Background: Type VII collagen is essential for skin stability as highlighted by related skin blistering diseases.
Results: A fusion protein comprising the cysteine-rich region and parts of the flanking domains forms a trimer upon oxidization.
Conclusion: The cysteine-rich region is an N-terminal cystine knot with novel topology.
Significance: The cystine knot is also found in type IX collagen, indicating a general principle.
Keywords: Collagen, Cysteine-mediated Cross-linking, Extracellular Matrix, Molecular Modeling, NMR, Skin, Dystrophic Epidermolysis Bullosa
Abstract
Collagens are a group of extracellular matrix proteins with essential functions for skin integrity. Anchoring fibrils are made of type VII collagen (Col7) and link different skin layers together: the basal lamina and the underlying connective tissue. Col7 has a central collagenous domain and two noncollagenous domains located at the N and C terminus (NC1 and NC2), respectively. A cysteine-rich region of hitherto unknown function is located at the transition of the NC1 domain to the collagenous domain. A synthetic model peptide of this region was investigated by CD and NMR spectroscopy. The peptide folds into a collagen triple helix, and the cysteine residues form disulfide bridges between the different strands. The eight cystine knot topologies that are characterized by exclusively intermolecular disulfide bridges have been analyzed by molecular modeling. Two cystine knots are energetically preferred; however, all eight disulfide bridge arrangements are essentially possible. This novel cystine knot is present in type IX collagen, too. The conserved motif of the cystine knot is CX3CP. The cystine knot is N-terminal to the collagen triple helix in both collagens and therefore probably impedes unfolding of the collagen triple helix from the N terminus.
Introduction
Collagens are an abundant class of proteins that fulfill important functions in an organism. Because collagens are extracellular matrix proteins, they are crucial for functions such as tissue integrity (1). A common characteristic of collagens is the repeating amino acid motif glycine-X-Y, with X very often proline and Y the posttranslational generated hydroxyproline. This unique amino acid pattern results in a left-handed helix, and three of these helices then form the right-handed collagen triple helix. The helices can undergo further assembly into fibrils and/or cross-linking events can take place. Cross-linking can occur intramolecular between different collagen strands or intermolecularly between different collagen triple helices after fibril formation. The cross-links are formed via disulfide bridges or in a lysyl oxidase-dependent manner, thus stabilizing the collagen structure (1).
Type VII collagen (Col7)3 is the major component of the anchoring fibrils. It links different skin layers, the basal lamina and the underlying connective tissue, together, thereby providing mechanical strength (2). Several related skin blistering diseases are described due to mutations in the gene of Col7 or due to the generation of autoantibodies against Col7. The acquired autoimmune skin blistering disease, epidermolysis bullosa acquisita, serves as a model system for autoimmune inflammation. Epidermolysis bullosa acquisita is well characterized with data ranging from genetic susceptibility (3, 4) and experimental mouse models (5, 6) to changes in metabolism (7). Genetic defects in Col7 result in the skin blistering disease dystrophic epidermolysis bullosa (DEB). Most mutations within Col7 are related to premature termination of the peptide chain or disturb the triple helical structure due to substitution of glycine residues (8).
There is a basic knowledge gap on structural information at an atomic level for Col7. Sequence comparison allowed determination of the domain architecture (9). The large central collagenous domain is flanked by two noncollagenous domains (NC). The subdomains of the NC1 domain have homology to the von Willebrand factor A (vWFA) or fibronectin III and mediate interactions with other extracellular proteins such as laminin-332 (10–12) and type I and IV collagen (10, 12–14), but interactions between neighboring Col7 domains have also been observed (15). Important for the function of Col7 is the formation of anchoring fibrils. Procollagen VII forms antiparallel dimers that are stabilized by the formation of disulfide bridges between the two triple helical monomers in the C-terminal region of the triple helix (16). Interestingly, a cysteine-rich region of unknown function is located at the transition of the last subdomain of NC1, the vWFA2 domain, and the triple helical collagenous domain (Fig. 1). This Col7 region also contains one of the rare missense mutations outside of the triple helical region of Col7 associated with DEB. We speculated that these cysteines are involved in stabilizing the collagen triple helix by preventing unfolding of the triple helix from the N terminus. To gain insight into the function of the cysteine-rich region, we investigated a synthetic peptide comprising this region. CD spectroscopy proved collagen triple helix formation, and the cysteines are within disulfide bridges as judged by NMR spectroscopy. Molecular modeling was employed to characterize different orientations of the disulfide bridges, i.e. different cystine knot topologies. The cysteine-rich region represents a novel cystine knot motif that is not restricted to Col7 only. A motif search identified a highly similar sequence in type IX collagen, a member of the FACIT collagen family (fibril-associated collagen with interrupted triple helices), that is found in cartilage, cornea, and vitreous (17). This novel cystine knot is in both collagens N-terminal to the triple helical region and thus probably impedes unfolding of the collagen triple helix.
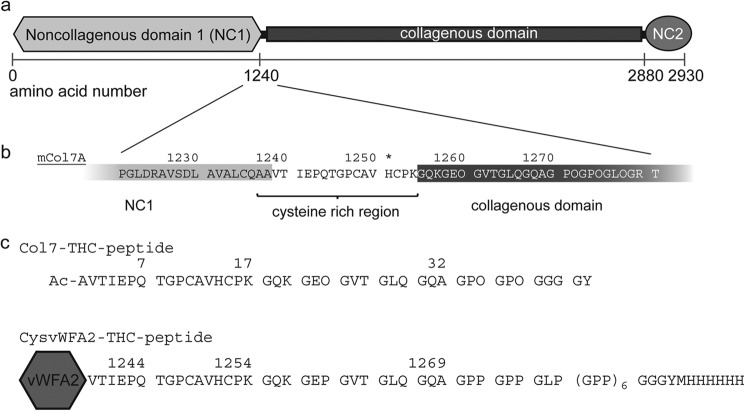
FIGURE 1.
