Background: Circadian gene expression in mammals is driven by rhythmic CLOCK·BMAL1 activities.
Results: Gm129 binds to the CLOCK·BMAL1 complex on DNA and represses its transcriptional activator activity to regulate the circadian gene expression phase.
Conclusion: Gm129 is a novel circadian clock modulator.
Significance: The discovery of Gm129 reveals a new regulatory component of circadian gene expression.
Keywords: Circadian, DNA-binding Protein, Gene Regulation, Signal Transduction, Transcription Repressor
Abstract
The mammalian circadian clock is a molecular oscillator composed of a feedback loop that involves transcriptional activators CLOCK and BMAL1, and repressors Cryptochrome (CRY) and Period (PER). Here we show that a direct CLOCK·BMAL1 target gene, Gm129, is a novel regulator of the feedback loop. ChIP analysis revealed that the CLOCK·BMAL1·CRY1 complex strongly occupies the promoter region of Gm129. Both mRNA and protein levels of GM129 exhibit high amplitude circadian oscillations in mouse liver, and Gm129 gene encodes a nuclear-localized protein that directly interacts with BMAL1 and represses CLOCK·BMAL1 activity. In vitro and in vivo protein-DNA interaction results demonstrate that, like CRY1, GM129 functions as a repressor by binding to the CLOCK·BMAL1 complex on DNA. Although Gm129−/− or Cry1−/− Gm129−/− mice retain a robust circadian rhythm, the peaks of Nr1d1 and Dbp mRNAs in liver exhibit a significant phase delay compared with control. Our results suggest that, in addition to CRYs and PERs, the GM129 protein contributes to the transcriptional feedback loop by modulating CLOCK·BMAL1 activity as a transcriptional repressor.
Introduction
Circadian clocks are endogenous timekeeping systems that govern the daily cycles of behavior and physiology in a variety of organisms (1, 2). In mammals, the 24-h circadian oscillator operates as a transcription-translation feedback loop involving both positive (transcriptional activators) and negative (transcriptional repressors) components. The transcriptional activators are CLOCK/NPAS2 and BMAL1, whereas Cryptochrome (CRY)3 (CRY1 and CRY2) and Period (PER1 and PER2) act as transcriptional repressors. In the transcription-translation feedback loop model, CLOCK·BMAL1 heterodimers bind to the promoter region of Per and Cry genes through E-box DNA regulatory sites and activate transcription. Upon transcriptional activation, gradually accumulated CRY and PER proteins abrogate their own transcription by repressing the activity of CLOCK·BMAL1 (3, 4). This feedback loop repeats every ∼24 h and results in rhythmic activities of the CLOCK·BMAL1 complex. Besides the promoters of Cry and Per genes, CLOCK·BMAL1 also binds to E-box elements in the promoter regions of thousands of clock-controlled genes, including transcription factors, such as the nuclear receptor Rev-erbα (Nr1d1), and the PAR domain bZIP transcription factors Dbp (12). These CLOCK·BMAL1 controlled transcription factors also exhibit high amplitude oscillation at the protein level in tissues. Thus, the CLOCK·BMAL1 complex and clock-controlled transcription factors drive massive transcriptional oscillation in various tissues in mammals (5).
As described above, the interactions between CLOCK·BMAL1 and E-box elements play a major role in circadian transcriptional regulation. To understand the direct targets of the CLOCK·BMAL1 complex and the molecular architecture of the circadian timing system, several studies have been conducted to identify the CLOCK·BMAL1 DNA-binding sites in vivo using ChIP-Seq analysis (6, 7). It was reported that, in liver, BMAL1 rhythmically bound to ∼2000 genomic targets (7). Based on these ChIP-Seq results, direct CLOCK·BMAL1 target genes were predicted and were ranked according to their binding strength. Interestingly, among the target genes with the highest BMAL1 chromatin binding strength, most are well studied circadian genes. The top 10 CLOCK·BMAL1 target genes include three core clock gene Per1, Per2, and Cry2 (8, 9); three nuclear receptors Rorc, Nr1d1, and Nr1d2 involved in the core clock regulation (10, 11), and three PAR domain bZIP transcription factors Dbp, Hlf, and Tef, which function as key genes in the output pathways (12). Only one gene remains uncharacterized, gene model 129 (Gm129), which encodes a novel protein with no predicted functional domains. Gm129 is also implicated as a direct CLOCK·BMAL1 target gene based upon its strong transcriptional oscillation in liver in a additional independent BMAL1 ChIP-seq analysis using NIH3T3 and WI38 cell lines (6).
Coincidently, a ChIP-seq and gene expression study in our group also found that Gm129 is a direct target gene of the BMAL1·CLOCK·CRY1 complex, and Gm129 exhibits high amplitude protein oscillation in mouse liver. These properties suggest that Gm129 might be an important factor in the molecular clock. Thus, we have conducted detailed molecular, biochemical, and genetic analysis to characterize the role of Gm129 in circadian clock regulation. In this study, we demonstrate that GM129 can directly interact with core clock proteins BMAL1 and PER2. GM129 also localizes in the nucleus and strongly represses CLOCK·BMAL1 transcriptional activity in a reporter gene assay. Additional in vitro and in vivo assays suggest that the GM129 protein directly interacts with the CLOCK·BMAL1 complex on DNA. In mouse liver, the absence of Gm129 extends the peak expression of Nr1d1 and Dbp. More importantly, in the Cry1−/− background, the Gm129 knock-out causes a significant phase shift of Nr1d1 and Dbp gene expression. We conclude that Gm129 functions as a novel transcriptional repressor in the mammalian clock transcription-translation feedback loop.
EXPERIMENTAL PROCEDURES
Mice
All animal procedures were in accordance with the National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee of the University of North Carolina, Chapel Hill. Gm129 heterozygote mice were purchased from the University of California, Davis, KOMP repository and bred to obtain Gm129−/− mice. Primers were designed as indicated (Fig. 5A and Table 1) for genotyping. Cry1−/−, Gm129−/− double knock-out mice were generated by crossing Cry1−/− (9) and Gm129−/− mice. All mice were maintained on a 12-h light/dark schedule.
FIGURE 5.
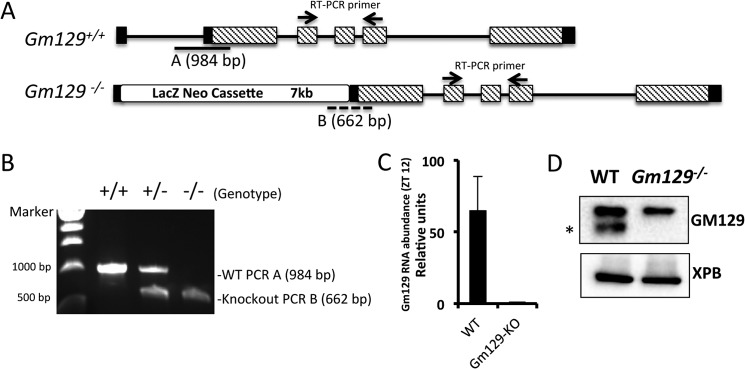
Generation of Gm129 knock-out mice. A, schematic representation of the WT (Gm129+/+) allele and targeted allele (Gm129−/−). Homologous recombination leads to the insertion of a 7-kb LacZ and Neomycin cassette between exon 1 and exon 2 in the WT Gm129 allele. Black lines represent introns. Black and dashed rectangles represent non-coding and coding exons, respectively. Solid line A and dashed line B locate the products of PCR genotyping. Two small arrows show the location of the primers used for RT-PCR (Table 1). B, primers were designed as indicated in Fig. 5A and Table 1 for genotyping. 984- and 662-bp PCR products were amplified using WT, heterozygote or knock-out mouse tail genomic DNA, respectively. C, Gm129 gene expression (normalized to Gapdh) in WT and Gm129−/− mice. The value of Gm129 expression in knock-out mice was set to 1. Error bars represent S.D. of 3 biological repeats. D, GM129 protein deficiency in Gm129−/− mouse liver. Liver nuclear extracts were prepared from wild-type (Gm129+/+) or homozygous Gm129−/− mutant mice at ZT12 and analyzed by SDS-PAGE followed by immunoblotting. The GM129 signal is marked by a black asterisk. XPB is a loading control.
