Abstract
Spinach (Spinacia oleracea L.) is widely known to be dioecious. However, monoecious plants can also occur in this species. Sex expression in dioecious spinach plants is controlled by a single gene pair termed X and Y. Our previous study showed that a single, incompletely dominant gene, which controls the monoecious condition in spinach line 03–336, should be allelic or linked to X/Y. Here, we developed 19 AFLP markers closely linked to the monoecious gene. The AFLP markers were mapped to a 38.2-cM chromosomal region that included the monoecious gene, which is bracketed between flanking markers with a distance of 7.1 cM. The four AFLP markers developed in our studies were converted into sequence-characterized amplified region (SCAR) markers, which are linked to both the monoecious gene and Y and are common to both populations segregating for the genes. Linkage analysis using the SCAR markers suggested that the monoecious gene (M) and Y are located in different intervals, between different marker pairs. Analysis of populations segregating for both M and Y also directly demonstrates linkage of the genes at a distance of ∼12 cM. The data presented in this study may be useful for breeding dioecious and highly male monoecious lines utilized as the pollen parents for hybrid seed production, as well as for studies of the evolutionary history of sexual systems in this species, and can provide a molecular basis for positional cloning of the sex-determining genes.
Keywords: dioecy, monoecy, spinach, Spinacia oleracea , sex chromosome
Introduction
Spinach (Spinacia oleracea L.) is an annual leafy vegetable that is rich in minerals and vitamins and is grown in >50 countries, mostly those with temperate climates (FAOSTAT, http://faostat3.fao.org/home/index.html). Spinach is a member of the subfamily Chenopodioideae (Chenopodiaceae) and is related to quinoa (Chenopodium quinoa Willd.), Swiss chard, table beet and sugar beet (Beta vulgaris L.) (Ware and McCollum, 1980; Kadereit et al., 2010; Fuentes-Bazan et al., 2012). Spinach is commonly known as a dioecious species with an even ratio of female to male individuals. However, certain lines and crosses produce monoecious individuals, among which the proportion of female to male (or hermaphrodite) flowers per plant varies widely (Janick and Stevenson, 1955b; Onodera et al., 2008, 2011).
The majority of spinach cultivars currently used in developed countries are F1 hybrids. Early hybrid seed production was carried out by removing male individuals before pollen shedding in dioecious lines used as seed parents. Currently, however, most hybrids are produced from seed parent lines that breed true for the highly female monoecious condition (Janick, 1998). Furthermore, current breeding programs are aimed at using highly male monoecious inbred lines as pollen parents (van der Vossen, 2004).
The progenies resulting from self-fertilization of male plants from the spinach cultivar Long Standing Bloomingdale, in which male plants occasionally bear hermaphroditic flowers with abnormal pistils producing seeds with poorly developed pericarps that appear almost as naked seeds, segregate for males and females. By contrast, self-fertilization of highly female monoecious plants produces female and/or monoecious progenies but never male progeny. These results suggest that male plants are heterogametic (XY) and females are homogametic in nature (XX). When female plants (XX) are pollinated with the pollen of YY plants from self-fertilized male plants, all-male progenies are produced (Janick and Stevenson, 1954). Examination of sex expression in various synthetic polyploids demonstrated that a single dose of Y causes the plant to be male regardless of the number of X chromosomes present (Janick and Stevenson, 1955c; Janick, 1956). Therefore, spinach has an active-Y system of sex determination rather than an X:autosome system. Trisomic analysis has placed the sex-determining allelic pair named X/Y on the longest chromosome pair (Ellis and Janick, 1960). Ellis and Janick (1960) showed that the chromosomes carrying the X and Y factors do not differ from each other morphologically. However, heteromorphic sex chromosomes were produced utilizing translocations on either end of chromosome 1 (Iizuka and Janick, 1966). A recent study using fluorescence in situ hybridization suggested that sex chromosomes may be differentiated by the loss of a 45S rDNA locus from Y-bearing members (Lan et al., 2006), while our fluorescence in situ hybridization analysis detected the same number of 45S rDNA loci in male and female plants of the spinach cultivar MAZERAN (TAKII Seed Co., Kyoto, Japan; unpublished data). The copy number of 45S rDNA on the chromosome may vary without relation to sex, as intraspecific copy number variation of ribosomal RNA genes has been observed in a wide range of plant species (Rogers and Bendich, 1987).
Janick and Stevenson (1955b) proposed, based on genetic analysis of a true-breeding monoecious line derived from the cultivar Nobel, that expression of the monoecious character is conditioned by a single, incompletely dominant factor named Xm, which is allelic to the gene for dioecism, X/Y. However, this single major factor alone is insufficient to explain completely the continuous range of monoecious types that are observed. Several lines of evidence indicate the involvement of (an) additional allele(s) and/or modifier(s) in monoecism (Janick and Stevenson, 1955b; Onodera et al., 2008, 2011). However, nothing further is known about monoecism in spinach, and the molecular basis of sex determination has not yet been described.
The sex-determining locus was previously localized to linkage group 3 of the spinach molecular map (Khattak et al., 2006). Onodera et al. (2011) subsequently developed 10 AFLP markers closely linked to Y, and AFLP markers and two known male-specific sequence-characterized amplified region (SCAR) markers (Akamatsu et al., 1998) were used to construct a high-density linkage map covering the X/Y locus. Our recent study showed that, as determined by Janick and Stevenson (1955b), a single, incompletely dominant gene is responsible for the monoecious character of breeding line 03–336 (Onodera et al., 2011), which was found to exhibit the most highly male monoecious condition among the true-breeding monoecious lines that we examined previously (Onodera et al., 2008 and unpublished data). However, when a male plant from a dioecious line (03–009) was crossed with an F1 monoecious hybrid of the cross between a monoecious plant from 03–336 and a female plant from 03–009, a large difference was found between the recombination frequencies of microsatellite marker SO4 with the male-determining gene (Y) and the monoecious gene; the recombination frequency between Y and SO4 is 1.6%, whereas recombination of the monoecious gene with SO4 occurs at a frequency of 12.6%. This finding led us to assume that these genes are located at different loci on the chromosomal region. However, this notion has not yet been confirmed by molecular evidence.
In this study, to narrow down the location of the monoecious gene, we developed 19 AFLP markers linked to the gene and constructed a regional linkage map. The present linkage map was subsequently compared with the other regional linkage map that indicates the position of the X/Y locus (Onodera et al., 2011) to elucidate the relationship between chromosomal locations of the genes for dioecism and monoecism. Furthermore, test-cross experiments combined with the use of sex-linked DNA markers were conducted to determine whether the sex-determining genes are located at a single locus.
