Abstract
Objective
Nicotine, the main addictive ingredient in tobacco, is metabolically inactivated to cotinine primarily by the hepatic enzyme CYP2A6. Substantial genetic variation in the CYP2A6 gene results in large variation in the rates of nicotine metabolism which in turn alters smoking behaviours (e.g., amount of cigarettes smoked, risk for dependence and success in smoking cessation). The goal of this study was to identify and characterize novel variants in CYP2A6.
Methods
The CYP2A6 gene from African American phenotypically slow nicotine metabolizers was sequenced and seven novel variants were identified [CYP2A6*39 (V68M), CYP2A6*40 (I149M), CYP2A6*41 (R265Q), CYP2A6*42 (I268T), CYP2A6*43 (T303I), CYP2A6*44 (E390K), CYP2A6*44 (L462P)]. Variants were introduced into a bi-cistronic cDNA expression construct containing CYP2A6 and P450 oxidoreductase (POR) and assessed for protein expression, enzymatic activity and stability as evaluated using western blotting and nicotine metabolism. Genotyping assays were developed and allelic frequencies were assessed in 534 African Americans.
Results
The variants displayed significantly lower protein expression (P<0.001) when compared with the wildtype as well as reduced metabolism of nicotine to cotinine when controlling for cDNA expression using POR (P<0.001). The variants also displayed reduced stability at 37ºC. Allelic frequencies ranged from 0.1-0.6% with a collective genotype frequency of 3.2%; the impact in vitro correlated significantly with in vivo activity (R2=0.40-0.48, P<0.05). Together, those with a novel variant had significantly lower nicotine metabolism in vivo than those without genetic variants (P<0.01).
Conclusion
Here we identified a number of novel variants with reduced/loss of CYP2A6 activity, increasing our understanding of CYP2A6 genetic variability.
Keywords: CYP2A6, genetic variation, nicotine metabolism, smoking, African Americans
INTRODUCTION
Nicotine is the main addictive compound in tobacco [1]. In the United States, approximately 19% of individuals over the age of 18 are smokers [2]. In vivo, approximately 80% of nicotine is inactivated to the primary metabolite cotinine (COT) in a reaction principally mediated by the enzyme CYP2A6 [3]. An association between genetic variation in CYP2A6 activity and numerous smoking behaviours has been demonstrated; slow metabolizers smoke fewer cigarettes, are less likely to be a current smoker, have lower dependence scores, reduced risk for lung cancer and better rates of smoking cessation [4-10]. In addition to nicotine, CYP2A6 is involved in the metabolism of other therapeutic agents including tegafur (an antineoplastic drug) [11], letrozole (an aromatase inhibitor used in the treatment of breast cancer) [12] and efavirenz [13], where variability in CYP2A6 is associated with altered drug levels, potentially impacting therapeutic response [14, 15].
CYP2A6 is the sole mediator of the conversion of cotinine to the metabolite trans-3’-hydroxycotinine (3HC) [16, 17]. The 3HC/COT metabolite ratio (also known as the nicotine metabolic ratio, NMR) is a reliable measure of in vivo CYP2A6 activity among regular smokers; it is stable over time, correlates with both the rate of nicotine clearance and nicotine metabolism to cotinine and altered by genetic variation in CYP2A6 [8, 16, 18-20]. Extensive genetic variability in CYP2A6 contributes to the large interindividual and interethnic variability in CYP2A6 activity and the NMR. There are 38 known genetic variants of CYP2A6 with the majority associated with reduce or loss of enzymatic function (http://www.cypalleles.ki.se/). Twin studies conducted to parse out the environmental and genetic influences on the NMR have shown substantial genetic contribution, much of which is not yet identified by known variants in CYP2A6 [21].