TABLE 1.
Primer sets used for ChIP assay
Plasmids
To generate pEGFPN1-Gm129, the Gm129 coding sequence was amplified by PCR from mouse fibroblast cDNA and inserted into the pEGFP-N1 vector. The pBABEpuro-Flag-HA-Gm129 plasmid was generated by inserting Gm129 and the FLAG tag sequence into the pBABEpuro vector backbone. The pFast-Flag-Gm129-Myc construct was generated by inserting the Gm129 coding sequence into the pFastbac1 (Invitrogen) vector with the FLAG tag sequence at the forward oligo and Myc tag sequences at the reverse oligo. Constructs for expressing mCLOCK, mBMAL1, mCRY1, and mPER2 were described previously (13).
Cell Lines
NIH3T3 cells expressing FLAG-GM129 were made by retrovirus infection. Briefly, the pBABEpuro-Flag-HA-Gm129 construct was co-transfected together with pVSVG and pCIHPZ into HEK293T to produce recombinant retrovirus particles. Then MEF cells were infected with the retrovirus and transfectants were selected in media containing puromycin for 2 weeks. Single colonies were picked and cultured for subsequent analysis. The Flp-In/FLAG-C1ORF51 cell line was generated according to the manufacturer's protocols using the Flp-In T-REx-293 system (Invitrogen).
Immunoblotting and Antibodies
Both mouse liver lysates and liver nuclear extracts were prepared for immunoblotting. To make mouse liver lysate, mouse liver samples were homogenized and lysed with lysis buffer (15 mm HEPES, pH 7.8, 250 mm NaCl, 1% Nonidet P-40, 10% glycerol, protease inhibitor mixture, from Roche Applied Science). Proteins from mouse liver nuclei were prepared according to the NUN procedure described previously (14). Anti-FLAG antibody was purchased from Sigma. Anti-CLOCK, anti-BMAL1 antibodies (Bethyl Lab), and anti-mPER2 antibody (ADI) and anti-Gm129 (Santa Cruz Biotechnology) were used in the immunoblotting assays. Anti-mCRY antibody was produced in our laboratory (13).
Immunoprecipitation
Insect cells or mammalian cells were collected and lysed in lysis buffer (50 mm Tris-Cl, pH 7.5, 150 mm NaCl, 10% glycerol, 1% Tween-20, 0.1% Nonidet P-40, protease inhibitor mixture, Roche Applied Science). Lysates were incubated with anti-FLAG M2-agarose beads (Sigma) for 4 h at 4 °C. Beads were washed 4 times with lysis buffer; bound proteins were eluted in 2× SDS-sample buffer and analyzed by immunoblotting.
Fluorescence Microscopy
For GFP fluorescence microscopy, HEK293T cells were cultured on poly-d-lysine 12-mm coverslips (BD Biosciences) placed in 6-well plates. Cells were transfected with pEGFP-N1 and pEGFPN1-Gm129 plasmids using Lipofectamine 2000 reagent (Invitrogen) at around 50% confluence. 48 h after transfection, cells were washed twice with PBS, fixed immediately with 3.7% methanol-free formaldehyde in PBS for 15 min, and washed twice with PBS. Coverslips were mounted with SlowFade Gold Antifade Reagent with DAPI (Invitrogen). Images were captured using a LeicaSP2 confocal microscope.
Chromatin Immunoprecipitation (ChIP) and ChIP Sequencing
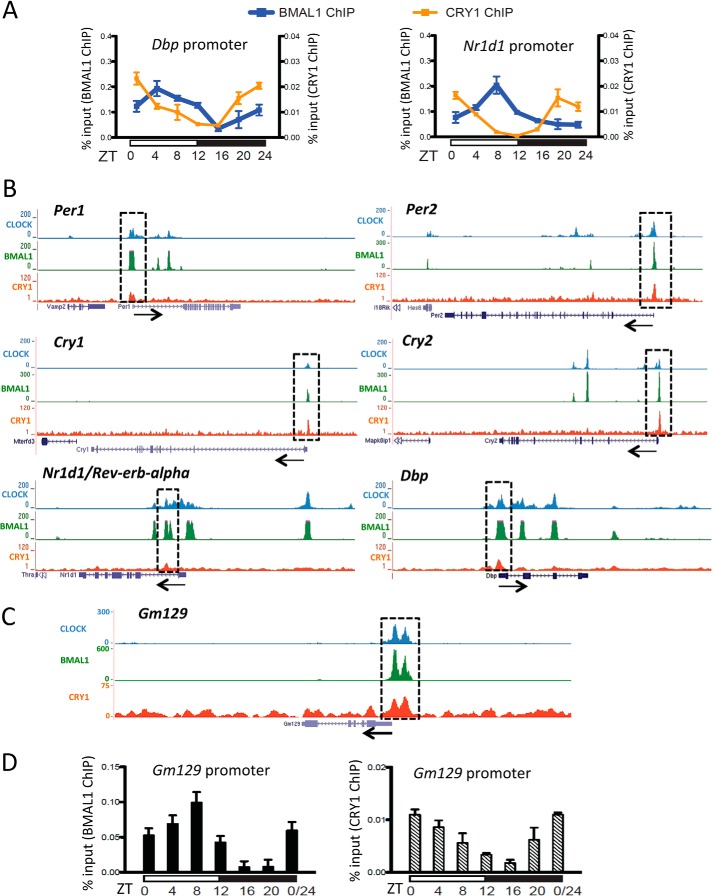
Primers used in ChIP-PCR are given in Table 1. ChIP-PCR assays using cell nuclear extracts were conducted as described previously (13). For ChIP-PCR and ChIP-Seq using mouse tissues, liver chromatin was prepared according to a previously published protocol (15) with minor modifications. Briefly, livers from mice were immediately homogenized and cross-linked in PBS buffer including 1% formaldehyde (5 ml/liver), and the homogenate was incubated for 15 min at 25 °C. Cross-linking reactions were stopped by the addition of 21 ml of ice-cold nuclei buffer (2.2 m sucrose, 125 mm glycine, 10 mm HEPES, pH 7.6, 15 mm KCl, 2 mm EDTA, 0.15 mm spermine, 0.5 mm spermidine, 0.5 mm DTT, 0.5 mm PMSF, and 10% glycerol). The homogenate was layered on top of 10 ml of cold nuclei buffer and centrifuged for 90 min at 24,000 rpm (100,000 × g) at 4 °C in a Beckmann SW27 rotor. The nuclei were washed with wash buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 2 mm EDTA) and stored at −80 °C. Cross-linked nuclei were then subjected to ChIP analysis as previously described (13). For ChIP-seq, ChIP DNA was ligated to single-end sequencing adapters and amplified. DNA fragments of 300–800 bp in size were purified and sequenced (50 base) on an Illumina Hi-seq2000 instrument (Illumina, CA). The reads were aligned to the mm9 mouse reference genome using Bowtie accepting up to 4 reportable alignments and two mismatches per read (16). In cases where more than a single alignment was possible, only the one with the highest mapping quality was reported using the “–best” option of Bowtie. To generate ChIP-Seq signal tracks, reads were extended in silico to 150 bp, and the number of reads overlapping with each base in the genome were counted and divided by the chromosome-wide average of per-base read coverage. The average signal over 50 bp windows was calculated and viewed using the University of California Santa Cruz (UCSC) genome browser (Fig. 1).
