Abstract
Understanding the mechanisms underlying diversification and speciation by introgressive hybridization is currently one of the major challenges in evolutionary biology. Here, the analysis of hybridization between two pairs of Iberian Leuciscinae provided new data on independent hybrid zones involving Achondrostoma oligolepis (AOL) and Pseudochondrostoma duriense (PDU), and confirmed the occurrence of hybrids between AOL and Pseudochondrostoma polylepis (PPO). A multilevel survey combining morphological, genetic and cytogenomic markers on a vast population screening successfully sorted the selected fishes as admixed. Results were similar in both AOL × PDU and AOL × PPO systems. Overall, hybrid morphotypes, cytogenomic data and genetic profiling indicated preferential backcrossing and suggested AOL as a major genomic contributor. Moreover, results implied AOL as more permissive to introgression than PDU or PPO. Although PDU- and PPO-like individuals appeared more resilient to genome modifications, AOL appeared to be more involved and affected by the ongoing hybridization events, as chromosomal translocations were only found in AOL-like individuals. All hybrids analysed evidenced extensive ribosomal DNA (rDNA) polymorphism that was not found in parental species, but usually seen falling within the range of possible parental combinations. Yet, transgressive phenotypes that cannot be explained by normal recombination, including more rDNA clusters than expected or the occurrence of syntenic rDNAs, were also detected. Present results proved rapid genomic evolution providing the genetic novelty for species to persist. In addition, although the ultimate consequences of such apparently extensive and recurrent events remain unknown, modern genome-wide methodologies are of great promise towards answering questions concerning the causes, dynamics and impacts of hybridization.
Keywords: Comparative genome hybridization, hybrid zones, introgression, rDNA polymorphism, Achondrostoma oligolepis , Pseudochondrostoma sp.
Introduction
Introgressive hybridization acts as a promoter of diversification and speciation in animals (Dowling and Secor, 1997; Mallet, 2005), and it was suggested to potentially induce more substantial genomic rearrangements and rapid genome repatterning than gene and/or genome duplications (reviewed in Mable, 2013). Hybrids are seen as triggers of transposition that may account for the genetic instability and extensive genome repatterning following hybridization (Fontdevila, 2005; Abbott et al., 2013). Chromosomes, allowing for a macro-examination of the genome, may afford for stronger evidences of hybridization and genome reorganization, given the right tools such as fluorescence in situ hybridization (FISH) with specific probes or comparative genomic hybridization (CGH) (Phillips and Reed, 1996; Dobigny and Yang, 2008).
Natural introgression is commonly found among freshwater fishes, particularly in Cyprinidae (Scribner et al., 2001), where several cases have been comprehensively evaluated (Gerber et al., 2001; Costedoat et al., 2007; Aboim et al., 2010; Broughton et al., 2011). The different outcomes witnessed (reviewed in Abbott et al., 2013) reinforce the opportunity of using cyprinid fish to search for genomic signatures of hybridization. Intergeneric hybrids involving the Iberian arched-mouth Achondrostoma oligolepis (AOL) and the straight-mouth nase Pseudochondrostoma duriense (PDU) were identified in the past (Steindachner, 1866; Almaça, 1965; Collares-Pereira and Coelho, 1983; but see Gante et al., 2004). These once congeneric chondrostomines (Robalo et al., 2007) are broadly sympatric (Figure 1) and seem to actively hybridize, contributing to the establishment and maintenance of several independent hybrid zones (HZs) with different levels of asymmetric bidirectional introgression (Gante et al., 2004; Aboim et al., 2010). In addition, hybridization between AOL and another straight-mouth nase Pseudochondrostoma polylepis (PPO) has long been suspected (Almaça, 1965; Collares-Pereira and Coelho, 1983), although it remained unexplored.
Figure 1.
Sampling locations ● across the distribution ranges of  AOL,
AOL,  PDU and
PDU and  PPO in the Iberian Peninsula. Sampling points: 1—Sousa River
(Douro basin), 2—Caima River (Vouga basin) representing AOL × PDU HZs;
3—Serra River (Vouga basin), 4—Mortágua River, 5—Alva River and
6—Ceira River (Mondego basin) representing AOL × PPO HZs.
PPO in the Iberian Peninsula. Sampling points: 1—Sousa River
(Douro basin), 2—Caima River (Vouga basin) representing AOL × PDU HZs;
3—Serra River (Vouga basin), 4—Mortágua River, 5—Alva River and
6—Ceira River (Mondego basin) representing AOL × PPO HZs.
The pure parental species are believed to have diverged around 11 million years ago (Aboim et al., 2010), representing distinct genetic and morphologic groups (Collares-Pereira and Coelho, 1983; Aboim et al., 2010). Earliest cytogenetic characterization confirmed great karyotype similarity (2n=50, 6–7 metacentric (m)+15–16 submetacentric (sm)+3 subtelo-acrocentric (st/a) chromosome pairs) (Pereira et al., 2009), and only recently diagnostic chromosome markers were designated for them (Pereira et al., 2012). PDU and PPO were characterized by a small and stable number of ribosomal DNA (rDNA) clusters, contrasting with the polymorphisms observed in AOL (Pereira et al., 2012). Still, there are few studies using in-depth molecular cytogenetic analyses on natural fish hybrids (Fujiwara et al., 1997; Zhu et al., 2006; Gromicho et al., 2006b; Hashimoto et al., 2012; Rampin et al., 2012).
Given the heterogeneous patterns of introgression previously found on HZs comprising these species (Gante et al., 2004; Aboim et al., 2010), present work integrates, for the first time, cytogenetic data with morphological and robust genetic profiling on AOL × PDU and putative AOL × PPO natural hybrids, in order to better understand the impact of the successful intergeneric hybridization process on such genomes. Using a multidisciplinary approach, we widened and reinforced previous analyses based on one or few feebler markers (Collares-Pereira and Coelho, 1983; Gante et al., 2004; Aboim et al., 2010), while testing for their value in depicting genome introgression in similar situations. Moreover, obtained results showed rapid genome modifications and instability not always explained by normal recombination, emphasizing the role of admixture events in generating genetic novelties for species (old or new) to thrive under particular spatial contexts.