While most CYP2A6 variants identified to date result in reduced activity, a substantial proportion of individuals without an established variant still exhibit slow nicotine metabolism, which may be due to novel genetic variation and/or environmental inhibitors. The goal of this study was to identify novel CYP2A6 variants by sequencing the CYP2A6 gene of individuals with unexplained slow activity. Novel variants were characterized in vitro after introduction to a bi-cistronic construct containing the cDNA of wildtype CYP2A6 and P450 oxidoreductase (POR) in E.coli and subsequent assessment of their impact on expression, activity and stability. New genotyping assays were developed and validated, and used to determine variant frequency. This study expands our knowledge of genetic variability in CYP2A6, improving our ability to investigate the impact of CYP2A6 activity on drug metabolism and smoking behaviours, with one goal being the use of this information to optimize smoking cessation treatments.
MATERIALS AND METHODS
Cloning and sequencing
To identify novel genetic variants, 32 individuals with low CYP2A6 activity (based on NMR) had the intronic, exonic and UTR regions of their CYP2A6 gene sequenced. Only individuals categorized as outliers (slow metabolizers outside one standard deviation from the group NMR mean) who were not homozygous for established CYP2A6 genetic variants were sequenced. A 10kb fragment spanning 1.4kb upstream and 8.5kb downstream of the CYP2A6 start site was amplified using a long polymerase chain reaction (PCR) assay. Long-PCR was carried out using the primers: 2A65Pr1F (forward) 5’ – ACC TAG ACT TAA TCT TCC CGT ATA C – 3'and 2A6R13 (reverse) 5’ – GCC TCC CAT AGT GCT ATA ATT AAC A – 3’ [22]. The conditions for the reaction were as follows: initial DNA denaturation at 95ºC for 2 min, 30 cycles of denaturation at 95ºC for 20 sec, annealing at 58ºC for 20 sec, elongation at 72°C for 3 min and a final elongation period at 72°C for 3 min. The resulting product was subcloned into a pCR-XL-TOPO plasmid (TOPO® XL PCR Cloning Kit; Invitrogen, Burlington, Canada) and alleles without established CYP2A6 variants were identified and sequenced at The Centre for Applied Genomics (Toronto, Canada). Sequencing was performed using a dual ABI 3730XL instrument (Life Technologies, Carlsbad, USA). Once a novel variant was identified, it was confirmed through the sequencing of at least three clones containing the novel variant allele followed by sequencing of the CYP2A6 gene directly from PCR amplified genomic DNA. Following the creation of a genotyping assay for each novel variant, DNA was sequenced from newly identified individuals positive for the variant allele.
Creation of CYP2A6 variant constructs
CYP2A6 novel non-synonymous single nucleotide polymorphisms (SNPs) were introduced, using primers found in supplementary table 1, into a bi-cistronic construct containing the cDNA of wildtype CYP2A6 and P450 oxidoreductase (POR) [23, 24] using a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Mississauga, Canada). A negative construct, in which a 802bp region of wildtype CYP2A6 cDNA was excised through restriction enzyme digest by BamHI, an established variant construct, CYP2A6*17, and the wildtype construct CYP2A6*1 were included as controls [24]. All constructs were sequenced to confirm the changes introduced.
Expression of variant constructs
Control and variant CYP2A6 cDNA constructs were transfected into DH5α competent cells (MAX Efficiency® DH5α Competent Cells, Invitrogen) and expressed. Membrane fractions were prepared as previously described [24] and protein concentrations were determined through the Bradford protein assay (Bio-Rad Laboratories, Mississauga, Canada). CYP2A6 and POR protein levels were determined by immunoblotting using polyclonal CYP2A6 (Polyclonal Anti-2A6 raised in mouse CAT# SAB1400063, Sigma-Aldrich, St. Louis, USA) and POR (Polyclonal Rabbit Anti-Human P450 Reductase CAT#AB1257, Millipore, Billerica, USA) antibodies, respectively. In addition, a standard curve using commercially available cDNA-expressed CYP2A6 protein (CAT# 456254; BD Biosciences, Mississauga, Canada) was used to determine absolute CYP2A6 protein levels. As expected POR expression was unaltered between the cDNA constructs (Supplementary Fig. 1), including the CYP2A6 negative construct, therefore the ratio of CYP2A6 protein levels to POR protein levels were used to evaluate changes in CYP2A6 expression for each variant relative to the wildtype construct. Ratios were derived from CYP2A6 and POR protein levels on Western blots within their individual linear ranges.