FIGURE 1.
Gm129 is a direct target of the CLOCK·BMAL1·CRY complex. A, rhythmic binding of BMAL1 (blue curve) and CRY1 (orange curve) to the Dbp and Nr1d1 promoter regions in mouse liver over one 12-h light/12-h dark cycle. Chromatin immunoprecipitation analysis was conducted using antibodies against BMAL1 and CRY1. Cross-linked chromatins were prepared from mouse liver tissues taken at 4-h intervals in a 24-h cycle. Error bars represent S.D. of 3 biological repeats. B, highly overlapping CLOCK, BMAL1, and CRY1 binding to the Per1, Per2, Cry1, Nr1d1, and Dbp gene. Gene tracks represent binding of CLOCK (blue), BMAL1 (green), and CRY1 (red) to various genes. Gray rectangles highlight co-occupied sites. The scale of the y axis (peak height) is adjusted for individual traces for visual clarity. Beneath each set of gene tracks is a representation of the intron/exon structure. Black arrow indicates the orientation of transcription. C, binding of the CLOCK·BMAL1·CRY1 complex to the promoter region of the Gm129 gene. D, rhythmic binding of BMAL1 (black bars) and CRY1 (dashed bars) to the Gm129 promoter region in mouse liver over one 12-h light/12-h dark cycle. Nuclear extracts were prepared from mouse liver tissues collected at 4-h intervals in a 24-h cycle and subjected to chromatin immunoprecipitation analysis using antibodies against BMAL1 and CRY1. Error bars represent S.D. of 3 biological repeats.
Reporter Gene Assay
Firefly luciferase was used as a reporter controlled by the mPer1 promoter sequence (17). Renilla luciferase was used as a transfection control. HEK 293T cells were co-transfected with the indicated amounts of plasmids using Lipofectamine® 2000 (Invitrogen). 36 h post-transfection, cells were collected, and luciferase activities were assayed using the Dual-Luciferase® Reporter Assay System according to the manufacturer's protocols (Promega).
Pulldown Assay with Immobilized DNA
Biotin-labeled E-box DNA or mutated E-box DNA (Table 1) was immobilized on magnetic streptavidin beads. Immobilized DNA was washed twice in reaction buffer (10 mm Tris-HCl, pH 7.9, 50 mm NaCl, 5 mm dithiothreitol, 10% glycerol, 0.01% Nonidet P-40, and 10 μg/ml of BSA) before setting up binding reactions. Reactions (200 μl) were prepared in reaction buffer with the indicated combinations of CLOCK·BMAL1 (10 nm), GM129 (10 nm), PER2 (10 nm), and CRY1 (10 nm) and 500 ng of immobilized DNA. After incubation for 1 h at 4 °C, DNA·protein complexes were collected on a magnet and washed three times with reaction buffer. The proteins associated with the DNA were analyzed by SDS-PAGE and immunoblotting.
RNA Extraction, Reverse Transcription and Quantitative Real-time RT-PCR
Mice of each genotype housed in light/dark (12:12) were sacrificed every 2 h starting from ZT 0 and livers were collected, washed with PBS, and immediately frozen in dry ice and stored in −80 °C until further use. RNA samples were prepared by TRIzol reagent (Invitrogen) and RNAeasy (Qiagen). Total RNA (500 ng) was reverse transcribed with oligo(dT)20 using SuperScript III (Invitrogen) according to the manufacturer's instructions. Quantitative real-time PCR assays were performed by using an ABI 7300 System (Applied Biosystems) and MaximaTM SYBR Green/ROX qPCR Master Mix (Fermentas) was used. Primers are listed in Table 1.
RESULTS
The Gm129 Gene Is a Direct Target of CLOCK·BMAL1·CRY1 and Oscillates in Mouse Liver
Previously we reported that CRY1 binds to CLOCK·BMAL1 on DNA by both in vitro EMSA and in vivo ChIP assays (13). To systematically examine binding of the native CLOCK·BMAL1·CRY1 core clock complex to CCG (clock-controlled gene) promoters in mouse tissue, we conducted ChIP assays using the liver to map the chromatin-binding sites of CLOCK, BMAL1, and CRY1. As shown in Fig. 1A, in agreement with a previously published report (18), both BMAL1 and CRY1 rhythmically bind to the Dbp and Nr1d1 promoters and reach their peak affinities at ZT6-ZT10 (for BMAL1) and ZT20-ZT24 (for CRY1) in ChIP assays. To simplify the experimental design and achieve the best signal-to-noise ratio for identifying chromatin-binding sites of the core clock protein complex, we conducted CLOCK and BMAL1 ChIP-seq using liver harvested at ZT8 and CRY1 ChIP-seq with liver harvested at ZT20. Fig. 1, B and C, show the representative samples of ChIP-seq data we obtained using anti-CLOCK, anti-BMAL1, and anti-CRY1 antibodies. As shown in Fig. 1B, CLOCK, BMAL1, and CRY1 all bind to promoters of the core clock repressive factors: Cry1, Cry2, Per1, and Per2. Besides these core components, these positive (CLOCK and BMAL1) and negative (CRY1) regulators also bind to promoters of the secondary/consolidating loop component Rev-Erbα/Nr1d1 as well as to the promoter of the clock-controlled transcription factor Dbp1.
Consistent with other published studies (6, 7, 19), our ChIP-seq results showed that the Gm129 gene has high affinity binding sites for CLOCK, BMAL1, and CRY1 in its promoter region (Fig. 1C). To further characterize this binding, we carried out ChIP-PCR assays with livers harvested at different time points to examine circadian rhythmicity of binding of the CLOCK·BMAL1·CRY1 complex to the Gm129 promoter. Fig. 1D shows ChIP-PCR data over the course of a diurnal cycle for the Gm129 gene. Clearly, both BMAL1 and CRY1 rhythmically occupy E-boxes in the Gm129 promoter with different binding phases.
