Background: Sequence requirements for triple-helical collagen to bind fibronectin are not fully understood.
Results: Fibronectin affinity was conferred on a recombinant bacterial collagen by incorporating specific human collagen II sequences.
Conclusion: The chimeric collagen defines the minimum collagen II sequence required for fibronectin affinity.
Significance: This system is useful to investigate sequence/activity relationships for triple-helical collagen and to generate scalable bioactive materials.
Keywords: Collagen, Extracellular Matrix Proteins, Fibronectin, Protein Chimeras, Recombinant Protein Expression, Binding, Gelatin, Triple Helix
Abstract
Interaction of collagen with fibronectin is important for extracellular matrix assembly and regulation of cellular processes. A fibronectin-binding region in collagen was identified using unfolded fragments, but it is not clear if the native protein binds fibronectin with the same primary sequence. A recombinant bacterial collagen is utilized to characterize the sequence requirement for fibronectin binding. Chimeric collagens were generated by inserting the putative fibronectin-binding region from human collagen into the bacterial collagen sequence. Insertion of a sufficient length of human sequence conferred fibronectin affinity. The minimum sequence requirement was identified as a 6-triplet sequence near the unique collagenase cleavage site and was the same in both triple-helix and denatured states. Denaturation of the chimeric collagen increased its affinity for fibronectin, as seen for mammalian collagens. The fibronectin binding recombinant collagen did not contain hydroxyproline, indicating hydroxyproline is not essential for binding. However, its absence may account, in part, for the higher affinity of the native chimeric protein and the lower affinity of the denatured protein compared with type II collagen. Megakaryocytes cultured on chimeric collagen with fibronectin affinity showed improved adhesion and differentiation, suggesting a strategy for generating bioactive materials in biomedical applications.
Introduction
Collagen, the most abundant protein in the human body, is distinguished by its signature triple-helical structure based on a (Gly-Xaa-Yaa)n repeating sequence (1). So far, 28 types of collagens have been described (2–4). The fibrillar type I collagen, which constitutes more than 90% of the collagen in the body, forms characteristic fibrils with an axial periodicity of 67 nm in tendon, bone, and skin. Type I collagen is a heterotrimer with two α1(I) chains and one α2(I) chain. Type II collagen forms fibrils with the same periodicity in cartilage and vitreous (5, 6), and is a homotrimer composed of three α1(II) chains, which show a high sequence homology to α1(I). Collagen fibrils provide physical support for cells and tissues, and also have a major impact on various cellular processes, such as adhesion, growth, migration, and differentiation. These biological functions are based on the interaction of collagen with various cell surface receptors and with other extracellular matrix proteins (7, 8). An understanding of how extracellular matrix molecules interact and influence each other can shed light on key aspects of extracellular matrix formation, remodeling, and tissue repair. One important example is the interaction between collagen and fibronectin, another abundant matrix protein implicated in cell proliferation as well as matrix assembly.
Fibronectin (Fn)3 is a large dimeric glycoprotein containing modular domains that mediate self-assembly, binding to cell surface receptors, and interactions with collagen and other extracellular matrix molecules (9, 10). A range of biochemical studies defined a 42-kDa domain containing six modules, 6FnI1–2FnII7–9FnI, to be the gelatin-binding domain (GBD) (11–13), due to its ability to strongly and specifically bind to denatured collagen (gelatin). Two separate fragments of the GBD, 6FnI1–2FnII7FnI and 8–9FnI, have been shown to independently bind denatured collagen, but with a decreased affinity (14–17). High resolution structures have been determined by NMR and x-ray crystallography for the 6FnI1–2FnII7FnI and 8–9FnI subfragments (15, 16), and recently a crystal structure was reported for the zinc-mediated dimer of the intact GBD as well (18).
The specific region within collagen responsible for binding to Fn and the nature of this interaction has been a long standing area of investigation. More than 30 years ago, it was demonstrated that Fn binds denatured collagen more tightly than the native triple-helical form (13, 19), and the binding site on the α1 chain of type I collagen was localized to a cyanogen bromide peptide that includes the collagenase (MMP-1) cleavage site (11, 20). Recently, important advances have been made in defining the interaction of denatured collagen peptides with Fn, using NMR and x-ray crystallography. Short synthetic (Gly-Xaa-Yaa)n peptides containing sequences from either the α1(I) or α2(I) chains have been employed to further define the required collagen sequence and mode of Fn binding (15–17). NMR monitoring of the perturbation of signals from individual residues within 8–9FnI or 6FnI1–2FnII7FnI modules showed that a 24-residue gelatin peptide (α1(I) Gly778-Gly799) bound tightly, and a crystal structure of the 8–9FnI module segment shows residues 784GLOGQRGER792 within the longer peptide form an anti-parallel β-strand that interacts with a β-sheet in 8FnI (15).
Although the binding of Fn to denatured collagen has been well characterized, the binding of Fn to native collagen has been more difficult to define. Native triple-helical collagen has been shown to interact with Fn by a range of techniques, including rotary shadowing (21), sucrose gradient centrifugation (21), and solid-state binding assays (19), but it is not clear if the same Gly-Xaa-Yaa sequence binds in the triple-helical versus the denatured states. Here, a recombinant bacterial collagen-like protein system is employed to characterize the amino acid sequences in native and denatured collagen required for Fn binding. A collagen-like protein found in Streptococcus pyogenes, which is readily expressed and easily modified in Escherichia coli, was previously shown to have a triple helix structure and stability similar to that of human fibrillar collagens, even though post-translationally formed hydroxyproline (Hyp) is absent (22–24). Chimeric collagen-like proteins were generated by inserting potential Fn binding sequences from human collagen type II between bacterial collagen modules. The binding of these chimeric constructs in their native and denatured states to Fn was investigated using solid-state binding assays to define the minimal required sequence. The results obtained from the recombinant bacterial collagens allowed comparison with previous studies done on mammalian collagen fragments as well as synthetic collagen-like peptides. Chimeric collagens with in vitro Fn binding ability were also shown to promote adhesion and differentiation of human megakaryocytes, suggesting these recombinant collagens could be developed as biomaterials for biomedical applications.
EXPERIMENTAL PROCEDURES
Molecular Cloning
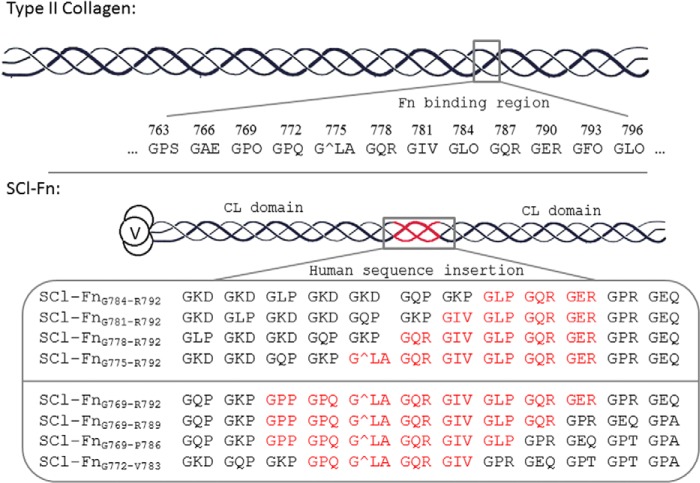
The protein sequence for the bacterial collagen SCl constructs was based on the Scl2.28 sequence from S. pyogenes with DNA codon optimized for E. coli expression. A His6 tag was introduced at the N terminus for purification purpose. The full DNA sequence encoding the SCl protein was synthesized at Biomatik Corp. (Wilmington, DE). Oligonucleotides encoding various lengths of the type II collagen Fn-binding region were designed and synthesized (Invitrogen). Annealed dsDNA were inserted between the two collagen-like (CL) domains of the SCl constructs through restriction sites XmaI and ApaI predesigned in the sequence. The final constructs containing the Fn-binding sites were cloned into the pColdIII vector (Takara Bio Inc.) through NdeI and XbaI restriction sites. All enzymes for cloning were purchased from New England Biolabs (Ipswich, MA). DNA sequencing to confirm fidelity was carried out at the Tufts Core Facility. The DNA constructs and their subsequent proteins were denoted as SCl, for Streptococcus collagen-like proteins, and those recombinant proteins containing type II collagen sequences predicted to binding Fn denoted as SCl-Fn. To define the length of the insertion of type II collagen, we use SCl-Fn#-#′, with # representing the starting residue and #′ representing the final residue of the inserted type II collagen sequence. For example, the putative GLPGQRGER sequence is located in the type II collagen triple-helix region from residue Gly784 to Arg792, and the recombinant bacterial VCLCL construct containing this sequence between the two CL domains is denoted as SCl-FnG784-R792.