Nicotine metabolism assay
In vitro metabolism of nicotine to cotinine was used to determine CYP2A6 activity as previously described [25]. A total of 10 pmol of CYP2A6 protein was used in a reaction mix (final volume of 500 μl) containing 50 mM Tris-HCl (pH 7.4) buffer, 20 pmol/ml expressed cytochrome b5 (CAT# P2252; Invitrogen), 1 mg of protein/ml human liver cytosol (added in excess) and nicotine (10 μM to 1000 μM). The reaction mix was pre-heated at 37°C for 2 minutes and metabolism was initiated by the addition of 1 mM NADPH (β-NADPH reduced tetrasodium salt, Sigma-Aldrich). The reaction duration was 15 minutes at 37°C and was stopped by the addition of 100 μl of NaCO3. High-pressure liquid chromatography (HPLC) was used to measure cotinine levels as previously described [26]. Nicotine kinetic parameters, Km and Vmax, were approximated by the software GraphPad Prism (Version 5.00 for windows; GraphPad Software, San Diego, USA) using the Michaelis-Menten equation.
Thermal stability assessment
Stability was assessed by incubating equal amounts of CYP2A6 protein (wildtype and variant) at 37°C for 3, 6, 9, 12 and 24 hours as time points. The 24 hour CYP2A6 protein aliquot incubation was initiated first followed by the incubation of each subsequent time stamped aliquot (i.e. 12 hour then 9 hour, etc.) with all incubations ending together 24 hours after the initial start time; levels of CYP2A6 protein and activity were measured immediately following the incubation (without freezing samples) by immunoblotting and metabolism of nicotine to cotinine as described above (using nicotine concentrations of 50 μM and 500 μM).
Genotyping
The CYP2A6 gene was assessed using three overlapping regions for initial PCR gene-specific amplification; each protocol using a specific set of primers (Supplementary Table 2). The reaction conditions for region 1 involved an initial DNA denaturation at 95°C for 2 min, 40 cycles of denaturation at 95°C for 20 sec, annealing at 55°C for 20 sec, elongation at 72°C for 1 min 15 sec and a final elongation period at 72°C for 3 min. Identical conditions were used for regions 2 and 3 except that the annealing temperature for region 2 was 60°C for 20 sec and elongation at 72°C was for 1 min and the annealing temperature was 50°C for 20 sec for region 3. Samples were genotyped using SYBR green (Power SYBR® Green PCR Master Mix; Life Technologies) using primers optimized for each variant (Supplementary Table 3) using the following conditions: initial denaturation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 sec and annealing/elongation at 65°C for 10 sec. Each sample, containing PCR region specific DNA and SYBR green master mix, was run in duplicate on a 384 well plate containing one common primer and either a wildtype or variant primer. Reactions were run and read by Applied Biosystems® ViiA™ 7 Real-Time PCR System (Life Technologies). Samples (N=10) were genotyped for 3524T>C using amplified DNA from both regions 1 and 2 first amplifications and for 5661G>A using amplified DNA from both from regions 2 and 3; genotyping results did not differ by first amplification region used. Baseline NMR was previously determined from each participant (N=534) from an African American smoking cessation study of light smokers (defined as smoking less than 10 cigarettes per day) [27, 28] and was used to evaluate the impact of the variants identified by genotyping on CYP2A6 activity.
Statistics
In vitro protein expression and catalytic activity of wildtype and variants were compared using a one way analysis of variance with a Bonferonni correction for post-hoc analysis. The NMR group mean of the CYP2A6 *1/*1 group was also compared to individual variant genotype group NMR means using a one way analysis of variance with a Bonferonni correction for post-hoc analysis. The NMR group mean of the combined variant group was compared to the CYP2A6 *1/*1 NMR group mean using an unpaired t-test.