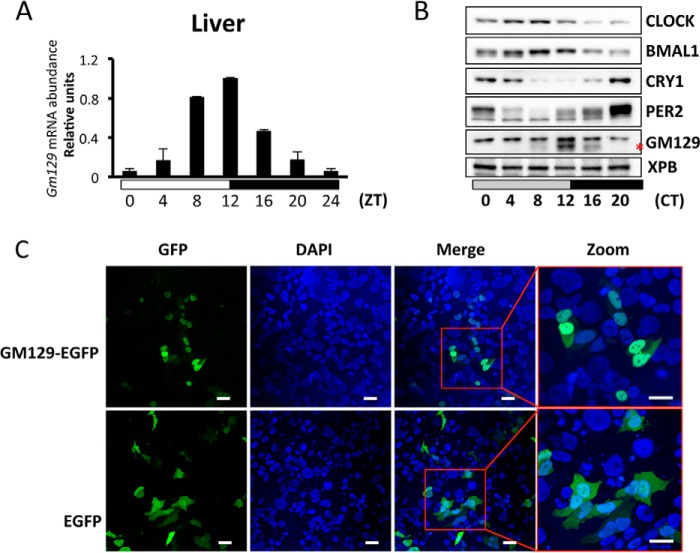
It was also reported that Gm129 exhibits circadian gene expression in the mouse liver (6). To confirm these results, time series quantitative PCR was conducted with mouse livers harvested over one diurnal cycle. Indeed, Gm129 mRNA levels show very high amplitude (∼17-fold difference between zenith and nadir) oscillation with a peak at ZT12 (Fig. 2A). Because in some cases a rhythmically expressed gene can produce a constitutively expressed protein (20), the GM129 protein level was also examined by time series immunoblotting of mouse liver extracts made over a circadian cycle (constant dark). As shown in Fig. 2B, GM129 protein also exhibits high amplitude oscillation in the mouse liver nucleus with a peak at CT12. Thus, all these results suggest that Gm129 is a direct target of the CLOCK·BMAL1·CRY1 complex and encodes a protein with high amplitude circadian oscillation.
FIGURE 2.
GM129 is a nuclear protein and oscillates in mouse liver. A, circadian oscillation of Gm129 mRNA in mouse liver. Mice were sacrificed at zeitgeber times (ZT) 4, 8, 12, 16, 20, and 24/0 h. The abundance of Gm129 mRNA was determined by quantitative RT-PCR and normalized to that of Gapdh. The maximal value was normalized to 1. Error bars represent S.D. of 2 biological repeats. The 0 to 24-h time point is plotted twice to emphasize the trend. B, oscillation of GM129 and core clock protein levels in liver nuclei. Mice were kept in constant dark for 24 h following 12-h dark/12-h light cycles. After 24 h in the dark, nuclear extracts were prepared from livers collected at 0, 4, 8, 12, 16, and 20 h in dim red light and GM129 was detected by Western blots. The XPB protein was used as a loading control. The GM129 signal is marked by a red asterisk. Note there is a nonspecific antibody cross-reactive band above the GM129 signal. The blot is representative of two experiments. C, subcellular localization of GM129-EGFP fusion protein. HEK293T cells were transfected with plasmid DNA expressing GM129-EGFP protein or EGFP alone. 48 h post-transfection, cells were fixed and visualized by fluorescence microscopy (GFP) and DAPI staining. Nuclear localization was confirmed by merging the GFP signal with the DAPI signal. EGFP alone is a control that localizes to both cytoplasm and nucleus. Bar = 10 μm.
GM129 Protein Localizes in the Nucleus and Interacts with PER2 and BMAL1
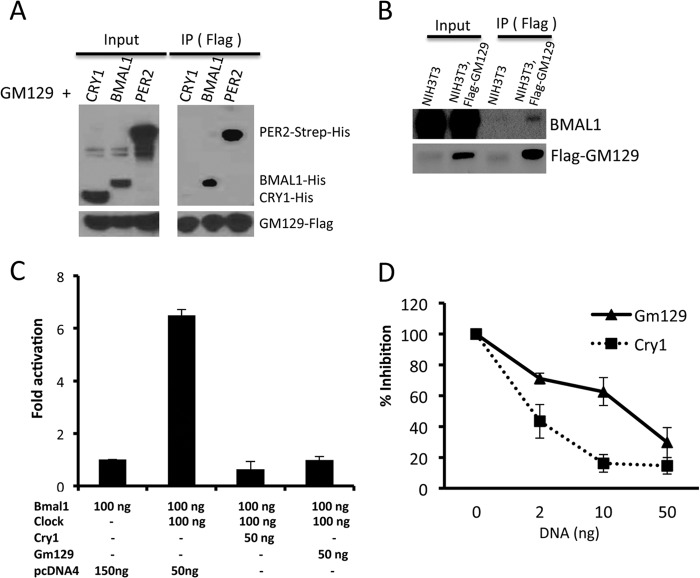
GM129 is a 375-amino acid protein with no analogs in the mouse genome, and it has no known protein motifs or domains to guide for a potential functional assignment. Thus, we decided to determine its subcellular localization to obtain clues about its molecular functions. To this end, we constructed a GFP fusion construct of GM129 and transfected HEK293T cells with this plasmid. We found that the GFP-GM129 protein largely localizes in the nucleus (Fig. 2C). In contrast, GFP alone exhibits both nuclear and cytoplasmic localization. Interestingly, the top 10 direct CLOCK·BMAL1 target genes with known function all encode nuclear proteins that function as either core clock components (PERs and CRYs) or circadian transcriptional factors that are responsible for output pathways (DBP, HLF, TEF, NR1D1, and NR1D2). Unlike circadian transcription factors in output pathways, core clock proteins normally interact with each other (21, 22). Thus, to further clarify the function of GM129, we decided to test protein-protein interactions between GM129 and core clock proteins using the baculovirus expression system. As shown in Fig. 3A, GM129 has a strong affinity to BMAL1 and PER2, but not CRY1. Additionally, GM129 binds to endogenous BMAL1 in an NIH3T3 cell line that stably expresses the FLAG-GM129 protein (Fig. 3B). Hence, nuclear localization and direct interaction with core clock proteins strongly support the hypothesis that GM129 is a novel circadian regulator.
FIGURE 3.
GM129 interacts with BMAL1 and PER2, and represses CLOCK/BMAL1-activated transcription. A, GM129 interacts with BMAL1 and PER2. Sf21 cells were co-infected with FLAG-GM129 baculovirus together with either PER2-Strep-His, BMAL1-His, or CRY1-His baculovirus. Cell extracts were prepared 48 h after infection and subjected to immunoprecipitation (IP) using FLAG resin. Immunoprecipitates were analyzed by Western blot. B, GM129 interacts with endogenous BMAL1 in an NIH3T3 cell line stably expressing FLAG-GM129. Immunoprecipitates obtained with FLAG resin from whole cell lysates were probed by immunoblotting. GM129 and BMAL1 were detected by anti-FLAG anti-BMAL1 antibody, respectively. C, HEK293 cells were transiently transfected with 25 ng of Per1 luciferase reporter construct, 2.5 ng of pBind (for normalization of transfection efficiency), and a total 250 ng of the indicated combinations of Clock, Bmal1, Gm129, and Cry1 expressing constructs together with pcDNA4. Cells were collected 48 h after transfection. Error bars represent S.D. from 3 experimental repeats. D, dose-response studies of inhibition of CLOCK·BMAL1-induced transcription by the GM129 and CRY1 proteins. Three amounts (2, 10, and 50 ng) of the Cry1 and Gm129 plasmids were used in the reporter gene assay. Experiments were done in triplicate and average values are plotted. Error bars represent standard deviation of 3 biological repeats. Note: x-axis is not set to scale.
