Abstract
The emerging availability of microsatellite markers from mammalian sex chromosomes provides opportunities to investigate both male- and female-mediated gene flow in wild populations, identifying patterns not apparent from the analysis of autosomal markers alone. Tammar wallabies (Macropus eugenii), once spread over the southern mainland, have been isolated on several islands off the Western Australian and South Australian coastlines for between 10 000 and 13 000 years. Here, we combine analyses of autosomal, Y-linked and X-linked microsatellite loci to investigate genetic variation in populations of this species on two islands (Kangaroo Island, South Australia and Garden Island, Western Australia). All measures of diversity were higher for the larger Kangaroo Island population, in which genetic variation was lowest at Y-linked markers and highest at autosomal markers (θ=3.291, 1.208 and 0.627 for autosomal, X-linked and Y-linked data, respectively). Greater relatedness among females than males provides evidence for male-biased dispersal in this population, while sex-linked markers identified genetic lineages not apparent from autosomal data alone. Overall genetic diversity in the Garden Island population was low, especially on the Y chromosome where most males shared a common haplotype, and we observed high levels of inbreeding and relatedness among individuals. Our findings highlight the utility of this approach for management actions, such as the selection of animals for translocation or captive breeding, and the ecological insights that may be gained by combining analyses of microsatellite markers on sex chromosomes with those derived from autosomes.
Keywords: sex-linked microsatellite, X chromosome, Y chromosome, STR, marsupial
Introduction
Microsatellites are versatile and informative genetic markers that have facilitated an unprecedented understanding of the genetic diversity present in natural populations. Consequently, autosomal microsatellite markers have become invaluable tools for molecular ecology (Guichoux et al., 2011). Additional insights have been gained by comparing microsatellite data to DNA sequence data from maternally inherited mitochondrial markers (Pope et al., 1996; Johnson et al., 2003), but the differing patterns of mutation and selection pressure may influence the inferences made (Sunnucks, 2000). Microsatellite markers from mammalian sex chromosomes can now provide further opportunities to investigate female- and male-mediated gene flow (Petit et al., 2002; Schaffner, 2004; Miller et al., 2010).
As a consequence of their different modes of inheritance, different patterns of variation are expected between genetic markers from autosomes and sex chromosomes (Petit et al., 2002; Ellegren, 2009). Several factors may lead to reductions in the genetic diversity of sex chromosomes relative to autosomes, particularly the smaller effective population sizes (Ne) of the sex chromosomes. In mammals, if the sex ratio is equal, there will be only three copies of the X chromosome and a single Y chromosome for every four copies of an autosome. Thus, over evolutionary time, the autosomes have more opportunities to accumulate mutations than the sex chromosomes and genetic variation on the different chromosomes will occur at the ratio of 1.0 to 0.75 to 0.25 for autosomal, X-linked and Y-linked markers, respectively (Charlesworth, 2009).
Ne can also vary within the genome as a consequence of selection and localised variation in recombination rates. For example, some regions of the genome (hotspots) will have disproportionately high recombination rates, whereas other genome regions undergo minimal recombination and are expected to have reduced Ne relative to the genome average (Nachman, 2002; Kaiser and Charlesworth, 2009). Data from human genome resequencing suggest that reductions in diversity, as a consequence of selection at sites linked to genes, have had a greater impact on the X chromosome than the autosomes (Gottipati et al., 2011), but this is not well understood in non-model species for which genomic data are not available. Mammalian Y chromosomes are haploid and, with the exception of pseudoautosomal regions in some but not all species (Page et al., 2005), undergo no recombination. Thus, the Y chromosome is expected to have one of the lowest effective population sizes of any genome region (Charlesworth, 2009), a characteristic that is also likely to be subject to variations in mating systems, for example, differences in reproductive success between the sexes. As a consequence, the Y chromosome will be especially sensitive to population bottlenecks, expansions or founder effects (Petit et al., 2002). Evidence from a range of mammalian species demonstrates a trend for reduced genetic variation on the Y chromosome (Hellborg and Ellegren, 2004; Lindgren et al., 2004; Meadows et al., 2004; Lawson Handley et al., 2006).
Similarly, the inheritance pattern of the X chromosome means that demographic factors and mating systems will differ in their effect on genetic diversity at X-linked relative to autosomal loci. For example, the autosomes spend a greater proportion of evolutionary time in the male germline than does the X chromosome and so variation on the autosomes will be more severely influenced by any male-specific bottleneck (Ellegren, 2009). The relative levels of genetic diversity at X-linked and autosomal loci have been shown to vary markedly even within a species, with humans providing a key example. The ratio of X to autosome diversity in humans is clearly different at regional scales than at a worldwide scale (Hammer et al., 2008; Ellegren, 2009; Keinan et al., 2009), indicating that understanding regional differences in selection, population structure and history will be important to understanding patterns of chromosome diversity (Casto et al., 2010).
Sex chromsome microsatellites have the potential to make significant contributions to wildlife management and conservation planning (Miller et al., 2010) by contributing a sex-specific perspective to population and conservation genetics. X- and Y-linked markers may improve identification of discrete genetic lineages within a population, contributing to efforts to maximise genetic diversity represented in individuals selected for relocation or captive breeding. Sex chromosome markers may also identify sex-specific effects such as skewed mating structure that will affect the way genetic variation is retained in small populations. Further, sex-linked microsatellites have the potential to illuminate patterns of sex-biased dispersal: previously their application to this field was limited by the poor availability of variable sex chromosome markers for non-model species (Prugnolle and de Meeus, 2002), but the application of sex-linked microsatellites to studies of sex-biased dispersal is becoming more common (Li and Merila, 2010; Yannic et al., 2012). Importantly, large-scale genome sequencing efforts targeting an ever-wider range of species (Genome 10K Community of Scientists, 2009) are likely to enable the development of sex chromosome markers for non-model organisms (Greminger et al., 2010) bringing the analysis of sex chromosomes for many mammals within grasp.
The tammar wallaby (Macropus eugenii) is the first marsupial species for which both X and Y chromosome microsatellites have been characterised (MacDonald et al., 2006, 2007), providing an opportunity to explore patterns of genetic diversity using markers from autosomes and both sex chromosomes. Tammar wallabies declined after European settlement of Australia, primarily as a consequence of hunting, habitat loss and introduced predators. All but one of the mainland populations are now extinct: the population around Adelaide was extinct by the 1920s and the Eyre Peninsula population was likely extinct by the 1970s. The only remaining mainland population occurs in southwest Western Australia and this has also declined and become fragmented during the 20th century, especially post-1960 (Tyndale-Biscoe, 2005). Despite this the species remains abundant on Kangaroo Island (in Spencer's Gulf, ∼130 km southwest of Adelaide, South Australia) and present in several isolated island populations off the Western Australian coast including the Abroholos Archipelago (near Geraldton), the Recherche Archipelago (near Esperance) and Garden Island (near Perth). These Western Australian island populations are genetically distinct from one another and display low levels of genetic diversity (Eldridge et al., 2004; Miller et al., 2011). The tammar wallaby is considered to have a promiscuous mating system with a male dominance hierarchy. In captivity, females mate with multiple males, with the dominant male siring approximately half of the offspring (Hynes et al., 2005; Miller et al., 2010). Thus, there is likely to be greater variation in reproductive success for males than for females in wild populations.
Here we investigate differences in genetic variation between autosomal and sex-linked microsatellites, focusing on two island populations of the tammar wallaby that have very different histories. Garden Island is <10 km2 and is home to an endemic population of an estimated 1800–2600 tammar wallabies (Brian Chambers, unpublished data) that was separated from the mainland populations by rising sea levels between 13 000 and 10 000 years ago (Tyndale-Biscoe, 2005). Previous work using autosomal microsatellites identified low genetic diversity and high levels of inbreeding in this population (Eldridge et al., 2004). In contrast, Kangaroo Island has a total area of around 3890 km2 (Inns, 1980) and holds the largest extant tammar wallaby population. Although the total size of the Kangaroo Island population is not well understood, tammars number in the hundreds of thousands at least: the species is considered overabundant and thousands of individuals are culled annually (Wright and Stott, 1999). High levels of polymorphism have been observed at autosomal microsatellite loci in Kangaroo Island tammar wallabies and, although no comprehensive population genetic study has yet been published for this population, significant geographic heterogeneity in allele frequencies is considered unlikely, given the area of the island, the mobility of the animals and the large population size (Taylor and Cooper, 1999).