Domain architecture of Col7 and investigated peptides/constructs. a, the Col7 monomer has a central collagenous domain that is flanked by two noncollagenous domains (NC1 and NC2). b, the transitional region between the NC1 domain and the collagenous domain contains two cysteines and is therefore named the cysteine-rich region. c, the amino acid sequences of the synthetic Col7-THC peptide and the CysvWFA2-THC peptide construct (murine forms) comprise that transitional cysteine-rich region and the N-terminal part of the collagenous domain. In the recombinant produced CysvWFA2-THC peptide, the peptide is fused with the last subdomain of NC1, the vWFA2, and a C-terminal His tag. O in the amino acid sequence represents hydroxyproline. One of the known recessive missense mutations in human Col7 is Y1250S (Ref. 37), variation ID in the Human Gene Mutation Database: CM014331) that corresponds to His-1251 in the murine protein. This residue is marked with an asterisk.
MATERIALS AND METHODS
Design of Col7-THC Peptide and Cloning of CysvWFA2-THC Peptide
The amino acid sequence of the investigated peptide (murine form) is given in Fig. 1. The Col7-THC peptide was synthesized by peptides&elephants (Potsdam, Germany). To allow quantification of peptide solutions, a tyrosine residue was introduced. This tyrosine and additional three glycine residues serving as linker were added at the C terminus. The N terminus was acetylated. All proline residues in the Y position of the GXY triplets were synthesized as hydroxyprolines. The predicted melting temperature of the triple helical part of the peptide is 9.7 °C (18).
To gain further insights into the disulfide bridge network, i.e. whether the disulfide bonds are within one peptide chain, or between different chains, a recombinant construct was used. This construct is termed CysvWFA2-THC peptide and comprises the vWFA2 domain, the linker region, and eight triplets of the Col7 collagenous domain (Fig. 1). Six additional GPP triplets were added to account for the loss of stability of the recombinant protein because posttranslational formation of hydroxyprolines contributes significantly to collagen triple helix stability. A polyhistidine tag was added for easier purification of the construct. The expected melting temperature of the triple helical region is 19.8 °C (18). The nucleotide sequence of CysvWFA2-THC peptide was codon-optimized, synthesized (Eurofins MWG Operon, Ebersberg, Germany), and cloned into the pTWIN1 vector (New England Biolabs). The amino acid sequence is given in the supplemental information. To verify that formation of disulfide bridges occurs between the different chains of the triple helical portion of CysvWFA2-THC peptide and not between different vWFA2 subdomains, an expression construct, containing only DNA encoding for CysvWFA2 in the pTWIN1 vector, was generated.
Protein Expression and Purification of CysvWFA2 and CysvWFA2-THC Peptide
CysvWFA2 and CysvWFA2-THC peptide have been expressed in Escherichia coli ER2566 as described previously for vWFA2 (15). Contrary to the previous expression construct, both proteins, CysvWFA2 and CysvWFA2-THC peptide, start with a cysteine. This causes premature intein cleavage of the intein and necessitated the development of new purification strategies. All chromatographic steps have been performed with an ÄKTA purifier (GE Healthcare) placed at 4 °C.
CysvWFA2 was purified by a combination of ammonium sulfate precipitation and cation exchange chromatography. All steps have been performed on ice or at 4 °C. Cells have been resuspended in 20 mm MES, pH 6, and lysed three times by a French press procedure or with a lysozyme/ultrasound treatment. Removal of cell debris was done by centrifugation. Solid ammonium sulfate was added to obtain 50% (m/v) saturation. Precipitated proteins were removed by centrifugation (30,000 × g, 30 min), and the ammonium sulfate concentration was raised to 70% (m/v). The precipitate was removed by centrifugation (30,000 × g, 30 min) dissolved in 20 mm MES, pH 6, and desalted with a Sephadex G-25 column (equilibrated with 20 mm MES, pH 6). Cation exchange chromatography was done with a Resource S (GE Healthcare) according to the manual. Protein elution was performed with 20 mm MES, 1 m NaCl, pH 6. CysvWFA2 eluted at 60 mm NaCl but still contained an impurity that was removed by a final size exclusion chromatography. A Superdex 200 10/300 GL column (GE Healthcare) was equilibrated with 10 mm sodium phosphate buffer, pH 7.4, and UV absorption was monitored at 214 nm because CysvWFA2 has a very low absorbance coefficient at 280 nm.
CysvWFA2-THC peptide was purified by a combination of nickel-nitrilotriacetic acid affinity chromatography (nickel-Sepharose fast flow, GE Healthcare) and cation exchange chromatography (Resource S, GE Healthcare). Purification was performed on ice or at 4 °C. Cells were resuspended in 20 mm sodium phosphate buffer, pH 8, containing 20 mm imidazole and lysed by sonification. After removal of cell debris by centrifugation, the lysate was applied to a nickel-Sepharose FF column. CysvWFA2-THC peptide was eluted by raising the imidazole concentration to 300 and 500 mm imidazole, respectively. Fractions containing CysvWFA2-THC peptide have been identified by SDS-PAGE and pooled. Buffer was exchanged to 20 mm MES, pH 6, and the sample was passed over a chitin matrix to remove potentially remaining uncleaved intein tag constructs and applied to a Resource S column. Elution was performed by increasing NaCl concentrations. CysvWFA2-THC peptide eluted in two peaks at ∼0.50 and 0.58 m NaCl. Fractions containing CysvWFA2-THC peptide were identified by SDS-PAGE and pooled.
Formation of interchain disulfide bridges in CysvWFA2-THC peptide has been performed by buffer exchange (ultrafiltration with molecular weight cut-off 10 kDa, 37 °C) to 20 mm HEPES, pH 8, containing 5 mm TCEP to ensure complete reduction of all cystines and dissociation of the collagen triple helix. After overnight incubation at 4 °C to allow for collagen triple helix formation, the TCEP concentration was lowered to ∼0.1 mm by buffer exchange (ultrafiltration, 2 °C). Cystine formation was started by the addition of a glutathione (oxidized form) stock solution in 20 mm HEPES, pH 8. Final glutathione concentration was 8.8 mm. Protein concentrations were determined by amino acid analysis as described in Ref. 19 without collagenase treatment.