Materials and methods
Plant materials and genetic crosses
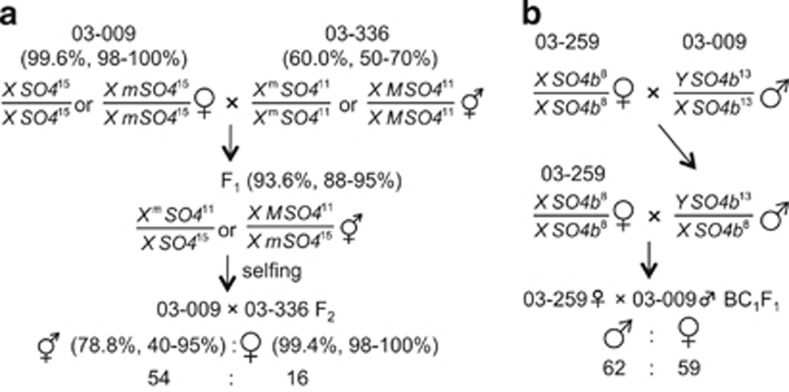
Seventy F2 plants from a cross between a female plant from dioecious line 03–009 and a plant from true-breeding monoecious line 03–336 were used to map the major factor determining the monoecious condition. This F2 population consisted of 54 monoecious and 16 female plants, which fit the Mendelian ratio 3:1 (Figure 1; Table 2 in Onodera et al., 2011). Femaleness classes (expressed as the index based on the percentage of female flowers per plant; Janick and Stevenson, 1955b) of the parental (03–009 and 03–336) F1 and F2 plants grown during the autumn–winter season of 2008/2009 are shown in Figure 1. F3 progeny plants from two F2 monoecious plants were grown in a growth chamber (LH-350S, Nippon Medical & Chemical Instruments Co. Ltd, Osaka, Japan) at 20 °C under an 8-h photoperiod during the first 3 weeks, at 25 °C under an 8-h photoperiod during the next three weeks and subsequently at 25 °C under a 24-h photoperiod.
Figure 1.
Crossing scheme to generate populations for mapping the monoecious gene (Xm or M) and the male-determining factor, Y. (a) 03–009 × 03–336 F2 plants segregated for the monoecious gene. Index of femaleness (expressed as the percentage of female flowers per plant) for the parental, F1 and F2 plants grown during the autumn–winter season of 2008/2009 are given as mean with range in parentheses (Onodera et al., 2011). The symbol ‘Xm' or ‘M' is used to refer to the monoecious gene, depending on the assumption of the presence or absence of its allelism to the gene pair (X/Y) for dioecism. (b) 03–259♀ × 03–009♂ BC1F1 plants segregated for Y (Onodera et al., 2011). Genotypes at sex-linked SSR loci (SO4 and SO4b) located in a 5′ flanking region of the Nir gene (accession ID, X17031.1; Back et al., 1991) are also indicated. Alleles of SO4 and SO4b loci were named after numbers of the repeated-motif sequences (TTG and TCA, respectively).
Table 2. Distribution of femaleness exhibited by the parental lines (03–009 and 03–336), F1, and the test-cross populations.
| Number of plants in femaleness classes (%) a | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Population
|
100
|
>95
|
>90
|
>75
|
>50
|
>25
|
>5
|
>0
|
0
|
Total
|
χ2
(1:1)b |
| Grown in 2009 | |||||||||||
| 03–009 | 4 | 4 | 8 | ||||||||
| 03–336 | 8 | 8 | |||||||||
| 03–009♀ × 03–336 F1 | 5 | 3 | 4 | 12 | |||||||
| Test-cross population Ac | 15 | 4 | 38 | 45 | 3 | 75 | 180 | 5.00d | |||
| Grown in 2010 | |||||||||||
| 03–009 | 10 | 14 | 24 | ||||||||
| 03–336 | 3 | 3 | 6 | ||||||||
| 03–009♀ × 03–336 F1 | 1 | 3 | 2 | 6 | |||||||
| Test-cross population Ac | 11 | 3 | 6 | 31 | 31 | 18 | 1 | 134 | 235 | 4.634d | |
| Test-cross population Bc | 7 | 1 | 3 | 23 | 32 | 5 | 70 | 141 | 0.007e | ||
| Grown in 2009+2010 | |||||||||||
| Test-cross population Ac | 26 | 3 | 6 | 35 | 69 | 63 | 4 | 209 | 415 | 0.022e | |
Femaleness classes are expressed as the percentage of female flowers per plant (Janick and Stevenson, 1955a).
χ2 test for a 1:1 ratio of male (0% femaleness class) to other sex types (monoecious (>0–>95% femaleness classes) and female (100% femaleness class)).
See MATERIALS AND METHODS for details.
Significant at the 0.01 level.
Significant at the 0.05 level.
A total of 121 backcross progeny plants (BC1F1) generated by a cross between all-female (true-breeding highly female monoecious) line 03–259 and dioecious line 03–009 (Onodera et al., 2011) were also used to map the male-determining genetic factor (Y). This population consisted of 62 male (0% female) and 59 female (>98% female) plants, which fit a ratio of 1:1 (Figure 1; Table 1 in Onodera et al., 2011). For the backcross, line 03–259 was used as the recurrent parent.
Table 1. SCAR markers derived from AFLP loci linked to the monoecious gene and Y.
| Marker | Source sequences for marker design | Polymorphism | Length of variant fragments (bp) 03–259/03–336/03–009 male/female |
|---|---|---|---|
| SP_0007 | E21M64a | Codominant | 161/161/179/179 |
| SP_0008 | 009_79-21Pb | Codominant | 271/271/274/274 |
| SP_0016 | 009_5-13Jb | Codominant | 336/336/401/401 |
| SP_0017 | 009_6-10Kb | Dominant | —/—/49/49 |
| SP_0022 | 009_8-16Nb | Codominant | 635/268/635/635 |
An AFLP marker developed in a previous study (Onodera et al., 2011).
BAC clones 009_79-21P, 009_5-13J, 009_6-10K and 009_8-16N were identified as positive for AFLP markers E28M48 (Onodera et al. 2011), E28M20-2, E27M31 and E28M31 (the present study), respectively.
Two test-cross populations (designated Test-cross population A and B) were prepared for allelism analysis of the genes for dioecism and monoecism. Test-cross population A was initiated by crossing a plant from 03–336 with a male plant from 03–009, and an F1 male plant was then used to pollinate a female plant from 03–009 (these crosses are labeled crosses 1 and 2 in Supplementary Figure 1). For Test-cross population B, a male plant was chosen from the family (03–009♀ × 03–336)-monoecious × 03–009♂ (crosses 3 and 4 in Supplementary Figure 1; see also Onodera et al., 2011). In test-cross B, segregation of the SSR marker SO4, which is linked to both the X/Y gene and the gene for monoecism (Onodera et al., 2011), was followed. In these families, the male-determining factor Y and the SO415 allele showed coupling, while the monoecious gene was associated with the SO411 allele, allowing a heterozygous SO415/ SO411 male plant to be chosen, which was also likely to be heterozygous for the gene for monoecy. This plant was test-crossed with a female plant from the 03–009 line (Cross 5, Supplementary Figure 1). Test-cross populations A and B were grown outdoors during two summer seasons (April–July of 2009 and 2010) and a single summer season (April–July 2010), respectively.