Our study provides an opportunity to investigate the insights to be gained from the inclusion of sex chromosome microsatellites. Based on our current knowledge of the two tammar wallaby populations studied, our expectations are that:
Genetic diversity will be higher in the Kangaroo Island population than the Garden Island population for markers from all three chromosomes, reflecting the differences between the islands in terms of area and population sizes and previously detected inbreeding in the Garden Island population.
If genetic diversity on a specific chromosome is predominantly influenced by the N e of that chromosome, then the theoretical expectation is that genetic diversity will be highest for markers from the autosome and lowest for Y-linked markers, at the ratio of 1.0 to 0.75 to 0.25 for autosomal, X-linked and Y-linked markers, respectively.
If, as anticipated for this species, reproductive success varies more for males than for females, then genetic diversity relative to autosomal loci will be lower than expected based on sex-specific differences in Ne at Y chromosome loci and/or higher than expected based on sex-specific differences in Ne at X-linked loci.
Materials and methods
Sampling and DNA extraction
Ear biopsies were taken from 78 male and 34 female tammar wallabies from the Stirling Naval Base on Garden Island in 2005–2006 (∼5 mm2 punch removed from mid-ear to allow placement of individual ear tags) and from 97 male and 102 female tammar wallabies from the northwestern end of Kangaroo Island in 2006 (∼10 mm2 clip removed from ear tip). All sampling was conducted with approval from animal ethics committees of either the University of Melbourne (Kangaroo Island) or the University of Western Australia and the Western Australian Department of Environment and Conservation (Garden Island). Tissues were stored in 80% ethanol and transferred to a −80 °C freezer as soon as possible after collection until DNA extraction. DNA was extracted from ear tissue samples using a salting out method. Tissues of ∼2 mm2 were incubated overnight at 55 °C in 330 μl tissue extraction buffer (40 mM Tris/HCl; 100 mM NaCl; 20 mM EDTA, pH 7.2; 0.6% SDS; 0.5 mg ml−1 Proteinase K). Following digestion, 150 μl of 7.5 M ammonium acetate was added. Samples were chilled at −80 °C for 25 min before centrifugation for 20 min at 13 000 r.p.m., 4 °C. Supernatant was transferred to a new tube and DNA was ethanol precipitated. DNA pellets were resuspended in 1 × TE buffer.
Microsatellite genotyping
PCR primer sequences for the 30 di- and tri-nucleotide microsatellites used in this study are provided in MacDonald et al. (2006, 2007). These markers include 10 Y chromosome loci, 9 X chromosome loci and 11 loci from chromosome 2 (Table 1). To allow fluorescent labelling of products, each forward primer was 5′-tailed with 19 bp of M13 sequence (5′-CACGACGTTGTAAAACGAC-3′) (Boutin-Ganache et al., 2001). PCRs of 10 μl total volume contained final concentrations of 1x PCR buffer (160 mM (NH4)2SO4, 670 mM Tris-HCl (pH 8.8 at 25 °C), 0.1% stabiliser: Bioline, London, UK), 2 mM MgCl2 (2.5 mM for locus Me2-123), 0.2 mM each dNTP, 500 mM betaine, 2.5 ng BSA, 0.4 μM WellRed dye-labelled M13 primer (Sigma-Aldrich, St Louis, MO, USA), 0.2 μM M13-tailed forward primer, 0.4 μM reverse primer, 0.3 U BioTaq Red Taq polymerase (Bioline) and ∼50 ng DNA. PCR conditions were: one cycle of 94 °C for 4 min; 36 cycles of 94 °C for 30 s, annealing temperature for 45 s, 72 °C for 1 min; and one cycle of 72 °C for 30 min. Amplified products were run on a Beckman Coulter CEQ 8000 with a 60–420 bp or 60–640 bp size standard (Beckman Coulter, Brea, CA, USA). Results were analysed using the CEQ 8000 Genetic Analysis System software Version 8.0.52 (Beckman Coulter). All genotypes were scored manually. To determine the importance of genotyping errors within this study, a total of 420 single-locus genotypes from seven loci were repeated from separate PCRs. Replicate genotypes were blind-scored and results of the two replicates were compared once all scoring had been completed. In all 420 cases, identical alleles were observed for each replicate, suggesting that mis-scored alleles do not represent a serious problem in this study.
Table 1. Genetic variation observed at 30 chromosome-specific microsatellites in two tammar wallaby populations.
| Locus a | Genbank b | BAC clone c |
Kangaroo Island
|
Garden Island
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Tm d | Ne | AR f | Ag | Ne | AR f | Ag | |||
| Me2-020 | EF105493 | MeAGI_170K8 | 58 | 336 | 1.0 | 1 | 212 | 1.0 | 1 |
| Me2-040h | EF105494 | MeAGI_389E8 | 58 | 196 | 2.0 | 2 | 94 | 2.0 | 2 |
| Me2-041 | EF105495 | MeAGI_389E8 | 58 | 88 | 7.0 | 7 | 114 | 3.0 | 3 |
| Me2-047 | EF105496 | MeAGI_389E8 | 60 | 342 | 9.6 | 11 | 144 | 4.0 | 4 |
| Me2-053 | EF105497 | MeAGI_206L23 | 56 | 332 | 1.0 | 1 | 134 | 1.0 | 1 |
| Me2-061 | EF105498 | MeAGI_206L23 | 58 | 330 | 7.5 | 8 | 176 | 2.6 | 3 |
| Me2-066 | EF105499 | MeAGI_206L23 | 58 | 350 | 2.9 | 3 | 194 | 2.0 | 3 |
| Me2-077h | EF105500 | MeAGI_458L18 | 58 | 312 | 3.0 | 3 | 142 | 1.9 | 2 |
| Me2-084 | EF105501 | MeAGI_458L18 | 58 | 328 | 15.0 | 19 | 196 | 2.0 | 3 |
| Me2-088 | EF105502 | MeAGI_458L18 | 58 | 314 | 6.9 | 8 | 202 | 1.5 | 2 |
| Me2-123 | EF105503 | MeAGI_458L18 | 58 | 250 | 1.0 | 1 | 188 | 1.0 | 1 |
| MeX-016 | EF105484 | MeAGI_162E23 | 58 | 238 | 1.0 | 1 | 96 | 1.0 | 1 |
| MeX-034 | EF105485 | MeAGI_583I21 | 58 | 274 | 6.2 | 8 | 88 | 2.0 | 2 |
| MeX-041 | EF105486 | MeAGI_583I21 | 60 | 260 | 1.0 | 1 | 75 | 1.0 | 1 |
| MeX-048 | EF105487 | MeAGI_583I21 | 56 | 118 | 2.9 | 3 | 116 | 2.0 | 2 |
| MeX-049 | EF105488 | MeAGI_583I21 | 58 | 224 | 2.6 | 3 | 105 | 1.0 | 1 |
| MeX-054 | EF105489 | MeAGI_583I21 | 60 | 119 | 7.9 | 8 | 85 | 3.0 | 3 |
| MeX-055 | EF105490 | MeAGI_583I21 | 60 | 234 | 5.2 | 6 | 61 | 1.0 | 1 |
| MeX-066 | EF105491 | MeAGI_583I21 | 60 | 188 | 3.0 | 3 | 79 | 3.9 | 4 |
| MeX-070 | EF105492 | MeAGI_583I21 | 58 | 160 | 7.7 | 8 | 97 | 1.8 | 2 |
| MeY-01 | DQ641481 | MeVIA_112D12 | 60 | 91 | 6.0 | 6 | 72 | 2.0 | 2 |
| MeY-03 | DQ641482 | MeVIA_112D12 | 60 | 83 | 3.0 | 3 | 66 | 3.0 | 3 |
| MeY-04 | DQ641483 | MeVIA_53A23 | 60 | 94 | 1.0 | 1 | 75 | 1.0 | 1 |
| MeY-06 | DQ641484 | MeVIA_53A23 | 60 | 82 | 1.0 | 1 | 57 | 1.0 | 1 |
| MeY-27i | DQ641485 | MeVIA_53A23 | 60 | ND | ND | ND | ND | ND | ND |
| MeY-28 | DQ641486 | MeVIA_112D12 | 60 | 84 | 6.0 | 6 | 67 | 2.0 | 2 |
| MeY-36 | DQ641487 | MeVIA_53A23 | 60 | 94 | 1.0 | 1 | 72 | 1.0 | 1 |
| MeY-37 | DQ641488 | MeVIA_80O22 | 60 | 95 | 2.0 | 2 | 75 | 1.0 | 1 |
| MeY-56 | DQ641489 | MeVIA_53A23 | 60 | 85 | 1.0 | 1 | 76 | 1.0 | 1 |
| MeY-57 | DQ641490 | MeVIA_53A23 | 60 | 83 | 1.0 | 1 | 73 | 2.0 | 2 |
Abbreviation: ND, not determined.