CD Measurements of the Col7-THC Peptide
To allow for triple helix formation, solutions of the Col7-THC peptide were incubated for 1 week on ice. CD spectra were recorded in 10 mm sodium phosphate buffer, pH 7.4, between 190 and 300 nm at 4 °C on a Jasco J-715A in quartz cuvettes (1-mm path length). Peptide concentrations ranged from 30 μm to 1.3 mm. Temperature-dependent measurements for the determination of the melting point of the collagen triple helix were performed between 4 and 80 °C with a heating rate of 20 °C per hour.
Resonance Assignment of Col7-THC Peptide
NMR spectra for resonance assignment of the peptide were recorded on a Bruker Avance 500 NMR spectrometer equipped with a CPTCI probe head. Resonance assignment was performed using the following spectra: 1H,1H TOCSY (total correlation spectroscopy), 1H,1H COSY, 1H,13C HSQC (heteronuclear single quantum correlation), and 1H,13C HMBC (heteronuclear multiple bond correlation) using standard Bruker pulse sequences at 281.2 K (8 °C). Peptide concentrations varied between 1.3 and 0.03 mm and were determined by UV absorbance of the tyrosine. Peptide samples were dissolved in 5 mm sodium phosphate buffer, pH 7.4 (pH meter reading), containing 10 or 100% (v/v) D2O and 3-(trimethylsilyl)propionate-d4 (TSP-d4) for spectral referencing. Chemical shifts were referenced in the indirect dimension via the Ξ scale (factor for 13C: 0.251449530). Processing of the spectra and resonance assignment were performed with Topspin 3.1 (Bruker).
Temperature dependence of the NMR spectra was investigated with a 0.72 mm peptide sample between 281.2 K (8 °C) and 310.2 K (37 °C) system temperature. Cystines were reduced by the addition of TCEP-d16 (5 mm final) to the NMR sample.
Molecular Modeling
Col7-THC peptide were modeled in the computer using PyMOL (20) and the GROMACS 4.5.6 program package (21) together with the CHARMM27-CMAP (22, 23) force field. Electrostatic interactions were treated by the particle mesh Ewald method (24); a time step of 2 fs was chosen for all molecular dynamics simulations. To reach an isothermal (NVT) ensemble, the weak coupling to an external temperature of 300 K and the removal of the center of mass motion were achieved by velocity rescaling (25).
Modeling of the Col7-THC peptide was performed in four steps. In the first one the crystal structure of a collagen-like peptide (26) (Protein Data Bank (PDB): 1CAG) was used to construct a triple helix. The peptide sequence was changed to PK GQK GEO GVT GLQ GQA GPO GPO GGG GY with the PyMOL mutagenesis wizard. For the resulting triple helix a steepest descent energy minimization and a subsequent molecular dynamics simulation run of 10 ns in the NVT ensemble were performed, which both left the triple helix structure preserved. In the second step the sequence CAVHC was linked to the N terminus of each strand, and again an energy minimization and a molecular dynamics simulation run of 10 ns were performed. In the third step eight replicas of the triple helix were defined by introducing eight different combinations of harmonic potentials that depend on the distances of the cysteine sulfur atoms. For each replica the harmonic potentials were chosen in a way to reach one of eight different models I–VIII that correspond to the possible topologies in supplemental Scheme 1. The equilibrium distances of the potentials were 0.23 nm, and the force constants were increased in nine logarithmic levels from a starting value of 5 MJ mol−1 nm−2 to a final value of 5000 MJ mol−1 nm−2. For each level and each replica a molecular dynamics simulation run of 10 ns in length was performed. In the last step the restraining potentials were discarded, and disulfide bonds were formed. The sequence AVTIEPQTGP was linked to the N terminus of each strand, and a final energy minimization was performed for each replica.
Database Search of the CX3C Motif
The cysteine-rich region seems to be important for Col7 function as it harbors one of the known missense mutations. To investigate whether the CX3C motif is present in other collagens, a motif search of the CX3C motif was performed with the GenomeNet Motif Search site.
RESULTS
For a better understanding of the triple helix formation of Col7 and the function of the cysteine-rich region of Col7, we employed a combination of CD spectroscopy, molecular modeling, and NMR spectroscopy.
Col7-THC Peptide Forms a Collagen Triple Helix
Formation of a collagen triple helix of the Col7-THC peptide was verified by the temperature and concentration dependence of the CD spectra (Fig. 2). The CD spectrum of Col7-THC peptide is typical for a collagen triple helix with a positive band at 230 nm. Temperature-dependent measurements show a sigmoid curve, with an inflection point at 12.6 °C that equals the melting temperature of Col7-THC peptide. The melting point is in good agreement with the prediction of 9.7 °C (18). 1H NMR spectra also show temperature-dependent changes that can be interpreted as triple helix formation (Fig. 2d).
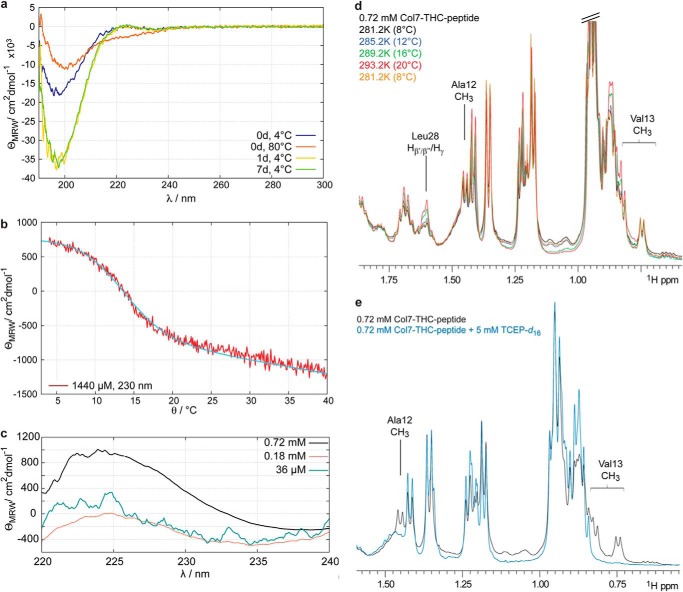
FIGURE 2.