GM129 Represses CLOCK and BMAL1-induced Transcription
Although GM129 strongly binds to BMAL1 and PER2 protein, it was not know whether this interaction can regulate the activity of the core clock protein complex. Interestingly, like GM129, CRY1 also binds to BMAL1 and PER2 and functions as a strong repressor of CLOCK·BMAL1. Thus, we reasoned that GM129 might function as a transcriptional regulator by interacting with BMAL1. To investigate GM129 function, a luciferase reporter gene assay was conducted in HEK293T cells using a Per1::Luc reporter (21). Coexpression of CLOCK and BMAL1 generate >6-fold induction of Luciferase activity compared with expressing BMAL1 alone (Fig. 3C). CRY1 is a positive control in this assay that efficiently represses CLOCK·BMAL1 activity. Importantly, the additional coexpression of either CRY1 or GM129 reduced the luciferase activity to the basal level (Fig. 3C). As shown in Fig. 3D, the repressor function of GM129 is also dose dependent, because as the amounts of Gm129 or Cry1 plasmids transfected were increased, there was increasing inhibition of CLOCK·BMAL1-activated transcription. In comparison with CRY1, it appears that GM129 is nearly equally efficient in repressing CLOCK·BMAL1 activity (Fig. 3D). Together with nuclear localization and protein-protein interaction results, repression of CLOCK·BMAL1-activated transcription by GM129 suggests a role in the negative arm of the transcription-translation feedback loop.
GM129 Interacts with the CLOCK·BMAL1 Complex on DNA Both in Vivo and in Vitro
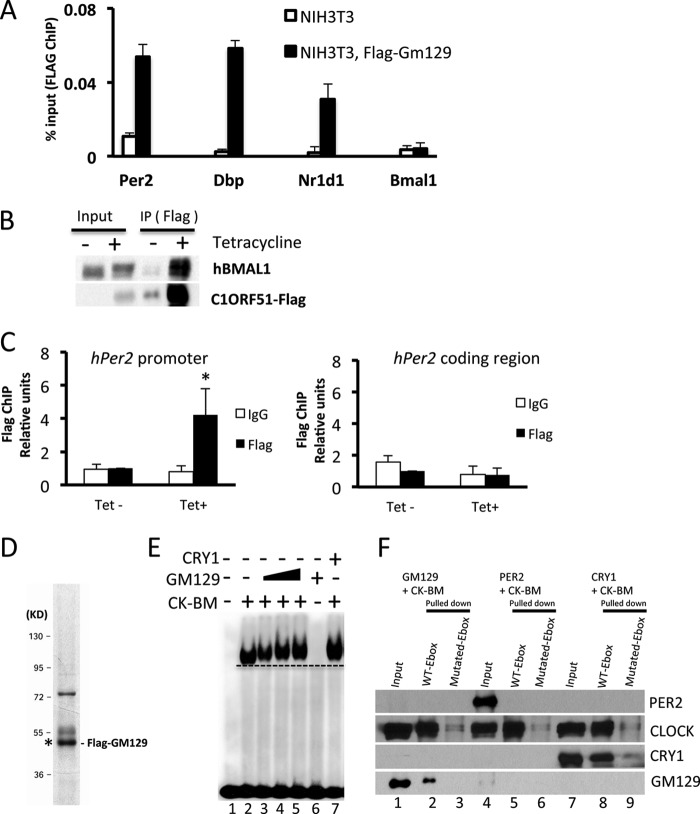
Although GM129 protein represses activity of the CLOCK·BMAL1 in the reporter gene assay, the detailed molecular mechanism is unclear. Generally, a protein can function as a repressor by two mechanisms: (a) the repressor can bind to the activator complex on DNA and interfere with its interaction with co-activator or with RNA polymerase II complex or (b) the repressor can directly dissociate the activator from DNA. Previously, we have reported that CRY1 binds to the CLOCK·BMAL1 complex on DNA in vitro and in vivo (13). Here, we have conducted parallel experiments to examine how GM129 functions as a repressor of CLOCK·BMAL1. The in vivo system that we used employed an NIH3T3 cell line that stably expresses FLAG-tagged GM129. A ChIP assay was conducted with these cells to measure the binding of GM129 to the E-box elements in the Per2, Dbp, Nr1d1, and Bmal1 promoters. As shown in Fig. 4A, FLAG-GM129 showed significant recruitment to the Per2, Dbp, and Nr1d1 E-box compared with the NIH3T3 controls. No significant recruitment was detected in the Bmal1 promoter that does not contain CLOCK·BMAL1-binding sites, which suggest the recruitment is specific and CLOCK·BMLA1 dependent. Equally important, FLAG-tagged C1ORF51, a human homolog of Gm129, also interacts with hBMAL1 and binds to the hPer2 promoter in HEK293R cells that stably expresses the FLAG-C1ORF51 protein, which suggests that the molecular function of the GM129 protein is evolutionarily conserved (Fig. 4, B and C).
FIGURE 4.
GM129 binds to CLOCK·BMAL1 on DNA both in vitro and in vivo. A, FLAG-GM129 binding to E-box DNA sequence in the Per2, Dbp, and Nr1d1 promoters was determined by ChIP assays followed by quantitative PCR. Cross-linked nuclear extracts were isolated from cells and subjected to ChIP assay with FLAG antibodies against FLAG-GM129. NIH3T3 cells and Bmal1 promoter region primers were used as controls for immunoprecipitation (IP) and PCR, respectively. Data are expressed as percentage of input. Error bars represent S.D. of 3 experiments. B, interaction of the human homolog of GM129 (C1ORF51) with endogenous hBMAL1 in extracts made from a 293R cell line stably expressing Flag-C1ORF51 under control of a tetracycline inducible promoter (Flp-In/FLAG-C1ORF51 cells). FLAG resin was used to pull down Flag-C1ORF51, and immunoprecipitates (IP) were analyzed by Western blot. Note that Flag-C1ORF51 protein is also enriched by FLAG resin without tetracycline due to the minor leaky expression. C, C1ORF51 binding to the E-box DNA sequence in the hPer2 promoter was determined by ChIP assays followed by quantitative PCR. Cross-linked nuclear extracts were isolated after a 24-h tetracycline induction and subjected to ChIP assay with FLAG antibodies against Flag-C1ORF51 or IgG. Upon tetracycline treatment, Flag-C1ORF51 showed significant recruitment to the hPer2 E-box compared with the IgG control and non-tetracycline controls (left panel, black asterisk: p < 0.05). No significant recruitment was detected in the hPer2 coding region (right panel). Data are expressed as percentage of input. Error bars represent S.D. of 3 experiments. D, Flag-GM129 protein was expressed using the Sf21/baculovirus system. After affinity chromatography purification, protein was analyzed by SDS-PAGE/Coomassie Blue staining. GM129 protein is marked with asterisk confirmed by immunoblotting. E, effects of GM129 on mobility of the CLOCK·BMAL1·E-box complex analyzed by EMSA. A radiolabeled 14-mer E-box duplex (1.5 nm) (M34, Table 1) (35) was incubated with purified BMAL1·CLOCK heterodimer (0.5 nm), and then increasing amounts of GM129 proteins (0.1, 0.5, and 1 nm) were added. Note the addition of GM129 results in slower migration of the protein·DNA complex (lanes 3–5). GM129 alone (1 nm) does not bind to DNA (lane 6). CRY1 (1 nm) was used as a positive control to show the shifted CLOCK·BMAL1·CRY1 complex on DNA. A dashed line parallel to the leading edge of the CLOCK·BMAL1·E-box complex is drawn to indicate the change in mobility of the protein·DNA bands upon addition of GM129 and CRY1. F, GM129 binds to the CLOCK·BMAL1·E-box determined by in vitro pulldown assays. The CLOCK·BMAL1 complex (10 nm) was mixed with 10 nm GM129, PER2, or CRY1 protein, then 500 ng of immobilized 30-mer WT E-box or mutated E-box duplex DNA was added for an additional 60-min incubation in binding buffer. Immobilized complexes were washed and then protein·DNA complexes were eluted and analyzed by Western blot. Proteins were detected by anti-CLOCK or anti-FLAG (GM129, PER2, and CRY1) antibodies.