Me2 loci are from chromosome 2, MeX loci are from the X chromosome and MeY loci are from the Y chromosome.
Genbank accession number.
BAC clone of origin for each locus.
PCR annealing temperature (°C).
Number of chromosomes analysed.
Allelic richness based on a minimum sample size of 82 chromosomes for Kangaroo Island samples and 57 chromosomes for Garden Island samples.
Number of alleles observed.
Null alleles detected at these loci in the Garden Island population.
This locus not scored because evidence of multiple alleles was observed in some males.
Population genetic analyses
Allelic richness, a measure of the number of alleles per locus, corrected for sample size, was calculated using FSTAT (Goudet, 2001) based on a minimum sample size of 82 chromosomes from Kangaroo Island and 57 chromosomes from Garden Island. The genotyping data collected comprised a mixture of haploid and diploid data. Haploid data can be analysed in FSTAT, but this requires the duplication of each haploid result to create a false homozygote. To determine allelic richness for all loci in one calculation, using data from both males and females, all data were analysed as if haploid, that is, diploid genotypes were split to make two false homozygotes from each genotype. For all other analyses, data were not modified and diploid data were analysed separately from haploid data as necessary. FSTAT was also used to evaluate linkage disequilibrium (LD) at polymorphic loci on each chromosome (assessed separately for haploid males and diploid females at X chromosome loci) and to test conformity to Hardy–Weinberg equilibrium for chromosomes X and 2. All analyses were performed separately for each population. MICRO-CHECKER (van Oosterhout et al., 2004) was used to identify loci that displayed evidence of null alleles.
We used GENALEX software version 6.5 (Peakall and Smouse, 2012), to calculate the observed heterozygosity (Ho), expected heterozygosity (He), Shannon's allele information index (sHA), Shannon's mutual information index (sHUA) and pairwise FST and ΦPT, from diploid X chromosome and autosomal data. We calculated haplotypic diversity (h), Shannon's allele information index (sHA) and Fixation index for haploid X chromosome and Y chromosome data as well as pairwise ΦPT for Y chromosome data. ΦPT allows estimation of population genetic differentiation from haploid or binary data as well as diploid data (Peakall et al., 1995). For X chromosome loci, GENALEX was also used to calculate He for male and female data together. From this mean He for autosomal and X chromosome data, and from mean h for Y chromosome data, we used the equation θ=0.5 * ((1/(1−He)2)−1) to estimate θ expected under the stepwise mutation model (Kimura and Ohta, 1975).
We estimated pairwise relatedness and inbreeding coefficients for individuals from each population using COANCESTRY (Wang, 2011). For these analyses, we used data from autosomal markers only. We selected the triadic likelihood estimator (TrioML) because this estimator is able to account for inbreeding. We used 1000 bootstrap simulations to generate 95% confidence limits around the expected difference in mean relatedness and inbreeding between random groups of individuals. The observed differences in mean relatedness and inbreeding coefficients between the Kangaroo Island and Garden Island populations, and between males and females within each population, were then compared with these simulations to test for significant differencies between observed groups of individuals.
STRUCTURE (Pritchard et al., 2000) was used to investigate genetic structure within each population for: (i) autosomal data only, (ii) X chromosome data only and (iii) all data combined, with Y chromosome haplotypes coded as a single locus in males and as missing data in females. Data input files included information on phase information for each genotype (haploid or diploid, with the second allele coded as missing data for haploid loci). The parameters used assumed admixture and correlated allele frequencies (Falush et al., 2003). To estimate the number of clusters, K, for each data set, 20 runs were conducted for each value of K from 1 to 7, each with a burnin of 10 000 repetitions followed by 150 000 MCMC repetitions. For each data set, using STRUCTURE HARVESTER (Earl and vonHoldt, 2012), we calculated the mean likelihood, LnP(K), and s.d. for each value of K, as well as ΔK, the second order rate of change of the likelihood with respect to K (Evanno et al., 2005). NETWORK version 4.6.1.1 (Bandelt et al. (1999); www.fluxus-engineering.com) was used to construct a Y chromosome haplotype network. We used the median-joining network algorithm with the parameter epsilon set to a value of 10. Knowledge of the mutation rates for these loci is inadequate, so perfect dinucleotide repeats (MeY-01, MeY-03 and MeY-57) were weighted w = 2 and imperfect repeats (MeY-28, MeY-37) were weighted w = 5, meaning that loci with lower expected mutation rates were assigned higher weights. We used the MP option (Polzin and Daneschmand, 2003) to identify and remove unnecessary median vectors and links.
Results
Characterising microsatellite variation
We found evidence of multiple alleles from some males at one Y-linked locus (MeY-27), suggesting that this locus or its primer binding sites may be duplicated. This locus was excluded from further study, leaving a total of 29 loci for analysis of which 19 and 17 loci were polymorphic among the Kangaroo Island and Garden Island animals, respectively. Of these, a total of 20 loci were polymorphic in one or more populations, with 16 polymorphic in both populations. Despite the similarity in numbers of polymorphic loci, there were substantial differences in the within locus variation observed among individuals in each population. The number of alleles per polymorphic locus ranged from two to 19 among Kangaroo Island wallabies, but only two to four alleles per locus were observed among Garden Island wallabies (Table 1). Null alleles were detected at two loci from chromosome 2 in the Garden Island population, with null allele frequencies estimated by MICRO-CHECKER (using Brookfield Estimator 1) as 0.04 for locus Me2-040 and 0.03 for locus Me2-077. No null alleles were detected at any locus in the Kangaroo Island population.
No deviations from Hardy–Weinberg equilibrium were detected at loci from chromosomes X and 2 in either population, although conformity to Hardy–Weinberg equilibrium could not be assessed for one bi-allelic locus (MeX-070) in the Garden Island population because its second allele was observed in only one male. Significant LD was detected between five pairs of autosomal loci and between four pairs of X chromosome loci: in each of these pairs both of the loci in question had been identified from the same BAC clone, suggesting a close physical proximity. Consequently, four autosomal loci, Me2-041, Me2-066, Me2-077 and Me2-088, were excluded from further population genetic analyses (only four loci were excluded because two loci already excluded, Me2-077 and Me2-088, constituted the fifth locus pair that displayed significant LD). In addition, three X-linked loci (MeX-048, MeX-049 and MeX-070) were excluded from further population genetic analyses (only three loci were excluded, because MeX-048 displayed significant LD with both MeX-054 and MeX-055). Significant LD was also detected between two pairs of Y-linked loci (between MeY-01 and MeY-28 and between MeY-01 and MeY-37) in the Kangaroo Island population but not in the Garden Island population. All Y chromosome microsatellites should be considered linked on this non-recombining chromosome, so the lack of significant LD for some pairs of Y-linked loci is most likely a reflection of low levels of variation and the number of rare alleles observed. In total, 13 loci comprising four autosomal (Me2-040, Me2-047, Me2-061 and Me2-084), 4 X-linked (MeX-034, MeX-054, MeX-055 and MeX-066) and 5 Y-linked (MeY-01, MeY-03, MeY-28, MeY-37 and MeY-57) loci were available for the comparative analyses.
Population genetics
Population genetic analyses revealed lower levels of gene diversity on Garden Island relative to Kangaroo Island across all chromosomes. We observed a mean allelic richness of 8.53 (Table 1), a mean of 9.5 alleles/locus and mean He of 0.64 (Table 2) for Kangaroo Island animals, and a mean allelic richness of 2.65 (Table 1), a mean of 2.75 alleles/locus and mean He of 0.19 (Table 2) for Garden Island animals, using four autosomal microsatellite loci. This result is in line with previous studies of the two islands using seven autosomal microsatellites which showed high polymorphism (mean alleles/locus 11.1; mean He 0.84) on Kangaroo Island (Taylor and Cooper, 1999) and relatively low genetic diversity (mean 3.7 alleles/locus; He 0.44) on Garden Island (Eldridge et al., 2004). For Kangaroo Island, the ratio of autosome to X to Y diversity (θ) was 1.0 to 0.367 to 0.190, whereas for Garden Island, this ratio was 1.0 to 2.14 to 0.14 (Table 3). We observed substantial genetic differentiation between the two populations, which was most pronounced at loci on the Y chromosome (Table 4). Fis was 0.29 (P=0.01) for autosomal loci and 0.33 (P=0.01) for X-linked loci (data from females only).