a, CD spectra of a 36 μm solution of Col7-THC peptide measured at different time points after sample preparation. Immediately after dissolution of the peptide, a CD spectrum is obtained that shows a negative band at ∼200 nm. Unfolding the peptide by increasing the temperature to 80 °C results in a CD spectrum that is typical for random coil structures. Triple helix formation is already achieved after incubation at 4 °C for 1 day. θMRW is the mean residue ellipticity. b, temperature-dependent changes of the CD signal at 230 nm allow the determination of the melting temperature of the collagen triple helix. The melting temperature has been determined to be 12.6 °C. c, formation of collagen triple helices is also concentration-dependent. At high peptide concentration a positive band at about 230 nm can be observed that is characteristic for collagen triple helix-forming peptides. d, detail of 500-MHz one-dimensional proton NMR spectra of a 0.72 mm peptide solution at different temperatures. Some resonances show temperature-dependent changes that are representative for unfolding of the triple helix (e.g. Leu-28). These changes are completely reversible (yellow curve). e, Val-13 and Ala-12 that are in between the two cysteine residues show two sets of resonances. Upon the addition of 5 mm TCEP-d16, which efficiently reduces disulfide bridges, one set of signals vanishes. These resonances are therefore sensitive to the formation of disulfide bridges.
Resonance Assignment of the Col7-THC Peptide
NMR experiments have been performed at 8 °C (281.2 K) to ensure formation of the collagen triple helix. At this temperature about 90% of the peptide is in the triple helical form. The peptide had to be chemically synthesized to allow incorporation of hydroxyproline residues; therefore, 13C is at natural abundance. Standard two-dimensional experiments were employed for resonance assignment. Due to the particular amino acid composition, i.e. ∼30% of the amino acids are glycine and 20% are proline residues, severe resonance overlap hampers complete resonance assignment. About 20% of the residues could be identified unambiguously (Table 1). The identified signals belong to residues that are within and outside the triple helical region. These residues can serve as well distributed probes, thereby allowing confirmation of the CD spectroscopic data.
TABLE 1.
Unambiguously assigned NMR resonances of Col7-THC peptide
| Number/amino acid | Chemical shifts in ppm |
||||
|---|---|---|---|---|---|
| HN | α | β | Others | ||
| Ac | CH3 group 2.025 (27.40) | ||||
| 1 | Ala | 8.368 | 4.302 (52.55) | 1.354 (19.32) | |
| 4 | Ile | 4.196 (60.78) | 1.841 (38.77) | γ CH2: 1.177 and 1.470 (27.19) | |
| 4 | Ile | δ CH3 0.858 (12.76) | |||
| 4 | Ile | γ CH3: 0.895 (17.49) | |||
| 5 | Glu | γ CH2: 2.291 (35.83) | |||
| 12 | Ala | (A) 8.557 | (A) 4.316 (52.54) | (A) 1.448 (20.20) | |
| 12 | Ala | (B)a 1.357 | |||
| 13 | Val | 4.042 (62.37) | 1.952 (32.26) | (A)a γ CH3: 0.739 (20.86) and 0.812 (20.47) | |
| 13 | Val | (B) γ CH3: 0.758 (20.87) and 0.837 (20.46) | |||
| 14 | His | 4.658 (56.01) | 3.084 (30.80) | δ H: 7.030 (119.97) | |
| 14 | His | ϵ H: 7.922 (138.21) | |||
| 27 | Gly | 3.979 (44.40) | |||
| 28 | Leu | 8.286 | 4.351 (55.67) | 1.604 (42.35) | γ CH: 1.602 (26.96) |
| 28 | Leu | δ CH3: 0.868 (23.34) and 0.920 (24.98) | |||
| 43 | Tyr | 4.402 (59.23) | 2.867 and 3.101 (39.48) | δ CH: 7.108 (133.38) | |
| 43 | Tyr | ϵ CH: 6.809 (118.05) | |||
a The residue shows two sets of resonances.
NMR Spectroscopy of Col7-THC Peptide
The melting point of the triple helix formed by Col7-THC peptide is 12.6 °C as determined by CD spectroscopy. Temperature-dependent NMR experiments show a transition that is in accordance with this value (Fig. 2d). For example resonances of the Leu-28 side chain increase in signal intensity with increasing temperature. These spectral changes are reversible.
The extracellular matrix is characterized by an oxidative environment and disulfide bridges are observed, e.g. type III collagen is stabilized by a cystine knot (27). The peptide was synthesized with free thiol groups. Resonances characteristic for the Cβ of cysteine residues are vanishing with time, rendering the formation of disulfide bridges very likely. This assumption is supported by the observation of two sets of resonances for Val-13. Val-13 is close to the cysteine residues of the peptide and lies outside the triple helical region. Because temperature-dependent experiments show no differences for Val-13, triple helix formation can be excluded as a cause (Fig. 2d). In contrast, upon the addition of TCEP, the characteristic resonances of Val-13 are vanishing, showing that the cysteines are indeed forming disulfide bridges. For the methyl group of Ala-12, the same changes are observed. Because two sets of resonances are observed for Val-13 but no peaks for the Cβ of free cysteines, we assume that one set of resonances belongs to interchain disulfide bridges and one set belongs to intrachain oxidized molecules, which is supported by a study of model peptides of the type III collagen cystine knot reporting a 70% yield for the homotrimer after air oxidation (27).
SDS-PAGE analysis of the peptide from the NMR sample under nonreducing conditions shows a major band that is absent under reducing conditions (Fig. 3a). The peptide is a monomer under reducing conditions and might diffuse out of the gel during staining. Under oxidizing conditions interchain disulfide bridges are present, and the higher molecular weight oligomers can be detected. Because peptides are known to potentially have very unusual behavior in SDS-PAGE analysis, the oligomerization state cannot be delineated from the apparent molecular weight.
