Background: Otub1 suppresses E2 UbcH5 to stabilize and activate p53.
Results: UbcH5 monoubiquitinates Otub1, and monoubiquitination-defective Otub1 mutants fail to inhibit UbcH5 and induce p53.
Conclusion: Monoubiquitination is critical for Otub1 to suppress UbcH5 and induce p53.
Significance: We report the discovery of a novel molecular mechanism underlying Otub1 suppression of E2 and activation of p53.
Keywords: Deubiquitination, p53, Ubiquitin, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitination, Deubiquitinating Enzymes, Otub1, UbcH5, Monoubiquitination
Abstract
Ovarian tumor domain-containing ubiquitin (Ub) aldehyde binding protein 1 (Otub1) regulates p53 stability and activity via non-canonical inhibition of the MDM2 cognate Ub-conjugating enzyme (E2) UbcH5. However, it is not clear how this activity of Otub1 is regulated in cells. Here we report that Otub1 is monoubiquitinated by UbcH5 in cells and in vitro, primarily at the lysine 59 and 109 residues. This monoubiquitination, in turn, contributes to the activity of Otub1 to suppress UbcH5. The lysine-free Otub1 mutant (Otub1K0) fails to be monoubiquitinated and is unable to suppress the Ub-conjugating activity of UbcH5 in vitro and the MDM2-mediated p53 ubiquitination in cells. Consistently, this mutant is unable to stabilize p53, induce apoptosis, and suppress cell proliferation. Overexpression of Otub1K0 inhibits DNA-damage induced apoptosis. Adding either Lys-59 or Lys-109 back to the Otub1K0 mutant restores the monoubiquitination of Otub1 and its function to stabilize and activate p53. We further show that UbcH5 preferentially binds to the monoubiquitinated Otub1 via Ub interaction with its backside donor Ub-interacting surface, suggesting that this binding interferes with the self-assembly of Ub-charged UbcH5 (UbcH5∼Ub) conjugates, which is critical for Ub transfer. Thus, our data reveal novel insights into the Otub1 inhibition of E2 wherein monoubiquitination promotes the interaction of Otub1 with UbcH5 and the function to suppress it.
Introduction
Ubiquitination-mediated proteasomal degradation plays a key role in the regulation of p53 protein stability and activity (1, 2). Under physiological conditions, p53 is tightly controlled at low levels, mainly by MDM2, a RING finger domain-containing ubiquitin (Ub)2 ligase (E3) (3, 4). MDM2 binds to the N-terminal transactivation domain of p53, directly suppressing p53 activity and mediating p53 ubiquitination and its subsequent proteasomal degradation (5–7). Diverse stress signals activate p53 by barricading MDM2-mediated p53 suppression, either disrupting their physical interaction (e.g. DNA damage) or directly suppressing MDM2 E3 activity toward p53 (e.g. oncogenic or ribosomal stress) (8, 9). Thus, MDM2-mediated p53 ubiquitination and degradation play a central role in controlling the stability and activity of p53.
Also important for the proper control of p53 dynamics is deubiquitination, a reverse process of ubiquitination mediated by deubiquitinating enzymes (Dubs) (10). p53 is regulated directly or indirectly by several ubiquitin-specific proteases (USPs), the largest Dub family of enzymes (10). For example, USP7 deubiquitinates and stabilizes p53, MDM2, and MDMX, an MDM2 homologue also required for the proper control of p53 levels and activity in cells (11, 12). USP7 preferentially deubiquitinates MDM2 under physiological conditions, whereas it deubiquitinates and stabilizes p53 in response to DNA damage (13, 14). DNA damage also activates USP10 to specifically deubiquitinate and stabilize p53 but not MDM2 and MDMX (15). USP29 has been shown to deubiquitinate and stabilize p53 in response to oxidative stress (16). USP42 appears to regulate p53 levels only during an early phase of the stress response (17). Also, USP2 deubiquitinates both MDM2 and MDMX (18, 19), whereas USP4 deubiquitinates ARF-BP1 (20), another ubiquitin ligase for p53. Consequently, both USP2 and USP4 destabilize p53 and inhibit its function. Of note, the USPs mentioned above regulate the p53 pathway via their deubiquitinating enzyme activity.
We recently identified ovarian tumor domain-containing Ub aldehyde binding protein 1 (Otub1), an ovarian tumor (OTU) family member Dub, as a novel positive regulator of p53 (21). Interestingly, Otub1 regulates p53 through non-canonical suppression of the ubiquitin-conjugating enzyme (E2) activity of UbcH5 (also called UbE2D), leading to the inhibition of MDM2-mediated p53 ubiquitination (21). Similarly, Otub1 inhibits the DNA damage-induced double strand break response by suppressing the Ubc13 (also called UbE2N)-mediated chromatin ubiquitination (22). Further, a recent yeast two-hybrid study revealed that Otub1 is a major Dub that interacts with the D and E classes of E2s as well as UbE2N (23), suggesting that Otub1 represents a unique Dub that mainly targets E2 enzymes. Mechanistically, it has been shown that Otub1 preferentially binds to the Ub-charged Ubc13 (Ubc13∼Ub) (22). The donor Ub binds to the N terminus of Otub1, and this is facilitated by a free Ub, which binds to a second Ub-binding site at the C terminus of Otub1, leading to its conformational change, which, in turn, promotes the donor Ub binding (24, 25). Consequently, this interaction blocks the Ub transfer from E2 to substrates (24, 25). However, how these structural observations relate, in a cellular context, to mediating E2 suppression activity is still not clear. It is also unknown whether Otub1 activity is regulated by posttranslational modification.
Here we report that Otub1 is monoubiquitinated by UbcH5, primarily at Lys-59 or Lys-109, and that this monoubiquitination is critical for the E2-suppressing activity of Otub1. A monoubiquitination-defective, lysine-free mutant of Otub1 (Otub1K0) is unable to suppress UbcH5 in vitro or inhibit MDM2-mediated p53 ubiquitination in cells. Consistently, Otub1K0 is unable to activate p53, induce apoptosis, and suppress cell proliferation, whereas adding either Lys-59 or Lys-109 back to Otub1K0 restores the function of Otub1 to regulate p53. We further show that UbcH5 preferentially binds to the monoubiquitinated Otub1 via its backside interaction with the Otub1-linked Ub. Together, these data reveal novel insights into the Otub1 regulation of E2 wherein monoubiquitination of Otub1 promotes its E2-suppressing activity.
EXPERIMENTAL PROCEDURES
Cell Culture, Plasmids, and Antibodies
Human p53-null lung non-small cell carcinoma H1299 and p53-proficient osteosarcoma U2OS cells were cultured in DMEM supplemented with 10% FBS, 50 units/ml penicillin, and 0.1 mg/ml streptomycin at 37 °C in a 5% CO2 humidified atmosphere, as described previously (21, 26). The GST-UbcH5c plasmid was constructed by inserting the full-length cDNA into the pGEX.4T.1 (Pharmacia) vector. The FLAG-Otub1 and His-Otub1 plasmids have been described previously (21). All Otub1 point mutant plasmids were constructed using site-directed mutagenesis (Stratagene). Other plasmids have been described previously (21). Anti-FLAG (M2, Sigma), anti-p21 (Ab-11, NeoMarkers), anti-p53 (DO-1, Santa Cruz Biotechnology), anti-MDM2 (SMP14, Santa Cruz Biotechnology), anti-Ub (Santa Cruz Biotechnology), and anti-V5 (Invitrogen) antibodies were purchased. Rabbit polyclonal anti-Otub1 antibodies were generated as described (21).
In Vitro Ubiquitination Assay
Recombinant His-Otub1 proteins were expressed in Escherichia coli and purified using the Ni-NTA purification method. The in vitro ubiquitination reactions were assembled in a total of 20 μl of reaction buffer containing recombinant UbE1 (0.025 μm, Boston Biochem), UbcH5 (0.4 μm, Boston Biochem), Ub (40 μm, WT or the I44A mutant, Boston Biochem), 50 mm Tris-HCl (pH 8.0), 5 mm MgCl2, 2 mm ATP, and 1 mm DTT in the absence or presence of 3.2 μm or the indicated concentrations of purified His-Otub1 at 37 °C for the indicated hours, as described previously (21). For detection of Otub1 monoubiquitination, the reaction was stopped by adding SDS sample buffer and analyzed by IB analysis with anti-Otub1 antibodies. For detection of the Ub-conjugating activity of UbcH5, the reaction was assayed by SDS-PAGE gel in non-reducing conditions, followed by IB analysis with anti-conjugated Ub (clone FK2, Millipore).