Materials and methods
Fish sampling
Information regarding the specimens used in this study is summarized in Table 1 (see also Figure 1 and Supplementary Material). A total of 200 individuals were captured by electrofishing in years 2007 and 2008 (see Supplementary Material). Genetic and morphological profiling was performed to retrieve potential hybrids for further analyses (see Supplementary Material). The successful cytogenetic characterization dictated sample size and selection of a total of 13 hybrid individuals from Douro (4), Vouga (6) and Mondego (3) basins (Table 1). Specimens were killed by an overdose of anesthetic MS-222, fixed in formalin and preserved in 70% ethanol for deposition in the Ichthyological Collection at MNHNC (Lisbon, Portugal). All manipulations were performed in accordance with Portuguese guidelines and regulations regarding animal welfare and experimentation.
Table 1. Specimens analysed in this study.
| Hybrid system | Basin | Tributary | Sampling point (Figure 1) | ID code | Sex | Capture date |
|---|---|---|---|---|---|---|
| AOLxPDU | Douro | Sousa River | 1 | cs3 | ♀ | 2008 |
| cs20 | ♂ | |||||
| cs19 | ♀ | |||||
| cs24 | ♂ | |||||
| Vouga | Caima River | 2 | cv23 | n.d. | 2007 | |
| cv26 | n.d. | |||||
| cv14 | ♂ | |||||
| cv21 | ♂ | |||||
| cv16 | ♂ | |||||
| AOLxPPO | Serra River | 3 | cv39 | ♂ | 2008 | |
| Mortágua River | 4 | zd20 | ♀ | |||
| Mondego | Alva River | 5 | zd52 | ♀ | 2007 | |
| Ceira River | 6 | zd61 | ♀ | |||
| Total | 13 |
Abbreviations: AOL, Achondrostoma oligolepis; ♀, female; ♂, male; n.d., not determined; PDU, Pseudochondrostoma duriense; PPO, Pseudochondrostoma polylepis.
Information on origin, sex and date of collection is given.
Morphological analysis
Upon recurrent backcrossing, hybrids tend to become indistinguishable from parental forms (Mallet, 2005). Representatives of Achondrostoma and Pseudochondrostoma genera exhibit clear morphological differences that are easily recognizable at first sight (Collares-Pereira, 1979; Coelho, 1985; see Supplementary Material), and intermediate forms of diagnostic traits can be used to distinguish hybrid morphotypes (Collares-Pereira and Coelho, 1983). Therefore, a specific set of morphological characters (shape and position of the mouth, presence of a corneous lower lip, number of scales in the lateral line and number of gill-rakers) were used in a primary approach of classification (see Supplementary Material). Specimens with an assortment of both parent-specific morphological characters and/or intermediate patterns were assigned as putative hybrids (Supplementary Table S1). Depending on their general phenotype and main parental characters' contribution, hybrids were classified and hereafter designated as AOL-like, PDU-like or PPO-like.
Genetic profiling
A considerable fraction of these hybrids cannot be reliably distinguished from pure parental species based on morphology alone (Aboim et al., 2010). For this reason, a genetic analysis on all 200 individuals was also included (see Supplementary Material), based on molecular markers previously developed for the parental species—mtDNA cyt b gene, 10 microsatellite loci (Aboim et al., 2010) and the single-copy RAG-1 nuclear gene (recombination activating gene 1) (Aboim et al., 2009). Representative sample sizes of each parental species per population/river/basin were considered, as well as the independent and heterogeneous nature of the already known HZs. Individuals from Caima River were characterized earlier by Aboim et al. (2010); the remaining ones were profiled using the same methods, and all of them were classified into one of the following classes: pure parental species, F1, F2, B × AOL, B × PDU or B × PPO backcrosses (Supplementary Table S2; see also Supplementary Material).
Cytogenetic analysis
Chromosome suspensions were obtained from standard kidney preparations and stored at −20 °C until further processing. Slides were thermally aged, digested with pepsin and RNase and immediately used for the following procedures. FISH experiments used 5S and 45S rDNA sequences (Pereira et al., 2012) and the (TTAGGG)n pool of sequences (Ijdo et al., 1991) as probes in either dual-colour or independent procedures. In addition, genomic DNA of AOL and PDU were used in the proportion of 1:1 for the CGH protocol. All probes were differentially labelled by nick translation with biotin-16-dUTP or digoxigenin-11-dUTP according to the manufacturer's recommendations. All in situ hybridizations were performed as detailed by Pereira et al. (2012), with the exception that CGH probes were allowed to pre-anneal (37 °C, 1 h). Slides selected for sequential analysis of constitutive heterochromatin (after FISH) were washed in 4 × SSC/0.01% Tween 20 (pH 7.0) and dehydrated in ethanol series. C-banding improved with DAPI counterstaining followed Pereira et al. (2009).
Chromosomes were observed under the fluorescence-equipped microscope Olympus AX70 coupled to a DP30VW (Olympus, Center Valley, PA, USA) CCD camera. Digital greyscale images were processed for contrast manipulation, over-layering and pseudo-coloration, affecting the whole image as one. At least 10 complete metaphases were analysed per individual. Karyotypes were arranged with chromosomes organized by decreasing sizes in three categories according to centromere position: m, sm and st/a (Levan et al., 1964).
Results
The combination of distinct methods for hybrid identification and classification detected signatures of introgression in all the 13 cytogenetically analysed individuals (Table 2 and Supplementary Material). All specimens presented diploid chromosome numbers of 2n=50 and grossly similar karyotypes. Molecular cytogenetic data were overall able to retrieve the non-pure origin of the individuals under analysis, even in cases for which morphology and/or genetic profiling were not proficient in detecting introgression (cv23, cv39, zd20, zd52 and zd61), with two exceptions (cs19 and cs20) (Table 2).