Table 2. Number of alleles (N), number of private alleles (Np), number of rare alleles (frequency<0.05) (Nr) observed (Ho) and expected (He) heterozygosities, haplotypic diversity (h) and Shannon's allele information index (sHA) for each chromosome for 13 chromosome-specific microsatellite loci in the Kangaroo Island and Garden Island tammar wallaby populations.
|
Kangaroo Island
|
Garden Island
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Np | Nr | Ho | He | h | sHA | N | Np | Nr | Ho | He | h | sHA | |
| Me2-040 | 2.00 | 0.00 | 1.00 | 0.076 | 0.093 | — | 0.282 | 2.00 | 0.00 | 1.00 | 0.000 | 0.043 | — | 0.154 |
| Me2-047 | 11.00 | 11.00 | 6.00 | 0.805 | 0.813 | — | 2.690 | 4.00 | 4.00 | 0.00 | 0.667 | 0.661 | — | 1.727 |
| Me2-061 | 7.00 | 5.00 | 1.00 | 0.825 | 0.809 | — | 2.529 | 3.00 | 1.00 | 2.00 | 0.053 | 0.052 | — | 0.200 |
| Me2-084 | 18.00 | 16.00 | 11.00 | 0.824 | 0.832 | — | 3.093 | 2.00 | 0.00 | 1.00 | 0.014 | 0.014 | — | 0.060 |
| Chrom. 2 mean | 9.50 | 8.00 | 4.75 | 0.633 | 0.637 | — | 2.149 | 2.75 | 1.25 | 1.00 | 0.183 | 0.192 | — | 0.535 |
| MeX-034—Fa | 5.00 | 5.00 | 3.00 | 0.179 | 0.212 | — | 0.707 | 2.00 | 2.00 | 0.00 | 0.500 | 0.455 | — | 0.934 |
| MeX-054—Fa | 5.00 | 4.00 | 1.00 | 0.643 | 0.643 | — | 1.821 | 3.00 | 2.00 | 0.00 | 0.611 | 0.568 | — | 1.352 |
| MeX-055—Fa | 4.00 | 4.00 | 0.00 | 0.537 | 0.651 | — | 1.726 | 1.00 | 1.00 | 0.00 | 0.000 | 0.000 | — | 0.000 |
| MeX-066—Fa | 3.00 | 3.00 | 1.00 | 0.317 | 0.328 | — | 0.870 | 3.00 | 3.00 | 1.00 | 0.278 | 0.245 | — | 0.682 |
| X chrom. mean—Fa | 4.25 | 4.00 | 1.25 | 0.419 | 0.459 | — | 1.281 | 2.25 | 2.00 | 0.25 | 0.347 | 0.317 | — | 0.742 |
| MeX-034—Mb | 4.00 | 4.00 | 2.00 | — | — | 0.163 | 0.539 | 2.00 | 2.00 | 0.00 | — | — | 0.320 | 0.722 |
| MeX-054—Mb | 8.00 | 6.00 | 4.00 | — | — | 0.706 | 2.170 | 3.00 | 1.00 | 0.00 | — | — | 0.604 | 1.457 |
| MeX-055—Mb | 6.00 | 6.00 | 2.00 | — | — | 0.652 | 1.848 | 1.00 | 1.00 | 0.00 | — | — | 0.000 | 0.000 |
| MeX-066—Mb | 3.00 | 3.00 | 1.00 | — | — | 0.349 | 0.857 | 3.00 | 3.00 | 0.00 | — | — | 0.494 | 1.224 |
| X chrom. mean—Mb | 5.25 | 4.75 | 2.25 | — | — | 0.467 | 1.354 | 2.25 | 1.75 | 0.00 | — | — | 0.354 | 0.851 |
| MeY-01 | 6.00 | 6.00 | 0.00 | — | — | 0.745 | 2.230 | 2.00 | 2.00 | 1.00 | — | — | 0.035 | 0.129 |
| MeY-03 | 3.00 | 2.00 | 2.00 | — | — | 0.089 | 0.312 | 3.00 | 2.00 | 2.00 | — | — | 0.070 | 0.258 |
| MeY-28 | 6.00 | 6.00 | 3.00 | — | — | 0.597 | 1.688 | 2.00 | 2.00 | 1.00 | — | — | 0.035 | 0.129 |
| MeY-37 | 2.00 | 2.00 | 0.00 | — | — | 0.239 | 0.580 | 1.00 | 1.00 | 0.00 | — | — | 0.000 | 0.000 |
| MeY-57 | 1.00 | 0.00 | 0.00 | — | — | 0.000 | 0.000 | 2.00 | 1.00 | 1.00 | — | — | 0.035 | 0.129 |
| Y chrom. mean | 3.60 | 3.20 | 1.00 | — | — | 0.334 | 0.962 | 2.00 | 1.60 | 1.00 | — | — | 0.035 | 0.129 |
X chromosome data from diploid females.
X chromosome data from haploid males.
Table 3. Theta ( θ ) and theta relative to chromosome 2 (Rel) for 13 chromosome-specific microsatellite loci in the Kangaroo Island and Garden Island tammar wallaby populations.
|
Kangaroo Island
|
Garden Island
|
|||||||
|---|---|---|---|---|---|---|---|---|
| C a | He | θ | Rel | C a | He | θ | Rel | |
| Me2-40 | 184 | 0.093 | 90 | 0.043 | ||||
| Me2-47 | 298 | 0.813 | 132 | 0.661 | ||||
| Me2-61 | 286 | 0.809 | 114 | 0.052 | ||||
| Me2-84 | 296 | 0.832 | 144 | 0.014 | ||||
| Chrom. 2 mean | 266 | 0.637 | 3.291 | 1.00 | 120 | 0.192 | 0.267 | 1.00 |
| MeX-34 | 170 | 0.212 | 30 | 0.455 | ||||
| MeX-54 | 116 | 0.643 | 49 | 0.568 | ||||
| MeX-55 | 170 | 0.651 | 40 | 0.000 | ||||
| MeX-66 | 174 | 0.328 | 45 | 0.245 | ||||
| X chrom. mean | 158 | 0.459 | 1.208 | 0.37 | 41 | 0.317 | 0.572 | 2.14 |
| MeY-01 | 65 | 0.745 | 56 | 0.035 | ||||
| MeY-03 | 65 | 0.089 | 56 | 0.070 | ||||
| MeY-28 | 65 | 0.597 | 56 | 0.035 | ||||
| MeY-37 | 65 | 0.239 | 56 | 0.000 | ||||
| MeY-57 | 65 | 0.000 | 56 | 0.035 | ||||
| Y chrom. mean | 65 | 0.334 | 0.627 | 0.19 | 56 | 0.035 | 0.037 | 0.14 |
Theta was calculated based on expected (He) heterozygosity for each chromosome, with haploid male and diploid female data combined for X chromosome loci.
Number of chromosomes included in this analysis for each locus.
Table 4. Genetic differentiation observed between tammar wallabies from the Kangaroo Island and Garden Island populations.
| FST | ΦPT | sHua |
Molecular variance
|
|||
|---|---|---|---|---|---|---|
| Among populations | Among individuals | Within individuals | ||||
| Chromosome 2 males and females | 0.29 (P=0.01) | 0.39 (P=0.01) | 0.59 | 29% | 20% | 51% |
| X chromosome females only | 0.54 (P=0.01) | 0.64 (P=0.01) | 0.82 | 54% | 15% | 31% |
| Y chromosome Males only | — | 0.93 (P=0.01) | 0.80 | 93% | 7% | — |
Complete genotypes at five variable Y chromosome microsatellites were obtained for 65 males from Kangaroo Island and 55 males from Garden Island (Table 5). A total of 17 distinct Y chromosome haplotypes were observed from these males in the Kangaroo Island population. In contrast, only four Y chromosome haplotypes were observed from Garden Island wallabies. Three of these haplotypes were observed in only a single individual each, meaning that the most common haplotype was observed in 52 out of 55 Garden Island males. No Y chromosome haplotypes were shared between the populations (Figure 1).
