domain of eEF3 (Fig. 1B), regions likely to play an important role in the ribosome interaction were identified based on sequence analysis and the cryo EM model of the eEF3-bound ribosome (Fig. 1A) (16). Residues 803–808, homologous to the DNA binding domain of the Sac7d protein of Sulfolobus acidocaldarius were proposed to interact with 5 S rRNA on the ribosome (Fig. 1C) (16). To test the significance of this region, a series of single and multiple alanine substitution mutants was prepared (Table 2) and transformed into a yeast strain expressing wild type eEF3 from a URA3 plasmid as the only copy. Mutants able to support growth in the absence of wild type eEF3 were identified by counter-selecting on 5-fluoroorotic acid plates.
TABLE 2.
Growth rate and paromomycin sensitivity of eEF3 chromodomain mutants
The eEF3 mutants are named after the amino acid residues substituted with alanine followed by their position in subscript. Column 2 shows the relative positions of the mutated residues within residues 803–808. Sensitivity to paromomycin was scored with respect to wild type as follows: +, identical to wild type; ++, slightly sensitive; +++, highly sensitive. NA denotes data not available due to inviability of the corresponding mutant.
| eEF3 mutants | Residues 803–808 | Doubling time | Paromomycin sensitivity |
|---|---|---|---|
| h | |||
| Wild type | R R K F K N | 1.58 ± 0.06 | + |
| R803A/R804A/K805A/F806A/K807A/N808A | A A A A A A | Inviable | NA |
| R803A/R804A/F806A | A A + A + + | Inviable | NA |
| R803A/K805A/N808A | A + A + + A | Inviable | NA |
| K805A/K807A/N808A | + + A + A A | 2.70 ± 0.03 | ++ |
| R804A/F806A/K807A | + A + A A + | 2.63 ± 0.04 | ++ |
| R803A/R804A | A A + + + + | Inviable | NA |
| R803A/F806A | A + + A + + | 4.01 ± 0.29 | +++ |
| R804A/F806A | + A + A + + | 1.87 ± 0.01 | + |
| R803A | A + + + + + | 2.13 ± 0.06 | + |
| R804A | + A + + + + | 1.81 ± 0.11 | + |
| F806A | + + + A + + | 1.67 ± 0.02 | + |
Block substitution of the six residues constituting the Sac7d homology region (803–808) with alanine resulted in an inviable mutant, supporting the notion that the chromodomain-like insertion is integral to the function of eEF3. Of the four triple mutants tested, R803A/R804A/F806A and R803A/K805A/N808A were also inviable, whereas the corresponding complementary mutants K805A/K807A/N808A and R804A/F806A/K807A exhibited a slow growth phenotype (Fig. 1D). The R803A/R804A mutant represented the minimal mutation required to completely inactivate eEF3 function in vivo.
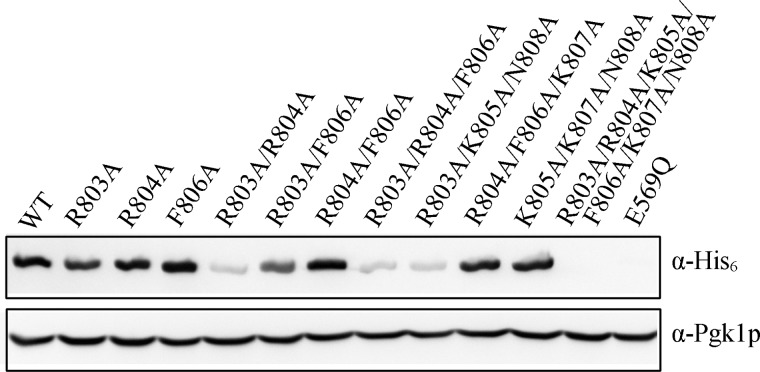
Mutations in essential genes may be inviable in vivo if either the steady state expression level of the mutant form is inadequate or if the protein is functionally impaired despite being expressed. His6-tagged chromodomain mutants were coexpressed in a strain expressing untagged wild type eEF3, and the levels of mutant protein were analyzed by a Western blot with anti-His6 antibody (Fig. 2). All the eEF3 mutants that were unable to support growth showed little to no expression in this system. This observation was not limited to mutations in the chromodomain-like insertion, as the expression of E569Q, an inviable mutant in the conserved Walker B motif of the ABC1 domain of eEF3 (unpublished observation), was also undetectable, ruling out a domain-specific effect. Protein expression levels, therefore, accounted for the inviability of the eEF3 mutants under study.
FIGURE 2.