Protein Purification
All SCl constructs in the pColdIII vector were expressed in E. coli BL21 strain, grown in 20 ml of LB medium with 100 μg/ml of ampicillin overnight at 37 °C. This starting culture was used to inoculate 500 ml of LB-ampicillin media in a shaking flask and grown at 37 °C to an A600 nm = 0.8. To induce protein expression, 1 mm isopropyl β-d-thiogalactopyranoside was added to the culture and the temperature was lowered to 22 °C. After 16 h induction, cells were harvested by centrifugation and resuspended in a His tag purification column binding buffer (20 mm sodium phosphate buffer, pH 7.4, 500 mm NaCl, 10 mm imidazole) containing 0.25 mg/ml of lysozyme and frozen at 80 °C until purification. The expression protocol was modified to decrease the level of truncated products in SCl-FnG769-R789 and SCl-FnG769-R792 expression by using HyperBrothTM (Athena Environmental Sciences, Inc., Baltimore, MD) instead of LB broth, and performing isopropyl β-d-thiogalactopyranoside induction for 4 h. Protease inhibitor mixture (Sigma) was also added to binding buffer during cell resuspension.
Purification of the His-tagged SCl-Fn was carried out by gravity-driven immobilized metal ion affinity chromatography with nickel ion binding and imidazole gradient elution. Frozen cells were thawed and further lysed by sonication. Cellular debris was removed by centrifugation at 8,228 × g at 4 °C. Supernatant containing the soluble target protein was loaded onto equilibrated purification columns packed with nickel-nitrilotriacetic acid beads (Qiagen, Valencia, CA) and run through the column by gravity. The column was then washed sequentially with 2 bed volumes of binding buffer, binding buffer plus 60 mm imidazole, and binding buffer plus 120 mm imidazole. His-tagged protein on the column was eluted by elution buffer (binding buffer plus 400 mm imidazole). Protein purity was checked by SDS-PAGE (NuPAGE® BisTris 4–12%, Invitrogen) and molecular weight was determined by MALDI-TOF MS. The concentration of protein was determined using an extinction coefficient of ϵ280 = 9970 m−1 cm−1 after dialysis into phosphate-buffered saline (PBS, pH 7.4). Chemicals used in all experiments are purchased from Sigma, unless otherwise indicated.
Circular Dichroism (CD) Spectroscopy
CD spectra were obtained on AVIV Model 420 CD spectrometer (AVIV Biomedical, Lakewood, NJ) using glass cuvettes with 1-mm path length. Protein solutions were equilibrated for at least 24 h at 4 °C before measurement. Wavelength scans were collected from 190 to 260 nm in 0.5-nm steps with a 4-s averaging time, 1.0-nm bandwidth and repeated three times. Temperature scans were monitored from 15 to 70 °C at 225 nm with a 10-s averaging time and 1.5-nm bandwidth. Samples were equilibrated for 2 min at each temperature, and the temperature was increased at an average rate of 0.1 °C/min.
Differential Scanning Calorimetry (DSC)
DSC was performed on a NANO DSC II model 6100 (Calorimetry Sciences Corp, Lindon, UT). Each sample was re-dialyzed against PBS overnight before measurement to collect the dialyzed buffer as reference in the experiment. Sample solutions were loaded at 0 °C into the cell and heated at a rate of 1 °C/min to 100 °C.
Mass Spectrometry (MS)
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectra were acquired on a Microflex LT system (Bruker Corporation, Billerica, MA) with 70% laser intensity using standard LP (linear positive) 60-kDa method provided by the software. MALDI matrix was prepared by saturating sinapinic acid in solution containing 50% (v/v) acetonitrile and 0.3% (v/v) trifluoroacetic acid. A 6-μl aliquot of a 1 mg/ml sample was mixed with 24 μl of matrix, 1 μl of solution was plated onto a 96-spot target plate and allowed to dry.
Trypsin Cleavage Assay
Purified SCl-Fn in PBS buffer was incubated with 0.01 mg/ml (430 nm) of trypsin at 25 °C for 30 min, 60 min, 180 min, and overnight. The reaction was stopped by adding phenylmethylsulfonyl fluoride (PMSF) to 1 mm. Cleavage was visualized on an SDS-PAGE.
Fibronectin Binding Assay
The binding of Fn to immobilized SCl-Fn were measured using a solid-state binding assay. 50 μl of 10 or 100 μg of SCl-Fn proteins were coated onto high binding 96-well assay plates (R&D Systems, Minneapolis, MN) overnight at 4 °C. Denatured samples were first incubated in a 90 °C water bath for 30 min before coating. Plates were blocked by 3% BSA for 1 h at RT. 50 μl of human plasma Fn (Millipore FC010–10MG) was subsequently added to each collagen-coated well at a concentration of 100 μg/ml and incubated for 2 h. For dose-response assay, series concentrations of Fn at 100, 60, 20, 6, 2, 0.6, and 0 μg/ml were used. For the gelatin inhibition assay, 100 μg/ml of fibronectin was mixed with 100 μg/ml of denatured collagen type II overnight at 4 °C before being incubated on the collagen-coated plates. Bound Fn on plates were detected by mouse anti-human Fn C-terminal antibody (or mouse anti-human Fn GBD antibody for competitive binding assay, Millipore MAB1935 and MAB1892) at a 1:1000 dilution for 1 h. Secondary anti-mouse HRP antibody (Santa Cruz) at 1:3000 dilution was incubated for 1 h and 50 μl of 3,30,5,50-tetramethylbenzidine solution (Invitrogen) was added for the colorimetric reaction. Plates were washed by 200 μl of PBST 6 times between every step. Color was allowed to develop at room temperature for 5 min and 50 μl of 1 n HCl was added to stop the reaction. A450 nm was recorded from the 96-well plate using a Spectra Max M2 plate reader (Molecular Devices, Sunnyvale, CA) for data analysis. Appropriate controls (bovine collagen type II and SCl with no insertion) were included using the same setup. All assays were performed in triplicates. Human cellular Fn (Sigma F2518) was also tested in the above mentioned binding assay, no obvious differences were observed compared with human plasma fibronectin, consistent with an early study (19). This is likely due to the fact that they share the same GBD.
Megakaryocyte Cell Cultures
Human umbilical cord blood was purchased from The Cord Blood Bank of New York (Long Island, NY). Megakaryocytes (MK) were differentiated as previously described (25). Briefly, mononuclear cells were separated by layering blood onto Lympholyte (<1077 g/ml, Cedarlane, Hornby, Canada). Cells were then washed twice in phosphate-buffered saline (PBS) and CD34+ cells were separated by the immunomagnetic bead selection technique (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions. CD34+ cells were then cultured in Stem Span medium (Stem-Cell Technologies, Vancouver, Canada), supplemented with 10 ng/ml of TPO, IL-6, and IL-11 (PeproTech, Rocky Hill, NJ) at 37 °C in a 5% CO2 fully humidified atmosphere. At the end of the culture (13 days), 1 × 105 cells were collected and plated per well of a 24-well tissue culture plate. The plates were pretreated with 100 μg/ml of filter-sterilized SCl-FnG769-R792 and SCl-FnG769-R789 solution overnight. The plate was washed twice with PBS and subsequently incubated with 25 μg/ml of human cellular Fn (Sigma F2518). The plate was washed twice with PBS before being seeded with mature human MK.
Data Analysis
All quantitative analyses were performed in triplicate. Results presented were based on the averages of data points and S.D. as error bars. The significance level was determined by a p value using two sample paired Student's t test between the means of two samples. Raw CD and MS data were processed and plotted in Origin 6.0 (MicroCal Inc.). Results of the binding assay were plotted in Excel 2013 (Microsoft Corporation) and Bmax and Kd values were calculated by fitting the plot to specific saturation binding equation: Y = Bmax × X/(Kd + X) in GraphPad Prism 6.0 (GraphPad Software Inc.) with the S.E. obtained from the curve fit.