Table 2. Summary table with the characterization of the AOL × PDU and AOL × PPO hybrids included in this study.
| ID code | Phenotype a | Genotype b |
Cytogenetic characterization
|
Corresponding figures | |||||
|---|---|---|---|---|---|---|---|---|---|
|
45S rDNA sites
|
5S rDNA sites
|
Syntenic units
|
|||||||
| Total | Chromosome distribution | Total | Chromosome distribution | Total | Chromosome distribution | ||||
| cs3 | AOL-like | Admixed | 4 | 3 sm+1 st/a | 7 | 4 st/a+2 sm+1 m | 1 | 1 st/a | Figure 4 |
| cs20 | AOL | Admixed | 6 | 4 sm+2 i sm | 8 | 4 st/a+2 sm+2 m | 0 | Figure 2a | |
| cs19 | AOL-like | Admixed | 6 | 6 sm | 8 | 4 st/a+2 sm+2 m | 0 | Figure 2b | |
| cs24 | AOL | Admixed | 7 | 4 sm+1 i sm+2 m | 8 | 4 st/a+2 sm+2 m | 2 | 2 m | Figure 2c |
| cv23 | PDU-like | PDU | 4 | 4 sm | 4 | 4 st/a | 0 | Figure 3a | |
| cv26 | PDU-like | Admixed | 4 | 4 sm | 6 | 4 st/a+1 sm+1m | 0 | Figure 3b | |
| cv14 | PDU-like | Admixed | 4 | 4 sm | 8 | 4 st/a+2 sm+2 m | 0 | Figures 3c and 5 | |
| cv21 | AOL-like | Admixed | 6 | 5 sm+1 st/a | 6 | 4 st/a+1 sm+1 m | 1 | 1 st/a | Figure 3d |
| cv39 | AOL | AOL | 6 | 3 sm+1 m+2 st/a | 8 | 4 st/a+2 sm+2 m | 3 | 1 m+2 st/a | Figure 3e |
| cv16 | AOL-like | Admixed | 8 | 6 sm+1 m+1 st/a | 8 | 4 st/a+2 sm+2 m | 2 | 1 m+1 st/a | Figure 3f |
| zd20 | PPO-like | PPO | 5 | 4 sm+1 i sm | 5 | 4 st/a+1 m | 0 | Figure 2d | |
| zd52 | AOL | AOL | 6 | 3 sm+2 i sm+1 m | 8 | 4 st/a+2 sm+2 m | 1 | 1 m | Figure 2e |
| zd61 | AOL-like | AOL | 6 | 4 sm+1 m+1 st/a | 8 | 4 st/a+2 sm+2 m | 2 | 1 m+1 st/a | Figure 2f |
| Parental species | AOL | AOL | 6 | 4 sm+2 i sm | 6–8 | 4 st/a+(1–2) sm+(1–2) m | 0 | Pereira et al., 2012 | |
| PDU | PDU | 3 | 3 sm | 4 | 4 st/a | 0 | |||
| PPO | PPO | 4 | 4 sm | 4 | 4 st/a | 0 | |||
Abbreviations: AOL, Achondrostoma oligolepis; i, interstitial; m, metacentric; PDU, Pseudochondrostoma duriense; PPO, Pseudochondrostoma polylepis; rDNA, ribosomal DNA; sm, submetacentric; st/a, subtelo-acrocentric.
Information on phenotype, genotype, number and chromosome location of rDNA clusters (chromosome type: m, sm and st/a; chromosome region: i=interstitial, otherwise terminal in the p arm) is given. General characterization of the respective parental species is included in the last rows. Discrete indicators of non-pure origin are highlighted in bold; the presence of at least one of these markers was considered enough to suspect admixture.
After detailed morphological analysis.
After detailed genetic profiling (see Supplementary Material).
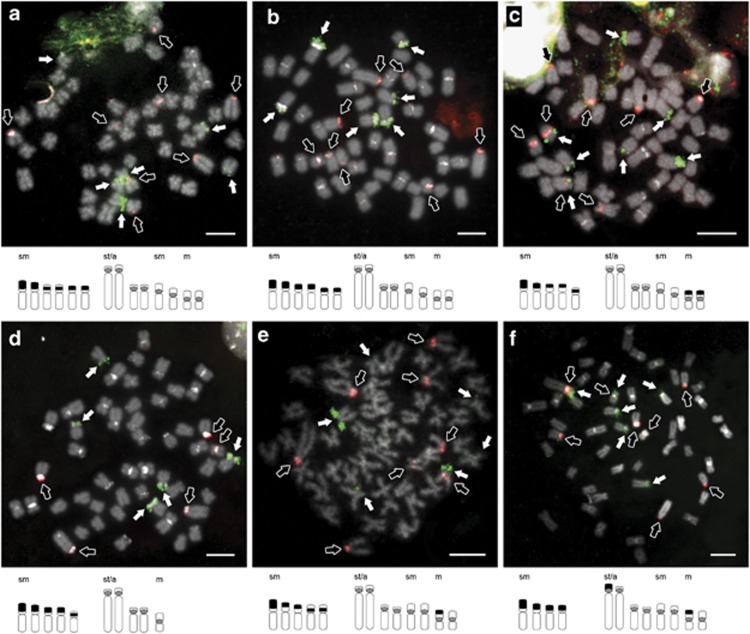
The simultaneous mapping of both rDNAs demonstrated new rDNA profiles and noticeable chromosome rearrangements when compared with parental genomes (Pereira et al., 2012). An assortment of combinations (number of clusters and chromosome location) was identified throughout the sample (Table 2; Figures 2, 3, 4a and b). Although 5S rDNA was mostly associated with pericentromeric regions, interstitial signals of 45S rDNA were often difficult to discriminate (Figures 2a and c–e). Numeric polymorphisms of both rDNA clusters ranged from four or five to eight FISH-positive signals, in agreement with parental profiles (Pereira et al., 2012). In general, PDU- (Figures 3a and c) and PPO-like genomes (Figure 2d) exhibited a lower number of signals, namely concerning 45S rDNA clusters. Genomes of AOL-like individuals revealed rDNA phenotypes with higher number of positive signals for both probes (Figures 2a–c, e, f3d–f and 4a, b), representing the most common combination of six clusters of 45S rDNA and eight of 5S rDNA (6/8). This profile was found in both AOL × PDU and AOL × PPO systems, even though it had not been observed in individuals from Caima River (Vouga basin). In addition, one AOL-like specimen from Sousa River (cs24) and another from Caima River (cv16) HZs evidenced unexpected rDNA phenotypes with seven (Figure 2c) and eight (Figure 3f) positive signals of 45S rDNA, respectively.