Table 5. Y chromosome microsatellite haplotypes observed from Kangaroo Island and Garden Island tammar wallabies.
| Haplotype | MeY-01 a | MeY-03 a | MeY-28 a | MeY-37 a | MeY-57 a | KI b | GI c |
|---|---|---|---|---|---|---|---|
| A | 340 | 303 | 349 | 179 | 358 | 2 | 0 |
| B | 340 | 303 | 352 | 179 | 358 | 7 | 0 |
| C | 340 | 303 | 358 | 179 | 358 | 6 | 0 |
| D | 342 | 303 | 352 | 179 | 358 | 1 | 0 |
| E | 342 | 303 | 358 | 179 | 358 | 8 | 0 |
| F | 342 | 303 | 361 | 179 | 358 | 1 | 0 |
| G | 342 | 305 | 352 | 179 | 358 | 1 | 0 |
| H | 344 | 301 | 352 | 181 | 358 | 1 | 0 |
| I | 344 | 303 | 352 | 179 | 358 | 17 | 0 |
| J | 344 | 303 | 352 | 181 | 358 | 8 | 0 |
| K | 346 | 303 | 352 | 179 | 358 | 2 | 0 |
| L | 346 | 303 | 355 | 179 | 358 | 2 | 0 |
| M | 348 | 301 | 358 | 179 | 358 | 1 | 0 |
| N | 348 | 303 | 358 | 179 | 358 | 1 | 0 |
| O | 348 | 303 | 364 | 179 | 358 | 3 | 0 |
| P | 350 | 303 | 358 | 179 | 358 | 1 | 0 |
| Q | 350 | 303 | 364 | 179 | 358 | 3 | 0 |
| R | 328 | 305 | 337 | 185 | 358 | 0 | 1 |
| S | 330 | 307 | 343 | 185 | 358 | 0 | 52 |
| T | 330 | 307 | 343 | 185 | 360 | 0 | 1 |
| U | 330 | 309 | 343 | 185 | 358 | 0 | 1 |
| 65 | 55 |
Allele size in base pairs for each locus.
Number of individuals with each haplotype in the Kangaroo Island population.
Number of individuals with each haplotype in the Garden Island population.
Figure 1.
Median-joining Y chromosome haplotype network showing haplotype diversity among males in the Kangaroo Island (yellow nodes) and Garden Island (blue nodes) populations. Node size is proportional to haplotype frequency. Black nodes indicate inferred median vectors.
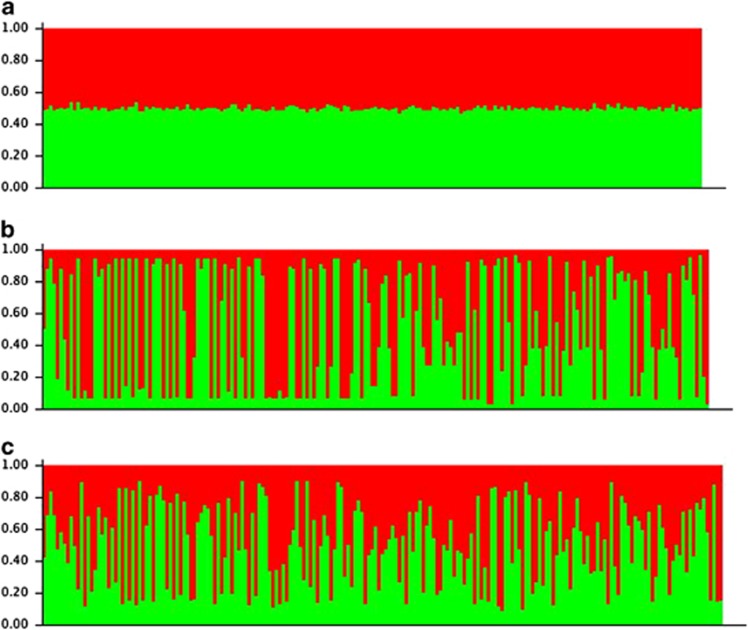
The number of genetic clusters identified within the Kangaroo Island population varied depending on the markers used (Table 6). For each data set, we first examined the mean LnP(K) values for each K, to identify those instances where K was clearly equal to 1 (where mean LnP(K) was essentially flat across all values of K). As ΔK cannot be calculated for K=1, these cases could not be evaluated using the method of Evanno et al. (2005). For the Garden Island population, we identified a single cluster as the most appropriate for all data sets, regardless of chromosome. This was also the case for the Kangaroo Island population using autosomal data alone. However, in the two other Kangaroo Island data sets, which used X-linked markers only, and autosomal, X- and Y-linked markers combined, we observed similar LnP(K) values for K=1 and K=2, with low s.d. values for K=2. In these data sets, using the ΔK approach, K=2 was identified as the most likely value for K (Figure 2). Given the sampling design of this study, in which all individuals have been sampled from a relatively small area, we do not believe that these results indicate that there are two genetically distinct sub-groups within the individuals sampled. Rather, these results reflect the detection of sex chromosome lineages reflecting the genealogies of the individuals sampled.
Table 6. Mean LnP(K), standard deviation in LnP(K) (s.d.), and ΔK observed in Structure analyses for K values of 1–7 in the Kangaroo Island (KI) and Garden Island (GI) populations.
| K=1 | K=2 | K=3 | K=4 | K=5 | K=6 | K=7 | |
|---|---|---|---|---|---|---|---|
| Kangaroo Island | |||||||
| Chrom. 2 | |||||||
| LnP(K) | −1998.98 | −2039.13 | −2064.15 | −2079.03 | −2053.60 | −2068.15 | −2089.08 |
| s.d. | 0.29 | 26.00 | 35.30 | 55.68 | 35.79 | 49.51 | 54.01 |
| ΔK | — | 0.58 | 0.29 | 0.72 | 1.12 | 0.13 | — |
| X chrom. | |||||||
| LnP(K) | −750.18 | −651.40 | −632.90 | −644.09 | −615.69 | −649.42 | −710.89 |
| s.d. | 0.18 | 2.16 | 7.39 | 12.99 | 5.86 | 8.33 | 19.58 |
| ΔK | — | 37.11 | 4.02 | 3.05 | 10.61 | 3.33 | — |
| All data | |||||||
| LnP(K) | −2900.35 | −2896.36 | −3123.38 | −3435.34 | −3668.60 | −3922.17 | −3772.11 |
| s.d. | 0.93 | 7.15 | 55.60 | 132.63 | 164.64 | 1211.92 | 524.85 |
| ΔK | — | 32.32 | 1.53 | 0.59 | 0.12 | 0.33 | — |
| Garden Island | |||||||
| Chrom. 2 | |||||||
| LnP(K) | −222.63 | −224.37 | −225.01 | −224.22 | −223.50 | −223.63 | −223.42 |
| s.d. | 0.07 | 1.09 | 1.31 | 1.20 | 0.75 | 1.23 | 0.80 |
| ΔK | — | 1.01 | 1.10 | 0.06 | 1.13 | 0.28 | — |
| X chrom. | |||||||
| LnP(K) | −185.02 | −190.33 | −187.86 | −186.46 | −185.41 | −185.62 | −185.19 |
| s.d. | 0.18 | 6.25 | 3.65 | 1.96 | 0.87 | 1.29 | 0.51 |
| ΔK | — | 1.25 | 0.29 | 0.18 | 1.44 | 0.49 | — |
| All data | |||||||
| LnP(K) | −424.01 | −444.36 | −439.70 | −436.73 | −430.29 | −428.42 | −428.33 |
| s.d. | 0.26 | 7.39 | 10.69 | 7.87 | 5.75 | 3.57 | 4.03 |
| ΔK | — | 3.38 | 0.16 | 0.44 | 0.80 | 0.50 | — |
Data were analysed from chromosome 2 only, from the X chromosome only and from all three chromosomes combined, with Y chromosome haplotypes coded as a single locus.
Figure 2.
Bar charts generated by the program STRUCTURE showing the assignment of individuals from the Kangaroo Island population to two genetic clusters: (a) data from chromosome 2 only; (b) data from the X chromosome only; (c) data from all three chromosomes combined. Each vertical line represents a different individual, colour-coded according to the proportion of its ancestry derived from each genetic cluster identified.