Note
The human collagen type II sequence used for the recombinant collagen design is based on UniProt entry P02458. Amino acids in the collagen domain are numbered starting from the beginning of the triple-helical region (residues 201–1214 of entry P02458), based on typical collagen notations. “O” or “Hyp” denotes 4-hydroxyproline in the text.
RESULTS
Design and Expression of Chimeric Bacterial Collagens
A collagen-like protein in S. pyogenes (Scl2.28) contains an N-terminal trimerization domain (V domain) followed by a CL domain with 79 Gly-Xaa-Yaa triplets (22, 26, 27). A construct in which the collagen domain was duplicated, VCLCL, was cloned and has been used in this study as the host for human collagen sequence insertions that may mediate binding to Fn. Both VCL and VCLCL constructs form homotrimers with a typical triple-helix structure and a thermal stability of 36–37 °C. These proteins are produced in high yields in E. coli using a cold-shock expression system (28), and were purified on a His tag column. Prolyl hydroxylase is not present in E. coli, so the expressed protein does not undergo any post-translational modification of Pro residues to Hyp.
The VCLCL construct, denoted as SCl, was modified by recombinant DNA technology to contain potential Fn binding sequences from human collagen, inserting (Gly-Xaa-Yaa)n sequences, where n = 3 to 8. Because the bacterial system produces homotrimers, sequences from the homotrimeric human α1(II) chain rather than the heterotrimeric type I collagen were inserted into the bacterial system (Fig. 1). The human collagen sequences were inserted between two tandem CL domains of bacterial collagen because this design was previously reported to be successful for conferring biological activity (24, 29). Recent studies by Erat et al. (15) indicated a peptide with residues 778–799 from the α1 chain of type I collagen bound tightly to Fn. These studies on denatured α1(I) collagen model peptides were used to guide the design of inserted human α1(II) sequences, because α1(II) and α1(I) are highly homologous in this region.
FIGURE 1.
Constructs of bacterial collagen-human collagen chimeras (SCl-Fn). Schematic diagram of type II collagen highlighting the putative fibronectin-binding region based on sequence homology with type I collagen (top panel), together with a diagram of the bacterial collagen constructs designed and produced (bottom panel). The constructs are named streptococcal collagen-like protein with Fn-binding site (SCl-Fn), supplemented with the starting and ending residue numbers of the inserted sequence in type II collagen. For example, the GLPGQRGER sequence is located in type II collagen triple-helix region from residue Gly784 to Arg792, thus it is denoted SCl-FnG784-V792. The MMP cleavage site in the sequence is marked by ∧.
Initially, the three triplets, GLPGQRGER, shown to participate in the β-sheet with the 8FnI module were inserted into the bacterial collagen construct (FnG784-R792) to see if this interacting sequence was sufficient for binding Fn. In further constructs, the human type II sequence was extended in the N-terminal direction to insert 4, 5, and 6 tripeptides, creating constructs SCl-FnG781-R792, SCl-FnG778-R792, and SCl-FnG775-R792. A second set of proteins was constructed to explore the requirements at the C terminus, inserting 4, 6, 7, or 8 tripeptides from the α1(II) chain: SCl-FnG772-V783, SCl-FnG769-P786, SCl-FnG769-R789, and SCl-FnG769-R792 (Fig. 1).
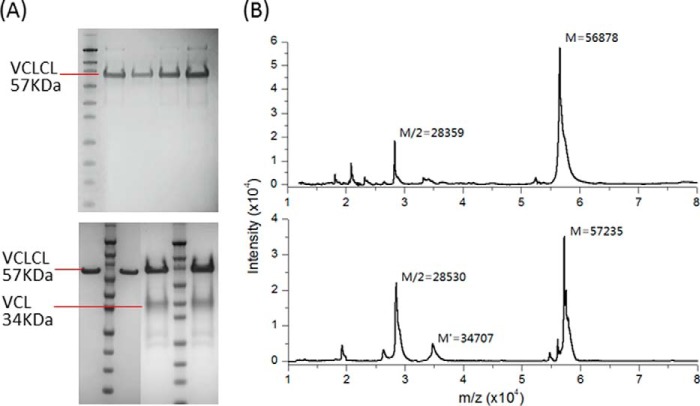
The recombinant proteins were expressed in E. coli and purified through a nickel-nitrilotriacetic acid column (immobilized metal ion affinity chromatography), yielding ∼50–100 mg/liter of chimeric collagen proteins. The purity of the proteins was confirmed by SDS-PAGE (Fig. 2A). All constructs showed one major band with the exception of the two proteins with the longest 7 and 8 triplet insertions, SCl-FnG769-R789 and SCl-FnG769-R792. On SDS-PAGE, these two proteins showed a major band that represent the full-length of SCl (VCLCL) constructs, together with one lower band corresponding to the size of VCL, suggesting that some portion had been degraded into VCL and CL fragments and/or that limited nutrients in the culture media caused premature termination of protein translation. The level of truncated fragments decreased after the protein production conditions were optimized (richer media: HyperBrothTM; shorter induction time; and protease inhibitors during bacteria lysis), but these shorter proteins were never fully eliminated. MALDI-TOF confirmed the identity of all purified proteins (Fig. 2B, Table 1).
FIGURE 2.
A, SDS-PAGE of SCl-Fn recombinant collagen constructs. Top panel from left to right: marker, SCl-FnG784-R792, SCl-FnG781-R792, SCl-FnG778-R792, and SCl-FnG775-R792. Bottom panel from left to right: SCl-FnG772-V783, marker, SCl-FnG769-P786, SCl-FnG769-R789, marker, and SCl-FnG769-R792. Lower bands in SCl-FnG769-R789 and SCl-FnG769-R792 represent truncated products with a single CL unit. Triple-helical proteins migrate slower than normal in SDS-PAGE, thus the Mr prediction from the protein standard is higher than the actual Mw. B, examples of MALDI-TOF mass spectra showing Mr of SCl-Fn. SCl-FnG775-R792, top panel; and SCl-FnG769-R792, bottom panel.
TABLE 1.
Biophysical characterization as well as Fn affinity of SCl-Fn constructs and bovine collagen control
The Kd values were obtained by fitting the binding curves to saturation binding equation: Y = Bmax × X/(Kd + X), which give an order of magnitude estimation to the relative Fn affinities of SCl-FnG775-R992, SCl-FnG769-R992 and bovine collagen II control in both native and denatured forms. Units for molecular mass is in kDa. The calculated Mw is based on amino acids sequence in the open reading frame of each construct. The actual Mw is acquired from MALDI-TOF results. Unit for MRE220 is deg × cm2 × dmol−1. The units for enthalpy is kJ × mol−1. Bmax is the maximum absorbance of the A450 value in arbitrary unit. The Kd value is in μg/ml. The Tm value is in °C.
| Mw measured (calculated) | CD |
DSCa |
Fn binding |
||||||
|---|---|---|---|---|---|---|---|---|---|
| MRE220 | Tm | Enthalpy | Tm | Bmax native | Bmax denatured | Kd native | Kd denatured | ||
| Bovine collagen II | (96.1) | 4306 | 41.0 | 0.74 ± 0.2 | 1.36 ± 0.1 | 13.22 ± 6.6 | 0.41 ± 0.1 | ||
| SCl | 55.2 (55.2) | 3502 | 36.8 | 4200 | 38.0 | ||||
| SCl-FnG784-R792 | 56.1 (56.2) | 2714 | 36.5 | 4100 | 37.9 | ||||
| SCl-FnG781-R792 | 56.3 (56.5) | 2220 | 36.2 | ||||||
| SCl-FnG778-R792 | 56.6 (56.8) | 1939 | 36.5 | ||||||
| SCl-FnG775-R792 | 56.9 (57.1) | 1918 | 36.2 | 3800 | 37.8 | 1.33 ± 0.1 | 1.51 ± 0.1 | 4.82 ± 1.0 | 1.27 ± 0.2 |
| SCl-FnG769-R792 | 57.2, 34.7 (57.6) | 1800 | 35.3 | 3700 | 37.1 | 1.26 ± 0.1 | 1.23 ± 0.1 | 5.04 ± 1.3 | 2.15 ± 0.6 |
| SCl-FnG769-R789 | 57.1, 34.5 (57.2) | 1987 | 35.5 | ||||||
| SCl-FnG769-P786 | 56.9 (56.9) | 2795 | 36.6 | ||||||
| SCl-FnG772-V783 | 56.2 (56.4) | 2952 | 36.5 | ||||||
a Blank cells in DSC columns indicate value not determined for that sample, blank cells in the Fn binding column indicate no strong binding has been observed for that sample.