Inviable mutations in the eEF3 Sac7d homology region reduce steady state protein levels. His6-eEF3 wild type and mutant plasmids were coexpressed along with untagged wild type eEF3, and the level of mutant proteins were analyzed by Western blot using anti-His6 antibody. E569Q is an eEF3 mutant in the ATP binding domain that also results in inviability. Pgk1p served as the loading control.
Of all viable mutants, the R803A/F806A double mutant exhibits the slowest growth (Fig. 1E), whereas the R803A single mutant was the only single mutant with a significant growth defect. In comparison, mutations at R804A and F806A alone or in combination have minimal effects on growth. Mutant strains were also tested for sensitivity toward paromomycin, a translation elongation inhibitor. Increased sensitivity to paromomycin was directly correlated with increased doubling times (Table 2), indicating that decreased growth rates of the mutants shown in Fig. 1, D and E, may be caused by defective translation elongation.
eEF3 R803A/F806A Has Compromised Translation Elongation
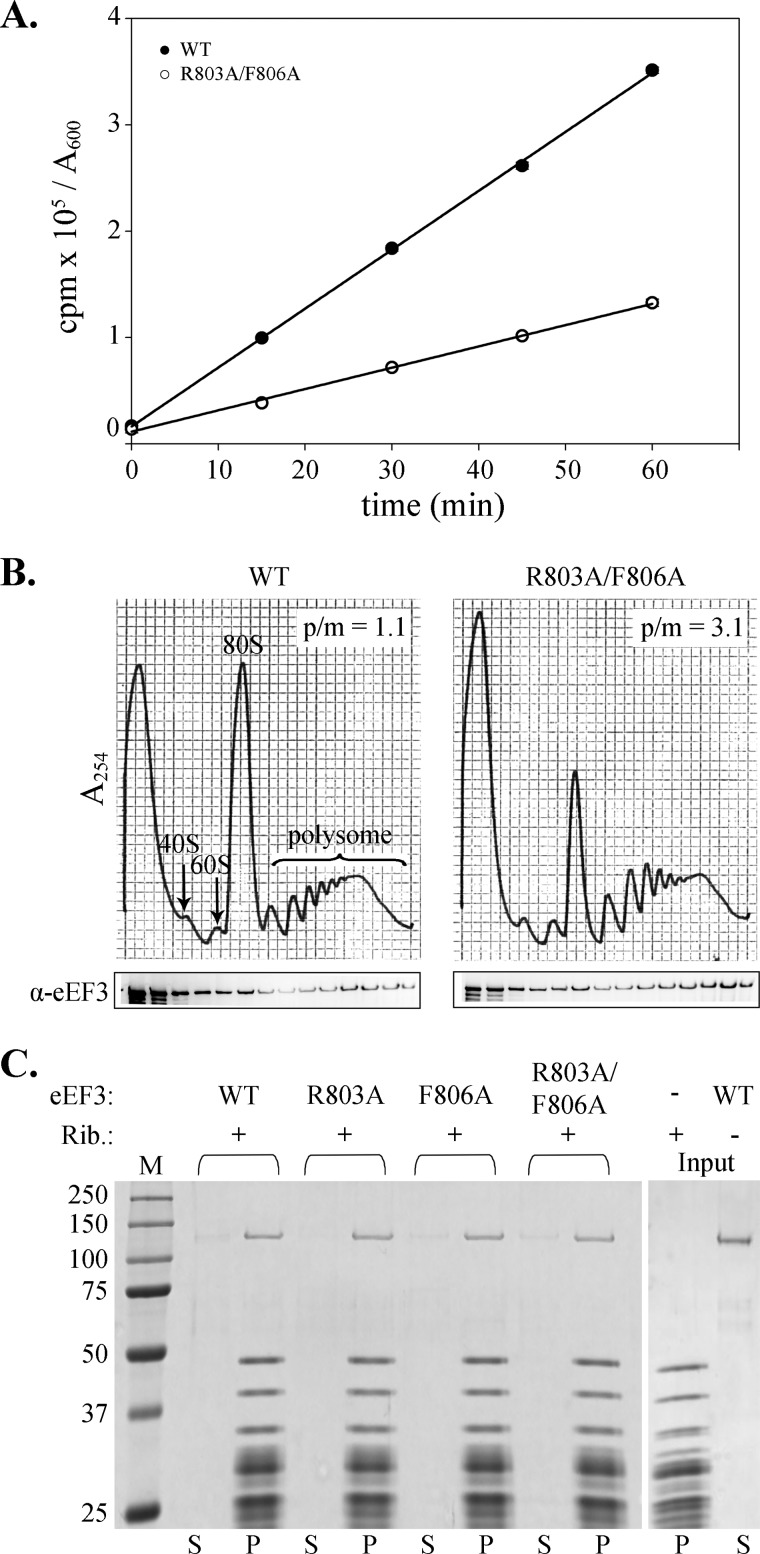
To determine how the mutations in the chromodomain-like insertion of eEF3 affects protein synthesis, [35S]methionine incorporation was determined in exponentially growing cells expressing either wild type eEF3 or the R803A/F806A mutant, the sickest of all mutants. Total protein synthesis by the slow-growing R803A/F806A strain was only 35% that of the wild type strain (Fig. 3A), which correlates well with the dramatically reduced growth rate observed in this mutant. To identify whether a specific step of translation is affected, polyribosome profiles of both wild type and R803A/F806A strains were analyzed. In the absence of cycloheximide, the R803A/F806A strain shows a reduced 80 S peak and a concurrent increase in polyribosome peaks (Fig. 3B) indicative of an elongation defect. The polyribosome to monosome ratio, calculated as the area under the curve, shows a roughly 3-fold difference between the wild type and mutant strain extracts. To test whether the defective translation elongation could be explained by a reduced binding of the mutant to the ribosome, the distribution of eEF3 across the polyribosome fractions was analyzed using Western blot analysis. Compared with a wild type strain, the relative eEF3 distribution between polyribosome and monosome fractions showed a 2.5-fold enrichment in extracts from the R803A/F806A strain, reflecting the increase in the polyribosomes observed in R803A/F806A.