Mean pairwise relatedness and inbreeding coefficients were significantly higher for the Garden Island population than the Kangaroo Island population (Table 7). Within Kangaroo Island, we detected no difference in the mean inbreeding coefficient between males and females, but we observed that mean relatedness was significantly higher among females than among males. Within Garden Island, we observed that both the mean relatedness and inbreeding coefficients were significantly higher among males than among females (Table 7).
Table 7. Mean relatedness (R) and inbreeding (I) coefficients calculated for Kangaroo Island (KI) and Garden Island (GI) tammar wallabies using COANCESTRY, including comparisons between the two populations and between males and females within each population.
| KI (1) vs GI (2) | KI males (1) vs KI females (2) | GI males (1) vs GI females (2) | ||||
|---|---|---|---|---|---|---|
| |
R |
I |
R |
I |
R |
I |
| n (1) | 18 528 | 193 | 4465 | 95 | 2850 | 76 |
| Mean (1) | 0.275 | 0.166 | 0.190 | 0.171 | 1.438 | 0.745 |
| Variance (1) | 0.908 | 0.048 | 0.080 | 0.050 | 0.708 | 0.104 |
| n (2) | 5995 | 110 | 4753 | 98 | 561 | 34 |
| Mean (2) | 1.502 | 0.732 | 0.211 | 0.134 | 0.451 | 0.476 |
| Variance (2) | 0.332 | 0.080 | 0.085 | 0.039 | 0.291 | 0.099 |
| Mean (1)−mean (2) | −1.227a | −0.566a | −0.022a | 0.037 | 0.986a | 0.269a |
| 5% Quantile | −0.016 | −0.068 | −0.009 | −0.047 | −0.064 | −0.114 |
| 95% Quantile | 0.016 | 0.073 | 0.010 | 0.052 | 0.072 | 0.108 |
n, number of pairwise comparisons (for relatedness estimates) or individuals (for inbreeding estimates) analysed for the first population (1) or the second population (2).
Statistically significant differences between populations or between the sexes, for which the difference between the two mean values tested fell outside the 95% confidence intervals generated during bootstrap simulations.
Discussion
This analysis of microsatellite variation in tammar wallabies demonstrates a clear genetic distinction between the two largest remaining populations of this species, a distinction that is apparent across autosomal loci as well as sex-linked loci. At the same time, variation among these suites of loci demonstrates differences that reflect the interaction between their different modes of inheritance and the different histories and characteristics of the populations. The combination provides a much broader picture of genetic variation in the two populations than would be obtained from the application of autosomal microsatellites alone.
We found lower levels of genetic diversity at autosomal loci in the Garden Island population than in the Kangaroo Island population. Our results agree with expections of higher genetic diversity on Kangaroo Island than Garden Island and are consistent with our observations of high levels of inbreeding and high relatedness coefficients for Garden Island wallabies. It is possible that a founder effect or an historic bottleneck, that occurred when rising sea levels isolated the Garden Island tammar wallabies from the mainland West Australian populations, may have contributed to the low levels of genetic diversity observed in this population (this study and Eldridge et al. (2004)).
We note that our mean He estimates for autosomal loci are lower than those obtained in previous studies (Taylor and Cooper, 1999; Eldridge et al., 2004). These differences could reflect variation in levels of diversity at the different loci used, but another possible explanation is that all tissue samples used by us were collected from a single region of each island, whereas the previous studies may have drawn on a greater number of collection sites, particularly on the much larger Kangaroo Island. Greater genetic diversity at autosomal loci in the earlier studies may therefore reflect greater heterogeneity in sampling locations. In addition, we were unable to score some loci in some individuals from Garden Island, despite successful amplification of other loci from the same DNA extractions. The markers we used were originally developed for Kangaroo Island tammar wallabies so this could reflect ascertainment bias when applying these markers to Garden Island samples. Such a bias would lead to reduced variation at these markers on Garden Island, however, we found evidence for null alleles (an indicator of ascertainment bias) at only two loci in the Garden Island population and our mean He estimates were also lower than those obtained in previous work for Kangaroo Island. Consequently, we do not believe that ascertainment bias alone can provide a comprehensive explanation for the low levels of diversity we observe in Garden Island tammar wallabies.
Genetic diversity is higher in the Kangaroo Island population than the Garden Island population for all measures of genetic diversity (allelic richness, Ho, He, haploid diversity and θ) on each of the three chromosomes. This difference is most pronounced on the Y chromosome, where over 90% of the males surveyed from Garden Island share the same haplotype, suggesting the predominance of a single, dominant Y chromosome lineage in this population. In fact, genetic variation is lowest for Y chromosome loci in both populations. Previous studies (Taylor and Cooper, 1999; Eldridge et al., 2004) observed lower genetic diversity for Garden Island than for Kangaroo Island using autosomal markers alone, we now observe the same trend using additional sex-linked markers.
Comparisons of genetic diversity among the different chromosomes reflect important population processes as well as differences in Ne among the chromosomes caused by their differing modes of inheritance. Genetic variation is also affected by the mutation rate, which influences the creation of new alleles. Mutation rates in sperm DNA were estimated to range from 1.5 × 10−2 to 2.2 × 10−3 mutations per locus per generation for three of the autosomal loci included in this study (MacDonald et al., 2011), but comparable data are not available for sex-linked loci, meaning that we cannot discount the possibility of mutation rate variation among chromosomes. One other factor that may influence variation in genetic diversity between different chromosomes is a male bias to the mutation process as a consequence of elevated cell divisions during spermatogenesis. However, this process is unlikely to have caused the difference we find between the two populations, because elevated cell division is likely to increase rather than decrease diversity at Y-linked loci (Miyata et al., 1987). Given that genetic diversity at Y-linked loci was reduced relative to diversity at X-linked and autosomal loci in both populations, any male mutation bias in tammar wallabies must be counteracted by even stronger factors acting to suppress Y chromosome diversity.
For Garden Island, the ratio of autosome to X to Y diversity (θ) is 1.0 to 2.14 to 0.14. Genetic diversity is lowest for the Y chromosome, and lower than expected relative to autosomal markers. However, diversity is higher for X-linked loci than for loci on chromosome 2. Higher than expected X to autosome ratios have been observed in several species (typically closer to 1:1 ratios, but X chromosome diversity was observed to be higher than autosomal diversity in some Drosophila melanogaster populations (Andolfatto, 2001)) and one mechanism proposed to explain this is an extreme skew in male reproductive success (Charlesworth, 2009; Ellegren, 2009). The ratio of X to autosome diversity is expected to increase when male Ne is much lower than female Ne because, assuming an equal sex ratio, the autosomes spend a greater proportion of evolutionary time in the male germline than does the X chromosome. Consequently, the autosomes have greater exposure to any bottleneck in the male germline caused by skewed male reproductive success. It is possible that our observations reflect such a male bottleneck in the Garden Island population, a possibility that is also supported by lower than expected Y chromosome diversity in this population. However, the ratio of X to autosome diversity we observe on Garden Island is much higher than might be expected based solely on variation in reproductive success and consequently may be influenced by other factors, or may reflect a lack of precision in our estimates of diversity among chromosomes. Given the small number of loci used in this study, the low number of alleles per locus, high levels of inbreeding and some evidence for an ascertainment bias, caution is required in the interpretation of these data. A genomic scale study, with many more markers and comprehensive sampling, may be required to resolve these questions in this population.