In Vivo Ubiquitination Assay
An in vivo ubiquitination assay under denaturing conditions was conducted in H1299 cells using an Ni-NTA pulldown method, as described previously (21, 26).
Transfection, IB Analysis, and Coimmunoprecipitation (Co-IP) Analysis
Cells were transfected with plasmids using TransIT®-LT1 reagents following the protocol of the manufacturer (Mirus Bio Corp.). Cells were harvested 36–48 h post-transfection and lysed in lysis buffer consisting of 50 mm Tris-HCl (pH 8.0), 0.5% Nonidet P-40, 1 mm EDTA, 150 mm NaCl, 1 mm PMSF, 1 mm DTT, 1 μg/ml pepstatin A, and 1 mm leupeptin. Equal amounts of clear cell lysate were used for IB analysis. Co-IP was conducted as described previously (21). Bound proteins were detected by IB analysis using antibodies as indicated in figure legends.
Generation of Tet-inducible Otub1K0 Expression Cell Lines
To generate Tet-inducible expression of Otub1K0, Otub1K0 cDNA was subcloned into the pcDNA4-TO (Invitrogen) vector to generate the pcDNA4-TO-FLAG-Otub1K0 plasmid. T-Rex-U2OS cells (Invitrogen) were transfected with pcDNA4-TO-FLAG-Otub1K0, followed by selection in culture medium containing 50 μg/ml of hygromycin and 100 μg/ml of zeocin for 2 weeks. Single colonies were isolated, expanded, and screened by IB analysis for Dox-induced expression using anti-FLAG antibodies.
RNAi
The 21-nucleotide siRNA duplexes with a 3′ dTdT overhang were synthesized by Dharmacon Inc. (Lafayette, CO). The siRNA sequence targeting the 3′ UTR of the Otub1 mRNA was GTGGTTGTAAATGGTCCTA (21). The control scramble RNA sequence has been described previously (21). These siRNA duplexes were introduced into cells using SilentFect lipid reagent (Bio-Rad) following the protocol of the manufacturer. The cells were analyzed 48 h after transfection.
RT Quantitative PCR Analysis
Total RNA was isolated from cells using Qiagen RNeasy mini kits (Qiagen, Valencia, CA). Reverse transcriptions were performed as described previously (21). Quantitative real-time PCR was performed on an ABI 7300 real-time PCR system (Applied Biosystems) using iTaqTM Universal SYBR Green Supermix (Bio-Rad) as described previously (21, 26). All reactions were carried out in triplicate. The relative gene expression was calculated using the ΔCτ method following the protocol of the manufacturer. The primers for p21, mdm2, bax, and GAPDH have been described previously (21, 26).
Cell Proliferation Assay
A cell proliferation assay was carried out on an IncuCyte system (Essen Bioscience), which allowed us to do kinetic, noninvasive imaging of cells in culture right inside the well controlled incubator environment. Tet-inducible expression cells were split into 12 well-plates with or without Dox treatment and monitored in the IncuCyte system for 48 h. A cell growth curve was calculated and presented as the percentage of confluence from every 2-h phase-contrast imaging. For further validation, we conducted colony formation assays. T-Rex-U2OS-FLAG-Otub1 or T-Rex-U2OS-FLAG-Otub1K0 cells were plated in the absence or presence of 2 μg/ml Dox for up to 2 weeks. The colonies were visualized by staining with crystal violet blue.
GST Fusion Protein Association Assays
GST-UbcH5c and its mutants were purified using glutathione-Sepharose 4B beads. GST fusion protein-protein association assays were conducted as described previously (27). After washing, bound proteins were analyzed using IB analysis with anti-Otub1 and anti-GST antibodies.
Mass Spectrometry
For identifying Otub1 lysine residue(s) subjected to monoubiquitination, monoubiquitinated Otub1 was generated using an in vitro ubiquitination reaction, followed by SDS-PAGE gel electrophoresis and colloidal blue gel staining. The monoubiquitinated Otub1 was excised and trypsinized, and then peptides were analyzed by liquid chromatography/tandem mass spectrometry using an LTQ Velos Pro linear ion trap (Thermo Scientific) to collect data-dependent MS/MS data, as described previously (28). To determine protein sequences, a UniProt Sprot human database (downloaded August 2011) was created and amended with sequence-reversed entries to estimate the false discovery rate (FDR). Peptides were identified using SEQUEST (version 28, rev. 12, Thermo Scientific) with trypsin cleavage specificity, a static C+57 modification (alkylation), and a variable modification of K+114 (ubiquitination). SEQUEST results were filtered to strict peptide and protein FDRs, estimated from the number of matches to sequence-reversed peptides, using PAW software (29). Independent FDR control for unmodified peptides and peptides with mass shifts of +114 assigned to Lys residues was performed, resulting in a 4.2% FDR for protein discovery (46 identifications to forward sequence proteins and two to reverse-sequenced proteins) and less than a 7.7% FDR for detection of ubiquitination sites (13 MS/MS spectra assigned to forward sequence peptides containing K+114 and none to reverse-sequence peptides from any entry in the database).
Flow Cytometry
U2OS-To-FLAG-Otub1 or U2OS-To-FLAG-Otub1K0 cells were treated with or without Dox (2 μg/ml) for 24 h. The cells were harvested and stained with propidium iodide (Sigma) staining buffer (50 μg/ml propidium iodide, 200 μg/ml RNase A, and 0.1% Triton X-100 in PBS) at 37 °C for 30 min. The cells were measured for DNA content using a BD Biosciences FACScan flow cytometer. Data were analyzed using the CellQuest software program.
Structural Modeling
All modeling was performed using the UBQ_Gp_LYX-Cterm application of the Rosetta 3.5 suite (30) using standard parameters according to the documentation. For modeling of the quaternary complex, the crystal structure 4DDI was used. UbcH5b and UbcH5b∼Ub were included using the “extra-bodies” option. A total of 500 models were created for either Lys-59 or Lys-109 ubiquitination. For modeling of mono-ubiquitinated Otub1, the crystal structure 2ZFY was used. A total of 200 models were created for either Lys-59 or Lys-109 ubiquitination. The resulting models were clustered, and the highest scoring structure of the most populated cluster was used for further analysis.
RESULTS
Otub1 Is Monoubiquitinated by UbcH5
During our investigation of Otub1 suppression of E2 activity in vitro using reconstituted ubiquitination reactions, we observed a slowly migrating Otub1 band recognized by both anti-Otub1 and anti-Ub antibodies. The molecular weight was approximately equal to an Otub1 attached to a single Ub, suggesting a monoubiquitination of Otub1. To test whether Otub1 is indeed modified by monoubiquitination in vitro, we incubated purified recombinant His-Otub1 protein in the presence of different combinations of E1, UbcH5, Ub, and ATP, followed by IB analysis. As shown in Fig. 1A, the putative monoubiquitinated Otub1 band was detected only when both E1 and UbcH5 were present (lane 5), suggesting that UbcH5 mediates the monoubiquitination of Otub1. This modification requires ATP and is time-dependent (Fig. 1B). To examine whether Otub1 is monoubiquitinated in cells, H1299 cells were transfected with FLAG-tagged Otub1 alone or together with V5-tagged Ub. The cell lysates were subjected to co-IP with anti-FLAG antibody, followed by IB analysis with anti-V5. As shown in Fig. 1C, the monoubiquitinated form of Otub1 was detected in cells transfected with both FLAG-Otub1 and V5-Ub, demonstrating that Otub1 is also monoubiquitinated in cells.
FIGURE 1.