AFLP analysis
AFLP analysis was carried out according to the method of Vos et al. (1995) with the following modifications: the digestion–ligation reaction was prepared in a total volume of 11 μl containing 1 μl of 10 × T4 Ligation buffer with ATP (New England BioLabs, Ipswich, MA, USA), 1 μl of 0.5 M NaCl, 0.5 μl of 1.0 mg ml−1 BSA, 0.1 μM EcoRI-Adapter, 1 μM MseI-Adapter, 1 unit EcoRI (New England BioLabs), 5 units MseI (New England BioLabs), 67 cohesive end units T4 DNA Ligase (New England BioLabs) and 0.5 μg DNA. The reaction was incubated for 2 h at 37 °C. The adaptor-ligated DNA was pre-amplified using primers with a single selective nucleotide. Selective amplification was performed using fluorescent dye-labeled primers (6-FAM, VIC and NED; Life Technologies, Carlsbad, CA, USA) with three nucleotide extensions (Supplementary Table 1). The PCR conditions for pre-amplification and selective amplification from the original method were employed (Vos et al., 1995). The products of selective amplification were run on an Applied Biosystems 3130 Genetic Analyzer (Life Technologies) and the obtained raw data files were analyzed with GeneMapper (Life Technologies). The AFLP markers obtained were named after the restriction enzymes used (E and M for EcoRI and MseI, respectively), followed by the code number assigned to the three-nucleotide extension (Supplementary Table 1; Onodera et al., 2011).
Bulked segregant analysis with AFLPs was performed to isolate sex-linked markers. The bulks were composed of pre-amplified AFLP templates from eight individual plants of the same sex, that is, monoecious or female, and were used for the subsequent selective amplification.
Development of SCAR and SSR markers used for linkage analysis
SCAR markers were designed from DNA sequences of sex-linked AFLP marker loci identified in a previous study (Onodera et al., 2011) and in the present study. The DNA sequence of AFLP marker locus E21M64 (Supplementary Table 2; Onodera et al., 2011) was directly determined from the AFLP products. The AFLP products were run on a polyacrylamide gel consisting of a stacking gel (6%, pH 6.8) and a separation gel (13%, pH 8.8) as described by Hori et al. (2003). The AFLP-gel was stained with 1 μl ml−1 SYBR Green I (Lonza, Basel, Switzerland) and the target AFLP band was then excised from the gel. The DNA fragments eluted from the gel piece were subjected to PCR with selective amplification primers appropriate for the target band. The amplified fragments were directly sequenced with an Applied Biosystems 3130 Genetic Analyzer.
DNA sequence information for AFLP marker loci, E28M48 (Supplementary Table 2; Onodera et al. 2011), E28M20-2, E27M31, and E28M31 (Supplementary Table 3) was obtained from BAC clones (Table 1). An arrayed genomic BAC library of 03–009 males (unpublished) was screened with the AFLP markers. The isolated BAC clones were used to determine the sequences of the markers and their flanking regions.
Primer pairs complementary to the five AFLP marker loci were designed for five SCAR markers (Table 1) and tested to see whether they amplified DNA fragments polymorphic between the parental lines (03–259, 03–336, and 03–009) of the mapping populations. SP_0007 produced a codominant variant between 03–259 and 03–009 and between 03–336 and 03–009 (Table 1 and Supplementary Figure 2). SP_0008 and SP_0016 produced codominant variants, while SP_0017 produced a dominant variant between 03–259 and 03–009 and between 03–336 and 03–009. In addition, SP_0022 gave a codominant variant only between 03–336 and 03–009 but no variant between 03–259 and 03–009. Sequence information for the SCAR marker primers is available upon request from Y Onodera (onodera@abs.agr.hokudai.ac.jp).
Primers for SSR marker SO4b (5′-CCACACTCCCTCACCGTTTGTC-3′ and 5′-GGTTGCTCTGTTTCTTGCAACTATG-3′) were designed based on the 5′ flanking sequence of the S. oleracea Nir gene (GenBank ID X17031) (Back et al., 1991) for nitrite reductase, which is linked to the sex-determining locus (Khattak et al., 2006; Onodera et al., 2011) (Supplementary Figure 3). The primer set successfully amplified variant alleles in the parental lines of the backcross population (03–259♀ × 03–009♂ BC1F1), and the allele from 03–009 was found to be linked in coupling phase to maleness.
Genotyping with SSR and SCAR markers
To genotype the mapping populations, PCR was carried out using primers for the SSR and SCAR markers developed in the present study. Amplification reactions were performed in total volumes of 10 μl containing 10 ng of template DNA, 0.2 μM of each primer, 0.2 mM of each dNTP and 5 μl of Go Taq Master Mix (Promega Corporation, Madison, WI, USA). Amplification was performed for 30 cycles after initial denaturation at 94 °C for 2 min. Each cycle consisted of 15 s at 94 °C, 30 s at the annealing temperature and 1 min at 72 °C, followed by 5 min at 72 °C prior to gel electrophoresis analysis.
SSR marker SO4 (Khattak et al., 2006) and male-specific DNA markers T11A and V20A (accession numbers E15132 and E15133, respectively) (Akamatsu et al., 1998) were also used for genotyping. Amplification reactions for the markers were performed as described by Onodera et al. (2008, 2011).
Map construction
The Mendelian segregation ratio for each marker used in the linkage analysis was first verified in the mapping populations. Linkage analysis of the markers was carried out using MAPMAKER/EXP3.0 (Lander et al., 1987) with a LOD threshold of 3.0. Map distances in centimorgans (cM) were calculated from recombination frequencies using the Kosambi function (Kosambi, 1944).
Results
Development and mapping of AFLP markers linked to the major factor determining the monoecious condition
In this study, gene notations follow the nomenclature proposed by Janick and Stevenson (1955b). X and Y refer to the gene pair for sex determination in dioecious lines, although these symbols are commonly used to refer to sex chromosomes. When referring to the major gene for monoecism, either the symbol Xm or M is used, depending on the assumption of the presence or absence of the gene's allelism to X/Y.
To find DNA markers linked to the monoecious gene (Xm or M) efficiently, DNAs from a 03–009♀ × 03–336 F2 population (Onodera et al., 2011) were subjected to bulked segregant analysis combined with the AFLP technique. Plants used for the female and monoecious bulks were selected for their femaleness and for their genotypes at sex-linked SSR locus SO4 (Onodera et al., 2011); eight female plants (100% femaleness) and eight highly male monoecious plants (40–70% femaleness) with SO415SO415 and SO411SO411 genotypes, respectively, were chosen from the F2 population. As in this population the Xm (or M) allele was closely linked in coupling to the SO411 allele, while X (or m) was closely linked to SO415, the chosen plants were most likely to be homozygous for the locus in question. Therefore, this bulked segregant analysis would likely reveal DNA markers linked in coupling and in repulsion phase to the monoecious gene (Xm or M).