Figure 2.
Simultaneous mapping of 5S (red probe, hollow arrows) and 45S rDNA (green probe, white
arrows) clusters in specimens with admixed origin from (a–c) Douro
(AOL-like) and (d–f) Mondego (one PPO- and two AOL-like) basins. Below each
metaphase is a partial (unscaled) ideogram of the chromosomes bearing 5S (grey circle
 ) and 45S rDNA (black
box ▪) gene clusters. Note the strong DAPI-positive bands at centromeres and other
heterochromatic regions (white). Bar=5 μm . A full color version of
this figure is available at the Heredity journal online.
) and 45S rDNA (black
box ▪) gene clusters. Note the strong DAPI-positive bands at centromeres and other
heterochromatic regions (white). Bar=5 μm . A full color version of
this figure is available at the Heredity journal online.
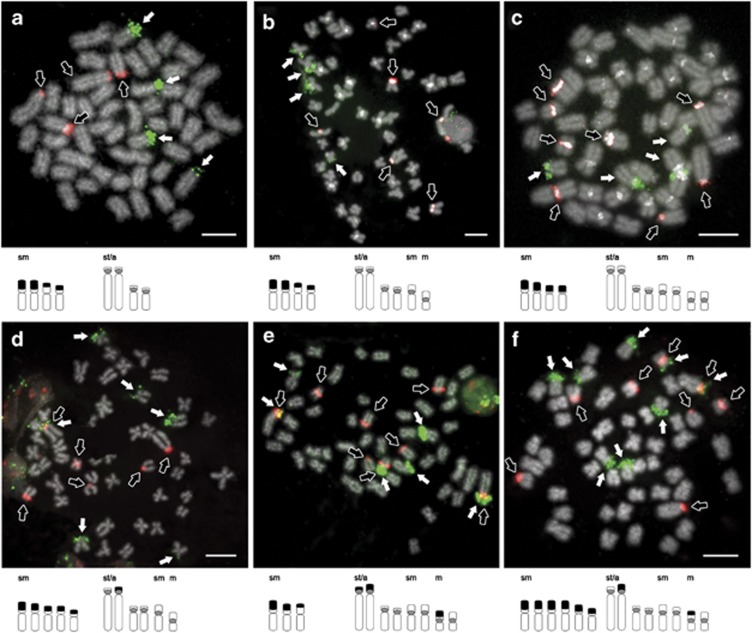
Figure 3.
Dual-colour FISH with specific 5S (red probe, hollow arrows) and 45S rDNA probes (green
probe, white arrows) in metaphases of (a–c) PDU-like and (d–f)
AOL-like specimens from Vouga basin. Below each metaphase is an (unscaled) ideogram of
the rDNA-bearing chromosomes (grey circle  =5S, black box ▪ =45S). Note the strong
centromeric DAPI-positive bands (white). Bar=5 μm. A full color version
of this figure is available at the Heredity journal online.
=5S, black box ▪ =45S). Note the strong
centromeric DAPI-positive bands (white). Bar=5 μm. A full color version
of this figure is available at the Heredity journal online.
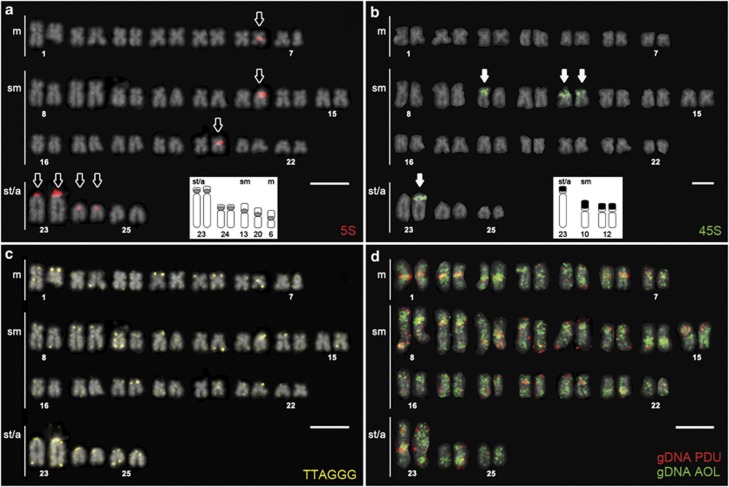
Figure 4.
Karyotype of a female AOL-like hybrid from Douro basin after (a) FISH with 5S
(red probe, hollow arrows) and (b) 45S rDNA (green probe, white arrows)
sequences, both including inset ideograms of the rDNA-bearing chromosomes (grey circle
 =5S, black box
▪ =45S); (c) FISH with telomeric (TTAGGG)n
repeats and (d) CGH using AOL and PDU genomic DNA as probes. Genetic admixture
can be retrieved from non-matching bands between homologue pairs after CGH, particularly
in marker chromosomes numbers 8, 10 and 23. Bar=5 μm. A full color
version of this figure is available at the Heredity journal online.
=5S, black box
▪ =45S); (c) FISH with telomeric (TTAGGG)n
repeats and (d) CGH using AOL and PDU genomic DNA as probes. Genetic admixture
can be retrieved from non-matching bands between homologue pairs after CGH, particularly
in marker chromosomes numbers 8, 10 and 23. Bar=5 μm. A full color
version of this figure is available at the Heredity journal online.
The first hints of genome reorganization were evidenced by the translocation of a big cluster of 45S rDNA to chromosome number 23 (Figures 2f, 3d–f and 4b) and/or by the presence of other syntenic associations of both 5S and 45S gene clusters (Figures 2c, e, f and 3e and f). Such translocations or associations were not observed in the parental species despite a relatively good sample coverage (N=15; Pereira et al., 2012), nor in PDU- or PPO-like specimens, but only in AOL-like hybrids (Table 2). Distinct syntenic combinations could be consistently recognized in one or two large st/a chromosomes and/or in a pair of small m chromosomes, although never exceeding three syntenic units per metaphase (Figure 3e). In addition, no rDNA profile or the incidence of translocations seemed to correlate with any particular genomic composition or with sex.