FIGURE 3.
Chromodomain mutations compromise translation elongation without affecting ribosome binding. A, exponentially growing wild type (● or R803A/F806A (○) eEF3 strains were monitored for total translation by measuring [35S]methionine incorporation for the indicated times. [35S]Methionine incorporation is expressed as cpm/A600 unit of cells. B, polyribosome extracts of wild type or R803A/F806A eEF3 strains were prepared in the absence of cycloheximide and analyzed by 7–47% sucrose density gradient centrifugation. Fractions were probed for the distribution of eEF3 by Western blot using anti-eEF3 antibody. C, eEF3 binding to ribosomes (Rib) was monitored in vitro by pelleting a reaction mixture containing 25 pm each of reconstituted 80 S ribosomes and various eEF3 mutants through a 20% sucrose cushion. The pellet (P, 20% of total) and unbound supernatant (S, 20% of total) were separated by SDS-PAGE and analyzed by Brilliant Blue staining.
Chromodomain-like Insertion Mutants Do Not Affect Ribosome Binding in Vitro
Because eEF3 is a ribosome-associated ATPase, mutations that reduce the elongation activity of eEF3 could affect its ribosome binding, ATP binding, or the ribosome-stimulated ATPase activity. To distinguish between these possibilities, the partial reactions of eEF3 were analyzed in vitro. In addition to the double mutant R803A/F806A, single mutants R803A and F806A were also analyzed to determine whether these mutations had a cumulative effect on the overall phenotype of R803A/F806A. The ability of eEF3 to bind 80 S ribosomes reconstituted from purified subunits was determined by sucrose cushion analysis (Fig. 3C). In the absence of ribosomes, eEF3 remains in the supernatant as expected after pelleting through a 20% sucrose cushion. In the presence of ribosomes, ∼80% of eEF3 pelleted with the ribosomes, demonstrating significant and stable binding between eEF3 and the ribosomes. No discernible differences were observed among the wild type and R803A, F806A, or R803A/F806A eEF3 mutants, which indicates that the mutations in the chromodomain-like insertion do not significantly affect the ability of eEF3 to interact with the 80 S ribosomes in vitro.