A total of 126 combinations of EcoRI- and MseI-selective-primers were screened using the monoecious and female bulks (Supplementary Table 1). We obtained ∼9000 AFLP peaks in total, with a range of ∼20–100 per primer combination. Of these combinations, 47 produced 55 peaks that were polymorphic between the bulks. Of these peaks, 28 were unique to the monoecious bulk, whereas the remaining 27 peaks were unique to the female bulk (Supplementary Figure 4). These 47 primer combinations were used to genotype the individuals of the bulks separately. Finally, 14 combinations produced polymorphic peaks with high intensity and reproducibility and yielded 19 AFLP markers. Ten of the markers are linked to the monoecious character in the coupling phase, while the remaining nine are linked to this character in the repulsion phase (Supplementary Table 3).
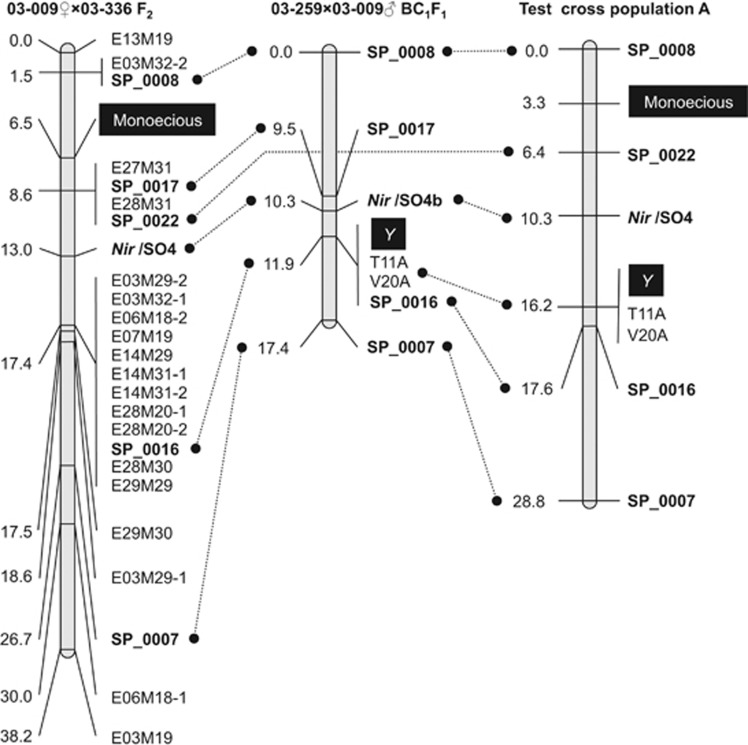
Using 70 F2 families of 03–009♀ × 03–336 (Figure 1; Onodera et al., 2011), the 19 AFLP markers and SO4 were mapped to a 38.2-cM region including the monoecious gene (Xm or M), with an average distance of 2.1 cM (ranging from 0–8.1 cM) between adjacent markers (Figure 2). However, more than half of the AFLP markers were mapped 17.4 cM from the origin of the map and formed a cluster in this small region. Based on the linkage map, the monoecious gene was located in a 7.1-cM interval between AFLP markers E03M32-2 and E27M31/E28M31. It is worth noting that E27M31 and E28M31 co-segregated completely in the 70 F2 plants.
Figure 2.
Genetic linkage maps of the chromosomal region covering the sex-determining genes. The linkage maps were constructed from three different populations, 03–009 × 03–336 F2, 03–259 × 03–009 BC1F1 and Test-cross population A (see Supplementary Figure 1). AFLP markers are indicated by ‘E' and ‘M' from the EcoRI/MseI selective primer pair (for example, E16M19) and SCAR markers are denoted by the prefix ‘SP'. In addition to the markers, SSR (SO4 and SO4b) on the 5′-flanking region of the Nir gene (Back et al., 1991) and male-specific DNAs (T11A and V20A; (Akamatsu et al., 1998) were also used for construction of the maps. Marker positions (cM) and names are written on the left and right sides of the map, respectively. Note that the linkage orders of SP_0008, Nir/SO4/SO4b, SP_0016, SP_0022 and SP_0007 are conserved across the populations.
Comparison of the two regional maps locating the positions of the loci for dioecism and monoecism
In our previous study, a regional linkage map locating the position of the X/Y locus (but not the monoecious gene) was established using 121 BC1F1 plants from the cross of 03–259♀ × 03–009♂, segregating for males and females in a 1:1 ratio (Figure 1; Onodera et al., 2011). The map comprises 10 AFLP markers and two male-specific SCAR markers (T11A and V20A; Supplementary Table 2). However, as none of these markers (and none of the markers mapped in the F2 families of 03–009♀ × 03–336) were transferable across the two mapping populations, a positional relationship between the sex-determining genes (Y and Xm or M) still remained unclear. To help elucidate the linkage order of the markers and the sex-determining genes mapped in the two mapping populations (Figures 1 and 2, Supplementary Table 2), the two (E21M64 and E28M48) and three (E28M20-2, E27M31, and E28M31) AFLP markers mapped in the BC1F1 and F2 families, respectively, were first converted into five SCAR markers (Table 1 and Supplementary Figure 2) common to both of the populations.
As expected, all of the SCAR markers (except SP_0022) were available across both populations; however, SP_0022 was available only for one population (03–009♀ × 03–336 F2) due to the lack of a variant between 03–259 and 03–009 (see MATERIALS AND METHODS for details). Furthermore, as SSR marker SO4 (mapped in the 03–009♀ × 03–336 F2 population), which is located in the 5′ flanking region of the Nir gene (accession ID X17031.1) (Back et al., 1991), does not vary between 03–259 and 03–009, an additional SSR marker (SO4b) that enables mapping of the Nir loci using the backcross population was also designed (see Supplementary Figure 3 and MATERIALS AND METHODS for details).
Seventy F2 families of 03–009♀ × 03–336 were genotyped for the five SCAR markers. The SCAR markers and Nir/SO4 were mapped to a 26.7-cM chromosomal region, where the monoecious gene was located in a 7.1-cM region between SP_0008 and SP_0017/SP_0022 (Figure 2). As expected, the linkage order of SP_0016, SP_0017 and SP_0022 in relation to Nir/SO4 and the monoecious gene agreed with that of the corresponding AFLP markers (E28M20-2, E27M31 and E28M31) shown in the original linkage map (Figure 2). Notably, SP_0008 (which originated from E28M48) (Supplementary Table 2) co-segregated completely with E03M32-2 in the F2 population. BAC genomic clone 009_79–21P (isolated with E28M48) was also identified as positive for E03M32-2, suggesting that there is physical proximity between SP_0008/E28M48 and E03M32-2.