Telomeric (TTAGGG)n repeats were mostly located at the terminal regions of each chromatid (Figure 4c) as expected. Interstitial telomeric sites, which have been documented as a ‘footprint' of recent chromosomal rearrangements (Phillips and Reed, 1996), were rarely detected but seemed to be present beneath the bigger units of 45S rDNA (numbers 10 and 23 in Figure 4c).
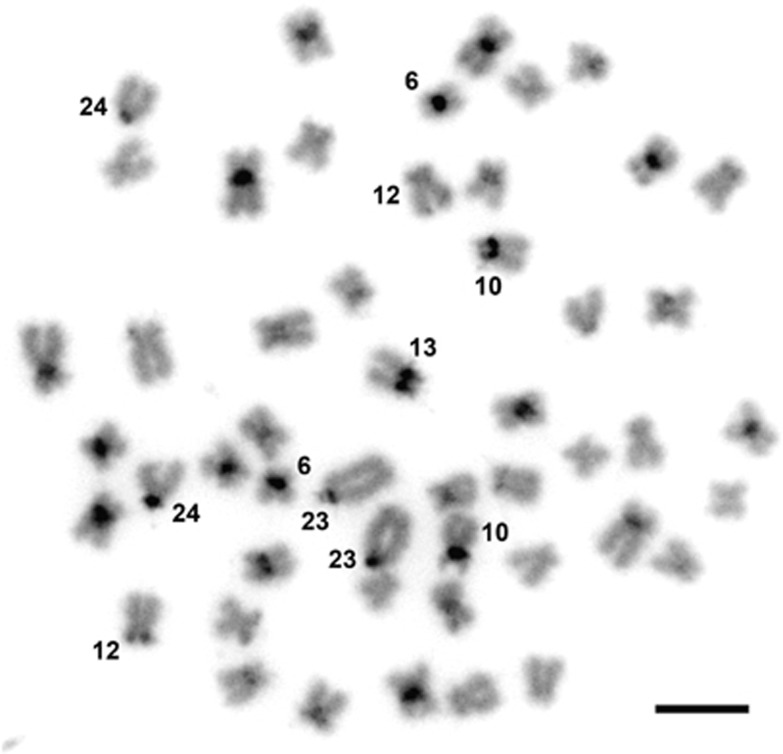
Similarly to the pure parental species (Pereira et al., 2009), constitutive heterochromatin (C-bands) was confined to most centromeres and very few other non-centromeric regions (Figure 5) that appeared as AT-rich DAPI-positive regions (Figures 2 and 3). Centromeric bands were also more pronounced in some pairs of chromosomes from each category (about 3 to 4 m, 10 sm and 2 st/a pairs) (Figure 5). After sequential analysis, C-bands were found to be intimately associated with 5S rDNA clusters but scarcely present in some of the 45S rDNA-bearing chromosomes (for example, number 12 in Figure 5). In addition, 45S rDNA chromosome regions appear as DAPI-negative and were hardly detected after C-banding (numbers 10 and 12 in Figure 5).
Figure 5.
C-banding improved with DAPI counterstaining on a metaphase of a male PDU-like hybrid from Vouga basin evidencing mostly centromeric bands of constitutive heterochromatin. Chromosome pairs numbers 6, 10, 12, 13, 23 and 24 are numbered for better paralleling with sequential banding results. Bar=5 μm.
CGH analyses provided good results (that is, hybridization patterns comparable between metaphases) for a single AOL-like specimen from Sousa River (Douro basin; cs3). Genomic DNAs of both pure parental species (AOL and PDU) were simultaneously hybridized on chromosomes of the hybrids, trying to quantify each specific contribution to the resultant genome. Hybridization signals were not uniform along the chromosomes, being strongest at specific regions where probes overlapped at least partially (Figure 4d), and seemed to match bands of constitutive heterochromatin (Figure 5). This result suggests that some of these bands may in fact be shared by both parental species (generally centromeric), most likely resultant from shared dispersed repetitive sequences and/or denoting common ancestry to some extent. Painted chromosomes could not be exclusively assigned to one or the other parental species; however, overall, chromosomes were mostly evidenced by the AOL genomic probe, suggesting a greater genomic identity to AOL, which is consistent with its morphotype and genotype (Table 2). However, it was possible to recognize a small number of species-specific bands from each parental species (single probe colour) either colocalized to the same chromosome and/or only present in one of the chromosome homologues (Figure 4d), revealing intergenomic recombination with PDU. Besides the obvious translocation of an rDNA segment to one of the large st/a chromosomes (Figure 4b), CGH results allowed the identification of additional possible translocations easily identifiable at the most heteromorphic pairs—perceptible in pair numbers 8, 10 and 11 (Figure 4d).
Discussion
Multilevel characterization of HZs
This investigation provided a multilevel survey on natural homoploid hybrids. New data on independent HZs involving AOL × PDU were endowed, while confirming the occurrence of AOL × PPO hybrids (Figure 1). Previous works strongly expressed the necessity of using multiple approaches when studying natural hybrids (Gerber et al., 2001; Scribner et al., 2001; Gante et al., 2004; Gromicho and Collares-Pereira, 2007; Mavárez and Linares, 2008; Lajbner et al., 2009), especially when second-generation crossing might be occurring, as it seems to be the case (Aboim et al., 2010). The combination of three different identification methods and a vast population screening data set allowed to successfully sort all the selected 13 individuals as admixed, whereas, if using each method independently, some hybrids were probably untraceable (Table 2 and see also Supplementary Material), as observed in Serra R. where hybrids were undetected by genetic markers (Supplementary Figure S1). Identifying hybrids on the basis of morphology alone may be challenging owing to ancestral polymorphisms, preferential backcrossing and intraspecific plasticity (Dowling and Secor, 1997; Mallet, 2005; Mavárez and Linares, 2008). Genetics might be more precise, but its resolution power is limited by the number of diagnostic markers available, their genomic coverage and also by intraspecific population structure. Chromosomes, on the other hand, provided herein solid evidences of hybridization, particularly significant given the gross chromosome homeology empirically estimated. In fact, observed translocations were crucial in distinguishing hybrid rDNA profiles that seemed to overlap those of pure parental species (Figures 2e, f, and 3d, e).