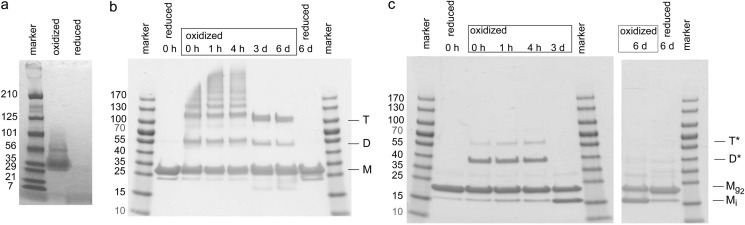
FIGURE 3.
a, SDS-PAGE (4–20%) analysis of a 1.4 mm solution of Col7-THC peptide. A major band is present for oxidative conditions but not for reducing conditions. The apparent molecular mass cannot be used to draw conclusions of the oligomerization state because peptides can have an unusual behavior in SDS-PAGE. b, SDS-PAGE (4–20%) analysis shows trimer formation of CysvWFA2-THC peptide. After the addition of glutathione, several additional bands at higher molecular mass are observed. The samples of day 3 and 6 show the presence of trimer (T), dimer (D), and monomer (M). Samples taken at 0 h and day 6 have also been analyzed under reducing conditions in the SDS-PAGE. CysvWFA2-THC peptide concentration was 10 μm, and sample loading is 7.5 μg of CysvWFA2-THC peptide per well. c, SDS-PAGE of a CysvWFA2 sample shows formation of oligomers upon the addition of oxidized glutathione (D*, T*), but after 3 days of incubation, only bands corresponding to monomers are present. The band of CysvWFA2 with an apparent molecular mass of 15 kDa represents the intramolecular disulfide bridge (Mi). Because the band at higher molecular mass is at the same height as the reduced proteins, this band is attributed to CysvWFA2 linked to two molecules of glutathione (Mg2). Oxidation was carried out with a 20 μm CysvWFA2 solution. 8 μg of CysvWFA2 are loaded per well.
Purification of CysvWFA2 and CysvWFA2-THC Peptide
The N-terminal cysteine of the expression constructs causes premature intein cleavage in E. coli, and thus affinity purification via the chitin-binding domain is not possible. New purification protocols had to be established.
CysvWFA2 was successfully purified by an initial ammonium sulfate precipitation followed by cation exchange chromatography and a final size exclusion chromatography. Purification of CysvWFA2-THC peptide was done by affinity chromatography via the polyhistidine tag; however, several other protein bands are present in the SDS-PAGE. Subsequent purification was performed by cation exchange chromatography at pH 6 (the theoretical pI of CysvWFA2-THC peptide is 7.36). At pH 6 half of the histidine side chains of the tag are protonated. Due to triple helix formation (the protein was purified at 4 °C), the three histidine tags are spatially close and form a positively charged area resulting in unexpectedly high salt concentrations necessary to elute the protein (see supplemental Fig. 1).
CysvWFA2-THC Peptide Forms Interchain Disulfide Bonds
Formation of interchain disulfide bridges in the CysvWFA2-THC peptide and subsequent SDS-PAGE should deliver the exact oligomerization state. SDS-PAGE behavior for this construct depends mainly on the vWFA2 domain. Reduction of cysteines of CysvWFA2-THC peptide was carried out at 37 °C. At this temperature the collagen triple helix is dissociated. Lowering the temperature to 4 °C allows for triple helix formation. Adding oxidized glutathione to the sample removes the remaining reducing agent TCEP, and the cysteines of CysvWFA2-THC peptide are oxidized, thus enabling formation of interchain disulfide bridges (Fig. 3b). In the beginning higher molecular weight species are also observed; however, finally a band with the molecular weight of the trimer appears in the SDS-PAGE, proving that the cysteines form interchain disulfide bridges. Disulfide bridge formation by air oxidation is inefficient, and only a very small fraction of trimers can be observed after 4 days (see supplemental Fig. 2).
The vWFA2 domain contains two cysteines that are located at the N and C terminus, respectively, and that have been shown to form a disulfide bridge (15). To exclude unspecific formation of oligomers due to these cysteines, CysvWFA2 was purified, and the cysteines were reduced and oxidized. SDS-PAGE analysis shows at the beginning of the oxidization reaction bands with molecular weights corresponding to dimer and trimer, but also a very weak band with a molecular weight of a tetramer is present. These bands disappear and thus represent unspecific formation of oligomers. After 3 days two bands remain that are assigned to CysvWFA2 linked to two molecules of glutathione and CysvWFA2 with an intramolecular disulfide bridge. The CysvWFA2 with an intramolecular disulfide bridge has an apparent molecular mass of 15 kDa (Fig. 3c).
Molecular Modeling of the Cystine Knot
Molecular modeling was employed to investigate whether certain cystine topologies are preferred. A model of the triple helical portion of Col7-THC peptide was energy-minimized before building the eight theoretically possible cystine knots. Molecular modeling shows that all topologies are essentially permitted but that they differ in their minimized energy (Fig. 4). Five models show a curvature within the collagen triple helix. For half of the models the collagen triple helix remains undisturbed. In model VII one strand of the collagen triple helix is moved out, and a short β-pleated sheet is formed. In model VI, the model with the highest energy, two disulfide bridges have very large dihedral angles that deviate substantially from the usually found value of about 90° (28). In model II, which has the second highest energy, the strands in the cystine knot are interlaced. Two models, namely model I and II, have a considerably lower energy when compared with the other models. The models are provided as supplemental File 2 (Overview of All Models).
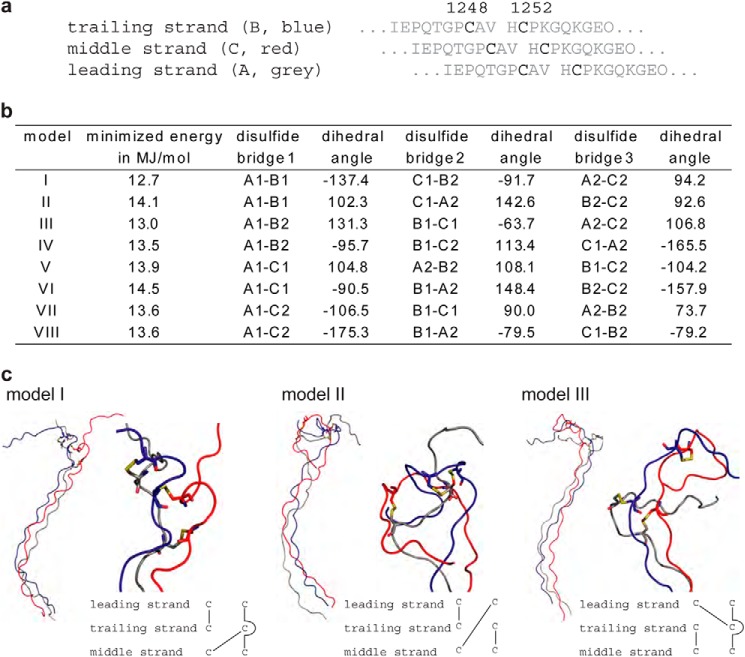
FIGURE 4.