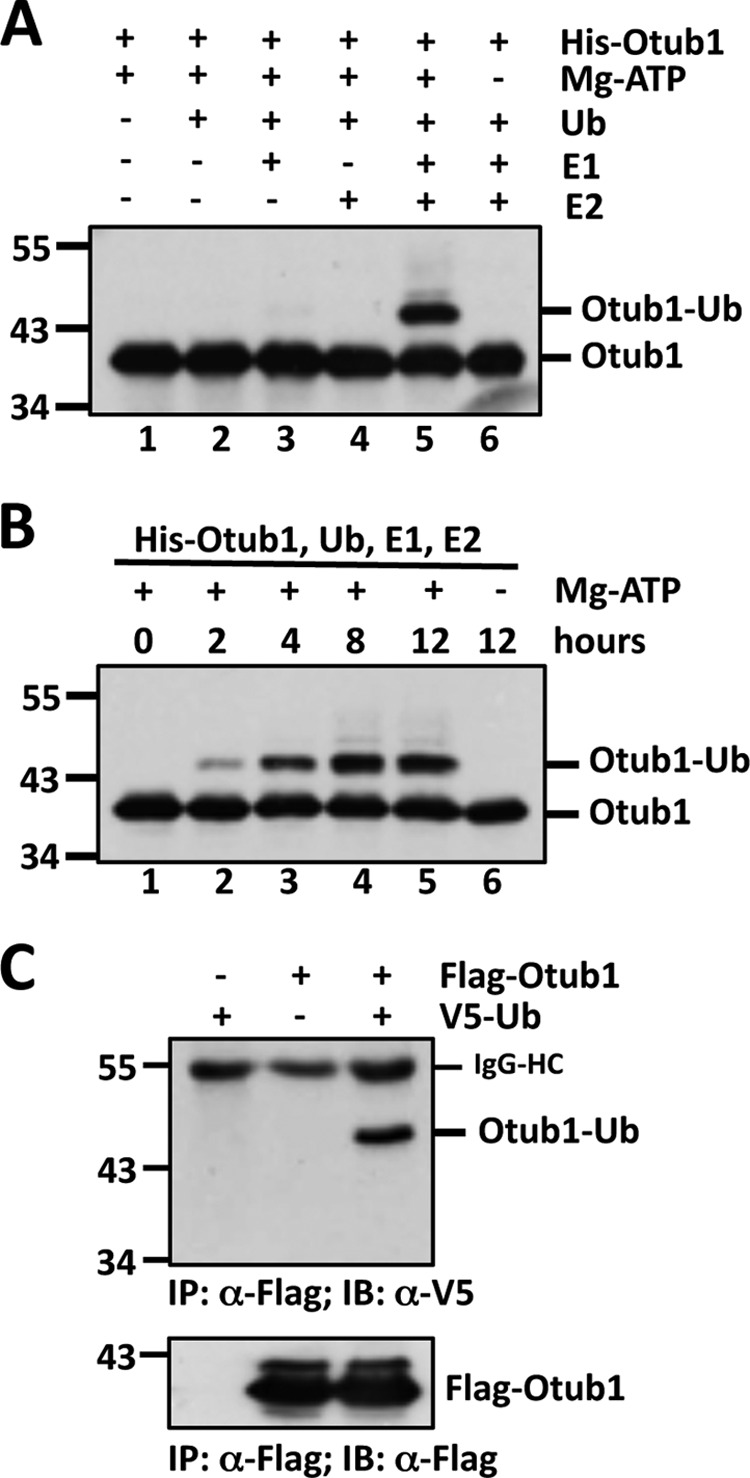
Otub1 is monoubiquitinated by UbcH5 in cells and in vitro. A and B, monoubiquitination of Otub1 by UbcH5 in vitro. His-Otub1 was incubated with different combinations of E1, UbcH5 (E2), Ub, and ATP, as indicated, for 8 h (A) or with E1, E2, and Ub in the presence or absence of ATP for different times, as indicated (B), followed by IB analysis with anti-Otub1. C, monoubiquitination of Otub1 in cells. H1299 cells transfected with FLAG-Otub1 with or without V5-Ub were subjected to co-IP with anti-FLAG antibody, followed by IB analysis with anti-V5 antibody (top panel) and anti-FLAG antibody (bottom panel). HC, heavy chain.
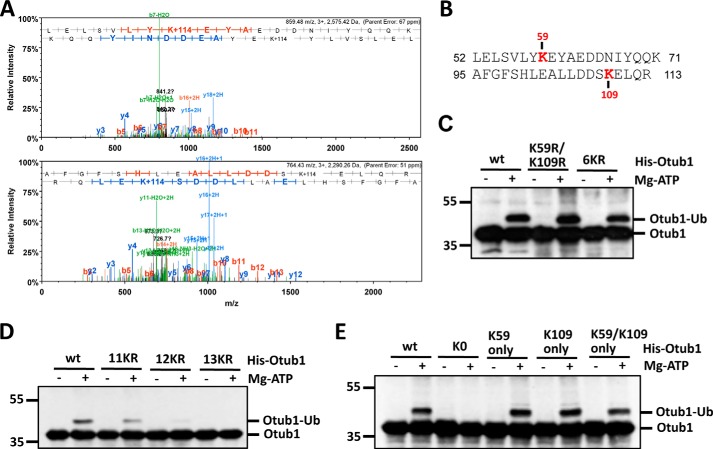
Lysines 59 and 109 Are Primary Monoubiquitination Sites
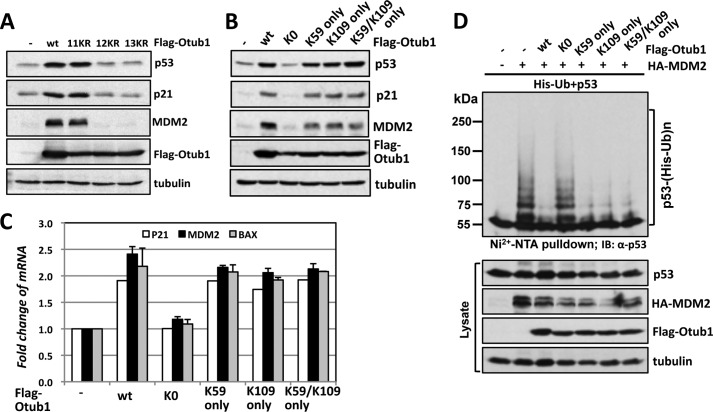
To identify which lysine residues are subjected to monoubiquitination, we performed a large volume of in vitro ubiquitination reactions using purified His-Otub1. The monoubiquitinated Otub1 was excised and in-gel trypsinized. The peptides were then subjected to mass spectrometry analysis. Two lysines (Lys-59 and Lys-109) were identified as putative Ub acceptor sites (Fig. 2, A and B). To validate whether Lys-59 or Lys-109 is indeed the monoubiquitination site, we generated Otub1 point mutants containing individual or both lysine residues mutated to arginine. Surprisingly, mutating a single lysine (K59R, K109R) (data not shown) or both (K59R/K109R) (Fig. 2C) did not abolish the monoubiquitination of Otub1, suggesting that Lys-59 and Lys-109 are either not primary ubiquitination sites or that monoubiquitin conjugation may take place in alternative lysine residues following the mutation of these primary lysines. To distinguish the two possibilities, we continued to mutate the rest of the lysines, initially focusing on lysines adjacent to Lys-59 and Lys-109, including 6KR (K59R/K109R/K71R/K73R/K77R/K78R) and 9KR (6KR+K115R/K120R/K122R), and found that no significant change in monoubiquitination was evident even when 11 of 17 lysines (11KR (9KR+K201R/K151R)) residues were replaced with arginine (Fig. 2, C and D, and data not shown). However, the level of monoubiquitinated Otub1 was drastically decreased when an additional Lys-188 residue was mutated (named 12KR) and completely abolished when both Lys-188 and Lys-213 were mutated (named 13KR) using 11KR as a template (Fig. 2D). These results led us to speculate that Lys-188 and/or Lys-213 might be the monoubiquitination sites. However, mutating individual (K188R, K213R) or both (K188R/K213R) failed to abolish the monoubiquitination of Otub1 (data not shown). Further, mutating four lysines (K188R/K213R/K59R/K109R) failed to abolish the monoubiquitination too (data not shown). These data suggest that Otub1 undergoes ubiquitination at alternative sites when primary sites are eliminated. This “alternative site theory” has been reported repeatedly in other studies (31–33). Thus, to further examine whether Lys-59 and/or Lys-109 are the primary sites, we went on to generate a lysine-free Otub1 (Otub1K0) where all 17 lysines are mutated to arginine and then mutated residue 59 or 109 or both back to Lys using Otub1K0 as a template (Otub1K59 only, Otub1K109 only, and Otub1K59/K109 only, respectively). As expected, Otub1K0 was unable to be monoubiquitinated by UbcH5 (Fig. 2E). Interestingly, adding individual lysine (Lys-59 or Lys-109) or both (Lys-59/Lys-109) back completely restored the monoubiquitination of Otub1K0 (Fig. 2E). These data strongly suggest that Lys-59 and Lys-109 are the primary monoubiquitination sites of Otub1.