The extent of hybridization
Hybridization may have multiple effects at different stages and spatial contexts (Gerber et al., 2001; Costedoat et al., 2007; Broughton et al., 2011; Abbott et al., 2013). The already surveyed HZs of AOL × PDU seem to behave differently (Aboim et al., 2010) with character displacement happening towards one or the other parental species (Collares-Pereira and Coelho, 1983; Gante et al., 2004; Aboim et al., 2010). Morphological similarities to one of the parental species are presumed to reflect its major genomic/genetic contribution during the introgression process (Mallet, 2005; Mavárez and Linares, 2008), and present data allude indeed to that presumption. Specifically, altogether, hybrid morphotypes (Supplementary Table S1), molecular cytogenetic data (Figures 2, 3, 4) and the genetic profiles (Supplementary Figure S1 and Supplementary Table S2) point to preferential backcrossing, as initially suspected (Gante et al., 2004). This may be correlated to both genetic attractiveness/compatibility and/or to the relative densities of the parental species within each HZ.
The conservative and widespread karyotype pattern of leuciscines (Ráb and Collares-Pereira, 1995) does not allow determining the directionality of introgression (but see Gante et al., 2004), nor discerning parental contributions to the hybrid genomes by means of conventional cytogenetic markers (that is, 2n, NF, karyotypes). Not even the major macrostructural differences between parental species (Pereira et al., 2009) could be unambiguously correlated to any hybrid morphotype or genotype. On the other hand, molecular cytogenetic results imply AOL to be more permissive to introgression—either more vulnerable or flexible (that is, prone to hybridize)—than each of the co-occurring and more widespread sister-species PDU or PPO. Irrespective of the hybrid system it engages, not only does AOL appear to be largely involved but also greatly affected by the ongoing hybridization events, as chromosomal translocations were only found in AOL-like individuals. Conversely, PDU- and PPO-like individuals appear to be more resilient to genome modifications, apparently retaining the original genome organization while simply inheriting rDNA-bearing chromosomes from each parent (Table 2). Backcrosses with PDU/PPO were also less frequently detected by the genetic assessment (Supplementary Figure S1 and Supplementary Table S2). Similarly, AOL-like morphotypes are strikingly more evident at first sight than PDU- or PPO-like morphotypes, owing to the detection of a corneous lower lip otherwise absent in Achondrostoma sp. These results seem conflicting with previous works on hybridizing cyprinids where distinct HZs represented different outcomes of independent hybridization processes (Costedoat et al., 2007; Aboim et al., 2010; Broughton et al., 2011), which means that either the environmental pressure is identical in the HZs considered here (unlikely) or that the endogenous (genetic) selection forces are in fact the strongest in action.
Hybrid rDNA polymorphisms—genetic novelties
In general, all hybrids evidenced extensive rDNA polymorphism that was not found in parental species (Table 2). Species-specific rDNA polymorphisms were recently characterized in European leuciscines (Gromicho et al., 2006a, 2006b; Kirtiklis et al., 2010; Pereira et al., 2012; Rossi et al., 2012), as well as in artificially reared hybrids of the Squalius alburnoides complex (Gromicho et al., 2006b). In the present study, each hybrid stood for a distinct rDNA profile, usually falling within the range of possible combinations of parental contributions (Supplementary Table S3) and reflecting their admixed nature. Exceptions included (i) two AOL-like individuals with more 45S rDNA clusters than expected (Figures 2c and 3f) and (ii) the occurrence of syntenic rDNAs. Both can be interpreted as transgressive—exceeding parental phenotypic values in either a negative or positive direction. Although less reported in animals, transgressive phenotypes are more common in interspecific hybrids than expected as a consequence of recombination between parental lines (Rieseberg et al., 1999). Still, present results document genetic novelties that cannot be explained by normal recombination between parental genomes. For example, translocations did not implicate homeologous chromosomes, as it would be expected in normal recombination. Instead, it involved chromosomes of even different classes (that is, usually, 45S rDNA-bearing chromosomes are sm, but after translocations occurred m and st/a chromosomes were also found carrying such clusters).
Considering the possible syntenic combinations observed in these hybrids, present results propose (i) higher ‘mobility' of 45S rDNA sequences, (ii) preferential ‘jumping' to chromosomes already bearing 5S rDNA with (iii) higher flexibility for incorporation (probably involving the non-transcribed spacers), resulting in (iv) repeatable rearrangements, and possibly in (v) erroneous pairing of the homeologous chromosomes inherited from each parent, as observed by Santos et al. (2002). Although the high number of 5S rDNA clusters was likely inherited from AOL (Pereira et al., 2012), the association of these clusters to heterochromatin (Figure 5) can eventually be correlated to mobile DNAs, which may as well account for their original dispersion throughout Achondrostoma spp. genomes (Pereira et al., 2012).
The number of studies in cyprinids using dual-FISH with rDNA probes is frankly growing, and colocalized ribosomal genes in leuciscines were described within Anaecypris, Leuciscus and Squalius genera (Gromicho et al., 2006a, 2006b; Kirtiklis et al., 2010; Rossi et al., 2012) but never in chondrostomine species (for example, parental species under focus; Pereira et al., 2012). The novel syntenic associations (Figures 2, 3, 4) are distinct evidences of genome instability and reshuffling, most likely driven by the hybridization events (Fontdevila, 2005; Abbott et al., 2013). It is rather probable that other regions of the genome might be experiencing the same processes as suggested by the CGH results (Figure 4d), even though no key interstitial telomeric sites were detected (Figure 4c). The ‘repeatability' of syntenic blocks, that is, different individuals bearing comparable 45S–5S rDNA associations, was earlier observed in both natural and synthetized hybrids of Helianthus sp., thus interpreted as a consequence of endogenous rather than ecological (exogenous) selection forces (reviewed in Fontdevila, 2005). This might also explain the ‘abnormal' recombination patterns previously discussed.