The Kangaroo Island population presents a contrasting situation to Garden Island. Here the ratio of autosome to X to Y diversity (θ) is 1.0 to 0.37 to 0.19. Thus, levels of genetic diversity vary among chromosomes in the order predicted by the rules of chromosomal inheritance, with the highest diversity on the autosome and the lowest on the Y chromosome, suggesting that variation in chromosomal Ne is an important determinant of genetic variation. We observe that both X-linked and Y-linked diversity are lower than the 1.0 to 0.75 to 0.25 ratio predicted based solely on chromosomal differences in Ne. This departure from expectations may reflect the relatively small number of loci from each chromosome used in this analysis. However, variation in Ne between chromosomes cannot be considered alone in its impact on levels of genetic variation. Demographic factors and population history will also influence the number of alleles transmitted between generations. These results provide little evidence for a strong skew in male reproductive success within the Kangaroo Island tammar wallaby population. If the majority of offspring were sired by a small number of dominant males we would expect to observe very low Y chromosome diversity and higher than expected X chromosome diversity. We also observe that relatedness is higher for females than for males. One explanation for this observation is a male bias to dispersal in the Kangaroo Island population. Sex-biased dispersal has traditionally been investigated through comparisons of bi-parentally inherited autosomal genetic variation with maternally inherited mitochondrial genetic variation, but to date sex chromosome markers have been used to address this topic in only a relatively small number of species. However, where they are available, Y chromosome markers, which provide a paternal perspective on population genetic variation, can be extremely useful. For example, in the bonobo (Pan paniscus), haplotype diversity has been shown to be significantly greater for mtDNA than for the Y chromosome, suggesting that dispersal in this species is highly female-biased (Eriksson et al., 2006). Male-biased dispersal has been recorded in several species of macropod, including eastern grey kangaroos, Macropus giganteus, (Zenger et al., 2003), brush-tailed rock wallabies, Petrogale penicillata (Hazlitt et al., 2004) and swamp wallabies, Wallabia bicolour (Paplinska et al., 2009). Here data from autosomal microsatellites suggest the occurrence of male-biased dispersal in the tammar wallaby, highlighting the need for further work on this topic in this model marsupial, for which X- and Y-linked microsatellites may prove to be valuable additional tools.
The utility of combining data from the different chromosomes can be seen through the results of our assignment analysis. All individuals from Kangaroo Island are assigned to a single genetic cluster when based on autosomal data alone. The incorporation of data from the Y chromosome, which clusters males by paternal lineage, and data from the X chromosome, which faces a different level of exposure to recombination, mutation and demographic factors than the autosomes, allows us to assign individuals to two genetic clusters, that reflect sex-chromosomal lineages within the Kangaroo Island population. Detailed information on genetic structure within populations and an understanding of sex-specific genetic lineages or skewed mating systems are important for conservation management where the aim is to preserve as much genetic variation as possible from a threatened population (Weeks et al., 2011). Our results demonstrate that sex-linked markers can provide information that complements data available from autosomal markers for application to conservation and management programs.
Given the importance of small offshore islands as refuges, both natural and artificial, for many Australian species (Eldridge et al., 2004; Miller et al., 2011), this work also highlights a potential need to monitor such island populations to ensure that genetic diversity is preserved across the whole genome. One aim of conservation programs is to preserve genetic diversity within threatened species, with the implication that this in turn will protect the evolutionary potential of those species. However, genetic diversity is rarely assessed at the genomic scale in conservation genetic studies, which are typically reliant on autosomal and mitochondrial markers. The mammalian Y chromosome is the only male-specific portion of the genome and carries genes crucial to male development and reproduction in most species investigated to date (Graves, 1995). The Y chromosome also has the lowest Ne and typically the lowest genetic diversity of all chromosomes, so the failure to specifically assess and conserve Y chromosome diversity seems an oversight that could have serious implications for the retention of male-specific diversity within threatened species. In this study, we observed only four Y chromosome haplotypes in the Garden Island population. Although we sampled only a relatively small proportion of males from one area of Garden Island, our sampling strategy was similar for the Kangaroo Island population where we observed 17 Y chromosome haplotypes. Other Western Australian island populations of the tammar wallaby may be in even greater need of management than Garden Island. For example, only three Y chromosome haplotypes were identified among male tammar wallabies from three different islands of the Houtman Abrolhos Archipelago using some of the markers included in the current study (Miller et al., 2011). Two of these islands share one of the Y chromosome haplotypes, which supports the inference from autosomal and mitochondrial data that tammar wallabies were introduced to North Island from West Wallabi Island. Thus, data from sex-linked markers have the capacity to contribute to management decisions such as the identification of management units and appropriate sources of animals for translocation or genetic rescue should one of the island populations become critically endangered or extinct. Future work may indicate circumstances where it is appropriate to consider extension of current management practices to specifically monitor genetic variation at sex chromosome markers and to augment threatened populations through sex-specific translocations, for example, to increase Y chromosome diversity within isolated island populations.
We anticipate that the availability of sex-linked markers from wildlife species will increase over the coming years. Here we have demonstrated, using only a relatively small number of loci, that the application of markers from the X and Y chromosomes can provide new insights into the tammar wallaby, which is arguably already one of the better-studied marsupial species (Hickford et al., 2012) and now has a genome sequence available (Renfree et al., 2011). This area has great potential for future research because the development of sex-linked markers in ‘genome-enabled' species (Thomson et al., 2010) and their subsequent application to genetic studies of these species and their close relatives is likely to be of value to both conservation genetics and wildlife management.
Data archiving
Data available from the Dryad Digital Repository: doi:10.5061/dryad.s76t7.
Acknowledgments
This work was supported by an Australian Research Council Discovery Grant (DP0211687) awarded to SDS and NNF. Professor Geoff Shaw, Dr Danielle Hickford and Dr Terry Fletcher assisted with sample collection and Rachel Walsh assisted with DNA extractions. We thank three anonymous reviewers, Niccy Aitken, William Sherwin, Marion Höhn, Dennis McNevin and participants in an Institute for Applied Ecology Science Writers' Workshops for helpful comments on earlier versions of this manuscript.
The authors declare no conflict of interest.
References
- Andolfatto P. Contrasting patterns of X-linked and autosomal nucleotide variation in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol. 2001;18:279–290. doi: 10.1093/oxfordjournals.molbev.a003804. [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. BioTechniques. 2001;31:24–28. [PubMed] [Google Scholar]
- Casto A, Li J, Absher D, Myers R, Ramachandran S, Feldman M. Characterization of X-Linked SNP genotypic variation in globally distributed human populations. Genome Biol. 2010;11:R10. doi: 10.1186/gb-2010-11-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resources. 2012;4:359–361. [Google Scholar]
- Eldridge MDB, Kinnear JE, Zenger KR, McKenzie LM, Spencer PBS. Genetic diversity in remnant mainland and ‘pristine' island populations of three endemic Australian macropodids (Marsupialia): Macropus eugenii, Lagorchestes hirsutus and Petrogale lateralis. Conserv Genet. 2004;5:325. [Google Scholar]
- Ellegren H. The different levels of genetic diversity in sex chromosomes and autosomes. Trends Genet. 2009;25:278–284. doi: 10.1016/j.tig.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Eriksson J, Siedel H, Lukas D, Kayser M, Erler A, Hashimoto C, et al. Y-chromosome analysis confirms highly sex-biased dispersal and suggests a low male effective population size in bonobos (Pan paniscus) Mol Ecol. 2006;15:939–949. doi: 10.1111/j.1365-294X.2006.02845.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome 10K Community of Scientists Genome 10K: a proposal to obtain whole-genome sequence for 10 000 vertebrate species. J Hered. 2009;100:659–674. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipati S, Arbiza L, Siepel A, Clark AG, Keinan A. Analyses of X-linked and autosomal genetic variation in population-scale whole genome sequencing. Nat Genet. 2011;43:741–743. doi: 10.1038/ng.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J.2001. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from: http://www.unil.ch/izea/softwares/fstat.html . Updated from Goudet (1995).
- Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes—an evolving understanding. BioEssays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Greminger MP, KrÜTzen M, Schelling C, Pienkowska Schelling A, Wandeler P. The quest for Y-chromosomal markers—methodological strategies for mammalian non-model organisms. Mol Ecol Resources. 2010;10:409–420. doi: 10.1111/j.1755-0998.2009.02798.x. [DOI] [PubMed] [Google Scholar]
- Guichoux E, Lagache L, Wagner S, Chaumeil P, LÉGer P, Lepais O, et al. Current trends in microsatellite genotyping. Mol Ecol Resources. 2011;11:591–611. doi: 10.1111/j.1755-0998.2011.03014.x. [DOI] [PubMed] [Google Scholar]
- Hammer MF, Mendez FL, Cox MP, Woerner AE, Wall JD. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 2008;4:e1000202. doi: 10.1371/journal.pgen.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlitt SL, Eldridge MDB, Goldizen AW. Fine-scale spatial genetic correlation analyses reveal strong female philopatry within a brush-tailed rock-wallaby colony in southeast Queensland. Mol Ecol. 2004;13:3621–3632. doi: 10.1111/j.1365-294X.2004.02342.x. [DOI] [PubMed] [Google Scholar]
- Hellborg L, Ellegren H. Low levels of nucleotide diversity in mammalian Y chromosomes. Mol Biol Evol. 2004;21:158–163. doi: 10.1093/molbev/msh008. [DOI] [PubMed] [Google Scholar]
- Hickford D, Frankenberg S, Renfree MB. The Tammar Wallaby, Macropus Eugenii: a Model Kangaroo For The Study Of Developmental And Reproductive Biology Emerging Model Organisms. Vol 2. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, USA; 2012. pp. 449–494. [DOI] [PubMed] [Google Scholar]
- Hynes EF, Rudd CD, Temple-Smith PD, Sofronidis G, Paris D, Shaw G, et al. Mating sequence, dominance and paternity success in captive male tammar wallabies. Reproduction. 2005;130:123–130. doi: 10.1530/rep.1.00624. [DOI] [PubMed] [Google Scholar]
- Inns RW.1980. Ecology of the Kangaroo Island wallaby, Macropus eugenii (Desmarest), in Flinders Chase National Park, Kangaroo Island. PhD thesis thesis, PhD thesis, University of Adelaide.