a, a model of the triple helical region of Col7-THC peptide was generated before introducing all eight possible disulfide bridge patterns. The three collagen strands in the model are referred to as leading, middle, and trailing strand (compare Ref. 42). b, disulfide pattern of the eight models and the dihedral angle of the disulfide bridge. A schematic representation is provided in the supplemental information. A, B, and C are the different strands, and 1 and 2 refer to Cys-1248 and Cys-1252, respectively. c, the cystine knots according to model I, II, and III are shown. An overview of the triple helical peptide is shown on the left-hand side, and an enlarged representation of the cystine knot is shown on the right-hand side. A schematic representation of the disulfide pattern is shown on bottom. From the eight imaginable topologies, five models show a curvature as depicted for models I to III.
The Cystine Knot Is Conserved, and the Consensus Sequence Is Also Present in Type IX Collagen
A database search showed that type IX collagen (Col9) also has this cystine knot consensus sequence that is N-terminal to the collagen triple helix (Fig. 5). Sequence comparisons of Col7 from different species and of Col7 and Col9 show a conserved CX3CP motif.
FIGURE 5.

Multiple sequence alignment of the cysteine-rich region of Col7 (a) and murine/human Col7 with the different isoforms of Col9 (b) relative to the CX3CP motif. Underlined sequences belong to triple helical domains. O represents hydroxyproline, but is omitted for clarity in the Col9 sequences.
DISCUSSION
Col7 is the major component of anchoring fibrils that are found at the dermal epidermal junction. Anchoring fibrils connect epidermis and dermis, and an impaired function causes skin blistering. A cysteine-rich region with unknown function is located at the transition of the N-terminal NC1 domain and the central collagen triple helix. A model peptide derived from the cysteine-rich region was investigated by a combination of CD spectroscopy, NMR spectroscopy, and molecular modeling. CD spectroscopy proved that the model peptide is able to form a collagen triple helix. NMR data showed that residues lying outside the triple helix undergo changes in chemical shift upon reduction of the cysteines, leading to the assumption that the cysteines are involved in formation of a cystine knot. Complete reduction of cysteines and reformation of the disulfide bridges in a fusion construct of the cysteine-rich region, the preceding vWFA2 domain, and a peptide of the succeeding triple helical domain of Col7 showed formation of trimer, proving that this cysteine-rich region represents a novel cystine knot. The consensus sequence of the cystine knot can be found in Col9. It was speculated for isolated Col9 that these cysteines form interchain disulfide bridges due to resistance to pepsin cleavage (29), indicating a relevance of this cystine knot in vivo. Molecular modeling of the theoretically possible cystine knot topologies indicates that all eight connectivities are allowed. The different models are energetically distinct, and two models are favored. Future studies will aim to obtain high resolution structural information of the cystine knot, and it will be of great interest to see whether these two topologies indeed occur in Col7.
Formation of collagen trimers occurs intracellularly. It was shown for type I collagen that the melting of the triple helix also occurs below body temperature (30) and that chaperones are required for collagen maturation (31). Trimerization domains ensure formation of triple helices. These domains are in general at the C terminus of collagens (32). As seen in type III collagen (Col3) or type XIX collagen (Col19), the formed triple helices are stabilized by C-terminal cystine knots (27, 33, 34). The Col3 cystine knot has recently been structurally characterized (35) and is also necessary for starting triple helix formation (36). However, this cystine knot is only correctly formed when it is C-terminal to the collagen triple helix (27). If the cysteines are placed N-terminal to the collagen helix, the cystine knot is virtually not formed (27). Interestingly, the cystine knot identified in Col7 is N-terminal to the collagen helix. Three amino acids are located between the cysteines that seem to provide the necessary steric requirements to allow for cystine knot formation. In Col7 the vWFA2 subdomain is preceding the collagenous domain and vWFA2 is interacting with other collagens. We hypothesize that the Col7 cystine knot prevents unfolding of the collagen triple helix from the N-terminal region and that the knot is not involved in triple helix formation as the cystine knot of Col3. The collagen triple helix of Col7 shows multiple interruptions with the first one after 13 GXY triplets. These interruptions point toward a desired flexibility of the anchoring fibrils. Because unfolding would impair the important function of Col7 for skin stability, unfolding is precluded by a cystine knot. The CX3CP cystine knot is located exclusively N-terminal to a collagen triple helix as seen in Col9 and Col7.
DEB is caused by mutations of the Col7 gene. Most mutations account for premature termination during translation resulting in truncated Col7 molecules (8). Only a few missense mutations within Col7 exist; most of them are within the triple helical region and are glycine substitutions. These mutations might affect folding of the triple helix and/or are located within binding sites of interacting proteins. Interestingly, there is a recessive missense mutation at the transition from the noncollagenous domain 1 to the triple helical collagenous domain. In combination with a mutation in the collagenous domain, this results in dystrophic nails, atrophic scars, and some large bullae (37). Because trimer formation of collagens occurs intracellularly, this mutation is probably affecting formation of the cystine knot. To access the structural impact of the mutation, it is important to know whether or not expression of the Col7 gene is allele-specific. This knowledge is also highly valuable for DEB regarding future therapies and a better understanding of disease manifestation. Most eukaryotic genes are expressed not allele-specific (e.g. Refs. 38 and 39). If expression of Col7 in a cell is not allele-specific, a mixture of collagen trimers, with a different number of strands bearing a mutation, is formed in the cell. Different trimers might show different stabilities and/or secretion properties. Therefore, observed differences in secretion of mutant Col7 molecules when compared with the wild type molecule in patient skin could be caused only by a subpopulation of the Col7 trimers. This renders therapy strategies for certain types of DEB on the basis of allele-specific repression of mutant Col7 mRNA highly interesting (40). If expression of Col7 is allele-specific, only one allele in a cell would be transcribed. In that case one would expect two classes of Col7 molecules, namely normal Col7 trimers and homotrimers of the mutated Col7 gene. A case report of a DEB patient supports allele-specific expression of Col7 because all mRNA transcripts originated from one allele (41). Consequently, depending on the expression condition of the Col7 gene in the organism, the missense mutation within the cystine knot might not only affect cystine knot formation but also lead to different cystine knot topologies.