Final remarks
The genomic instability herein documented proved rapid and traceable genomic evolution. Genetic novelty is thus generated for selection to operate and for species to persist under newly imposed stressful conditions (environmental and/or genomic) (Dowling and Secor, 1997; Gerber et al., 2001; Seehausen, 2004; Mallet, 2005; Abbott et al., 2013; Mable, 2013). In this case study, ecological barriers between hybrids and parental species might be altered and/or degraded, so that gene flow back to pure species never actually ceases. However, the ultimate consequences of such apparently extensive and recurrent events to these populations and species remain unknown, and the probability of fixation of the resulting rearrangements depends on the demography of the hybrid swarms (Gerber et al., 2001; Seehausen, 2004; Costedoat et al., 2007; Broughton et al., 2011; Abbott et al., 2013). Usually disregarded (reviewed in Costedoat et al., 2007), these interactions might ultimately result in speciation or extinction faster than expected. Coupling the study of these hybrid complexes with modern genome-wide approaches (Twyford and Ennos, 2012) will allow for the exploration of evolutionarily relevant processes and answering questions inherent to hybridization, its causes, dynamics and impacts.
Data archiving
Data deposited in the Dryad repository: doi:10.5061/dryad.5m121.
Acknowledgments
This work was supported by Centro de Biologia Ambiental (PEst-OE/BIA/UI0329/2011) and the Portuguese Fundação para a Ciência e Tecnologia (PhD Grant SFRH/BD/44980/2008 awarded to CSAP; Post-doc Grant SFRH/BPD/26850/2006 awarded to MAA). Fish samples were collected with permits provided by the Portuguese agency Instituto da Conservação da Natureza e das Florestas (ICNF). We thank the anonymous reviewers for their insightful comments that helped improving the manuscript. This paper is part of the publication series ‘Chromosome studies of European cyprinid fishes'.
Author contributions
CSAP performed all morphological and cytogenetic analyses and drafted the manuscript. MAA performed all genetic analyses and co-drafted the manuscript. PR and MJCP designed the study and co-drafted and revised the text. All authors participated in the discussion of the results and read and agreed to the final version of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, et al. Hybridization and speciation. J Evol Biol. 2013;26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- Aboim M, Cunha C, Coelho MM. Redistribution of the geographical ranges of the Iberian cyprinid genus Pseudochondrostoma based on a phylogenetic analysis: implications for the historical rearrangements of the north-western Iberian drainages. J Fish Biol. 2009;74:1337–1346. doi: 10.1111/j.1095-8649.2009.02189.x. [DOI] [PubMed] [Google Scholar]
- Aboim M, Mavárez J, Bernatchez L, Coelho MM. Introgressive hybridization between two Iberian endemic cyprinid fish: a comparison between two independent hybrid zones. J Evol Biol. 2010;23:817–828. doi: 10.1111/j.1420-9101.2010.01953.x. [DOI] [PubMed] [Google Scholar]
- Almaça C. Contribuition à la connaissance des poissons des eaux intérieures du Portugal (in French) Rev Fac Cienc Univ Lisboa. 1965;13:225–262. [Google Scholar]
- Broughton RE, Vedala KC, Crowl TM, Ritterhouse LL. Current and historical hybridization with differential introgression among three species of cyprinid fishes (genus Cyprinella) Genetica. 2011;139:699–707. doi: 10.1007/s10709-011-9578-9. [DOI] [PubMed] [Google Scholar]
- Coelho MM. The straight-mouth Portuguese Chondrostoma Agassiz, 1835. Taxonomic position and geographic distribution of Ch. polylepis Steindachner, 1865 and Ch. willkommii Steindachner, 1866, with the description of a new subspecies Ch. polylepis duriensis. Arq Mus Bocage. 1985;3:13–38. [Google Scholar]
- Collares-Pereira MJ. On the systematic position of the Iberian Cyprinidae forms arcasi (Steindachner, 1866) and macrolepidotus (Steindachner, 1866) of the genus Rutilus. Arq Mus Bocage. 1979;6:361–393. [Google Scholar]
- Collares-Pereira MJ, Coelho MM. Biometrical analysis of Chondrostoma polylepis × Rutilus arcasi natural hybrids (Osteichthyes-Cypriniformes-Cyprinidae) J Fish Biol. 1983;23:495–509. [Google Scholar]
- Costedoat C, Pech N, Chappaz R, Gilles A. Novelties in hybrid zones: crossroads between population genomic and ecological approaches. PLoS One. 2007;2:e357. doi: 10.1371/journal.pone.0000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobigny G, Yang F. Comparative cytogenetics in the genomics era: cytogenomics comes of age. Chromosome Res. 2008;16:1–4. doi: 10.1007/s10577-008-1213-y. [DOI] [PubMed] [Google Scholar]
- Dowling TE, Secor CL. The role of hybridization and introgression in the diversification of animals. Annu Rev Ecol Syst. 1997;28:593–619. [Google Scholar]
- Fontdevila A. Hybrid genome evolution by transposition. Cytogenet Genome Res. 2005;110:49–55. doi: 10.1159/000084937. [DOI] [PubMed] [Google Scholar]
- Fujiwara A, Abe S, Yamaha E, Yamazaki F, Yoshida MC. Uniparental chromosome elimination in the early embryogenesis of the inviable salmonid hybrids between masu salmon female and rainbow trout male. Chromosoma. 1997;106:44–52. doi: 10.1007/s004120050223. [DOI] [PubMed] [Google Scholar]
- Gante HF, Collares-Pereira MJ, Coelho MM. Introgressive hybridisation between two Iberian Chondrostoma species (Teleostei, Cyprinidae) revisited: new evidence from morphology, mitochondrial DNA, allozymes and NOR-phenotypes. Folia Zool. 2004;53:423–432. [Google Scholar]
- Gerber A, Tibbets C, Dowling T. The role of introgressive hybridization in the evolution of the Gila robusta complex (Teleostei: Cyprinidae) Evolution. 2001;55:2028–2039. doi: 10.1111/j.0014-3820.2001.tb01319.x. [DOI] [PubMed] [Google Scholar]