Structural and Stability Characterization of the Chimeric Proteins
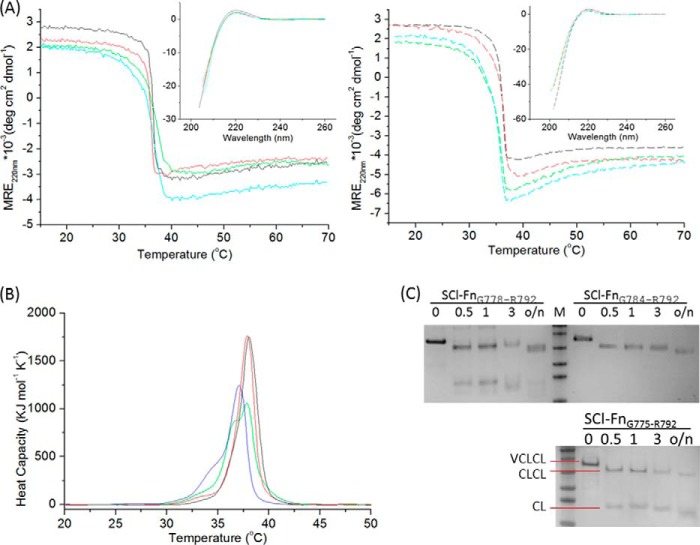
The CD spectra of all chimeric proteins showed a characteristic maximum near 220 nm, indicating that they adopted a triple-helix conformation (Fig. 3A, Table 1). The constructs with the longest insertions generally had somewhat lower MRE220 nm values, which could reflect the presence of denatured fragments or a distortion of the triple-helix. The thermal stability of the triple-helix was very similar for all constructs. Monitoring the MRE220 nm versus temperature indicated a sharp thermal transition, with a Tm near 36.5 °C for all chimeric proteins, except those with the longest 7 and 8 tripeptide insertions, where Tm values were slightly lower (35.5 and 35.2 °C). The DSC profile of the SCl control showed a sharp transition near 38 °C, with a calorimetric enthalpy of 4200 kJ/mol (Fig. 3B, Table 1). The insertion of 3 tripeptides, GLPGQRGER, in SCl-FnG784-R792 gave a DSC profile with the same Tm value and only a slight decrease in enthalpy. However, when 6 or 8 triplets were inserted, the enthalpies were lowered by ∼400–500 kJ/mol. For both of these proteins, SCl-FnG775-R792 and SCl-FnG769-R792, a shoulder of lower temperature was visible in addition to the main peak, consistent with a small fraction of truncated or denatured protein. The position of the major DSC thermal transition for SCl-FnG769-R792 also dropped ∼1 °C compared with the control, similar to the CD Tm results. All DSC transitions were at higher temperatures than the CD Tm values because of the faster heating rate (30).
FIGURE 3.
Structural and thermal analysis of SCl-Fn recombinant collagens. A, circular dichroism temperature scans showing the Tm values of SCl-FnG784-R792 (solid black), SCl-FnG781-R792 (solid red), SCl-FnG778-R792 (solid green), and SCl-FnG775-R792 (solid cyan) in the left panel and SCl-FnG772-V783 (dash black), SCl-FnG769-P786 (dash red), SCl-FnG769-R789 (dash green), and SCl-FnG769-R792 (dash cyan) in the right panel. The CD wavelength spectra of these proteins are shown in the inset. Insets share the same y axis label with the parent figures. B, differential scanning calorimetry of SCl-FnG775-R792 (green), SCl-FnG769-R792 (blue), and SCl-FnG784-R792 (red) as well as SCl control without insertion (black) showing heat capacity as a function of temperature. Areas under the curve indicate the calorimetric enthalpy. C, trypsin digestion of SCl-FnG775-R792, SCl-FnG778-R792, and SCl-FnG784-R792 on a time course of 0.5, 1, and 3 h and overnight (>16 h).
Trypsin digestion was performed on three of the collagen constructs, SCl-FnG775-R792, SCl-FnG778-R792, SCl-FnG784-R792, to ascertain if the inserted human sequences maintained a tight triple-helix (Fig. 3C). The V domain of SCl was cleaved off quickly, yielding a slightly lower band on SDS-PAGE, corresponding to the size of the CL-CL unit. With trypsin digestion over long time periods there appears to be a gradual digestion from the ends, leading to slightly smaller bands. SCl-FnG775-R792 and SCl-FnG778-R792 proteins with 5 triplet and 6 triplet insertions, respectively, showed partial cleavage at the insertion site, yielding a lower band corresponding to the size of a single CL domain. In contrast, SCl-FnG784-R792 with the 3 triplet insertion was not susceptible at the insertion site, with no single CL unit detectable on the gel.
Minimum Amino Acid Sequence Requirement for Fibronectin Binding
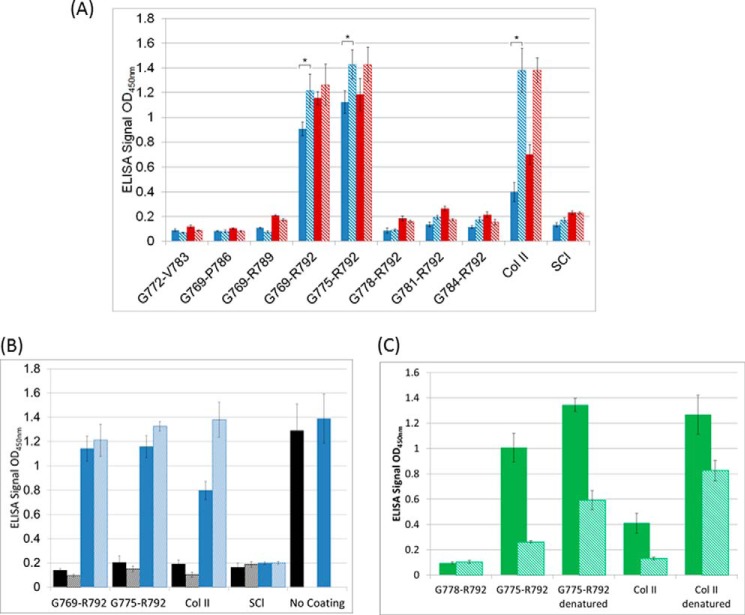
Solid-state binding assays were used to measure soluble Fn binding to solid-phase collagen adsorbed on a plastic surface. Such a solid-phase assay may mimic Fn interactions with insoluble collagen fiber bundles in the extracellular matrix (31, 32). These binding assays were carried out on all 8 chimeric bacterial collagen constructs, as well as type II collagen and the control SCl, to determine the minimum amino acid sequence for binding of Fn to native recombinant collagens. The collagen samples were coated on ELISA plates and binding was assessed for two different Fn concentrations (Fig. 4A). The binding assays were performed at room temperature, and because all of the triple-helical proteins had Tm values greater than 30 °C, these assays represent binding of Fn to native triple-helical collagen. Fn did not show any binding to the original SCl protein, indicating there is no binding site within the original bacterial collagen CL sequence. In the first set of sequences, Fn did not bind to SCl-FnG784-R792, SCl-FnG781-R792, and SCl-FnG778-R792, indicating the inserted sequences were not sufficient for binding. However, Fn did bind to SCl-FnG775-R792. In the second set of recombinant proteins with a progressive increase of insertion length at the C terminus, SCl-FnG772-V783, SCl-FnG769-P786, and SCl-FnG769-R789 did not bind Fn, whereas SCl-FnG769-R792 did bind. These studies indicated that the 775GLA triplet on the N terminus and the 790GER triplet on the C terminus were required for Fn binding, bracketing the essential sequence. These results define the minimal requirement for Fn binding to human type II collagen as the 6-triplet sequence, GLAGQRGIVGLPGQRGER, in a triple-helix context. Native bovine type II did bind to Fn using the same binding assay, but the signal observed was weaker than seen for the two recombinant collagens SCl-FnG775-R792 and SCl-FnG769-R792.