The cysteine-rich region of Col7 forms a cystine knot of novel topology. The cystine knot has a consensus sequence of CX3CP that is also found in Col9. Identification of the cystine knot not only provides the basis for a better understanding of collagen triple helix stabilization in Col7, it also opens the possibility to introduce an N-terminal cystine knot in collagen model peptides where a C-terminal cystine knot is undesired.
Supplementary Material
Acknowledgments
We acknowledge Jürgen Brinckmann for critical reading of the manuscript and helpful discussions. We thank Florian Flachsenberg for technical assistance. Jürgen Brinckmann and Heiko Steenbock are acknowledged for the amino acid analysis.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) Excellence Cluster “Inflammation at Interfaces” (DFG 306/1).

This article contains supplemental text, Scheme 1, Figs. 1 and 2, and an overview.
- Col7
- type VII collagen
- Col9
- type IX collagen
- Col3
- type III collagen
- vWFA2
- von Willebrand factor A domain 2 of Col7
- DEB
- dystrophic epidermolysis bullosa
- NC
- noncollagenous domain
- TCEP
- tris(2-carboxyethyl)phosphine
- TSP-d4
- 3-(trimethylsilyl)propionate-d4.
REFERENCES
- 1. Eyre D., Wu J.-J. (2005) Collagen cross-links. in Collagen (Brinckmann J., Notbohm H., Müller P. K., eds) pp. 207–229, Springer, Berlin [Google Scholar]
- 2. Gupta R., Woodley D. T., Chen M. (2012) Epidermolysis bullosa acquisita. Clin. Dermatol. 30, 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gammon W. R., Heise E. R., Burke W. A., Fine J.-D., Woodley D. T., Briggaman R. A. (1988) Increased frequency of HLA-DR2 in patients with autoantibodies to epidermolysis bullosa acquisita antigen: Evidence that the expression of autoimmunity to type VII collagen is HLA class II allele associated. J. Investig. Dermatol. 91, 228–232 [DOI] [PubMed] [Google Scholar]
- 4. Ludwig R. J., Recke A., Bieber K., Müller S., Marques Ade C., Banczyk D., Hirose M., Kasperkiewicz M., Ishii N., Schmidt E., Westermann J., Zillikens D., Ibrahim S. M. (2011) Generation of antibodies of distinct subclasses and specificity is linked to H2s in an active mouse model of epidermolysis bullosa acquisita. J. Invest. Dermatol. 131, 167–176 [DOI] [PubMed] [Google Scholar]
- 5. Sitaru C., Mihai S., Otto C., Chiriac M. T., Hausser I., Dotterweich B., Saito H., Rose C., Ishiko A., Zillikens D. (2005) Induction of dermal-epidermal separation in mice by passive transfer of antibodies specific to type VII collagen. J. Clin. Invest. 115, 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iwata H., Bieber K., Tiburzy B., Chrobok N., Kalies K., Shimizu A., Leineweber S., Ishiko A., Vorobyev A., Zillikens D., Köhl J., Westermann J., Seeger K., Manz R., Ludwig R. J. (2013) B cells, dendritic cells, and macrophages are required to induce an autoreactive CD4 helper T cell response in experimental epidermolysis bullosa acquisita. J. Immunol. 191, 2978–2988 [DOI] [PubMed] [Google Scholar]
- 7. Schönig S., Recke A., Hirose M., Ludwig R. J., Seeger K. (2013) Metabolite analysis distinguishes between mice with epidermolysis bullosa acquisita and healthy mice. Orphanet J. Rare Dis. 8, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dang N., Murrell D. F. (2008) Mutation analysis and characterization of COL7A1 mutations in dystrophic epidermolysis bullosa. Exp. Dermatol. 17, 553–568 [DOI] [PubMed] [Google Scholar]
- 9. Christiano A. M., Rosenbaum L. M., Chung-Honet L. C., Parente M. G., Woodley D. T., Pan T.-C., Zhang R. Z., Chu M.-L., Burgeson R. E., Uitto J. (1992) The large non-collagenous domain (NC-1) of type VII collagen is amino-terminal and chimeric. Homology to cartilage matrix protein, the type III domains of fibronectin and the A domains of von Willebrand factor. Hum. Mol. Genet. 1, 475–481 [DOI] [PubMed] [Google Scholar]
- 10. Brittingham R., Uitto J., Fertala A. (2006) High-affinity binding of the NC1 domain of collagen VII to laminin 5 and collagen IV. Biochem. Biophys. Res. Commun. 343, 692–699 [DOI] [PubMed] [Google Scholar]
- 11. Chen M., Marinkovich M. P., Jones J. C. R., O'Toole E. A., Li Y.-Y., Woodley D. T. (1999) NC1 domain of type VII collagen binds to the β3 chain of laminin 5 via a unique subdomain within the fibronectin-like repeats. J. Invest. Dermatol. 112, 177–183 [DOI] [PubMed] [Google Scholar]
- 12. Chen M., Marinkovich M. P., Veis A., Cai X., Rao C. N., O'Toole E. A., Woodley D. T. (1997) Interactions of the amino-terminal noncollagenous (NC1) domain of type VII collagen with extracellular matrix components. J. Biol. Chem. 272, 14516–14522 [DOI] [PubMed] [Google Scholar]
- 13. Wegener H., Leineweber S., Seeger K. (2013) The vWFA2 domain of type VII collagen is responsible for collagen binding. Biochem. Biophys. Res. Commun. 430, 449–453 [DOI] [PubMed] [Google Scholar]
- 14. Villone D., Fritsch A., Koch M., Bruckner-Tuderman L., Hansen U., Bruckner P. (2008) Supramolecular interactions in the dermo-epidermal junction zone. J. Biol. Chem. 283, 24506–24513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leineweber S., Schönig S., Seeger K. (2011) Insight into interactions of the von-Willebrand-factor-A-like domain 2 with the FNIII-like domain 9 of collagen VII by NMR and SPR. FEBS Lett. 585, 1748–1752 [DOI] [PubMed] [Google Scholar]