FIGURE 2.
Otub1 is monoubiquitinated by UbcH5 primarily at Lys-59 and Lys-109. A, identification of the monoubiquitination sites by mass spectrometry. The MS/MS spectrum (+3 ion) of Otub152–71 indicating ubiquitination at residue Lys-59 (top panel) and the MS/MS spectrum (+3 ion) of Otub195–113 indicating ubiquitination at residue Lys-109 (bottom panel) are shown. Matched b-series fragment ions are indicated in red, matched y-ion series fragment ions are indicated in blue, and matched fragment ions (both y and b) undergoing loss of water are indicated in green. B, summary of the peptides containing Ub-modified lysines (Lys-59 and Lys-109) identified by LC-MS/MS analysis. C and D, Otub1wt, Otub1K59R/K109R, Otub16KR, and Otub111KR, but not Otub112KR and Otub113KR, were monoubiquitinated by UbcH5 in vitro. WT Otub1 or its mutants, as indicated, were incubated with E1, E2, and Ub in the presence or absence of ATP, followed by IB analysis using anti-Otub1. E, Otub1wt, Otub1K59 only, Otub1K109 only, and Otub1K59/K109 only, but not Otub1K0, were monoubiquitinated by UbcH5 in vitro. WT Otub1 or its mutants, as indicated, were incubated with E1, E2, and Ub in the presence or absence of ATP, followed by IB analysis using anti-Otub1.
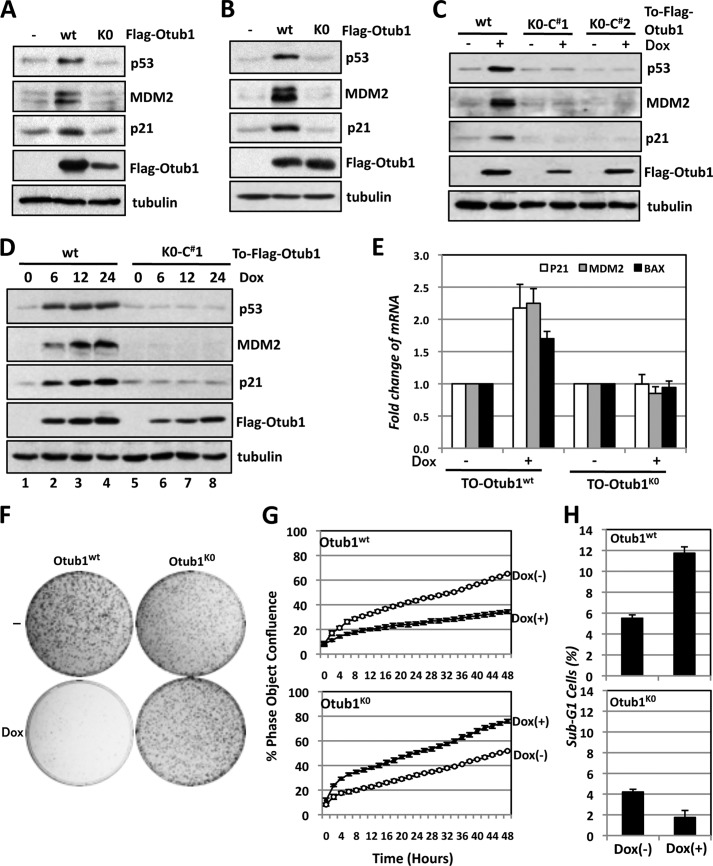
Lysine-free Otub1 Does Not Activate p53
To test whether monoubiquitination affects the function of Otub1 to regulate the stability and activity of p53, we took advantage of the monoubiquitination-defective Otub1K0 mutant. As shown in Fig. 3A and as expected (21), overexpression of WT Otub1 significantly induced the levels of p53 as well as its targets p21 and MDM2 in p53-proficient U2OS cells. However, overexpression of Otub1K0 failed to do so. Of note, we observed that the expression of Otub1K0 mutant protein is consistently lower than that of Otub1wt. To test whether the defective effect of Otub1K0 is due to this lower level of expression, Otub1wt and the Otub1K0 mutant plasmids were transfected into U2OS cells at different ratios. As shown in Fig. 3B, Otub1wt and Otub1K0 proteins were expressed at similar levels when the plasmids were transfected at a 1:6 ratio. Under this condition, Otub1K0 still failed to induce p53 and its target genes, whereas Otub1wt potently induced and activated p53 (Fig. 3B). Therefore, the inability of Otub1K0 to induce p53 is not due to its lower levels of expression relative to Otub1wt. Also, Dox-induced expression of Otub1K0 in two representative U2OS-TO-FLAG-Otub1K0 clones failed to induce p53 and its targets p21 and MDM2, whereas induced expression of WT Otub1 drastically induced p53 and its target proteins (Fig. 3C) and the levels of p21, bax, and mdm2 mRNA in U2OS cells (Fig. 3E). A time course study of induced Otub1 expression further confirmed that Otub1K0 failed to induce and activate p53 (Fig. 3D), even when its expression level was similar to that of Otub1wt (Fig, 3D, compare lane 8 to lane 2). Consistently, in contrast to the expression of Otub1wt, induced expression of Otub1K0 failed to suppress cell proliferation, as determined by both colony formation assays (Fig. 3F) and measuring of cell confluence over time in culture using the IncuCyte system (Fig. 3G). Instead, we consistently observed that overexpression of Otub1K0 slightly promoted cell proliferation (Fig. 3G) and increased the size of colonies (Fig. 3F). Because overexpression of Otub1wt also induces apoptosis (21), we next examined whether the lysine-free Otub1 could fail to do so. As shown in Fig. 3H, overexpression of Otub1wt significantly induced apoptosis, as evidenced by the marked increase in sub-G1 populations, whereas overexpression of Otub1K0 slightly inhibited apoptosis. Together, these data suggest that the lysine-free Otub1 fails to induce and activate p53 and is unable to induce apoptosis as well as suppress cell proliferation. Instead, it may act as a dominant-negative mutant in cells.
FIGURE 3.
Lysine-free Otub1 fails to induce and activate p53. A and B, Otub1K0 does not induce p53. U2OS cells were transfected with equal amounts (3 μg) of the FLAG-Otub1wt or FLAG-Otub1K0 plasmid (A) or 0.5 μg of the FLAG-Otub1wt or 3 μg of the FLAG-Otub1K0 plasmid (B), followed by IB analysis. C, induced expression of Otub1K0 fails to induce p53. Two representative U2OS-TO-FLAG-Otub1K0 clones and the U2OS-TO-FLAG-Otub1wt stable cell line were cultured in the absence or presence of 2 μg/ml of Dox for 24 h. The cell lysates were assayed by IB analysis. D, time course study of the p53 induction by Otub1wt but not Otub1K0. U2OS-TO-FLAG-Otub1wt or U2OS-TO-FLAG-Otub1K0 clone 1 (C#1) was cultured in the presence of 2 μg/ml of Dox and harvested at different time points, as indicated, followed by IB analysis. E, Otub1K0 does not induce p53 activity. U2OS-TO-FLAG-Otub1K0 or U2OS-TO-FLAG-Otub1wt cells were treated with or without Dox for 24 h, followed by RT-qPCR detection of p21, mdm2, and bax mRNA normalized to the expression of GAPDH. F, Otub1K0 does not inhibit cell proliferation. U2OS-TO-FLAG-Otub1wt or U2OS-TO-FLAG-Otub1K0 cells were cultured in the presence or absence of Dox for up to 3 weeks. The colonies were visualized by staining with crystal violet blue. G, T-Rex-U2OS-FLAG-Otub1wt or T-Rex-U2OS-FLAG-Otub1K0 cells were cultured in the presence or absence of 2 μg/ml doxycycline. The cell confluence was measured over time using the IncuCyte system. H, Otub1K0 does not induce apoptosis. U2OS-TO-FLAG-Otub1wt or U2OS-TO-FLAG-Otub1K0 cells were cultured with or without Dox for 24 h. The cells were stained with propidium iodide, followed by flow cytometry analysis. The average percentages of cells in sub-G1 are shown.