FIGURE 4.
Solid-phase binding assay of SCl-Fn recombinant collagen to soluble human plasma fibronectin. A, Fn binding affinity of all SCl-Fn constructs in both triple-helical (solid) and denatured (hatched) states. Two concentrations of fibronectin were used, 10 (blue) and 100 μg/ml (red). The binding signals of native SCl-FnG769-R792 and SCl-FnG775-R792 are statistically different from their respective signals in the denatured state, as evaluated using a paired t test; an asterisk (*) is used to indicate pairs where the means are statistically different (p < 0.05). B, binding of SCl-FnG769-R792 and SCl-FnG775-R792 to Fn prevented the recognition of Fn by the anti-Fn GBD antibody (black). A non-competitive anti-Fn C-terminal antibody (blue) was used to show the presence of Fn in ELISA plates with no competition to the gelatin-binding domain. Both native (solid) and denatured states (hatched) were tested. The beginning and ending residues of the inserted sequence are used to represent each SCl-Fn construct. Bovine collagen type II and recombinant collagen SCl with no insertion were used as control. C, soluble gelatin (denatured collagen type II) could inhibit the binding between collagen and fibronectin. When denatured collagen type II was incubated together with Fn in a collagen-coated plate, the binding signal (hatched green) observed were lower than the ones without gelatin inhibition (solid green).
To investigate if the minimal required Fn binding sequence was the same in the denatured and native states of collagen, solid-state binding assays were performed after incubating all recombinant collagen samples at 90 °C for 30 min. Again, Fn bound only to constructs SCl-FnG775-R792 and SCl-FnG769-R792 in the denatured form (Fig. 4A). This indicated that the minimum amino acid sequence requirement for Fn binding was the same for denatured and native collagens. The binding was also assayed in reverse by immobilizing Fn on the plate and monitoring the binding of soluble SCl-Fn through anti-His tag antibody. Similar results were observed indicating the same sequence requirement for Fn affinity (data not shown), but we were unable to compare this result with bovine collagen type II due to the difficulty in getting His-tagged animal collagen and the lack of an appropriate antibody to detect both animal and bacterial collagen.
To confirm the GBD of Fn was responsible for binding the recombinant collagens, a competitive binding assay was performed that utilized an anti-Fn GBD antibody, as well as an antibody to the distant C terminus of Fn. Fibronectin that bound to plates coated with SCl-FnG775-R792, SCl-FnG769-R792, or type II collagen was detectable by an antibody to the C terminus of Fn, but could not be identified by an antibody to the Fn GBD domain (Fig. 4B). This observation was consistent with the GBD on Fn being occupied by the recombinant collagen proteins or type II collagen, such that the site was no longer accessible to this specific antibody targeting the GBD through a steric exclusion mechanism. Thus, the interaction between SCl-Fn chimeric proteins and Fn was a site-specific interaction that involved the GBD of Fn.
To investigate whether this specific interaction between bacterial collagen and Fn could be disrupted by soluble gelatin, a gelatin inhibition assay was performed by premixing Fn with denatured collagen type II before incubating on collagen-coated plates. The results indicated that soluble gelatin serves as an effective inhibitor for binding between Fn and immobilized collagen. The inhibition effect was more dramatic on recombinant collagen and native collagen II as compared with coated denatured collagen type II (Fig. 4C). This may imply that denatured collagen type II has a lower dissociation constant (Kd) that makes it more effectively compete with recombinant collagen or native collagen type II for the binding site on Fn.
Quantitation of Fibronectin Binding Affinity for Native and Denatured Recombinant Chimeras and Type II Collagen
The two recombinant constructs that bound Fn were SCl-FnG775-R792 and the construct with the same insert plus two additional N-terminal triplets, SCl-FnG769-R792. The Fn binding affinity of these two recombinant constructs, as well as type II collagen, was studied in both the native and denatured states at varying concentrations of Fn, and typical saturation kinetics were observed for all curves (Fig. 5). Fn bound more tightly to SCl-FnG775-R792 and SCl-FnG769-R792 in their denatured states than in their native triple-helical form, demonstrating a preference of Fn for the denatured state in the context of bacterial collagen. The two chimeric proteins showed similar binding in the native state, but the shorter insertion bound somewhat more tightly in the denatured form (Table 1).
FIGURE 5.
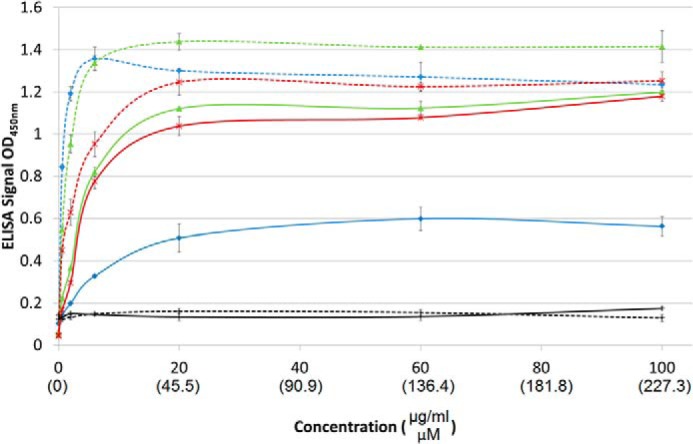
Dose-response of Fn binding to collagens adsorbed onto ELISA plates: SCl-FnG775-R792 (green), SCl-FnG769-R792 (red), bovine collagen type II (blue), and SCl with no insertion (black). Collagen in a native triple-helical state is represented by solid lines, whereas denatured collagen is represented by dashed lines.
The difference in Fn binding between native versus denatured forms was more striking for collagen type II than for the recombinant proteins. In the native triple-helical form, SCl-FnG775-R792 and SCl-FnG769-R792 bound more efficiently than native bovine collagen type II (Table 1). However, after denaturation, type II collagen showed a dramatic increase in affinity, so that its Kd is now 3–5 times lower than the denatured SCl-FnG775-R792 and SCl-FnG769-R792 values. The higher Fn affinity seen for denatured type II collagen compared with the chimeric collagens could reflect additional Fn-binding sites within mammalian collagen (17, 33, 34).
SCl-Fn Promote Cell Adhesion and Growth
The solid-state binding assays reported here demonstrated that insertion of residues α1(II) Gly775–Arg792 within the bacterial collagen context conferred the ability of these proteins to bind to Fn. To investigate whether this Fn binding would be active in a biological context, the influence on megakaryocte (MK) maturation and proplatelet formation mediated by SCl-Fn was assayed. Human mature MK were plated on SCl-FnG769-R789, which did not show binding in the solid-state binding assay, and SCl-FnG769-R792, which did exhibit Fn binding. The recombinant collagens were coated on tissue culture plastic followed by incubation with 25 μg/ml of human cellular Fn. As shown in Fig. 6, MK adhered preferentially to SCl-FnG769-R792 as compared with the SCl-FnG769-R789-coated plate. Controls were performed by seeding MK on uncoated or bovine type II collagen-coated plate wells, showing, respectively, low and high MK adhesion. At least three Fn binding integrins are expressed by MK (α4β1, α5β1, and αIIβ3) (35) and previous data demonstrated a pivotal role for these integrins in regulating MK development and interaction with the bone marrow extracellular matrix environment (25, 36, 37). The observed increase in MK adhesion and proplatelet formation on SCl-FnG769-R792 is consistent with its higher Fn affinity in solid-phase binding assays.
FIGURE 6.