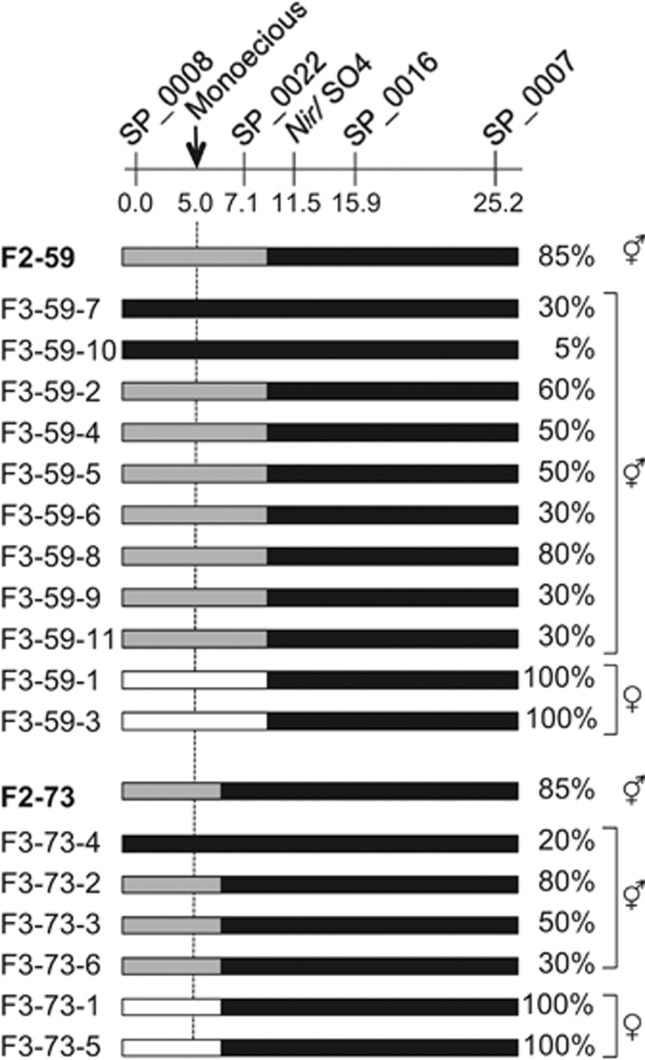
Two highly female monoecious plants (F2-59 and F2-73, both with 85% female flowers) chosen from the F2 plants were self-pollinated. As shown in Figure 3, F2-59 is heterozygous at the SP_0008 and SP_0022 loci but homozygous for 03–336 alleles at Nir/SO4, SP_0016 and SP_0007 and is most likely to be heterozygous for the monoecious gene. F2-73 is heterozygous at the SP_0008 locus but homozygous for 03–336 alleles at four other marker loci (SP_0022, Nir/SO4, SP_0016, and SP_0007); this plant may be either homozygous or heterozygous for the monoecious gene. The F3 families F2-59 and F2-73 (6 and 11 plants, respectively) both segregated for monoecious (5–80% female) and 100% female plants, suggesting that both of the F2 plants selected for test-crossing are heterozygous for the monoecious gene. As expected, the nine and four monoecious F3 plants (Figure 3) from F2-59 and F2-73, respectively, carried 03–336 alleles at the five marker loci. On the other hand, the female progenies (F3-59-1 and -3) from F2-59 did not have 03–336 alleles at SP_0022 and SP_0008 loci, but they carried the alleles at Nir/SO4, SP_0016 and SP_0007. Similarly, the female progenies (F3-73-1 and -5) from F2-73 were homozygous for 03–009 alleles at the SP_0008 locus, but carried 03–336 alleles at the SP_0022, Nir/SO4, SP_0016 and SP_0007 loci in the homozygous state. Taken together, the results from the analysis of the F2 and F3 progenies suggest that the monoecious gene is located in the interval between SP_0008 and SP_0022 rather than between SP_0022 and SP_0007.
Figure 3.
Graphical genotypes and femaleness of F3 families derived from two selections (F2-73 and F2-59) in 03–009 × 03–336 F2. Black bars represent homozygous 03–336 segments, gray bars indicate heterozygous 03–009/03–336 regions, and white bars represent homozygous 03–009 segments. F3 families are denoted by the prefix ‘F3'. Indexes of femaleness (expressed as the percentage of female flowers per plant) are indicated to the right of the bars. Marker positions (cM) and names are indicated above the bars.
Next, 121 BC1F1 progeny plants from the cross of 03–259♀ × 03–009♂ were genotyped for the markers SP_0007, SP_0008, SP_0016, SP_0017 and Nir/SO4b. These markers, as well as two male-specific markers (T11A and V20A), mapped to a 17.4-cM chromosomal region including the X/Y locus (Figure 2). Notably, SP_0016, along with T11A and V20A, co-segregated with Y in this mapping population. The linkage order of SP_0007 and SP_0008 relative to the male-specific markers and the X/Y locus was consistent with that of the corresponding AFLP markers (E21M64 and E28M48) in the previously reported linkage map (Supplementary Table 2; Onodera et al., 2011).
As shown in Figure 2, across the two mapping populations, the linkage order of the common markers SP_0008, SP_0017, Nir (SO4/SO4b), SP_0016, and SP_0007 was conserved. However, the chromosomal region covered by the markers includes the loci for dioecism (X/Y) and monoecism (Xm or M), which were mapped to different marker intervals; Y is located between Nir and SP_0007, while the monoecious gene was placed between flanking markers SP_0008 and SP_0017. The mapping data suggest that X/Y and the monoecious gene are more likely to be located at different loci in the chromosomal region than at an identical locus.
Allelism test between the genes for dioecism and monoecism
As in a previous study (Janick and Stevenson, 1955b), we analyzed two sets of sibships (designated as Test-cross population A and B; see Supplementary Figure 1 and MATERIALS AND METHODS for details) from crosses between the dioecious line (03–009) and the true-breeding monoecious line (03–336) to test two hypotheses: (1) that the monoecious gene is an allele of X/Y and (2) that the monoecious gene is not allelic but is linked to X/Y. According to the nomenclature used in the previous study, based on the first hypothesis, the genotype of 03–336 is XmXm and that of 03–009 is XY (male) or XX (female) (Onodera et al., 2011). Based on the second hypothesis, the genotype of 03–336 is XXMM and that of 03–009 is XYmm (male) or XXmm (female). Prior to presenting the following results, it is also necessary to mention that, assuming the first hypothesis is correct, Y is dominant to Xm and Xm is incompletely dominant to X (Janick and Stevenson 1955b; Onodera et al., 2011). Based on the second hypothesis, M would be the gene with incompletely dominant action and Y and M would be epistatic to M and X, respectively (Onodera et al., 2011).