Chromodomain-like Insertion Mutants Have Reduced ATPase Activity
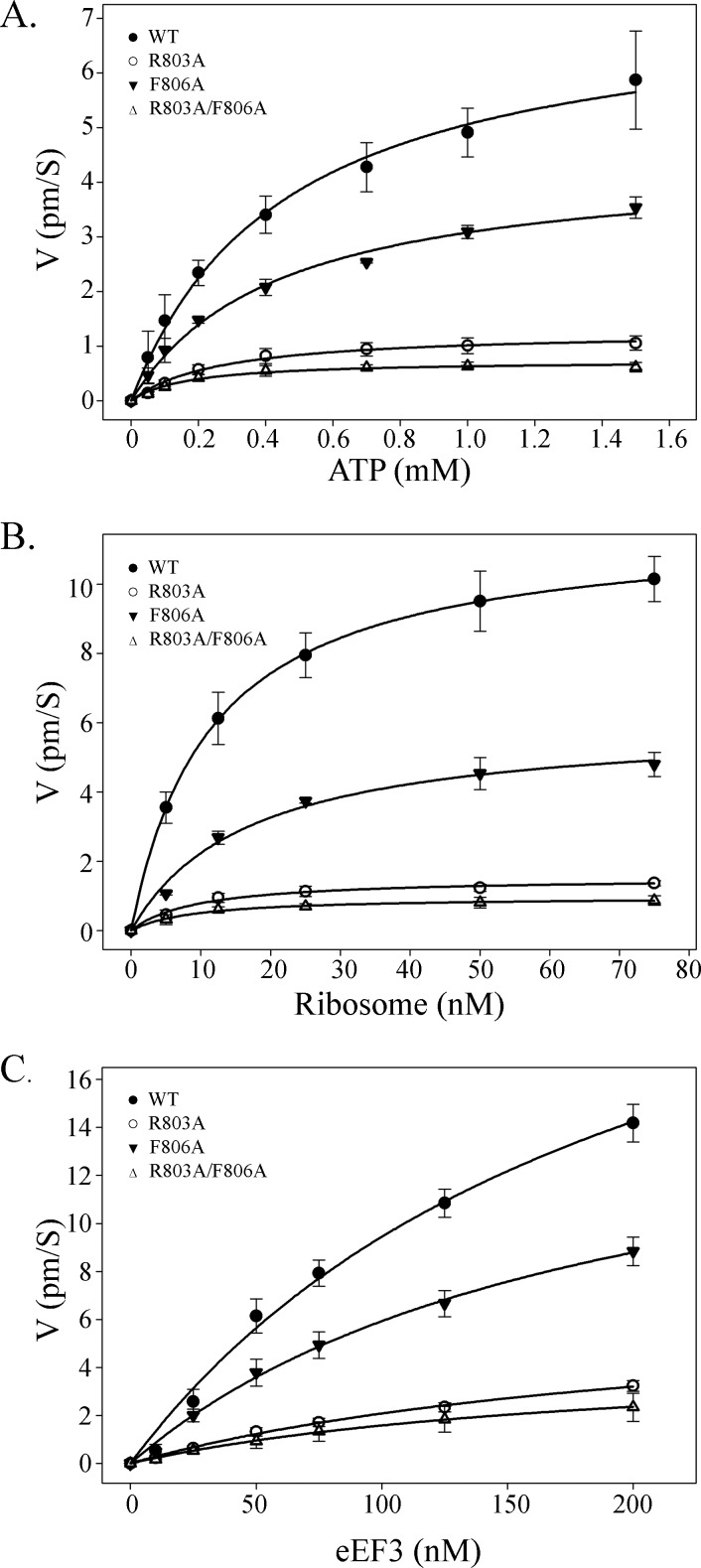
In the absence of any observable defects in ribosome binding by the various eEF3 mutants, we sought to determine whether the phenotypic effects of the mutants could be explained by differences in their ribosome-stimulated ATPase activities. Although the chromodomain-like insertion is not modeled to directly bind to ATP, it could potentially exert an indirect effect on the ATP binding and hydrolysis functions of eEF3. To test what partial reactions may be influenced by the chromodomain-like insertion, the ribosome-stimulated ATPase activities of the wild type as well as the R803A, F806A, and R803A/F806A eEF3 mutants were analyzed under conditions of varying concentrations of ATP, ribosomes, or eEF3. First, the dependence of ribosome-stimulated ATPase activity of various eEF3 mutants on varying concentrations of ATP was determined (Fig. 4A). Wild type eEF3 had the highest ATPase activity followed by F806A, R803A, and R803A/F806A mutants. The ATPase activity response to increasing concentrations of the substrate ATP followed a Michaelis-Menten curve. For each eEF3 mutant the curve was fitted to an equation describing ligand binding with one-site saturation using Sigmaplot, and the Km·ATP was determined. Under the conditions tested, the Km·ATP was found to be highest for the wild type at 453.9 μm followed by F806A with 433.6 μm, R803A with 259.8 μm, and R803A/F806A with 168.6 μm (Table 3).
FIGURE 4.