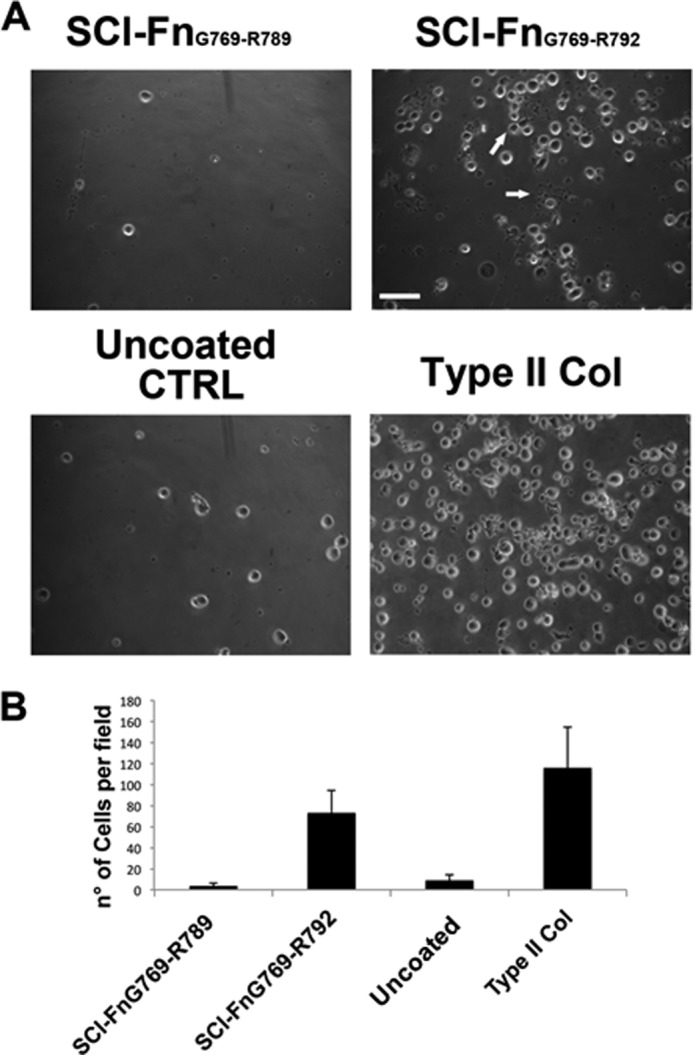
Megakaryocytes cultured on different SCl-Fn recombinant collagens. A, human mature MK were plated on two different SCl-Fn constructs: SCl-FnG769-R789, which did not bind Fn, and SCl-FnG769-R792, which did bind Fn. SCl-Fn-coated plates were preincubated with human cellular fibronectin prior to cell plating. After 16 h, MK showed higher adhesion and proplatelet formation on SCl-FnG769-R792 (right panel) compared with SCl-FnG769-R789 (left panel). In parallel controls were performed by seeding MK on uncoated and bovine type II collagen-coated plates. Arrows show proplatelet bearing MK. Scale bar = 100 μm. B, adherent cells were counted by phase-contrast microscopy (magnification ×40). Reported results are the mean ± S.D. of at least 10 randomly chosen fields in three independent experiments.
DISCUSSION
Collagen sequences from the putative fibronectin-binding region of human collagen type II were included within a triple-helical bacterial collagen context, expressed as recombinant proteins, and studied for their ability to bind Fn. The S. pyogenes bacterial collagen Scl2.28 forms a stable triple-helical structure and has no known biological interactions or activities (38). Previous studies show the insertion of human collagen sequences within the recombinant triple-helical protein conferred biological activity similar to that found in human collagens. For example, recombinant Scl2.28 proteins with inserted integrin binding sequences (GFPGER, GFPGEN, and GLPGER) were shown to bind cell surface integrins in vitro and to mediate cell binding (23, 39), whereas insertion of the GRPGKRGKQGQK sequence led to heparin binding (40). Incorporation of 6 triplets from the human collagen type III MMP cleavage site led to digestion by MMP-1 at the same site as in native type III collagen (29). All of these collagen sequences only have biological activity when in a triple helix context. In contrast, Fn is known to bind to denatured collagen even stronger than native collagen. The bacterial collagen system offers an opportunity to define Fn binding to native and denatured recombinant proteins containing different lengths of human collagen sequences proposed to interact with Fn.
Recent studies on the binding of various unfolded (Gly-Xaa-Yaa)n peptides to Fn are extended here to include residues in a recombinant triple-helix context, allowing definition of the minimum sequence requirement for Fn binding in both the native and denatured states. The original bacterial collagen SCl (VCLCL) without genetic modification could not bind Fn, but insertion of sufficient lengths of the amino acid sequence from the human collagen α1(II) chain, near the collagenase cleavage site, conferred Fn binding activity. Solid-state binding assays on the chimeric collagen showed that the 6-triplet type II collagen sequence, 775GLAGQRGIVGLPGQRGER792, was the minimal requirement for Fn binding in both triple-helical and denatured conditions. Although residues GLPGQRGER appear to be key in interacting with the 8FnI module in the crystal structure (15), full-length Fn did not bind to the SCl-FnG784-R792 protein, indicating these three triplets may be necessary but are not sufficient for binding to the triple-helical or denatured states in our system. Extending the required 6 triplet sequence two triplets further in the N-terminal direction did not increase the binding to Fn. It is likely that these 6 triplets also represent the binding site in the α1(1) chain, which differs from the α1(II) sequence in only two residues in this region, L776I and I782V, showing conservative changes in these positions.
The collagen sequence defined here as the minimum for Fn binding falls in a region known to be a ligand binding hot spot in fibrillar collagens (41). Within the required 6-tripeptide Fn binding sequence, the three triplets in GLAGQRGIV that precede GLPGQRGER include the well characterized G∧LA collagenase cleavage site. Human collagenases, such as MMP-1 and MMP-13, cleave between Gly775 and Leu776 and divide collagen chains into ¼ and ¾ fragments (42, 43). A recent study indicated that Leu residues at positions 776 and 785 were critical for MMP activity through direct interaction with the MMP catalytic and hemopexin domains, respectively (44). Because the minimum Fn binding sequence defined here overlaps with the key residues for MMP cleavage, MMP and Fn could compete for binding at this site.
It is well known that Fn binds more tightly to denatured collagen than to native triple-helical collagen (19), and this preference for the unfolded state was retained in the chimeric bacterial collagens studied here. There is much interest in how Fn can bind to collagen in both the denatured as well as the native states, and whether both binding abilities play a physiological role. In our system, the same GLAGQRGIVGLPGQRGER sequence was found to be required for Fn binding to gelatin or native triple-helical collagen. It is not clear how Fn binds to a sequence within the tightly packed native collagen, which shows resistance to digestion by most enzymes and a lack of immunogenicity. If the GLPGQRGER sequence is to participate in a β-sheet with the 8FnI module, as suggested from the crystal structure of the 8–9FnI module with peptide Gly778–Gly799 (15), this sequence would have to be pulled out from the supercoil of the triple-helix and extended from the 2.9-Å rise/residue of a triple-helix to the 3.4-Å rise/residue of a β strand. This is not an obvious structural transition because the sequence GLPGQRGER is predicted to have a relatively high propensity for triple-helix formation (45) and a relatively low propensity for a β strand (46). However, a range of observations suggests that this may be a flexible region, which is able to adopt an alternative equilibrium conformation. There is only one imino acid residue within the entire 6 triplet region (6%), and no Pro/Hyp within the first 3 triplets, compared with an overall 20% imino acid content within type I/II collagens. The importance of this imino acid deficiency is illustrated by an early study on transgenic mouse models showing that introduction of a Pro at a residue immediately before or after the Gly775 collagenase cleavage site (changing Gln-Gly775-Leu-Ala to either Pro-Gly775-Leu-Ala or Gln-Gly775-Pro-Ala) interferes with both MMP cleavage and Fn binding (47). In addition, two of the first three triplets, GLA and GIV, have a very low triple-helix propensity (45). Computational studies indicate this region has an alternative “vulnerable” conformation in equilibrium with the triple-helical state (48, 49), which is susceptible to collagenases. It is possible that a vulnerable conformation in this relatively loose triple helix region at the MMP cleavage site is transiently extended C terminally to unwind the GLPGQRGER sequence, so it can participate in the Fn β-sheet structure. Any alternative state must exist on a fast time scale in equilibrium with a well folded triple helix. Consistent with this hypothesis, the recombinant chimeric collagens with longer insertions do show partial susceptibility to trypsin at the human sequence insertion site, even though a fraction of the protein remained trypsin resistant after 24 h.