- Gromicho M, Coelho M, Alves M, Collares-Pereira MJ. Cytogenetic analysis of Anaecypris hispanica and its relationship with the paternal ancestor of the diploid-polyploid Squalius alburnoides complex. Genome. 2006;49:1621–1627. doi: 10.1139/g06-121. [DOI] [PubMed] [Google Scholar]
- Gromicho M, Collares-Pereira MJ.2007The evolutionary role of hybridization and polyploidy in an Iberian cyprinid fish—a cytogenetic reviewIn: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG (eds)Fish Cytogenetics Science Publishers: Enfield, NH, USA; 41–67. [Google Scholar]
- Gromicho M, Coutanceau J-P, Ozouf-Costaz C, Collares-Pereira MJ. Contrast between extensive variation of 28S rDNA and stability of 5S rDNA and telomeric repeats in the diploid-polyploid Squalius alburnoides complex and in its maternal ancestor Squalius pyrenaicus (Teleostei, Cyprinidae) Chromosome Res. 2006;14:297–306. doi: 10.1007/s10577-006-1047-4. [DOI] [PubMed] [Google Scholar]
- Hashimoto D, Ferguson-Smith M, Rens W, Prado F, Foresti F, Porto-Foresti F. Cytogenetic mapping of H1 histone and ribosomal RNA genes in hybrids between catfish species Pseudoplatystoma corruscans and Pseudoplatystoma reticulatum. Cytogenet Genome Res. 2012;360:1–5. doi: 10.1159/000345299. [DOI] [PubMed] [Google Scholar]
- Ijdo JW, Wells RA, Baldini A, Reeders ST. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991;19:4780. doi: 10.1093/nar/19.17.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirtiklis L, Porycka K, Borón A, Coutanceau J-P, Dettai A. Use of the chromosomal co-location of the minor 5S and the major 28S rDNA as a cytogenetic marker within the genus Leuciscus (Pisces, Cyprinidae) Folia Biol. 2010;58:245–249. doi: 10.3409/fb58_3-4.245-249. [DOI] [PubMed] [Google Scholar]
- Lajbner Z, Šlechtová V, Šlechta V, Svátora M, Berrebi P, Kotlík P. Rare and asymmetrical hybridization of the endemic Barbus carpathicus with its widespread congener Barbus barbus. J Fish Biol. 2009;74:418–436. doi: 10.1111/j.1095-8649.2008.02098.x. [DOI] [PubMed] [Google Scholar]
- Levan A, Fredga K, Sandberg A. Nomenclature for centromeric positions on chromosomes. Hereditas. 1964;52:201–220. [Google Scholar]
- Mable BK. Polyploids and hybrids in changing environments: winners or losers in the struggle for adaptation. Heredity. 2013;110:95–96. doi: 10.1038/hdy.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Mavárez J, Linares M. Homoploid hybrid speciation in animals. Mol Ecol. 2008;17:4181–4185. doi: 10.1111/j.1365-294x.2008.03898.x. [DOI] [PubMed] [Google Scholar]
- Pereira C, Neto A, Collares-Pereira MJ. Cytogenetic survey of species of two distinct genera of Iberian nases (Cyprinidae, Leuciscinae) that hybridize extensively in nature. I. Evidence of a similar and conserved chromosome pattern with some few species-specific markers at macro-structural level. Genetica. 2009;137:285–291. doi: 10.1007/s10709-009-9379-6. [DOI] [PubMed] [Google Scholar]
- Pereira C, Ráb P, Collares-Pereira MJ. Chromosomes of European cyprinid fishes: comparative cytogenetics and chromosomal characteristics of ribosomal DNAs in nine Iberian chondrostomine species (Leuciscinae) Genetica. 2012;140:485–495. doi: 10.1007/s10709-013-9697-6. [DOI] [PubMed] [Google Scholar]
- Phillips RB, Reed KM. Application of fluorescence in situ hybridization (FISH) techniques to fish genetics: a review. Aquaculture. 1996;140:197–216. [Google Scholar]
- Rampin M, Bi K, Bogart J, Collares-Pereira MJ. Methodological improvements on genomic in situ hybridization (GISH) for identifying parental chromosome sets in hybrids of fish species with small genome size. Comp Cytogenet. 2012;6:287–300. doi: 10.3897/CompCytogen.v6i3.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L, Archer M, Wayne R. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- Robalo JI, Almada VC, Levy A, Doadrio I. Re-examination and phylogeny of the genus Chondrostoma based on mitochondrial and nuclear data and the definition of 5 new genera. Mol Phylogenet Evol. 2007;42:362–372. doi: 10.1016/j.ympev.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Rossi AR, Milana V, Hett AK, Tancioni L. Molecular cytogenetic analysis of the Appenine endemic cyprinid fish Squalius lucumonis and three other Italian leuciscines using chromosome banding and FISH with rDNA probes. Genetica. 2012;140:469–476. doi: 10.1007/s10709-012-9695-0. [DOI] [PubMed] [Google Scholar]
- Ráb P, Collares-Pereira MJ. Chromosomes of European cyprinid fishes (Cyprinidae, Cypriniformes): a review. Folia Zool. 1995;44:193–214. [Google Scholar]
- Santos VH, Foresti F, Oliveira C, Almeida-Toledo LF, Toledo-Filho SA, Bernardino G. Synaptonemal complex analysis in the fish species Piaractus mesopotamicus and Colossoma macropomum, and in their interspecific hybrid. Caryologia. 2002;55:73–79. [Google Scholar]
- Scribner KT, Page KS, Bartron ML. Hybridization in freshwater species: a review of case studies and cytonuclear methods of biological inference. Rev Fish Biol Fish. 2001;10:293–323. [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Steindachner F. Icthyologischer Bericht über eine nach Spanien und Portugal unternommene Reise (Zweit Fortsetzung) (in German) Sitzungsb Kais Akad Wiss. 1866;54:6–27. [Google Scholar]
- Twyford AD, Ennos RA. Next-generation hybridization and introgression. Heredity. 2012;108:179–189. doi: 10.1038/hdy.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu HP, Ma DM, Gui JF. Triploid origin of the gibel carp as revealed by 5S rDNA localization and chromosome painting. Chromosome Res. 2006;14:767–776. doi: 10.1007/s10577-006-1083-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.