Mutations in eEF3 chromodomain-like insertion severely compromise ribosome-stimulated ATPase activity. The ribosome stimulation of ATPase activity by wild type (●), R803A (○), F806A (▾), or R803A/F806A (▵) mutants were analyzed by measuring the amount of ATP hydrolysis under varying conditions. The variable was the amount of ATP (0–1.5 mm) with 10 nm ribosomes and 50 nm wild type or mutant eEF3 (A), ribosomes (0–75 nm) with 50 nm wild type or mutant eEF3 and 1 mm ATP (B), or eEF3 (0–200 nm) with 10 nm ribosomes and 1 mm ATP (C) in the reaction. A graph of the reaction velocity (pm ATP hydrolyzed/s) was plotted against increasing concentrations of the indicated variable while keeping the other two constant.
TABLE 3.
Summary of kinetic parameters of ribosome stimulated ATPase activity of eEF3
| Variables | Kinetic parameter | Wild type | R803A | F806A | R803A/F806A |
|---|---|---|---|---|---|
| ATP, 0–1.5 mm | |||||
| Ribosome, 10 nm | Km·ATP (mm) | 0.45 ± 0.08 | 0.26 ± 0.05 | 0.43 ± 0.04 | 0.17 ± 0.03 |
| eEF3, 50 nm | |||||
| ATP, 1 mm | Km·Ribosome (nm) | 11.46 ± 1.42 | 9.39 ± 1.74 | 16.85 ± 2.22 | 8.97 ± 2.34 |
| Ribosome, 0–75 nm | |||||
| eEF3, 50 nM | Kcat·eEF3 (min−1) | 70.11 ± 2.52 | 9.18 ± 0.46 | 36.11 ± 1.61 | 5.81 ± 0.40 |
| ATP, 1 mm | Km·eEF3 (nm) | 206.9 ± 27.0 | 230.7 ± 31.9 | 187.1 ± 25.1 | 193.9 ± 73.9 |
| Ribosome, 10 nm | |||||
| eEF3, 0–200 nm | Kcat·Ribosome (min−1) | 869.1 ± 69.8 | 206.4 ± 18.2 | 510.5 ± 40.8 | 139.2 ± 32.0 |
To characterize the requirement of the ribosome in the reaction, the ATPase activity response of the different eEF3 mutants (50 nm each) in response to increasing concentrations of ribosomes in the presence of 1 mm ATP were determined and plotted as before (Fig. 4B). The intrinsic ATPase activity of either the wild type or any of the mutant eEF3 tested was below the detection range of the assay. The ribosome-stimulated processivity of eEF3 for ATP in the reaction is given by the turnover number (Kcat.eEF3). The turnover number as determined from the curves indicate that wild type eEF3 has maximum activity with a Kcat of 70 min−1. eEF3 F806A had a slower ATPase activity with a measured Kcat of 36 min−1. Both R803A and R803A/F806A mutants had severely compromised activities as reflected by turnover numbers of 9 and 6 min−1, respectively. The Km·ribosome for the reactions are all in the similar range, which indicates that wild type eEF3 as well as the various mutants showed similar dependence on the ribosome for the stimulation of the ATPase activity of eEF3. It also supports the observation that the reduced activity of chromodomain-like insertion mutants is not due to significantly altered ribosome interaction. Finally, complementary experiments were also carried out wherein the amount of eEF3 was varied in the presence of 10 nm ribosome and 1 mm ATP (Fig. 4C). The curves obtained in these reactions barely started to plateau even at 200 nm eEF3, which points to eEF3 being a limiting factor even at these high concentrations relative to the ribosome. The Kcat·ribosomes, which is a measure of eEF3 turnover by the ribosomes, estimated from the curves were an order of magnitude larger than the corresponding Kcat.eEF3 values (Table 3) at 869 min−1 for wild type, 511 min−1 for F806A, 206 min−1 for R803A, and 139 min−1 for R803A/F806A. A significantly higher processivity for ribosomes compared with eEF3 indicates that the ribosome-associated step(s) of the ATPase reaction is significantly faster than the remaining partial reactions, which are involved in the recycling of eEF3 to form the next active ribosome-eEF3-ATP complex.
DISCUSSION
The absolute requirement for eEF3 in translation elongation by fungal ribosomes, an otherwise highly conserved process from bacteria to mammals, remains an intriguing question with the potential to be exploited to target pathogenic fungi (28). In the present investigation we explored the role of a unique chromodomain-like insertion in the ABC2 domain of eEF3, which positioned proximal to the 5 S rRNA in the ribosome. The eEF3 chromodomain has previously been suggested to undergo a conformational switch upon ribosome binding and help facilitate its function as a dynamic E-site factor (16).