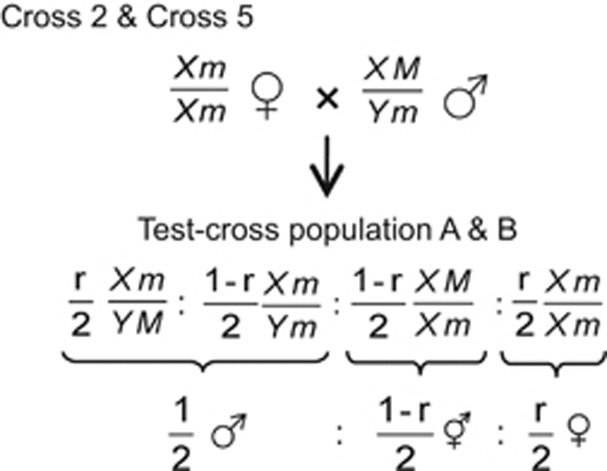
As shown in Figure 4, if the monoecious gene and X/Y are located on the identical locus (the first hypothesis), no recombination could occur between the genes (recombination frequency, r=0); thus, male and monoecious would segregate in equal proportions in Test-cross population A. On the other hand, if the monoecious gene is not allelic but is linked to X/Y (the second hypothesis), recombination could occur between the genes (0.5>r>0), and subsequently the test-cross (Cross 2, Figure 4 and Supplementary Figure 1) would produce progenies segregating for all three types (male, monoecious and female).
Figure 4.
Expected sex ratio in Test-cross population A and B. Assuming X/Y and M/m are located on an identical locus (no recombination occurs between X/Y and M/m; r=0), both test-cross populations can segregate at a ratio of one male to one monoecious. Alternatively, if X/Y and M/m are located on different loci (0<r<0.5), the crosses (Cross 2 and Cross 5) would give progenies segregating into three sex forms, that is, male, monoecious and female. See Supplementary Figure 1 for details of Cross 2 and Cross 5.
Part of Test-cross population A (180 individuals) was grown and evaluated for femaleness in 2009. The remaining portion of the progenies (235 individuals) was grown in 2010. In both years, the parental lines and their F1 plants (03–336 × 03–009♀) were also analyzed as control samples. Segregation ratios for the first and second sets of the test-cross populations are shown in Table 2. In both cases, all three types, including male (0% femaleness class; Table 2), monoecious (>0–>95% femaleness classes) and female (100% femaleness class) segregated in the population, with male plants accounting for approximately half of the population and with the monoecious segregants outnumbering females. The monoecious plants from 03–336, 03–336 × 03–009♀F1 and Test-cross population A grown in 2010 tended to have higher values for femaleness than those grown in 2009, as shown in Table 2. The variation between the years may have been due to an environmental effect, because sex expression in monoecious plants is readily influenced by temperature and photoperiod conditions (Janick and Stevenson, 1955a; Onodera et al., 2008). It should also be mentioned that T11A completely co-segregated with Y in the 415 individuals examined to date (data not shown). These results support the second hypothesis, which states that the monoecious gene is not allelic but is linked to X/Y. Based on the segregation ratio, the recombination frequency between M and Y was calculated to be 12.6±2.3%.
Like Test-cross population A, Test-cross population B also segregated for all three types, that is, male (0% femaleness class), monoecious (>0–>95% femaleness classes) and female (100% femaleness class), with male plants accounting for approximately half of the population and with the monoecious segregants outnumbering females. Moreover, T11A completely co-segregated with Y in the 141 individuals examined to date (data not shown). As shown in Figure 4, these results can be explained using the second hypothesis that the monoecious gene (M) is not allelic but is linked to X/Y. Based on the sex ratio of the population, the recombination frequency between M and Y was calculated to be 9.9±3.5%.
To locate both M and Y in relation to the four sex-linked SCAR markers (SP_0007, SP_0008, SP_0016 and SP_0022), Nir/SO4 and two male-specific markers (T11A and V20A), Test-cross population A and B, were genotyped for the markers. As shown in Figure 2 and Supplementary Figure 5, the linkage order of all seven markers and the genes in question was consistent across the two populations; T11A and V20A co-segregate with Y, which is closely or completely linked to SP_0016 and is located between Nir/SO4 and SP_0007, and M was mapped between SP_0008 and SP_0022, illustrating that the genes for dioecism and monoecism are not allelic but are linked to each other. It is also worth mentioning that the results are also consistent with the mapping data described in the preceding section; the linkage order of SP_0007, SP_0008, SP_0016, and SP_0022/SP_0017 and Nir was conserved across all four populations examined in the present study, and the locations of Y and M relative to the markers were also consistent among the populations (Figure 2 and Supplementary Figure 5).
Discussion
In this study, the genes for dioecism (Y) and monoecism (M) were located at different loci in a chromosomal region based on allelism tests, and mapping and linkage analysis were performed with the help of molecular markers. This is the first report to provide molecular evidence for the positional relationship between the genes for monoecism and dioecism in spinach. Janick and Stevenson (1955b) previously reported that the monoecious character in spinach is controlled by one major gene, Xm, which is an allele of the X/Y factor pair. This conclusion differs slightly from the current results. This discrepancy may be due to a difference in the observed recombination frequencies of the two sex-determining loci (X/Y and M/m) across the segregating populations used for the present versus the previous study. The limited sizes of the populations may lead to a bias that would produce different observed recombination frequencies, as different recombination estimates between populations have been reported in many crops, for example, grapevine (Doligez et al., 2006) and apple (N'Diaye et al., 2008). However, there may be heterogeneous recombination rates of the two sex-determining loci (X/Y and M/m) between the populations. Significant differences in recombination rates have also been observed in some plant species (Fatmi et al., 1993; Doligez et al., 2006). Furthermore, we cannot exclude the possibility that the monoecious gene examined in the present study is not identical to that observed in the previous study; confirmation of this difference awaits further study.
Recent breeding programs for hybrid spinach have tended to favor the use of the highly male monoecious lines as pollen parents over dioecious lines, as continuous self-fertilizations enabled by the monoecious condition enable the efficient production of highly homozygous lines that breed true for favorable traits. The line 03–336 used in this study bears a (relatively) high proportion of male flowers and is, therefore, highly useful as a source of the desirable monoecious character. The monoecious character was conditioned by M, which was mapped to a 7.1-cM interval between the codominant markers SP_0008 and SP_0022. If 100 F2 plants were selected using both SP_0008 and SP_0022, most of them would be expected to have the MM genotype, with a bit error rate of ∼1% (see Koh et al., 1996). Using this marker set may serve as an efficient alternative to traditional phenotypic evaluation for this character.
Most angiosperm species are co-sexual (monoecious and hermaphrodite), whereas dioecy, found in only 6% of flowering plants, is taxonomically distributed among >40% of angiosperm families (Renner and Ricklefs, 1995; Charlesworth, 2002, 2013). This low representation of dioecy suggests that the co-sexual condition is the ancestral sexual system in angiosperms, whereas dioecy evolved from the ancestral system both repeatedly and recently. This holds true for dioecy in subfamily Chenopodioideae, to which spinach belongs. In this subfamily, the co-sexual condition is the predominant sexual system and dioecy is scattered among several tribes, suggesting the recent and multiple origin of dioecy in this subfamily (Kadereit et al., 2010; Fuentes-Bazan et al., 2012). In this context, it is reasonable to assume that sex chromosomes (XY) in spinach may have evolved recently from autosomes in an ancestral species with the co-sexual condition.