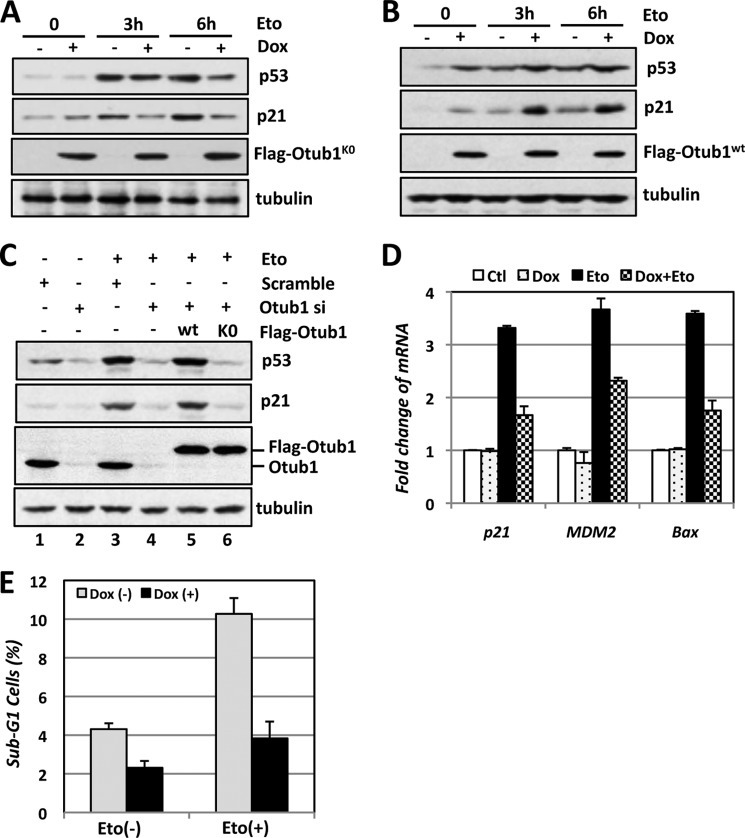
Lysine-free Otub1 Attenuates p53 Activation in Response to DNA Damage
We have shown previously that Otub1 plays a critical role in DNA damage-induced p53 activation (21). Therefore, we next asked whether Otub1K0 affects p53 signaling in response to DNA damage. To this end, U2OS-TO-FLAG-Otub1K0 cells were cultured with or without Dox, followed by treatment of etoposide (Eto) for different durations. As shown in Fig. 4A, overexpression of Otub1K0 significantly attenuated the induction of p53 and p21 proteins. However, this was not the case in U2OS-TO-FLAG-Otub1wt cells. Instead, overexpression of Otub1wt enhanced the induction of p53 and p21 proteins by Eto treatment (Fig. 4B). To further confirm the role of Otub1K0 in attenuating p53 activation in response to DNA damage, we performed siRNA knockdown and rescue experiments. As shown in Fig. 4C, although transfection with FLAG-Otub1wt completely rescued p53 induction by Eto treatment in cells transfected with Otub1 siRNA targeting the 3′ UTR of the Otub1 mRNA that is not present in the FLAG-Otub1 expression plasmid (Fig. 4C, compare lane 5 to lane 4), FLAG-Otub1K0 clearly failed to do so (lane 6). RT-qPCR assays showed that the induction of p21, mdm2, and Bax mRNA by Eto treatment was reduced significantly by overexpression of Otub1K0 (Fig. 4D). Consistently, overexpression of Otub1K0 suppressed Eto-induced apoptosis in cells (Fig. 4E). These results suggest that Otub1K0 attenuates p53 activation in response to DNA damage.
FIGURE 4.
Lysine-free Otub1 attenuates p53 activation in response to DNA damage. A and B, Otub1K0, but not Otub1wt, attenuates p53 activation following Eto treatment. U2OS-TO-FLAG-Otub1K0 cells (A) and U2OS-TO-FLAG-Otub1wt cells (B) were cultured in the absence or presence of Dox for 24 h, followed by treatment with Eto (20 μm) or control dimethyl sulfoxide for the indicated times, followed by IB analysis. C, reintroduction of Otub1wt, but not the Otub1K0 mutant, rescues the p53 response following DNA damage in cells with Otub1 knockdown. U2OS cells transfected with control, the FLAG-Otub1wt or FLAG-Otub1K0 plasmid, and scrambled or Otub1 siRNA targeting the 3′ UTR of the Otub1 mRNA are shown, as indicated. The cells were treated with Eto (20 μm) for 6 h before harvesting 48 h after siRNA transfection, followed by IB analysis. D and E, Otub1K0 attenuates p53 activation in response to DNA damage. U2OS-TO-FLAG-Otub1K0 cells were cultured in the absence or presence of Dox for 24 h, followed by treatment with Eto (20 μm) or control (Ctl) dimethyl sulfoxide for 6 h, and assayed by RT-qPCR (D) and cell cycle profile analysis for sub-G1 phase cells (E).
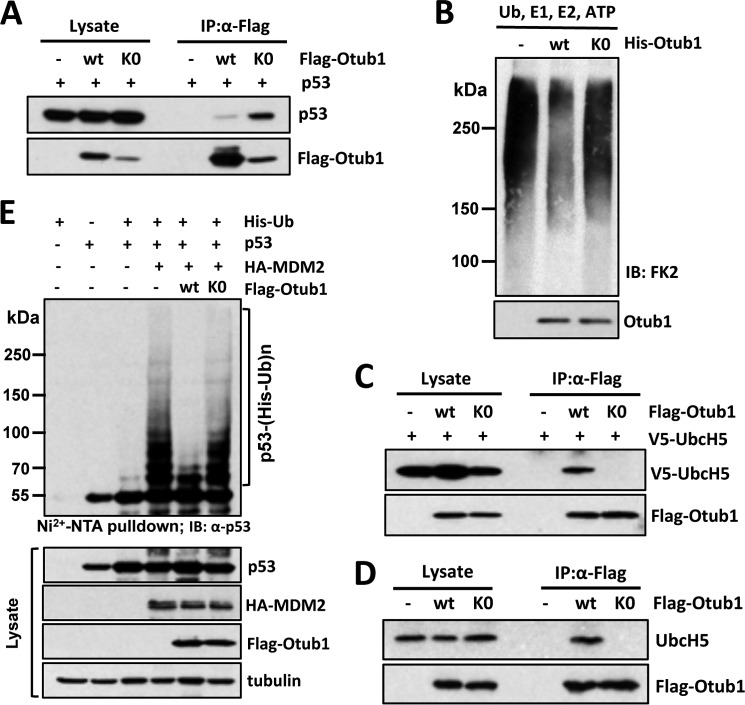
Lysine-free Otub1 Fails to Suppress E2 in Vitro
To determine why Otub1K0 fails to induce p53 activity, we first tested the physical interaction between Otub1 and p53. However, co-IP assays clearly showed that Otub1K0 bound to p53 much more strongly than Otub1wt (Fig. 5A). This effect is reminiscent of the Otub1D88A mutant, which does not induce and activate p53 (21). We then examined whether Otub1K0 fails to suppress E2 because Otub1 exerts its regulatory function on p53 or DNA damage response via a non-canonical mechanism (21, 22). In vitro reactions were assembled using recombinant E1, UbcH5, and Ub in the absence or presence of His-tagged Otub1wt or Otub1K0, followed by IB analysis with anti-conjugated Ub antibody (FK2). As expected, Otub1wt dramatically suppressed the Ub-conjugating activity of UbcH5 to form poly-Ub chains. However, Otub1K0 failed to do so (Fig. 5B). Also, Otub1K0 did not interact with either exogenous or endogenous UbcH5, as determined by co-IP assays in cells either transfected with equal amounts of the FLAG-Otub1wt and FLAG-Otub1K0 plasmids (data not shown) or with the plasmids at a 1:6 ratio to obtain equal protein expression (Fig. 5, C and D). Consistently, Otub1K0 failed to suppress MDM2-mediated p53 ubiquitination in cells (Fig. 5E). Together, these results revealed that the monoubiquitination-defective Otub1K0 does not suppress E2 and, thus, fails to induce p53 stabilization and activation.