The in vivo interactions between GM129 and the Per2 promoter could be achieved either directly through the CLOCK·BMAL1 complex or through other proteins. Here, we used a defined in vitro system to elaborate on these protein-DNA interactions. The CLOCK·BMAL1 complex and FLAG-GM129 were purified using anti-FLAG affinity chromatography and baculovirus expression system as described previously (13). Purified GM129 is shown in Fig. 4D. Then, we investigated the effects of GM129 on the CLOCK·BMAL1·E-box ternary complex using a electromobility shift assay (EMSA). As shown in Fig. 4E, CLOCK·BMAL1 binds to the E-box specifically (lane 2), and addition of GM129 protein retards the mobility of the CLOCK·BMAL1·E-box complex indicating the binding of GM129 to the ternary complex (Fig. 4E, lanes 3–5). CRY1 is a positive control that can also shift the CLOCK·BMAL1·E-box complex (lane 7) (13). GM129 on its own does not bind to E-box under the conditions used (lane 6), indicating its binding to the ternary CLOCK·BMAL1·E-box complex through interaction with CLOCK·BMAL1.
Next, the interactions of GM129 with CLOCK·BMAL1·E-box was tested by an in vitro DNA pulldown assay to further confirm the conclusion from the EMSA experiments. In this assay, we used a 30-bp fragment encompassing a non-canonical E-box (CACGTT, Table 1) in the mouse Per2 promoter, which was shown to bind to the CLOCK·BMAL1 complex (14, 23). Purified GM129 and other clock proteins were incubated with either wild-type or mutated E-box DNA immobilized on streptavidin-coupled magnetic beads. The beads were washed, and the DNA-bound proteins were analyzed by Western blot as shown in Fig. 4F. Clearly, GM129 binds strongly to the CLOCK·BMAL1 complex on wild-type E-box DNA substrate (lane 2) but not to mutated E-box (lane 3). In agreement with previously published results (13), PER2 protein (negative control) does not bind to the CLOCK·BMAL1 complex on DNA (lane 5). CRY1 (positive control) also showed strong interaction with the CLOCK·BMAL1·E-box (lane 8). Overall, these data unambiguously show that GM129 tightly binds to CLOCK·BMAL1 on E-box DNA. Together with the reporter gene assay, our biochemical analysis suggest that GM129 appears to repress transcription as a consequence of binding stably to the CLOCK·BMAL1·E-box complex.
Gm129 Gene Deficiency Affects the Expression of Clock-controlled Genes
If the repressor function of GM129 is essential and cannot be compensated by other genes, depletion of Gm129 in mice would affect the circadian rhythm rhythmicity. To test this, Gm129tm1e(KOMP)Wtsi heterozygous mice (hereafter Gm129+/−) were obtained from the NIH Knock-out Mouse Project (KOMP) Repository. The targeting strategy for the knock-out is summarized in Fig. 5A (24). Gm129−/− homozygous mice were obtained by mating these heterozygotes. Genotyping was done by PCR as shown in Fig. 5B. Fig. 5, C and D, show that Gm129 mRNA and protein are not expressed in knock-out mice, thus the Gm129−/− is a null allele. Heterozygote intercrosses produced progeny of each genotype in the expected Mendelian frequency, indicating that homozygous animals are viable. Overall, homozygous Gm129−/− mice had no gross developmental abnormalities.
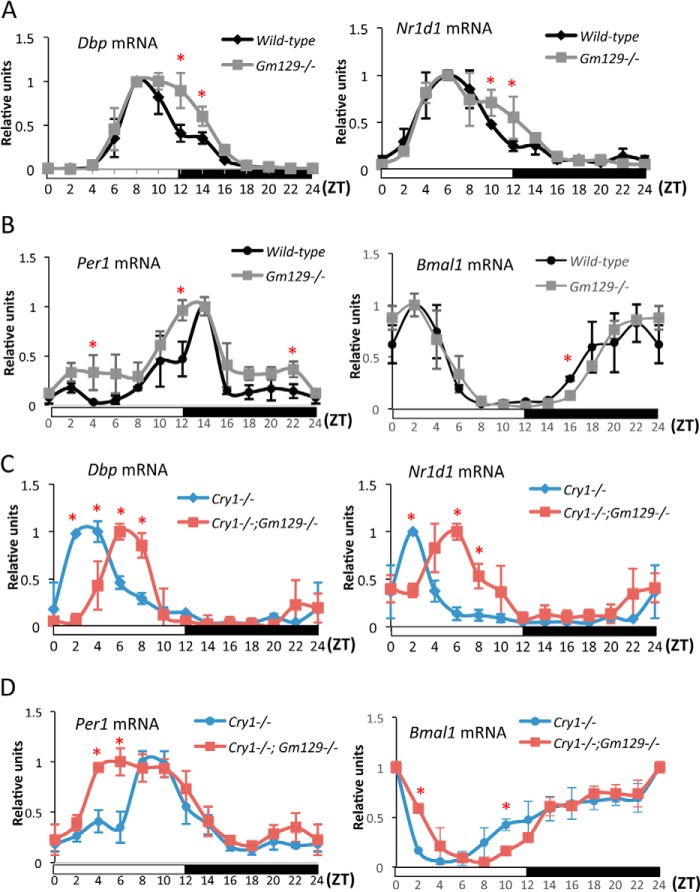
To determine the effects of GM129 disruption on behavioral rhythmicity, we monitored wheel-running activity under constant darkness (DD) for 1 week, following an initial 14 days in a 12-h light/12-h dark (LD) cycle. Although the animals appear robustly rhythmic, we were not powered to detect minor period length differences (data not shown). The circadian oscillator controls not only behavior, but also gene transcription in the SCN and peripheral tissue. Hence, it was of interest to determine the effects of Gm129 deficiency on circadian regulation of gene transcription. Thus, we analyzed the expression pattern of canonical clock genes Dbp, Nr1d1, Per1, and Bmal1 in mouse liver samples collected every 2 h for 24 h. In agreement with the behavioral analysis, the rhythmic expression of these clock genes was retained in Gm129−/− mice compared with wild-type (WT) mice (Fig. 6, A and B). However, the peaks of Dbp, Nr1d1, and Per1 gene expression persists longer in the Gm129−/− mouse liver between ZT10 and ZT14. Bmal1 expression is also slightly altered at ZT16. These effects are likely caused by absence of the GM129 transcriptional repressor that exhibits a peak expression level at ZT12.
FIGURE 6.