Heteromorphic sex chromosome pairs (XY) have been found in male plants of white campion (Silene latifolia), hemp (Cannabis sativa) and sorrel (Rumex acetosa). However, it is not unusual for homomorphic sex chromosome pairs to be observed in dioecious angiosperms such as spinach, asparagus (Asparagus officinalis), papaya (Carica papaya) and poplar (Populus trichocarpa) (Ming et al., 2011). It is generally accepted that the evolution of heteromorphic sex chromosomes involves the suppression of recombination around a sex determination locus on homomorphic proto-sex chromosomes and the subsequent expansion of a non-recombination region associated with genetic degradation (Bergero and Charlesworth, 2009; Bachtrog, 2011). The lethality of YY genotypes was reported in several dioecious plants (white campion and papaya), suggesting that their Y chromosomes had already undergone genetic degradation. By contrast, YY genotypes are viable in spinach and asparagus. In these plants, Y may preserve the genetic integrity but may differ from X only in its sex determination genes (Janick and Stevenson, 1954; Ming et al., 2011). It is reasonable to assume that in spinach and asparagus, sex chromosomes may represent the most recent evolutionary state, considering the homomorphism and the widespread occurrence of recombination in their sex chromosomes (Ellis and Janick, 1960; Khattak et al., 2006; Telgmann-Rauber et al., 2007; Onodera et al., 2011).
A notable feature of spinach is that examining the monoecious gene can provide clues to the evolutionary pathway to dioecy. A recent phylogenetic study revealed that Spinacia species are placed as a sister lineage to the taxon comprising polygamo-monoecious species with both hermaphrodite and unisexual flowers on the same plant (Kadereit et al., 2010). Given that cosexuality is the ancestral sex form in the taxon, based on the genetic model of Charlesworth and Charlesworth (1978), dioecy in spinach can be assumed to have evolved via at least two mutations at closely linked loci, that is, one mutation (male-sterility) giving females and the other increasing the maleness (that is, suppressing the femaleness) of the ancestral co-sexual form. These mutations may have provided a proto-male-determining factor, which produced ‘initial males (or inconstant males)' with a monoecious condition to some degree. Next, an additional mutation(s) at the ‘monoecious gene' may have produced an allele that promotes maleness, making the initial males have a fully (or almost fully) male phenotype with a low frequency of female flowers. If this is the case, it may be reasonable to assume that the sex form brought about by M represents the ‘initial male', because M functions as a male-promoting/female-suppressing factor, while it is less effective than Y. The monoecious character in 03–336 is inherited in an incompletely dominant manner (Onodera et al., 2008), and an increase in M dosage brings about highly male monoecious conditions (shift towards maleness); MM plants tended to exhibit lower femaleness than Mm plants in the 03–009 × 03–336 F2 population used in the present study (Haseda et al., in preparation). When grown in the summer, 03–336 plants often exhibit a phenotype close to that of male plants and produce fewer female flowers than are observed in the winter season (Onodera et al., 2008), suggesting that M can function almost equivalently to Y at high temperatures. Nevertheless, based on our current results, M cannot simply be interpreted as an ancestral allele of Y; instead, M can be assumed to represent a ‘relic' of the putative ancestral Y allele, which has arisen by duplication, as linked duplicated genes are prevalent in plant genomes (Messing et al., 2004; Moore and Purugganan, 2005).
A remarkable finding of the current study is that the male-specific DNA markers T11A and V20A (Akamatsu et al., 1998) completely co-segregated with Y in the three populations segregating for the gene (677 plants in total). Akamatsu et al. (1998) also observed that there was no recombinant plant between the male-specific markers and Y in 667 progeny plants from a sib-cross of dioecious spinach cultivar Atlas (Sakata Seed Corporation, Yokohama, Japan). The lack of observed recombination (r=0.00–0.22%, 95% CI) may signify severe or complete recombination suppression around the sex-determining locus, as is widely observed in sex chromosomes in other dioecious plants. This notion seems reasonable, as dioecy in angiosperms must have evolved from cosexuality through at least two mutations, which should be closely linked, and suppressed recombination between these loci is favorable for the maintenance of a stable dioecious population (Charlesworth, 2002, 2013).
The AFLP-based linkage map constructed in this study located only one (M) of the sex-determining loci. However, AFLP marker E28M20-2 was converted into SCAR marker SP_0016, which was completely or tightly linked to Y in the three populations; this marker most likely represents a site very close to the X/Y locus. Interestingly, E28M20-2 was mapped to a marker cluster region, suggesting that the sex-determining locus may be located in the vicinity of a centromere, because association of marker-clusters with centromeric regions has been reported in many plants, that is, tomato, potato (Tanksley et al., 1992) and maize (Castiglioni et al., 1999). Indeed, the proximity of the sex-determining region to the centromere was found in the Y chromosome of papaya (Yu et al., 2007).
In conclusion, the series of spinach molecular linkage maps constructed in our present and previous studies, along with spinach genomic BAC libraries (unpublished), may be helpful for future studies focusing on cloning the spinach sex-determining loci as well as identifying the chromosomal location of the loci by fluorescence in situ hybridization analysis, which, in turn, would further elucidate the evolutionary relationship between dioecism and monoecism in spinach and molecular features of the sex chromosomes at the most recent evolutionary stage.
Data archiving
Data available from the Dryad Digital Repository: doi:10.5061/dryad.155df.
Acknowledgments
We are grateful to Tohoku Seed Co., Ltd. (Utsunomiya, Tochigi, Japan) for providing the spinach breeding lines used in this study. We appreciate the technical assistance provided by Mrs H Yokomoto. This work was supported in part by JSPS KAKENHI Grant Number 23580001, MEXT KAKENHI Grant Number 221S0002, Suhara Memorial Foundation and Takeda Science Foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Akamatsu T, Suzuki T, Uchimiya H.1998. Sakata No Tane:KK: Japan.