FIGURE 5.
Lysine-free Otub1 fails to interact with UbcH5 and suppress UbcH5 activity in vitro and MDM2-mediated p53 ubiquitination in cells. A, Otub1K0 binds to p53. H1299 cells transfected with p53 together with Otub1wt or Otub1K0 were subjected to co-IP using anti-FLAG antibodies, followed by IB analysis. B, Otub1K0 does not suppress UbcH5-dependent ubiquitin chain formation. The in vitro ubiquitination reactions were conducted in the presence of E1, E2, Ub, and ATP in the absence or presence of His-tagged Otub1wt or Otub1K0, as indicated. The reactions were assayed by IB analysis using anti-conjugated Ub antibody (clone FK2) (top panel). C and D, Otub1K0 does not interact with UbcH5 in cells. H1299 cells transfected with 0.25 μg of the FLAG-Otub1wt plasmid or 1.5 μg of the FLAG-Otub1K0 plasmid alone (D) or together with 1.5 μg of V5-UbcH5 (C) were subjected to co-IP with anti-FLAG antibodies, followed by IB analysis. E, Otub1K0 does not inhibit MDM2-mediated p53 ubiquitination in cells. H1299 cells transfected with indicated plasmids were subjected to pulldown using Ni-NTA beads under denaturing conditions, followed by IB analysis. The ubiquitinated species of p53 are indicated.
Monoubiquitination of Otub1 Correlates with Its Activity to Induce p53
To determine whether monoubiquitination is critical for Otub1 to stabilize and activate p53, we analyzed a series of Otub1 mutants for their activity to induce and activate p53. We found that the monoubiquitination-competent 11KR mutant Otub1 was able to induce p53 and its targets p21 and MDM2 in cells, whereas the monoubiquitination-defective Otub1 mutants (12KR and 13KR) failed to do so (Fig. 6A). Interestingly, adding either Lys-59 or Lys-109 or both back to Otub1K0 restored the function of Otub1 to induce p53 and its targets p21 and MDM2 (Fig. 6B). An RT-qPCR assay also showed that Otub1K59 only and Otub1K109 only, but not Otub1K0, induced the expression of p21, mdm2, and bax mRNAs (Fig. 6C). Consistently, the Otub1K59 only and Otub1K109 only mutants drastically suppressed MDM2-mediated p53 ubiquitination as efficiently as Otub1wt in cells (Fig. 6D). Thus, monoubiquitination of Otub1 plays a critical role in the function of Otub1 to regulate p53.
FIGURE 6.
Monoubiquitination of Otub1 is required for its activity to induce and activate p53. A, monoubiquitination-competent but not monoubiquitination-defective mutants induce p53. U2OS cells were transfected with Otub1wt or its mutants (11KR, 12KR, and 13KR) followed by IB analysis. B and C, adding Lys-59 or Lys-109 back to Otub1K0 restores Otub1 function to regulate p53. U2OS cells were transfected with Otub1wt, Otub1K0, Otub1K59 only, Otub1K109 only, or Otub1K59/K109 only mutants, followed by IB analysis (B) and RT-qPCR (C) analysis. D, Lys-59-only or Lys-109-only Otub1 suppresses MDM2-mediated p53 ubiquitination in cells. H1299 cells transfected with His-Ub and p53 in the presence of the indicated plasmids were subjected to Ni-NTA pulldown under denaturing conditions, followed by IB analysis. The ubiquitinated species of p53 are indicated.
UbcH5 Preferentially Binds to Monoubiquitinated Otub1
To further elucidate the molecular insights into the role of monoubiquitination in the function of Otub1, we reasoned that monoubiquitination may promote the association of Otub1 with UbcH5 because Otub1K0 fails to interact with UbcH5 (Fig. 5, C and D) and UbcH5 makes extensive contacts with Ub (34). It is likely that monoubiquitination could increase the binding of Otub1 to an E2 through Ub-E2 interaction. Indeed, when the in vitro ubiquitination reaction mixture containing monoubiquitinated and unmodified Otub1 was incubated with GST-UbcH5, we found that UbcH5 preferentially bound to the monoubiquitinated over the unmodified form of Otub1 (Fig. 7A). These results suggest that monoubiquitination of Otub1 promotes Otub1-UbcH5 binding, thereby suppressing the Ub-conjugating activity of the UbcH5.
FIGURE 7.
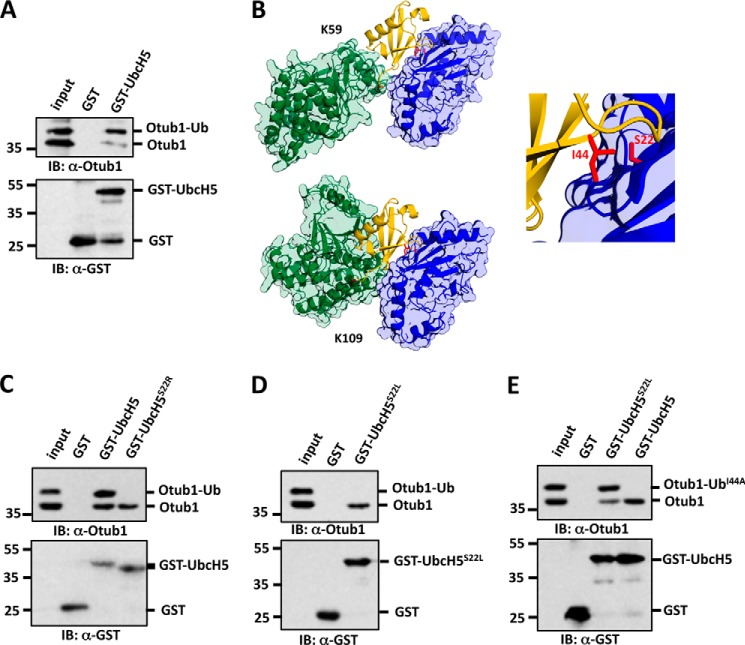
UbcH5 binds preferentially to monoubiquitinated Otub1 through backside UbcH5-Ub interaction. A, UbcH5 binds preferentially to the monoubiquitinated Otub1. His-Otub1 was subjected to an in vitro ubiquitination reaction as in Fig. 1. The reaction mixture containing both monoubiquitinated and unmodified Otub1 was then incubated with GST alone or GST-UbcH5c immobilized onto GSH beads. After washing, bead-bound proteins were assayed by IB analysis. B, modeling of the Ub-Otub1/UbcH5b interaction through docking of Ub linked to either Lys-59 (top left panel) or Lys-109 (bottom left panel) with the backside of UbcH5b. Otub1, UbcH5b, and Ub are colored green, blue, and yellow, respectively. Lys-59 and Lys-109 of Otub1, Ile-44 of Ub, and Ser-22 of UbcH5 are indicated in red. The enlarged view in the right panel shows the canonical Ub Ile-44 interacting with Ser-22 on UbcH5. C and D, UbcH5S22R and UbcH5S22L do not bind to monoubiquitinated Otub1. The in vitro ubiquitination reaction mixture containing both monoubiquitinated and unmodified Otub1 was incubated with GST alone, GST-UbcH5c, GST-UbcH5S22R, or GST-UbcH5S22L immobilized onto GSH beads. After washing, bead-bound proteins were assayed by IB analysis. E, mutating Ser-22 to Leu rescues the binding defect of UbcH5 with Otub1-UbI44A. His-Otub1 was subjected to an in vitro ubiquitination reaction using recombinant UbI44A. The reaction mixture containing both monoubiquitinated (Otub1-UbI44A) and unmodified Otub1 was incubated with GST alone, GST-UbcH5c, or GST-UbcH5S22L immobilized onto GSH beads. After washing, bead-bound proteins were assayed by IB analysis.