There is only one Pro in the 6 triplets needed for Fn binding, at position 786 in Gly-Leu-Pro786, and it is located in the Y position, where it normally undergoes hydroxylation to Hyp in type I collagen (50). The bacterially expressed recombinant collagen, which bound Fn did not undergo any post-translational modification and thus did not contain Hyp at position 786. These results indicate that Hyp is not essential for collagen or gelatin binding to Fn, just as it was found not to be essential for collagen binding to integrin or heparin or for MMP-1 cleavage. However, it is possible that the absence of Hyp may be responsible for some differences in Fn affinity observed between the recombinant collagen and type II collagen, because Hyp is known to affect the triple helix structure and stability and to form direct interactions with some proteins and receptors (51, 52). It is expected that the absence of Hyp786 will decrease local stability and rigidity, and in the context of Fn binding, a looser triple helix should lead to tighter binding. This is consistent with the ability of trypsin to digest the recombinant constructs with 5 and 6 tripeptides, but not type II collagen, and the apparent tighter binding of the native triple-helical bacterial constructs with 5–6 tripeptides compared with type II collagen.
Because SCl-Fn was produced in a E. coli system, the scalability and easy modification of the recombinant system also raise the possibility for this chimeric protein to be a novel biomaterial. Previously, the potential to obtain collagenous proteins with controllable biological activity modules using the bacterial collagen system has been demonstrated, leading to improved human mesenchymal stem cell growth in two- and three-dimensional cultures (24). To further demonstrate SCl-Fn as a suitable bioactive material, human megakaryocytes were cultured on SCl-Fn in the presence of Fn. SCl-Fn could effectively immobilize soluble Fn in the cell culture and subsequently support the attachment and differentiation of megakaryocytes. Fibronectin has been known to play a key role in supporting hemopoiesis (53), megakaryocyte development, and platelet release (54). It has been previously demonstrated that megakaryocytes are involved in the process of fibronectin fibrillogenesis and that, in turn, fibronectin plays a pivotal role in regulating megakaryocyte-type I collagen interaction in the bone marrow environment (37). Thus, SCl-Fn, as a Fn binding biomaterial, could support megakaryocyte function with the potential for bone marrow modeling and platelet production.
In conclusion, Fn binding affinity was conferred on bacterial collagen by incorporating a minimum of 6 triplets of human collagen type II sequence from residue Gly775–Arg792. We demonstrated the potential of this bacterial collagen system as a model to study collagen and Fn binding, and its practical use in promoting cell attachment and differentiation. Defining this fibronectin binding sequence and demonstrating its function in the recombinant system can help guide future recombinant protein design to build biomaterials with selective Fn binding ability.
Acknowledgments
We thank Dr. Jean Schwarzbauer from the Department of Molecular Biology, Princeton University, for valuable reagents and suggestions; Dr. John Ramshaw from the Commonwealth Scientific and Industrial Research Organisation (CSIRO) of Australia for helpful discussions; and Dr. David Wilbur, Tufts University Chemistry Department, for allowing us to access the MALDI-TOF MS equipment.
This work was supported, in whole or in part, by National Institutes of Health Grant EB011620 (to B. B. and D. K.).
- FN
- fibronectin
- GBD
- gelatin-binding domain
- Hyp
- hydroxyproline
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DSC
- differential scanning calorimetry
- MK
- megakaryocytes
- CL
- collagen-like
- SCl-Fn
- streptococcal collagen-like protein with Fn-binding site.
REFERENCES
- 1. Brodsky B., Ramshaw J. A. (1997) The collagen triple-helix structure. Matrix Biol. 15, 545–554 [DOI] [PubMed] [Google Scholar]
- 2. Kielty C. M., Grant M. E. (2002) The Collagen family: structure, assembly and organization in the extracellular matrix. In Connective Tissue and Its Heritable Disorders: Molecular, Genetic, and Medical Aspects (Royce P. M., Steinmann B., eds) 2nd Ed., pp. 158–222, Wiley-Liss, New York [Google Scholar]
- 3. Veit G., Kobbe B., Keene D. R., Paulsson M., Koch M., Wagener R. (2006) Collagen XXVIII, a novel von Willebrand Factor A domain-containing protein with many imperfections in the collagenous domain. J. Biol. Chem. 281, 3494–3504 [DOI] [PubMed] [Google Scholar]
- 4. Shoulders M. D., Raines R. T. (2009) Collagen structure and stability. Annu. Rev. Biochem. 78, 929–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kadler K. E., Baldock C., Bella J., Boot-Handford R. P. (2007) Collagens at a glance. J. Cell Sci. 120, 1955–1958 [DOI] [PubMed] [Google Scholar]
- 6. Piez K. A., Reddi A. H. (1984) Extracellular Matrix Biochemistry, Elsevier, New York [Google Scholar]
- 7. Leitinger B., Hohenester E. (2007) Mammalian collagen receptors. Matrix Biol. 26, 146–155 [DOI] [PubMed] [Google Scholar]
- 8. Di Lullo G. A., Sweeney S. M., Korkko J., Ala-Kokko L., San Antonio J. D. (2002) Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J. Biol. Chem. 277, 4223–4231 [DOI] [PubMed] [Google Scholar]
- 9. Schwarzbauer J. E., DeSimone D. W. (2011) Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb. Perspect. Biol. 3, pii: a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hynes R. (1985) Molecular biology of fibronectin. Annu. Rev. Cell Biol. 1, 67–90 [DOI] [PubMed] [Google Scholar]
- 11. Dessau W., Adelmann B. C., Timpl R. (1978) Identification of the sites in collagen α-chains that bind serum anti-gelatin factor (cold-insoluble globulin). Biochem. J. 169, 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Engvall E., Ruoslahti E. (1977) Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int. J. Cancer 20, 1–5 [DOI] [PubMed] [Google Scholar]
- 13. Ingham K. C., Brew S. A., Isaacs B. S. (1988) Interaction of fibronectin and its gelatin-binding domains with fluorescent-labeled chains of type I collagen. J. Biol. Chem. 263, 4624–4628 [PubMed] [Google Scholar]
- 14. Katagiri Y., Brew S. A., Ingham K. C. (2003) All six modules of the gelatin-binding domain of fibronectin are required for full affinity. J. Biol. Chem. 278, 11897–11902 [DOI] [PubMed] [Google Scholar]
- 15. Erat M. C., Slatter D. A., Lowe E. D., Millard C. J., Farndale R. W., Campbell I. D., Vakonakis I. (2009) Identification and structural analysis of type I collagen sites in complex with fibronectin fragments. Proc. Natl. Acad. Sci. U.S.A. 106, 4195–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Erat M. C., Schwarz-Linek U., Pickford A. R., Farndale R. W., Campbell I. D., Vakonakis I. (2010) Implications for collagen binding from the crystallographic structure of fibronectin 6FnI1–2FnII7FnI. J. Biol. Chem. 285, 33764–33770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Erat M. C., Sladek B., Campbell I. D., Vakonakis I. (2013) Structural analysis of collagen type I interactions with human fibronectin reveals a cooperative binding mode. J. Biol. Chem. 288, 17441–17450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Graille M., Pagano M., Rose T., Ravaux M. R., van Tilbeurgh H. (2010) Zinc induces structural reorganization of gelatin binding domain from human fibronectin and affects collagen binding. Structure 18, 710–718 [DOI] [PubMed] [Google Scholar]
- 19. Engvall E., Ruoslahti E., Miller E. J. (1978) Affinity of fibronectin to collagens of different genetic types and to fibrinogen. J. Exp. Med. 147, 1584–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kleinman H. K., McGoodwin E. B., Martin G. R., Klebe R. J., Fietzek P. P., Woolley D. E. (1978) Localization of the binding site for cell attachment in the α1(I) chain of collagen. J. Biol. Chem. 253, 5642–5646 [PubMed] [Google Scholar]