Effects of disruption of the Gm129 gene on circadian gene expression in mouse liver. A and B, circadian expression of Dbp, Nr1d1, Per1, and Bmal1 in liver from wild-type (black curve) and Gm129−/− (gray curve) mice analyzed by quantitative RT-PCR. C and D, circadian expression of Dbp, Nr1d1, Per1, and Bmal1 in Cry−/− (blue curve) and Cry−/−Gm129−/− (red curve) mouse livers analyzed by quantitative RT-PCR. Liver samples were collected every 2 h from mice that were housed under 12-h light/12-h dark conditions. Dbp and Nr1d1 mRNA levels were first normalized to Gapdh expression. Then, the maximal value of gene expression in the day/night cycle was normalized to 1. Error bars represent S.D. of two biological repeats. Red asterisk indicates significantly altered Dbp, Nr1d1, Per1, and Bmal1 transcription in livers from Gm129−/− and Cry−/−Gm129−/− mice compared with control (p < 0.05).
Lack of Gm129 in Cry1 Knock-out Mice Delays the Phase of Nr1d1 and Dbp Expression
Because Gm129−/− mice do not show any significant alteration in their circadian rhythm, we suspect that GM129 functions in a redundant manner with another protein in mice. It was previously reported that, except for Bmal1 (25), deficiencies of single core clock regulator genes normally do not abolish circadian rhythms. For example, Cry1−/− and Cry2−/− single knock-out mice have stable circadian rhythms with altered periods (9), but Cry−/− double knock-out mice are totally arrhythmic. In addition, Per1 and Per2 genes also have redundant roles in circadian regulation (8, 26). Therefore, we decided to combine the Gm129 mutation with another core clock gene mutation that possibly compensates for GM129 function. Although Gm129 does not have analogs in the mouse genome, our protein-protein interaction assay and reporter gene assay suggest that the Gm129 gene has a functionally redundant role with Cryptochrome as a transcriptional repressor on CLOCK·BMAL1. Hence, we decided to make Cry1−/−Gm129−/− double knock-out mice to examine the roles of Gm129 in circadian regulation. Dbp, Nr1d1, Per1, and Bmal1 rhythmic gene expression in the liver was used as readouts of the circadian rhythm. Results are shown in Fig. 6, C and D, which is aligned under and may be compared with Fig. 6, A and B. In agreement with a previous report, the Cry1 mutation alone causes an advanced phase shift (∼4 h) of Dbp (18), Per1 and Nr1d1 (black curve and blue curve). Most importantly, Cry−/−Gm129−/− mice exhibit robust, but significantly phase-delayed Dbp and Nr1d1 circadian gene expression (about 2–4 h phase shift) compared with Cry−/− mice (Fig. 6C). Per1 gene expression exhibits a broader peak between ZT4 and ZT10 compared with control (Fig. 6D, left panel). These results are consistent with the finding in Gm129−/− mice, in which peak transcription of Dbp, Nr1d1, and Per1 persists longer than in WT mice between ZT12 and ZT14. Equally important, rhythmic Bmal1 expression that is under control of the NR1D1 protein (3, 10) is also affected by the Gm129 knock-out mutation in Cry1−/− mice (Fig. 6D, right panel). Thus, we reasoned that in WT mice, the GM129 phase modulator function is largely masked by CRY1 that is a more potent repressor than GM129. However, in the absence of CRY1, GM129 has a stronger role in regulating CLOCK·BMAL1 activity. Thus, our genetic analyses suggest that Gm129 is a circadian clock modulator rather than an essential core component.
DISCUSSION
In the circadian transcription-translation feedback loop model, CRY and PER transcriptional repressors are essential components of the feedback loop. Besides CRY and PER, there are additional non-essential components of the core clock machinery that can repress CLOCK·BMAL1 activity. These proteins include the helix-loop-helix transcription factors, such as Differentially Expressed in Chondorocytes-1 and -2 (DEC1 and DEC2) and Inhibitor of DNA Binding 2 (ID2). These proteins can inhibit CLOCK·BMAL1 activities possibly by interfering with formation of the CLOCK·BMAL1 complex (27–29). DEC1 protein was also reported to delay the phases of Per1, Dbp, and Nr1d1 in cells after serum synchronization (30). Another non-essential repressor is CLOCK-interacting Protein, Circadian (CIPC), which can interact with CLOCK and inhibits CLOCK·BMAL1 transcriptional activity presumably by disrupting the binding between CLOCK and its transcriptional coactivators (31). Here, we demonstrated that similar to Cryptochrome, GM129 is a novel transcriptional repressor of the CLOCK·BMAL1 protein complex that interacts with CLOCK·BMAL1 on DNA.
It was surprising to find that the effect of GM129 on CLOCK·BMAL1 activity in the reporter gene assay is almost as strong as CRY1, and that the mechanism of GM129 is similar to that of the CRYs in that it binds to CLOCK·BMAL1·E-box complexes. However, important differences must exist between the function of CRYs and GM129 because GM129 is unable to compensate for the absence of Crys in Cry1−/−Cry2−/− double knock-out mice, which are arrhythmic (9). Our results also suggest that, unlike CRY1, GM129 is a non-essential CLOCK·BMAL1 repressor that is not required for circadian rhythm generation, but functions as a modulator to influence the phase of CLOCK·BMAL1 controlled transcriptional oscillation. We noticed that, in addition to BMAL1, GM129 also interacts with PER2 in our immunoprecipitation assays. Further investigation on these protein-protein interactions might help us to understand the differences between GM129 and Cryptochromes. It is also conceivable that Gm129 might be essential for the function of some peripheral clocks when isolated cells are analyzed.
Our finding of Gm129 was driven by the hypothesis that mining ChIP-seq data for genes with high affinity CLOCK·BMAL1-binding sites in their promoter will identify important factors in the circadian clock regulation. In addition to Gm129, other genes also contain high affinity CLOCK·BMAL1-binding sites in their promoter region with uncharacterized functions in circadian physiology. Investigations on these genes could introduce more new players in molecular clock regulation.
Gm129 may also contribute to non-clock pathways in addition to its function in the core clock machinery. Many circadian transcriptional regulators have roles in multiple pathways. It was previously reported that both PER and CRY can directly interact with nuclear receptors to influence related signaling pathways (32–34). Thus, to understand the function of the GM129 protein in circadian physiology, it will be important to identify the binding partners of GM129 and map its genome-wide binding sites in future studies.
In conclusion, our findings indicate a physiological role for Gm129 in the transcriptional regulation of clock-controlled genes. GM129 protein levels show very strong oscillation with a peak at CT12, 8 h earlier than the peak of PER and CRY transcriptional repressors (peak CT20-CT24) (Fig. 2B). Thus, GM129 protein can repress CLOCK·BMAL1 activity in a different time windows compared with CRY and PER (Fig. 7). Together with other essential and non-essential CLOCK·BMAL1 repressors, GM129 contributes to the precision of the molecular clock and fine-tunes regulation of rhythmic gene expression.
FIGURE 7.
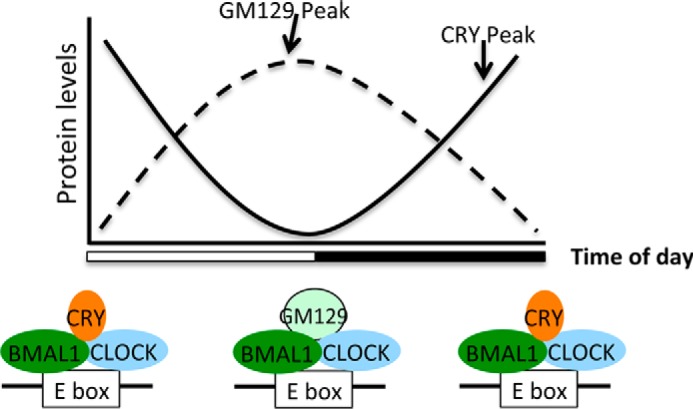
Model for the role of GM129 in modulating CLOCK·BMAL1 activities. Both GM129 and Cryptochromes could repress he transcription activator CLOCK·BMAL by similar mechanisms but at different times. Cryptochromes reach their peak protein expression levels at ZT20-ZT24, and the GM129 protein has peak expression at ZT12. Thus, GM129 and CRYs regulate the molecular clock in two separate time windows.