Monoubiquitinated Otub1 Binds to UbcH5 via Ub Interaction with the Backside Ub-interacting Surface of UbcH5
How does monoubiquitination promote Otub1-E2 interaction? Biochemical (22) and structural (24, 25) studies have shown that Otub1 preferentially binds to Ub-charged E2 (E2∼Ub). Because UbcH5 preferentially binds to monoubiquitinated Otub1 (Otub1-Ub) (Fig. 7A), we reasoned that monoubiquitination at Lys-59 or Lys-109 may stabilize the Otub1-UbcH5 interaction by forming a quadruple molecular complex containing Otub1-Ub·UbcH5∼Ub via direct interaction of Otub1-linked Ub with UbcH5∼Ub within the Otub1·UbcH5∼Ub complex characterized previously (24). Thus, we modeled the quaternary complex between UbcH5b∼Ub and Otub1-Ub using Rosetta software (30). After extensive sampling of the conformational space available to Ub, we found that Ub linked to Lys-109 was unable to form a direct interaction with UbcH5b∼Ub (data not shown). Ub linked to Lys-59 formed interactions with UbcH5b∼Ub in a small subsets of sampled conformations (data not shown). Because ubiquitination of either Lys-59 or Lys-109 has a similar effect to regulate p53, we suggest that Ub linked to Otub1 does not stabilize Otub1-UbcH5 interaction through direct interactions within the characterized Otub1·UbcH5∼Ub complex (24).
To further understand how monoubiquitination promotes Otub1-UbcH5 interaction, we modeled the structures of monoubiquitinated Otub1 alone. Interestingly, for monoubiquitination at either Lys-109 or Lys-59, the most frequently sampled poses exposed the canonical Ile-44 interface of Ub, potentially allowing for the interaction of Ub with the “backside” of UbcH5 against the catalytic Cys known previously for donor Ub binding (34, 35). When we superimposed the Ub linked to Otub1 with Ub (charged to another UbcH5) bound to the backside of UbcH5b (PDB code 3A33), we found no steric clashes between UbcH5b and Otub1. This suggested that the Ub covalently linked to Otub1 could facilitate the Otub1-UbcH5 interaction through binding to the backside Ub-interacting surface of UbcH5 (Fig. 7B). This backside interaction of UbcH5 with Ub involves key residues, including Ser-22 on UbcH5 and Ile-44 on Ub (Fig. 7B, right panel) (34). To test this possibility, we first examined whether mutating Ser-22 on UbcH5 to Arg (UbcH5S22R) could affect this backside UbcH5-Ub interaction because this mutation has been shown to disrupt the Ub-UbcH5 interaction (34). As shown in Fig. 7C, although wild-type GST-UbcH5 bound to the monoubiquitinated Otub1, the GST-UbcH5S22R mutant failed to bind. Similarly, mutating Ser-22 to Leu (UbcH5S22L) also disrupted the binding between UbcH5 and the monoubiquitinated Otub1 (Fig. 7D). These mutants generate a steric clash with Ub Ile-44 because of the larger side chain of Arg or Leu compared with Ser (34, 36). Next, we asked whether mutating Ile-44 of Ub to Ala with a smaller side chain could also affect the binding. The mutant Ub-I44A (UbI44A) can still be used for Otub1 monoubiquitination in vitro (Fig. 7E). Using the reaction mixture containing Otub1 monoubiquitinated with UbI44A (Otub1-UbI44A) for GST pulldown assays, we found that wild-type GST-UbcH5 indeed failed to interact with Otub1-UbI44A (Fig. 7E, last lane). Interestingly, the I44A mutation relieves the steric clash generated by the S22L mutation through complementation, resulting in the rescued binding between UbcH5S22L and Otub1-UbI44A (Fig. 7E). Together, these results demonstrate that monoubiquitinated Otub1 interacts with the backside Ub-interacting surface of UbcH5 through the covalently linked Ub. This backside binding could potentially cripple the self-assembly of UbcH5∼Ub conjugates thought to be critical for ubiquitin transfer to substrates and poly-Ub chain formation (34, 35), thereby suppressing the Ub-conjugating activity of UbcH5.
DISCUSSION
Otub1 has recently emerged as a unique Dub that binds to and inhibits several classes of E2s, including Ubc13 and UbcH5s (21–25). By doing so, Otub1 regulates the stability and activity of a number of proteins, including p53 (21), histones (22), SMAD2/3 (37), and gene related to anergy in lymphocytes (GRAIL) (38), independently of its deubiquitinating enzyme activity. Otub1 also regulates several other substrates via its canonical deubiquitinating enzyme activity, such as the cellular inhibitor of apoptosis (c-IAP) (39), estrogen receptor α (40), and tumor necrosis factor receptor-associated factors (TRAFs) 3 and 6 (41). Together, Otub1 plays an important role in diverse biological processes, including DNA damage response, immune response, inflammation, apoptosis, and signal transduction.
In this study, we discovered a novel mechanism for the Otub1 suppression of E2. We found that Otub1 is monoubiquitinated by UbcH5 in cells and in vitro. UbcH5, in turn, binds preferentially to the monoubiquitinated Otub1 (Otub1-Ub). Structural modeling and biochemical analysis revealed that this binding occurs through the extensive contacts between the β1-β3 backside Ub-binding surface of UbcH5 and the canonical “Ile-44 surface” of Ub covalently linked to Otub1 (Fig. 7B). Mutating either Ser-22 on UbcH5 to Arg or Leu or Ile-44 on Ub to Ala abolished the interaction between UbcH5 and the ubiquitinated Otub1 (Fig. 7, C–E) because of the creation of a steric clash in the Ub-E2 interface. Remarkably, this steric clash can be compensated by mutating both Ser-22 of UbcH5 to Leu and Ile-44 of Ub to Ala, resulting in the rescued binding between UbcH5S22L and the UbI44A covalently linked to Otub1 (Otub1-UbI44A). Consequently, this backside E2 interaction with monoubiquitinated Otub1 would interfere with the binding of a charged donor Ub to the same site of a second E2, which is critical for the Ub transfer and Ub chain elongation during the Ub-conjugating reaction (34, 36, 42).
This novel mechanism could exist in parallel with a mechanism described previously wherein Otub1 binds to E2∼Ub (24, 25) and they act in concert to potentiate the function of Otub1 to suppress E2. Previous structural studies have shown that binding of a free Ub to the distal Ub-binding site on Otub1 causes a conformational change of Otub1 to form an N-terminal Ub-binding helix, allowing for the binding of the donor Ub (24, 25). This Otub1 binding blocks the interaction of the donor Ub with another E2 and the attack on the thioester bond by an acceptor Ub and also suppresses Ub transfer (34, 36, 42). Future studies are warranted to address how the two distinct mechanisms could interplay, timely and spatially, and whether the mechanism described here is also regulated by the cellular concentration of free Ub or the molecular ratio of charged versus uncharged E2. Alternatively, monoubiquitinated Otub1 could bind to Ub-charged E2 to form a quadruple inhibitory complex (Otub1-Ub·E2∼Ub) through the backside UbcH5-Ub interaction. This quadruple complex would then form an infinite spiral, as in the case of the self-assembly of E2∼Ub conjugates (34, 35) through the donor Ub interaction with Otub1. This complex could, therefore, disrupt the assembly of E2∼Ub conjugates and suppress the Ub transfer and efficient polyubiquitination of substrates (34, 35). In this scenario, it is important to test whether monoubiquitination of Otub1 could result in a conformational change to promote the donor Ub binding to Otub1. Thus, future characterization of the structure of the inhibitory Otub1-Ub·UbcH5∼Ub complex would provide further insights into how Otub1 suppresses E2.