- Johnson JA, Toepfer JE, Dunn PO. Contrasting patterns of mitochondrial and microsatellite population structure in fragmented populations of greater prairie-chickens. Mol Ecol. 2003;12:3335–3347. doi: 10.1046/j.1365-294x.2003.02013.x. [DOI] [PubMed] [Google Scholar]
- Kaiser VB, Charlesworth B. The effects of deleterious mutations on evolution in non-recombining genomes. Trends Genet. 2009;25:9–12. doi: 10.1016/j.tig.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat Genet. 2009;41:66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ohta T. Distribution of Allelic Frequencies in a Finite Population under Stepwise Production of Neutral Alleles. Proc Natl Acad Sci USA. 1975;72:2761–2764. doi: 10.1073/pnas.72.7.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Handley LJ, Hammond RL, Emaresi G, Reber A, Perrin N. Low Y chromosome variation in Saudi-Arabian hamadryas baboons (Papio hamadryas hamadryas) Heredity. 2006;96:298–303. doi: 10.1038/sj.hdy.6800803. [DOI] [PubMed] [Google Scholar]
- Li M-H, Merila J. Genetic evidence for male-biased dispersal in the Siberian jay (Perisoreus infaustus) based on autosomal and Z-chromosomal markers. Mol Ecol. 2010;19:5281–5295. doi: 10.1111/j.1365-294X.2010.04870.x. [DOI] [PubMed] [Google Scholar]
- Lindgren G, Backström N, Swinburne J, Hellborg L, Einarsson A, Sandberg K, et al. Limited number of patrilines in horse domestication. Nat Genet. 2004;36:335–336. doi: 10.1038/ng1326. [DOI] [PubMed] [Google Scholar]
- MacDonald A, Sarre S, FitzSimmons N, Aitken N. Determining microsatellite genotyping reliability and mutation detection ability: an approach using small-pool PCR from sperm DNA. Mol Genet Genomics. 2011;285:1–18. doi: 10.1007/s00438-010-0577-9. [DOI] [PubMed] [Google Scholar]
- MacDonald AJ, Sankovic N, Sarre SD, FitzSimmons NN, Wakefield MJ, Graves JAM, et al. Y chromosome microsatellite markers identified from the tammar wallaby (Macropus eugenii) and their amplification in three other macropod species. Mol Ecol Notes. 2006;6:1202–1204. [Google Scholar]
- MacDonald AJ, Sarre SD, FitzSimmons NN, Graves JAM. Chromosome-specific microsatellites from the tammar wallaby X chromosome and chromosome 2. Mol Ecol Notes. 2007;7:1063–1066. [Google Scholar]
- Meadows JRS, Hawken RJ, Kijas JW. Nucleotide diversity on the ovine Y chromosome. Anim Genet. 2004;35:379–385. doi: 10.1111/j.1365-2052.2004.01180.x. [DOI] [PubMed] [Google Scholar]
- Miller EJ, Eldridge MDB, Herbert CA.2010Dominance and paternity in the tammar wallabyIn: Coulson G, Eldridge MDB (eds)Macropods: The Biology of Kangaroos, Wallabies and Rat-kangaroos CSIRO Publishing; 77–86. [Google Scholar]
- Miller EJ, Eldridge MDB, Morris KD, Zenger KR, Herbert CA. Genetic consequences of isolation: island tammar wallaby (Macropus eugenii) populations and the conservation of threatened species. Conserv Genet. 2011;12:1619–1631. [Google Scholar]
- Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harbor Symp Quant Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- Nachman MW. Variation in recombination rate across the genome: evidence and implications. Curr Opin Genet Dev. 2002;12:657–663. doi: 10.1016/s0959-437x(02)00358-1. [DOI] [PubMed] [Google Scholar]
- Page J, Berrios S, Parra MT, Viera A, Suja JA, Prieto I, et al. The program of sex chromosome pairing in meiosis is highly conserved across marsupial species: implications for sex chromosome evolution. Genetics. 2005;170:793–799. doi: 10.1534/genetics.104.039073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paplinska JZ, Eldridge MDB, Cooper DW, Temple-Smith PDM, Renfree MB. Use of genetic methods to establish male-biased dispersal in a cryptic mammal, the swamp wallaby (Wallabia bicolor) Aust J Zool. 2009;57:65–72. [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse PE, Huff DR. Evolutionary implications of allozyme and RAPD variation in diploid populations of dioecious buffalograss Buchloë dactyloides. Mol Ecol. 1995;4:135–147. [Google Scholar]
- Petit E, Balloux F, Excoffier L. Mammalian population genetics: why not Y. Trends Ecol Evol. 2002;17:28–33. [Google Scholar]
- Polzin T, Daneschmand SV. On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 2003;31:12–20. [Google Scholar]
- Pope LC, Sharp A, Moritz C. Population structure of the yellow-footed rock-wallaby Petrogale xanthopus (Gray, 1854) inferred from mtDNA sequences and microsatellite loci. Mol Ecol. 1996;5:629–640. doi: 10.1111/j.1365-294x.1996.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, de Meeus T. Inferring sex-biased dispersal from population genetic tools: a review. Heredity. 2002;88:161–165. doi: 10.1038/sj.hdy.6800060. [DOI] [PubMed] [Google Scholar]
- Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, Belov K, et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011;12:R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner SF. The X chromosome in population genetics. Nat Rev Genet. 2004;5:43–51. doi: 10.1038/nrg1247. [DOI] [PubMed] [Google Scholar]
- Sunnucks P. Efficient genetic markers for population biology. Trends Ecol Evol. 2000;15:199–203. doi: 10.1016/s0169-5347(00)01825-5. [DOI] [PubMed] [Google Scholar]
- Taylor AC, Cooper DW. Microsatellites identify introduced New Zealand tammar wallabies (Macropus eugenii) as an ‘extinct' taxon. Anim Conserv. 1999;2:41. [Google Scholar]
- Thomson RC, Wang IJ, Johnson JR. Genome-enabled development of DNA markers for ecology, evolution and conservation. Mol Ecol. 2010;19:2184–2195. doi: 10.1111/j.1365-294X.2010.04650.x. [DOI] [PubMed] [Google Scholar]
- Tyndale-Biscoe H. Life of Marsupials. CSIRO Publishing: Collingwood, Australia; 2005. [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Wang J. COANCESTRY: A program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resources. 2011;11:141–145. doi: 10.1111/j.1755-0998.2010.02885.x. [DOI] [PubMed] [Google Scholar]
- Weeks AR, Sgro CM, Young AG, Frankham R, Mitchell NJ, Miller KA, et al. Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evol Appl. 2011;4:709–725. doi: 10.1111/j.1752-4571.2011.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M, Stott P.1999. Rural Industries Research and Development Corporation, Australia.
- Yannic G, Basset P, Büchi L, Hausser J, Broquet T. Scale-specific sex-biased dispersal in the Valais shrew unveiled by genetic variation on the Y chromosome, autosomes and mitochondrial DNA. Evolution. 2012;66:1737–1750. doi: 10.1111/j.1558-5646.2011.01554.x. [DOI] [PubMed] [Google Scholar]
- Zenger KR, Eldridge MDB, Cooper DW. Intraspecific variation, sex-biased dispersal and phylogeography of the eastern grey kangaroo (Macropus giganteus) Heredity. 2003;91:153–162. doi: 10.1038/sj.hdy.6800293. [DOI] [PubMed] [Google Scholar]