We analyzed the functional implications of the chromodomain-like insertion on the interaction of eEF3 with the ribosome and its associated ATPase activity. Using mutational analysis we show that this domain is integral to the function of eEF3 (Table 2), as a six-amino acid alanine block substitution of the Sac7d homology region within the chromodomain-like insertion abrogated its function in vivo. It is interesting to note that all the mutations that fail to complement the function of the wild type eEF3 protein are poorly expressed (Fig. 2). Although we cannot rule out reduced mRNA levels as a potential cause for this down-regulation, it seems unlikely given the specificity for only some mutants. Most likely this relates to protein turnover either triggered by defective folding or functional inactivity, implicating a close involvement of protein surveillance and turnover machinery with the translational apparatus.
Several viable mutants in the chromodomain-like insertion of eEF3 were isolated, of which R803A/F806A shows the most severe growth defect. In agreement with previous reports of eEF3 mutants in the ABC domains or the hinge between the domains affecting translation (7, 14), R803A/F806A was also shown to have a significant defect in total protein synthesis (Fig. 3A) due to reduced translation elongation as evidenced by the accumulation of polyribosomes in the cell (Fig. 3B). Thus the chromodomain-like insertion is an integral part of eEF3 required not only for stable expression but also for its function in translation elongation. A potential explanation for reduced translation elongation is that changes in this domain may affect the ribosome binding by eEF3. However, both the distribution of eEF3 across the polyribosomes (Fig. 3B) as well as the steady state binding of eEF3 mutants to reconstituted 80 S ribosomes (Fig. 3C) argues that these mutations do not alter the ability of eEF3 to bind to the ribosomes.
To explain the reduced translation, we tested the ribosome-stimulated ATPase activity of the various mutants in vitro. The Km·ATP values obtained are in the similar range as the previously reported values for eEF3 from Saccharomyces carlsbergensis at 120 μm (29). The ATPase activity of the different mutants correlated with their growth rate, with wild type eEF3 exhibiting the highest ATPase stimulation by the ribosomes. To explore the probable mechanism of the ribosome stimulation of the eEF3 ATPase activity, the ATP hydrolysis data from the multiple assays, differing in concentrations of ATP, eEF3, or ribosomes, were globally fit into minimal mechanistic models using KinTek Global Kinetic Explorer. The two models that best fit our data differed in the initial binding of eEF3 to either the ribosome (ribosome model) or ATP (ATP model), and the kinetic parameters estimated for these models are given in Table 4. According to the ribosome model, eEF3 binds to the ribosome in the apo-state. After this, ATP binding to eEF3 occurs on the ribosome as has been previously proposed (16). This is identical to the published switch model for ABC transporters wherein substrate binding triggers ATP binding (30). Consistent with this model, standard filter binding assays, which can measure radiolabeled nucleotide binding for other elongation factors, failed to show ATP binding by isolated eEF3 (data not shown). Once the ribosome-EF3-ATP ternary complex is formed, the ribosome stimulates the ATP hydrolysis in a fast step and releases eEF3-ADP, which is followed by the dissociation of ADP in a significantly slower rate. The faster rate for ribosome-stimulated ATP hydrolysis step is supported by the processivity (as measured by Kcat) of the ribosome being an order of magnitude higher than that of eEF3. Mutations in the chromodomain-like insertion seem to adversely affect the hydrolysis and, to a larger extent, the ADP release steps of the reaction.
TABLE 4.
Predicted rate constants from global fitting of ATPase activity of eEF3 into multiple models (R, ribosome; E, eEF3; A, ATP; D, ADP)
| Reaction mechanism models | Predicted kinetic constants |
|||
|---|---|---|---|---|
| Wild type | R803A | F806A | R803A/F806A | |
| Ribosome model | ||||
| R + E ↔ R·E, Kdl | 1.86 μm | 2.23 μm | 1.97 μm | 2.27 μm |
| R·E + A↔ R·E·A, Kd2 | 2.78 μm | 3.15 μm | 3.23 μm | 3.91 μm |
| R·E·A → R + E·D + Pi, k3 | 753 s−1 | 162 s−1 | 474 s−1 | 123 s−1 |
| E·D → E + D, k4 | 75 s−1 | 8.8 s−1 | 32.4 s−1 | 5.6 s−1 |
| ATP model | ||||
| E + A ↔ E·A, Kdl | 1.29 mm | 0.90 mm | 1.05 mm | 0.96 mm |
| E·A + R↔ R·E·A, Kd2 | 0.024 μm | 0.024 μm | 0.028 μm | 0.024 μm |
| R·E·A → R + E·D + Pi, k3 | 722 s−1 | 157 s−1 | 455 s−1 | 129 s−1 |
| E·D → E + D, k4 | 94.5 s−1 | 10.5 s−1 | 42 s−1 | 6.5 s−1 |
An alternate model (the ATP model; Table 4) that fits the data equally well differs in the order of addition of the various components to form the active ribosome-eEF3-ATP species. In contrast to the ribosome model, ATP binding by eEF3 precedes the ribosome binding step in the ATP model. However this model predicts that the eEF3 binding to ATP is an unfavorable reaction with the dissociation constant in the low millimolar range, which is unlikely in the cellular context. In contrast, the ribosome model predicts a strong interaction between eEF3 and ribosome as well as the subsequent ATP binding step as supported by the low micromolar range dissociation constants. Moreover, eEF3 can bind to reconstituted ribosomes in vitro even in the absence of ATP (data not shown), suggesting that the ribosome model might reflect the cellular conditions better than the ATP model.