- 21. Lwebuga-Mukasa J. S., Madri J. A., Albert J., Furthmayr H. (1984) Studies on the interaction of human plasma-fibronectin with native type I calf skin collagen molecules using the rotary shadowing technique. Coll. Relat. Res. 4, 95–110 [DOI] [PubMed] [Google Scholar]
- 22. Xu Y., Keene D. R., Bujnicki J. M., Höök M., Lukomski S. (2002) Streptococcal Scl1 and Scl2 proteins form collagen-like triple helices. J. Biol. Chem. 277, 27312–27318 [DOI] [PubMed] [Google Scholar]
- 23. Seo N., Russell B. H., Rivera J. J., Liang X., Xu X., Afshar-Kharghan V., Höök M. (2010) An engineered alpha1 integrin-binding collagenous sequence. J. Biol. Chem. 285, 31046–31054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. An B., DesRochers T. M., Qin G., Xia X., Thiagarajan G., Brodsky B., Kaplan D. L. (2013) The influence of specific binding of collagen silk chimeras to silk biomaterials on hMSC behavior. Biomaterials 34, 402–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Balduini A., Pallotta I., Malara A., Lova P., Pecci A., Viarengo G., Balduini C. L., Torti M. (2008) Adhesive receptors, extracellular proteins and myosin IIA orchestrate proplatelet formation by human megakaryocytes. J. Thromb. Haemost. 6, 1900–1907 [DOI] [PubMed] [Google Scholar]
- 26. Mohs A., Silva T., Yoshida T., Amin R., Lukomski S., Inouye M., Brodsky B. (2007) Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline. J. Biol. Chem. 282, 29757–29765 [DOI] [PubMed] [Google Scholar]
- 27. Yoshizumi A., Yu Z., Silva T., Thiagarajan G., Ramshaw J. A., Inouye M., Brodsky B. (2009) Self-association of Streptococcus pyogenes collagen-like constructs into higher order structures. Protein Sci. 18, 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qing G., Ma L. C., Khorchid A., Swapna G. V., Mal T. K., Takayama M. M., Xia B., Phadtare S., Ke H., Acton T., Montelione G. T., Ikura M., Inouye M. (2004) Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 22, 877–882 [DOI] [PubMed] [Google Scholar]
- 29. Yu Z., Visse R., Inouye M., Nagase H., Brodsky B. (2012) Defining requirements for collagenase cleavage in collagen type III using a bacterial collagen system. J. Biol. Chem. 287, 22988–22997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Persikov A. V., Xu Y., Brodsky B. (2004) Equilibrium thermal transitions of collagen model peptides. Protein Sci. 13, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh P., Carraher C., Schwarzbauer J. E. (2010) Assembly of fibronectin extracellular matrix. Annu. Rev. Cell Dev. Biol. 26, 397–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Akiyama S. K., Yamada K. M. (1983) Fibronectin in disease. Monogr. Pathol. 24, 55–96 [PubMed] [Google Scholar]
- 33. Ingham K. C., Brew S. A., Migliorini M. (2002) Type I collagen contains at least 14 cryptic fibronectin binding sites of similar affinity. Arch. Biochem. Biophys. 407, 217–223 [DOI] [PubMed] [Google Scholar]
- 34. Guidry C., Miller E. J., Hook M. (1990) A second fibronectin-binding region is present in collagen α chains. J. Biol. Chem. 265, 19230–19236 [PubMed] [Google Scholar]
- 35. Fox N. E., Kaushansky K. (2005) Engagement of integrin α4β1 enhances thrombopoietin-induced megakaryopoiesis. Exp. Hematol. 33, 94–99 [DOI] [PubMed] [Google Scholar]
- 36. Mazharian A., Thomas S. G., Dhanjal T. S., Buckley C. D., Watson S. P. (2010) Critical role of Src-Syk-PLCγ2 signaling in megakaryocyte migration and thrombopoiesis. Blood 116, 793–800 [DOI] [PubMed] [Google Scholar]
- 37. Malara A., Gruppi C., Rebuzzini P., Visai L., Perotti C., Moratti R., Balduini C., Tira M. E., Balduini A. (2011) Megakaryocyte-matrix interaction within bone marrow. New roles for fibronectin and factor XIII-A. Blood 117, 2476–2483 [DOI] [PubMed] [Google Scholar]
- 38. Caswell C. C., Barczyk M., Keene D. R., Lukomska E., Gullberg D. E., Lukomski S. (2008) Identification of the first prokaryotic collagen sequence motif that mediates binding to human collagen receptors, integrins α2β1 and α11β1. J. Biol. Chem. 283, 36168–36175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cosgriff-Hernandez E., Hahn M. S., Russell B., Wilems T., Munoz-Pinto D., Browning M. B., Rivera J., Höök M. (2010) Bioactive hydrogels based on designer collagens. Acta Biomater. 6, 3969–3977 [DOI] [PubMed] [Google Scholar]
- 40. Peng Y. Y., Stoichevska V., Schacht K., Werkmeister J. A., Ramshaw J. A. (2013) Engineering multiple biological functional motifs into a blank collagen-like protein template from Streptococcus pyogenes. J. Biomed. Mater. Res. Part A, 10.1002/jbm.a.34898 [DOI] [PubMed] [Google Scholar]
- 41. Sweeney S. M., Orgel J. P., Fertala A., McAuliffe J. D., Turner K. R., Di Lullo G. A., Chen S., Antipova O., Perumal S., Ala-Kokko L., Forlino A., Cabral W. A., Barnes A. M., Marini J. C., San Antonio J. D. (2008) Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J. Biol. Chem. 283, 21187–21197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Visse R., Nagase H. (2003) Matrix metalloproteinases and tissue inhibitors of metalloproteinases. Structure, function, and biochemistry. Circ. Res. 92, 827–839 [DOI] [PubMed] [Google Scholar]
- 43. Han S., Makareeva E., Kuznetsova N. V., DeRidder A. M., Sutter M. B., Losert W., Phillips C. L., Visse R., Nagase H., Leikin S. (2010) Molecular mechanism of type I collagen homotrimer resistance to mammalian collagenases. J. Biol. Chem. 285, 22276–22281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Manka S. W., Carafoli F., Visse R., Bihan D., Raynal N., Farndale R. W., Murphy G., Enghild J. J., Hohenester E., Nagase H. (2012) Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1. Proc. Natl. Acad. Sci. U.S.A. 109, 12461–12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Persikov A. V., Ramshaw J. A., Brodsky B. (2005) Prediction of collagen stability from amino acid sequence. J. Biol. Chem. 280, 19343–19349 [DOI] [PubMed] [Google Scholar]
- 46. Sen T. Z., Jernigan R. L., Garnier J., Kloczkowski A. (2005) GOR V server for protein secondary structure prediction. Bioinformatics 21, 2787–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dzamba B. J., Wu H., Jaenisch R., Peters D. M. (1993) Fibronectin binding site in type I collagen regulates fibronectin fibril formation. J. Cell Biol. 121, 1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nerenberg P. S., Salsas-Escat R., Stultz C. M. (2008) Do collagenases unwind triple-helical collagen before peptide bond hydrolysis? Reinterpreting experimental observations with mathematical models. Proteins 70, 1154–1161 [DOI] [PubMed] [Google Scholar]
- 49. Stultz C. M., Edelman E. R. (2003) A structural model that explains the effects of hyperglycemia on collagenolysis. Biophys. J. 85, 2198–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fietzek P. P., Rexrodt F. W., Hopper K. E., Kühn K. (1973) The covalent structure of collagen. 2. The amino-acid sequence of α1-CB7 from calf-skin collagen. Eur. J. Biochem. 38, 396–400 [DOI] [PubMed] [Google Scholar]
- 51. Perret S., Eble J. A., Siljander P. R., Merle C., Farndale R. W., Theisen M., Ruggiero F. (2003) Prolyl hydroxylation of collagen type I is required for efficient binding to integrin α1β1 and platelet glycoprotein VI but not to α2β1. J. Biol. Chem. 278, 29873–29879 [DOI] [PubMed] [Google Scholar]
- 52. Asselin J., Knight C. G., Farndale R. W., Barnes M. J., Watson S. P. (1999) Monomeric (glycine-proline-hydroxyproline)10 repeat sequence is a partial agonist of the platelet collagen receptor glycoprotein VI. Biochem. J. 339, 413–418 [PMC free article] [PubMed] [Google Scholar]
- 53. Jiang F., Jia Y., Cohen I. (2002) Fibronectin- and protein kinase C-mediated activation of ERK/MAPK are essential for proplateletlike formation. Blood 99, 3579–3584 [DOI] [PubMed] [Google Scholar]
- 54. Matsunaga T., Fukai F., Kameda T., Shide K., Shimoda H., Torii E., Kamiunten A., Sekine M., Yamamoto S., Hidaka T., Kubuki Y., Yokokura S., Uemura M., Matsuoka A., Waki F., Matsumoto K., Kanaji N., Ishii T., Imataki O., Dobashi H., Bandoh S., Shimoda K. (2012) Potentiated activation of VLA-4 and VLA-5 accelerates proplatelet-like formation. Ann. Hematol. 91, 1633–1643 [DOI] [PubMed] [Google Scholar]