- Bachtrog D. Plant sex chromosomes: a non-degenerated Y. Curr Biol. 2011;21:R685–R688. doi: 10.1016/j.cub.2011.08.027. [DOI] [PubMed] [Google Scholar]
- Back E, Dunne W, Schneiderbauer A, Deframond A, Rastogi R, Rothstein S. Isolation of the spinach nitrite reductase gene promoter which confers nitrate inducibility on GUS gene expression in transgenic Tobacco. Plant Mol Biol. 1991;17:9–18. doi: 10.1007/BF00036801. [DOI] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24:94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Castiglioni P, Ajmone-Marsan P, van Wijk R, Motto M. AFLP markers in a molecular linkage map of maize: codominant scoring and linkage group distribution. Theoret Appl Genet. 1999;99:425–431. doi: 10.1007/s001220051253. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. Model for evolution of dioecy and gynodioecy. Am Nat. 1978;112:975–997. [Google Scholar]
- Charlesworth D. Plant sex determination and sex chromosomes. Heredity. 2002;88:94–101. doi: 10.1038/sj.hdy.6800016. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Plant sex chromosome evolution. J Exp Bot. 2013;64:405–420. doi: 10.1093/jxb/ers322. [DOI] [PubMed] [Google Scholar]
- Doligez A, Adam-Blondon A, Cipriani G, Laucou V, Merdinoglu D, Meredith C, et al. An integrated SSR map of grapevine based on five mapping populations. Theoret Appl Genet. 2006;113:369–382. doi: 10.1007/s00122-006-0295-1. [DOI] [PubMed] [Google Scholar]
- Ellis JR, Janick J. The chromosomes of Spinacia oleracea. Am J Bot. 1960;47:210–214. [Google Scholar]
- Fatmi A, Poneleit C, Pfeiffer T. Variability of recombination frequencies in the Iowa stiff stalk synthetic (Zea mays L) Theoret Appl Genet. 1993;86:859–866. doi: 10.1007/BF00212613. [DOI] [PubMed] [Google Scholar]
- Fuentes-Bazan S, Mansion G, Borsch T. Towards a species level tree of the globally diverse genus Chenopodium (Chenopodiaceae) Mol Phylogenet Evol. 2012;62:359–374. doi: 10.1016/j.ympev.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Hori K, Kobayashi T, Shimizu A, Sato K, Takeda K, Kawasaki S. Efficient construction of high-density linkage map and its application to QTL analysis in barley. Theoret Appl Genet. 2003;107:806–813. doi: 10.1007/s00122-003-1342-9. [DOI] [PubMed] [Google Scholar]
- Iizuka M, Janick J. The synthesis of heteromorphic sex chromosomes in spinach. J Hered. 1966;57:182–184. [Google Scholar]
- Janick J. Inheritance of Sex in tetraploid spinach. Proc Am Soc Hortic Sci. 1956;66:361–363. [Google Scholar]
- Janick J.1998Hybrids in horticultural cropsIn: Lamkey KR, Staub JE (eds)Concepts and Breeding of Heterosis in Crop Plants Crop Science Society of America; Crop Science Society of America Special Publication pp45–56. [Google Scholar]
- Janick J, Stevenson E. A genetic study of the heterogametic nature of the staminate plant in spinach. Proc Am Soc Hortic Sci. 1954;63:444–446. [Google Scholar]
- Janick J, Stevenson E. Environmental influences on sex expression in monoecious lines of spinach. Proc Am Soc Hortic Sci. 1955;65:416–422. [Google Scholar]
- Janick J, Stevenson E. Genetics of the monoecious character in spinach. Genetics. 1955;40:429–437. doi: 10.1093/genetics/40.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janick J, Stevenson E. The effects of polyploidy on sex expression in spinach. J Hered. 1955;46:150–156. [Google Scholar]
- Kadereit G, Mavrodiev E, Zacharias E, Sukhorukov A. Molecular phylogeny of Atripliceae (Chenopodioideae, Chenopodiaceae): implications for systematics, biogeography, flower and fruit evolution, and the origin of C-4 photosynthesis. Am J Bot. 2010;97:1664–1687. doi: 10.3732/ajb.1000169. [DOI] [PubMed] [Google Scholar]
- Khattak J, Torp A, Andersen S. A genetic linkage map of Spinacia oleracea and localization of a sex determination locus. Euphytica. 2006;148:311–318. [Google Scholar]
- Koh HJ, Heu MH, McCouch SR. Molecular mapping of the ge(S) gene controlling the super-giant embryo character in rice (Oryza sativa L) Theoret Appl Genet. 1996;93:257–261. doi: 10.1007/BF00225754. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distance from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Lan T, Zhang S, Liu B, Li X, Chen R, Song W. Differentiating sex chromosomes of the dioecious Spinacia oleracea L. (spinach) by FISH of 45S rDNA. Cytogenet Genome Res. 2006;114:175–177. doi: 10.1159/000093335. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, et al. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations (vol 1 pg 174, 1987) Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Messing J, Bharti A, Karlowski W, Gundlach H, Kim H, Yu Y, et al. Sequence composition and genome organization of maize. Proc Natl Acad Sci USA. 2004;101:14349–14354. doi: 10.1073/pnas.0406163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, Bendahmane A, Renner S, Merchant S, Briggs W, Ort D. Sex chromosomes in land plants. Ann Rev Plant Biol. 2011;62:485–514. doi: 10.1146/annurev-arplant-042110-103914. [DOI] [PubMed] [Google Scholar]
- Moore R, Purugganan M. The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol. 2005;8:122–128. doi: 10.1016/j.pbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- N'Diaye A, de Weg W, Kodde L, Koller B, Dunemann F, Thiermann M, et al. Construction of an integrated consensus map of the apple genome based on four mapping populations. Tree Genet Genomes. 2008;4:727–743. [Google Scholar]
- Onodera Y, Yonaha I, Masumo H, Tanaka A, Niikura S, Yamazaki S, et al. Mapping of the genes for dioecism and monoecism in Spinacia oleracea L.: evidence that both genes are closely linked. Plant Cell Rep. 2011;30:965–971. doi: 10.1007/s00299-010-0998-2. [DOI] [PubMed] [Google Scholar]
- Onodera Y, Yonaha I, Niikura S, Yamazaki S, Mikami T. Monoecy and gynomonoecy in Spinacia oleracea L.: morphological and genetic analyses. Sci Hortic. 2008;118:266–269. [Google Scholar]
- Renner S, Ricklefs R. Dioecy and its correlates in the flowering plants. Am J Bot. 1995;82:596–606. [Google Scholar]
- Rogers S, Bendich A. Ribosomal-RNA genes in plants—variability in copy number and in the intergenic spacer. Plant Mol Biol. 1987;9:509–520. doi: 10.1007/BF00015882. [DOI] [PubMed] [Google Scholar]
- Tanksley S, Ganal M, Prince J, Devicente M, Bonierbale M, Broun P, et al. High-density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telgmann-Rauber A, Jamsari A, Kinney MS, Pires JC, Jung C. Genetic and physical maps around the sex-determining M-locus of the dioecious plant asparagus. Mol Genet Genomics. 2007;278:221–234. doi: 10.1007/s00438-007-0235-z. [DOI] [PubMed] [Google Scholar]
- van der Vossen H,A,M.2004Spinacia oleracea LIn: Grubben G,J,H, Denton OA (eds)PROTA 2: Vegetables/Légumes PROTA: Wageningen, Netherlands [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, et al. AFLP—a new technique for DNA-fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware GW, McCollum JP.1980Spinach Producing Vegetable Crops3rd ednInterstate Printers & Publishers: Danville, CA, USA; 437–449. [Google Scholar]
- Yu Q, Hou S, Hobza R, Feltus FA, Wang X, Jin W, et al. Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Mol Genet Genomics. 2007;278:177–185. doi: 10.1007/s00438-007-0243-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.