Our work supports the hypothesis that ATPase activity of eEF3 is important for its biological activity and that the Sac7d homology region of the chromodomain-like insertion plays a key role in its function. The chromodomain-like insertion most likely exerts an allosteric effect on the nucleotide binding pocket of eEF3, affecting hydrolysis as well as subsequent release of ADP, as predicted by the models. A multiple sequence alignment of eEF3 sequences from across different representative fungi shows that this domain shares significant similarity. An analysis of sequence conservation within 10 residue stretches in this domain suggests that the Sac7d homology loop had the highest similarity along with residues 836–845. The latter stretch interestingly lies close to the Sac7d loop in the crystal structure of eEF3, strongly supporting a functional role in the activity of eEF3. Unlike the ABC domains, which are highly conserved and ubiquitous, the chromodomain-like insertion provides a uniquely specific site for the design of new broad spectrum anti-fungal drugs. Although the novel features of fungal ribosomes that necessitate the requirement for eEF3 in translation elongation remain unclear, future work may target fungal translation by modifying the ATPase activity of eEF3 through its unique chromodomain-like insertion.
Acknowledgments
We thank Dr. Smita Patel for assistance with the kinetic analysis of eEF3 activity and thoughtful comments on the models. We also thank Dr. Paul Copeland and the members of the Kinzy laboratory for critical reading of the manuscript, helpful comments, and discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM57483.
- aa-tRNA
- aminoacyl-tRNA
- eEF3
- eukaryotic translation elongation factor 3
- ABC
- ATP binding cassette.
REFERENCES
- 1. Hinnebusch A. G., Dever T. E., Asano K. (2007) in Translational Control in Biology and Medicine (Sonenberg N., Hershey J. W. B., Mathews M. B., eds.) pp. 225–268, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Merrick W. C., Nyborg J. (2000) in Translational Control of Gene Expression (Sonenberg N., Hershey J. W. B., Mathews M. B., eds.) pp. 89–126, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 3. Valente L., Kinzy T. G. (2003) Yeast as a sensor of factors affecting the accuracy of protein synthesis. Cell. Mol. Life Sci. 60, 2115–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carvalho M. D., Carvalho J. F., Merrick W. C. (1984) Biological characterization of various forms of elongation factor 1 from rabbit reticulocytes. Arch. Biochem. Biophys. 234, 603–611 [DOI] [PubMed] [Google Scholar]
- 5. Skogerson L., Moldave K. (1968) Characterization of the interaction of aminoacyltransferase II with ribosomes. Binding of transferase II and translocation of peptidyl transfer ribonucleic acid. J. Biol. Chem. 243, 5354–5360 [PubMed] [Google Scholar]
- 6. Qin S. L., Xie A. G., Bonato M. C., McLaughlin C. S. (1990) Sequence analysis of the translational elongation factor 3 from Saccharomyces cerevisiae. J. Biol. Chem. 265, 1903–1912 [PubMed] [Google Scholar]
- 7. Anand M., Chakraburtty K., Marton M. J., Hinnebusch A. G., Kinzy T. G. (2003) Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J. Biol. Chem. 278, 6985–6991 [DOI] [PubMed] [Google Scholar]
- 8. Anand M., Balar B., Ulloque R., Gross S. R., Kinzy T. G. (2006) Domain and nucleotide dependence of the interaction between Saccharomyces cerevisiae translation elongation factors 3 and 1A. J. Biol. Chem. 281, 32318–32326 [DOI] [PubMed] [Google Scholar]
- 9. Hollenstein K., Dawson R. J., Locher K. P. (2007) Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418 [DOI] [PubMed] [Google Scholar]
- 10. Deeley R. G., Westlake C., Cole S. P. (2006) Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol. Rev. 86, 849–899 [DOI] [PubMed] [Google Scholar]
- 11. Decottignies A., Goffeau A. (1997) Complete inventory of the yeast ABC proteins. Nat. Genet. 15, 137–145 [DOI] [PubMed] [Google Scholar]