Acknowledgments
We thank Dr. John Hogenesch for communicating data prior to publication and Dr. Christopher Selby for critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM31082.
The data sets reported in this paper have been deposited in the GEO database with code GSE53828.
- CRY
- Cryptochrome
- PER
- Period
- ZT
- zeitgeber times.
REFERENCES
- 1. Bell-Pedersen D., Cassone V. M., Earnest D. J., Golden S. S., Hardin P. E., Thomas T. L., Zoran M. J. (2005) Circadian rhythms from multiple oscillators. Lessons from diverse organisms. Nat. Rev. Genet. 6, 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Young M. W., Kay S. A. (2001) Time zones. A comparative genetics of circadian clocks. Nat. Rev. Genet. 2, 702–715 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L. (2008) The genetics of mammalian circadian order and disorder. Implications for physiology and disease. Nat. Rev. Genet. 9, 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reppert S. M., Weaver D. R. (2002) Coordination of circadian timing in mammals. Nature 418, 935–941 [DOI] [PubMed] [Google Scholar]
- 5. Hughes M. E., DiTacchio L., Hayes K. R., Vollmers C., Pulivarthy S., Baggs J. E., Panda S., Hogenesch J. B. (2009) Harmonics of circadian gene transcription in mammals. PLoS Genet. 5, e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hatanaka F., Matsubara C., Myung J., Yoritaka T., Kamimura N., Tsutsumi S., Kanai A., Suzuki Y., Sassone-Corsi P., Aburatani H., Sugano S., Takumi T. (2010) Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol. Cell. Biol. 30, 5636–5648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rey G., Cesbron F., Rougemont J., Reinke H., Brunner M., Naef F. (2011) Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 9, e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng B., Albrecht U., Kaasik K., Sage M., Lu W., Vaishnav S., Li Q., Sun Z. S., Eichele G., Bradley A., Lee C. C. (2001) Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694 [DOI] [PubMed] [Google Scholar]
- 9. Vitaterna M. H., Selby C. P., Todo T., Niwa H., Thompson C., Fruechte E. M., Hitomi K., Thresher R. J., Ishikawa T., Miyazaki J., Takahashi J. S., Sancar A. (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc. Natl. Acad. Sci. U.S.A. 96, 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cho H., Zhao X., Hatori M., Yu R. T., Barish G. D., Lam M. T., Chong L. W., DiTacchio L., Atkins A. R., Glass C. K., Liddle C., Auwerx J., Downes M., Panda S., Evans R. M. (2012) Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takeda Y., Jothi R., Birault V., Jetten A. M. (2012) RORγ directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 40, 8519–8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gachon F., Olela F. F., Schaad O., Descombes P., Schibler U. (2006) The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 4, 25–36 [DOI] [PubMed] [Google Scholar]
- 13. Ye R., Selby C. P., Ozturk N., Annayev Y., Sancar A. (2011) Biochemical analysis of the canonical model for the mammalian circadian clock. J. Biol. Chem. 286, 25891–25902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Asher G., Reinke H., Altmeyer M., Gutierrez-Arcelus M., Hottiger M. O., Schibler U. (2010) Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142, 943–953 [DOI] [PubMed] [Google Scholar]
- 15. Ripperger J. A., Schibler U. (2006) Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374 [DOI] [PubMed] [Google Scholar]
- 16. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 18. Stratmann M., Stadler F., Tamanini F., van der Horst G. T., Ripperger J. A. (2010) Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by CRYPTOCHROME1. Genes Dev. 24, 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koike N., Yoo S. H., Huang H. C., Kumar V., Lee C., Kim T. K., Takahashi J. S. (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338, 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reddy A. B., Karp N. A., Maywood E. S., Sage E. A., Deery M., O'Neill J. S., Wong G. K., Chesham J., Odell M., Lilley K. S., Kyriacou C. P., Hastings M. H. (2006) Circadian orchestration of the hepatic proteome. Curr. Biol. 16, 1107–1115 [DOI] [PubMed] [Google Scholar]
- 21. Griffin E. A., Jr., Staknis D., Weitz C. J. (1999) Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286, 768–771 [DOI] [PubMed] [Google Scholar]
- 22. Shearman L. P., Sriram S., Weaver D. R., Maywood E. S., Chaves I., Zheng B., Kume K., Lee C. C., van der Horst G. T., Hastings M. H., Reppert S. M. (2000) Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 23. Yoo S. H., Ko C. H., Lowrey P. L., Buhr E. D., Song E. J., Chang S., Yoo O. J., Yamazaki S., Lee C., Takahashi J. S. (2005) A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 2608–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., Jackson D., Severin J., Biggs P., Fu J., Nefedov M., de Jong P. J., Stewart A. F., Bradley A. (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bunger M. K., Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., Simon M. C., Takahashi J. S., Bradfield C. A. (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bae K., Jin X., Maywood E. S., Hastings M. H., Reppert S. M., Weaver D. R. (2001) Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525–536 [DOI] [PubMed] [Google Scholar]
- 27. Grechez-Cassiau A., Panda S., Lacoche S., Teboul M., Azmi S., Laudet V., Hogenesch J. B., Taneja R., Delaunay F. (2004) The transcriptional repressor STRA13 regulates a subset of peripheral circadian outputs. J. Biol. Chem. 279, 1141–1150 [DOI] [PubMed] [Google Scholar]
- 28. Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K. (2002) Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419, 841–844 [DOI] [PubMed] [Google Scholar]
- 29. Hou T. Y., Ward S. M., Murad J. M., Watson N. P., Israel M. A., Duffield G. E. (2009) ID2 (Inhibitor of DNA Binding 2) is a rhythmically expressed transcriptional repressor required for circadian clock output in mouse liver. J. Biol. Chem. 284, 31735–31745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakashima A., Kawamoto T., Honda K. K., Ueshima T., Noshiro M., Iwata T., Fujimoto K., Kubo H., Honma S., Yorioka N., Kohno N., Kato Y. (2008) DEC1 modulates the circadian phase of clock gene expression. Mol. Cell. Biol. 28, 4080–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao W. N., Malinin N., Yang F. C., Staknis D., Gekakis N., Maier B., Reischl S., Kramer A., Weitz C. J. (2007) CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat. Cell Biol. 9, 268–275 [DOI] [PubMed] [Google Scholar]
- 32. Lamia K. A., Papp S. J., Yu R. T., Barish G. D., Uhlenhaut N. H., Jonker J. W., Downes M., Evans R. M. (2011) Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480, 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmutz I., Ripperger J. A., Baeriswyl-Aebischer S., Albrecht U. (2010) The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 24, 345–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grimaldi B., Bellet M. M., Katada S., Astarita G., Hirayama J., Amin R. H., Granneman J. G., Piomelli D., Leff T., Sassone-Corsi P. (2010) PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 12, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hogenesch J. B., Gu Y. Z., Jain S., Bradfield C. A. (1998) The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. U.S.A. 95, 5474–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]