- 16. Colombo M., Brittingham R. J., Klement J. F., Majsterek I., Birk D. E., Uitto J., Fertala A. (2003) Procollagen VII self-assembly depends on site-specific interactions and is promoted by cleavage of the NC2 domain with procollagen C-proteinase. Biochemistry 42, 11434–11442 [DOI] [PubMed] [Google Scholar]
- 17. Diab M., Wu J. J., Eyre D. R. (1996) Collagen type IX from human cartilage: a structural profile of intermolecular cross-linking sites. Biochem. J. 314, 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Persikov A. V., Ramshaw J. A. M., Brodsky B. (2005) Prediction of collagen stability from amino acid sequence. J. Biol. Chem. 280, 19343–19349 [DOI] [PubMed] [Google Scholar]
- 19. Brinckmann J., Hunzelmann N., Kahle B., Rohwedel J., Kramer J., Gibson M. A., Hubmacher D., Reinhardt D. P. (2010) Enhanced fibrillin-2 expression is a general feature of wound healing and sclerosis: potential alteration of cell attachment and storage of TGF-β. Lab. Invest. 90, 739–752 [DOI] [PubMed] [Google Scholar]
- 20. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.3, Schrödinger, LLC, New York [Google Scholar]
- 21. Hess B., Kutzner C., Spoel D. v. d., Lindahl E. (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 22. MacKerell A. D., Bashford D., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 23. MacKerell J. A. D., Feig M., Brooks C. L., III. (2004) Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comp. Chem. 25, 1584–1604 [DOI] [PubMed] [Google Scholar]
- 24. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 25. Bussi G., Donadio D., Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 [DOI] [PubMed] [Google Scholar]
- 26. Bella J., Eaton M., Brodsky B., Berman H. M. (1994) Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science 266, 75–81 [DOI] [PubMed] [Google Scholar]
- 27. Barth D., Kyrieleis O., Frank S., Renner C., Moroder L. (2003) The role of cystine knots in collagen folding and stability, part II. Conformational properties of (Pro-Hyp-Gly)n model trimers with N- and C-terminal collagen type III cystine knots. Chemistry 9, 3703–3714 [DOI] [PubMed] [Google Scholar]
- 28. Katz B. A., Kossiakoff A. (1986) The crystallographically determined structures of atypical strained disulfides engineered into subtilisin. J. Biol. Chem. 261, 15480–15485 [PubMed] [Google Scholar]
- 29. van der Rest M., Mayne R., Ninomiya Y., Seidah N. G., Chretien M., Olsen B. R. (1985) The structure of type IX collagen. J. Biol. Chem. 260, 220–225 [PubMed] [Google Scholar]
- 30. Leikina E., Mertts M. V., Kuznetsova N., Leikin S. (2002) Type I collagen is thermally unstable at body temperature. Proc. Natl. Acad. Sci. U.S.A. 99, 1314–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ishida Y., Nagata K. (2011) Hsp47 as a collagen-specific molecular chaperone. Methods Enzymol. 499, 167–182 [DOI] [PubMed] [Google Scholar]
- 32. Boudko S. P., Engel J., Bächinger H. P. (2012) The crucial role of trimerization domains in collagen folding. Int. J. Biochem. Cell Biol. 44, 21–32 [DOI] [PubMed] [Google Scholar]
- 33. Boudko S. P., Engel J., Bächinger H. P. (2008) Trimerization and triple helix stabilization of the collagen XIX NC2 domain. J. Biol. Chem. 283, 34345–34351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boudko S. P., Engel J. (2004) Structure formation in the C terminus of type III collagen guides disulfide cross-linking. J. Mol. Biol. 335, 1289–1297 [DOI] [PubMed] [Google Scholar]
- 35. Boudko S. P., Engel J., Okuyama K., Mizuno K., Bächinger H. P., Schumacher M. A. (2008) Crystal structure of human type III collagen Gly991-Gly1032 cystine knot-containing peptide shows both 7/2 and 10/3 triple helical symmetries. J. Biol. Chem. 283, 32580–32589 [DOI] [PubMed] [Google Scholar]
- 36. Bächinger H. P., Bruckner P., Timpl R., Prockop D. J., Engel J. (1980) Folding mechanism of the triple helix in type-III collagen and type-III pN–collagen. Eur. J. Biochem. 106, 619–632 [DOI] [PubMed] [Google Scholar]
- 37. Ryoo Y.-W., Kim B.-C., Lee K.-S. (2001) Characterization of mutations of the type VII collagen gene (COL7A1) in recessive dystrophic epidermolysis bullosa mitis (M-RDEB) from three Korean patients. J. Dermatol. Sci. 26, 125–132 [DOI] [PubMed] [Google Scholar]
- 38. Levesque M. J., Ginart P., Wei Y., Raj A. (2013) Visualizing SNVs to quantify allele-specific expression in single cells. Nat. Methods 10, 865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hansen C. H., van Oudenaarden A. (2013) Allele-specific detection of single mRNA molecules in situ. Nat. Methods 10, 869–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pendaries V., Gasc G., Titeux M., Tonasso L., Mejía J. E., Hovnanian A. (2012) siRNA-mediated allele-specific inhibition of mutant type VII collagen in dominant dystrophic epidermolysis bullosa. J. Invest. Dermatol. 132, 1741–1743 [DOI] [PubMed] [Google Scholar]
- 41. Gardella R., Barlati S., Zoppi N., Tadini G., Colombi M. (2000) A -96C→T mutation in the promoter of the collagen type VII gene (COL7A1) abolishing transcription in a patient affected by recessive dystrophic epidermolysis bullosa. Hum. Mutat. 16, 275. [DOI] [PubMed] [Google Scholar]
- 42. Bella J. (2010) A new method for describing the helical conformation of collagen: Dependence of the triple helical twist on amino acid sequence. J. Struct. Biol. 170, 377–391 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.