- 12. Dasmahapatra B., Skogerson L., Chakraburtty K. (1981) Protein synthesis in yeast II. Purification and properties of elongation factor 1 from Saccharomyces cerevisiae. J. Biol. Chem. 256, 10005–10011 [PubMed] [Google Scholar]
- 13. Uritani M., Miyazaki M. (1988) Characterization of the ATPase and GTPase activities of the elongation factor 3 (EF-3) from yeasts. J. Biochem. 103, 522–530 [DOI] [PubMed] [Google Scholar]
- 14. Yang H., Hamada K., Terashima H., Izuta M., Yamaguchi-Sihta E., Kondoh O., Satoh H., Miyazaki M., Arisawa M., Miyamoto C., Kitada K. (1996) A point mutation within each of two ATP-binding motifs inactivates the functions of elongation factor 3. Biochim. Biophys. Acta 1310, 303–308 [DOI] [PubMed] [Google Scholar]
- 15. Triana-Alonso F. J., Chakraburtty K., Nierhaus K. H. (1995) The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem. 270, 20473–20478 [DOI] [PubMed] [Google Scholar]
- 16. Andersen C. B., Becker T., Blau M., Anand M., Halic M., Balar B., Mielke T., Boesen T., Pedersen J. S., Spahn C. M., Kinzy T. G., Andersen G. R., Beckmann R. (2006) Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 443, 663–668 [DOI] [PubMed] [Google Scholar]
- 17. Andrade M. A., Bork P. (1995) HEAT repeats in the Huntington's disease protein. Nat. Genet. 11, 115–116 [DOI] [PubMed] [Google Scholar]
- 18. Brehm A., Tufteland K. R., Aasland R., Becker P. B. (2004) The many colours of chromodomains. Bioessays 26, 133–140 [DOI] [PubMed] [Google Scholar]
- 19. Smith M. W., Meskauskas A., Wang P., Sergiev P. V., Dinman J. D. (2001) Saturation mutagenesis of 5 S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 8264–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burke D., Dawson D., Stearns T. (2000) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Ito H., Fukuda Y., Murata K., Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carr-Schmid A., Valente L., Loik V. I., Williams T., Starita L. M., Kinzy T. G. (1999) Mutations in elongation factor 1β, a guanine nucleotide exchange factor, enhance translational fidelity. Mol. Cell. Biol. 19, 5257–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Esposito A. M., Mateyak M., He D., Lewis M., Sasikumar A. N., Hutton J., Copeland P. R., Kinzy T. G. (2010) Eukaryotic polyribosome profile analysis. J. Vis. Exp. 10.3791/1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shin B. S., Dever T. E. (2007) Molecular genetic structure-function analysis of translation initiation factor eIF5B. Methods Enzymol. 429, 185–201 [DOI] [PubMed] [Google Scholar]
- 25. Moreno J. M., Kildsgaard J., Siwanowicz I., Mortensen K. K., Sperling-Petersen H. U. (1998) Binding of Escherichia coli initiation factor IF2 to 30S ribosomal subunits. A functional role for the N terminus of the factor. Biochem. Biophys. Res. Commun. 252, 465–471 [DOI] [PubMed] [Google Scholar]
- 26. Johnson K. A. (2009) Fitting enzyme kinetic data with KinTek Global Kinetic Explorer. Methods Enzymol. 467, 601–626 [DOI] [PubMed] [Google Scholar]
- 27. Johnson K. A., Simpson Z. B., Blom T. (2009) Global kinetic explorer. A new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 387, 20–29 [DOI] [PubMed] [Google Scholar]
- 28. Sturtevant J. (2002) Translation elongation-3-like factors. Are they rational antifungal targets? Expert. Opin. Ther. Targets 6, 545–553 [DOI] [PubMed] [Google Scholar]
- 29. Miyazaki M., Uritani M., Kagiyama H. (1988) The yeast peptide elongation factor 3 (EF-3) carries an active site for ATP hydrolysis which can interact with various nucleoside triphosphates in the absence of ribosomes. J. Biochem. 104, 445–450 [DOI] [PubMed] [Google Scholar]
- 30. Higgins C. F., Linton K. J. (2004) The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11, 918–926 [DOI] [PubMed] [Google Scholar]
- 31. Leshin J. A., Heselpoth R., Belew A. T., Dinman J. (2011) High throughput structural analysis of yeast ribosomes using hSHAPE. RNA Biol. 8, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]