Functionally, we showed that monoubiquitination of Otub1 is critical for Otub1 to suppress UbcH5 and stabilize and activate p53. This is evident from observations showing that the monoubiquitination-defective mutants, including Otub1K0, failed to suppress E2 and induce p53, whereas the monoubiquitination-competent mutants, such as Otub111KR, Otub1K59 only, and Otub1K109 only, induced p53 as efficiently as Otub1wt (Figs. 3 and 6). Thus, our study further emphasizes the emerging function of Otub1 as a potent E2 inhibitor and demonstrates a novel mechanism whereby monoubiquitination of Otub1 promotes its function to suppress E2 and activate p53. This novel mechanism could be implicated in targeting E2 for cancer therapy.
This work was supported, in whole or in part, by NCI, National Institutes of Health grants R00 CA127134 and R01 CA160474 (to M. S. D.). This work was also supported by Department of Defense Grant W81XWH-10-1029 (to M. S. D.).
- Ub
- ubiquitin
- USP
- ubiquitin-specific protease
- Dub
- deubiquitinating enzyme
- Ni-NTA
- nickel-nitrilotriacetic acid
- IB
- immunoblot
- IP
- immunoprecipitation
- Dox
- doxycycline
- RT-qPCR
- quantitative RT-PCR
- FDR
- false discovery rate
- Eto
- etoposide.
REFERENCES
- 1. Devine T., Dai M. S. (2013) Targeting the ubiquitin-mediated proteasome degradation of p53 for cancer therapy. Curr. Pharm. Des. 19, 3248–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kruse J. P., Gu W. (2009) Modes of p53 regulation. Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275, 8945–8951 [DOI] [PubMed] [Google Scholar]
- 4. Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 5. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 6. Honda R., Tanaka H., Yasuda H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 7. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 8. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y., Lu H. (2009) Signaling to p53. Ribosomal proteins find their way. Cancer Cell 16, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 11. Li M., Chen D., Shiloh A., Luo J., Nikolaev A. Y., Qin J., Gu W. (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653 [DOI] [PubMed] [Google Scholar]
- 12. Meulmeester E., Maurice M. M., Boutell C., Teunisse A. F., Ovaa H., Abraham T. E., Dirks R. W., Jochemsen A. G. (2005) Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol. Cell 18, 565–576 [DOI] [PubMed] [Google Scholar]
- 13. Cummins J. M., Rago C., Kohli M., Kinzler K. W., Lengauer C., Vogelstein B. (2004) Tumour suppression. Disruption of HAUSP gene stabilizes p53. Nature 428, 1 p following 486 [DOI] [PubMed] [Google Scholar]
- 14. Li M., Brooks C. L., Kon N., Gu W. (2004) A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13, 879–886 [DOI] [PubMed] [Google Scholar]
- 15. Yuan J., Luo K., Zhang L., Cheville J. C., Lou Z. (2010) USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140, 384–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu J., Chung H. J., Vogt M., Jin Y., Malide D., He L., Dundr M., Levens D. (2011) JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 30, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hock A. K., Vigneron A. M., Carter S., Ludwig R. L., Vousden K. H. (2011) Regulation of p53 stability and function by the deubiquitinating enzyme USP42. EMBO J. 30, 4921–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allende-Vega N., Sparks A., Lane D. P., Saville M. K. (2010) MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene 29, 432–441 [DOI] [PubMed] [Google Scholar]
- 19. Stevenson L. F., Sparks A., Allende-Vega N., Xirodimas D. P., Lane D. P., Saville M. K. (2007) The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 26, 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X., Berger F. G., Yang J., Lu X. (2011) USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J. 30, 2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun X. X., Challagundla K. B., Dai M. S. (2012) Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 31, 576–59222124327 [Google Scholar]
- 22. Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y. C., O'Donnell L., Kumakubo A., Munro M., Sicheri F., Gingras A. C., Natsume T., Suda T., Durocher D. (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 [DOI] [PubMed] [Google Scholar]
- 23. Zulkifle N. (2013) Systematic yeast two-hybrid analysis of human E2 ubiquitin-conjugating enzyme and deubiquitin (DUB) protein interaction. Int. J. Biol. Chem. 7, 1–14 [Google Scholar]
- 24. Juang Y. C., Landry M. C., Sanches M., Vittal V., Leung C. C., Ceccarelli D. F., Mateo A. R., Pruneda J. N., Mao D. Y., Szilard R. K., Orlicky S., Munro M., Brzovic P. S., Klevit R. E., Sicheri F., Durocher D. (2012) OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 45, 384–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wiener R., Zhang X., Wang T., Wolberger C. (2012) The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483, 618–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun X. X., DeVine T., Challagundla K. B., Dai M. S. (2011) Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. J. Biol. Chem. 286, 22730–22741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan F. F., Pratt E. B., Chen P. C., Wang F., Skach W. R., David L. L., Shyng S. L. (2010) Role of Hsp90 in biogenesis of the β-cell ATP-sensitive potassium channel complex. Mol. Biol. Cell 21, 1945–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilmarth P. A., Riviere M. A., David L. L. (2009) Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Infor. 2, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baker R., Lewis S. M., Sasaki A. T., Wilkerson E. M., Locasale J. W., Cantley L. C., Kuhlman B., Dohlman H. G., Campbell S. L. (2013) Site-specific monoubiquitination activates Ras by impeding GTPase-activating protein function. Nat. Struct. Mol. Biol. 20, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Denuc A., Bosch-Comas A., Gonzàlez-Duarte R., Marfany G. (2009) The UBA-UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition. PLoS ONE 4, e5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dimova N. V., Hathaway N. A., Lee B. H., Kirkpatrick D. S., Berkowitz M. L., Gygi S. P., Finley D., King R. W. (2012) APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat. Cell Biol. 14, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ju D., Xie Y. (2006) Identification of the preferential ubiquitination site and ubiquitin-dependent degradation signal of Rpn4. J. Biol. Chem. 281, 10657–10662 [DOI] [PubMed] [Google Scholar]
- 34. Sakata E., Satoh T., Yamamoto S., Yamaguchi Y., Yagi-Utsumi M., Kurimoto E., Tanaka K., Wakatsuki S., Kato K. (2010) Crystal structure of UbcH5b∼ubiquitin intermediate. Insight into the formation of the self-assembled E2∼Ub conjugates. Structure 18, 138–147 [DOI] [PubMed] [Google Scholar]
- 35. Brzovic P. S., Lissounov A., Christensen D. E., Hoyt D. W., Klevit R. E. (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 [DOI] [PubMed] [Google Scholar]
- 36. Saha A., Lewis S., Kleiger G., Kuhlman B., Deshaies R. J. (2011) Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Herhaus L., Al-Salihi M., Macartney T., Weidlich S., Sapkota G. P. (2013) OTUB1 enhances TGFβ signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat. Commun. 4, 2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soares L., Seroogy C., Skrenta H., Anandasabapathy N., Lovelace P., Chung C. D., Engleman E., Fathman C. G. (2004) Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat Immunol. 5, 45–54 [DOI] [PubMed] [Google Scholar]
- 39. Goncharov T., Niessen K., de Almagro M. C., Izrael-Tomasevic A., Fedorova A. V., Varfolomeev E., Arnott D., Deshayes K., Kirkpatrick D. S., Vucic D. (2013) OTUB1 modulates c-IAP1 stability to regulate signalling pathways. EMBO J. 32, 1103–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stanisi V., Malovannaya A., Qin J., Lonard D. M., O'Malley B. W. (2009) OTU Domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) α and affects ERα transcriptional activity. J. Biol. Chem. 284, 16135–16145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li S., Zheng H., Mao A. P., Zhong B., Li Y., Liu Y., Gao Y., Ran Y., Tien P., Shu H. B. (2010) Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 285, 4291–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wickliffe K. E., Lorenz S., Wemmer D. E., Kuriyan J., Rape M. (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]