RESULTS
Novel CYP2A6 variants identified
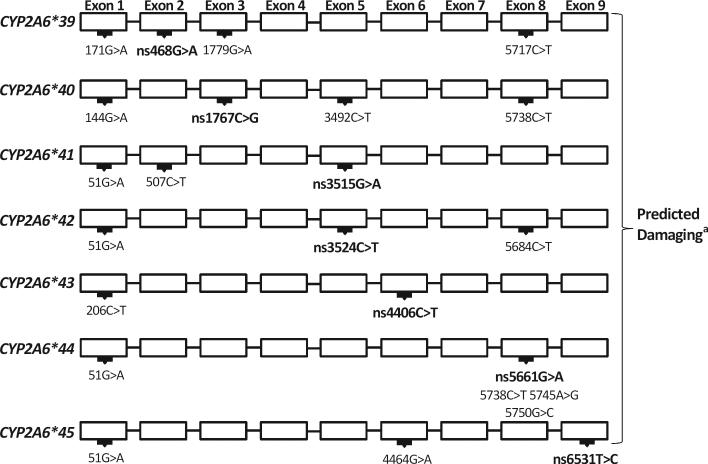
Using the NMR as an indicator of CYP2A6 activity, individuals with low CYP2A6 activity had their CYP2A6 gene sequenced. Based on this approach, seven novel non-synonymous genetic variants, CYP2A6*39 (468G>A, V68M), CYP2A6*40 (1767C>G, I149M), CYP2A6*41 (3515G>A, R265Q), CYP2A6*42 (3524T>C, I268T), CYP2A6*43 (4406C>T, T303I), CYP2A6*44 (5661G>A, E390K) and CYP2A6*45 (6531T>C, L462P) (Fig. 1) were identified which had not been previously characterized for impact on CYP2A6. A phenotype prediction software, Polyphen-2 [29], indicated that each variant was damaging to CYP2A6 (Fig. 1). Furthermore, there is high amino acid conservation at the location of these variants across numerous species (Table 1) suggesting their importance to enzyme function.
Figure 1.
Representation of exonic variation in individuals with an identified novel variant. Bolded genomic changes indicate non-synonymous SNPs predicting the corresponding amino acid alteration. aThe prediction software PolyPhen2 determined each variant to be damaging to CYP2A6 structure or function. PolyPhen2 uses a scaling system to determine variant impact. In short, scores <0.5 are determined to be benign, scores between 0.5 and 2.0 are possibly damaging and scores >2.0 are probably damaging (Ramensky et al., 2002). Genomic changes are relative to the CYP2A6 ATG start site using the reference sequence NG_008377.1. The protein sequence P11509.3 was used as an amino acid reference.
Table 1.
Novel variants exist in highly conserved locations among different species
| CYP2A6*39 V68M | CYP2A6*40 I149M | CYP2A6*41 R265Q | CYP2A6*42 I268T | CYP2A6*43 T303I | CYP2A6*44 E390K | CYP2A6*45 L462P | |
|---|---|---|---|---|---|---|---|
| Humana | --GPVFT-- | --ERIQE-- | --SPRD--- | ---FIDS-- | --GGTET-- | --GTEVF-- | --FRLKS-- |
| Gorillab | --GPVFT-- | --ERIQE-- | --SPRD--- | ---FIDS-- | --GGTET-- | --GTEVF-- | --FRLKS-- |
| Macaquec | --GPVFT-- | --ERIQE-- | --SPRD--- | ---FIDS-- | --AGTET-- | --GTEVF-- | --FRFKS-- |
| Moused | --GPVFT-- | --ERIQE-- | --SPRD--- | ---FIDS-- | --AGTET-- | --GTEVF-- | --FHFKS-- |
| Rate | --GPVFT-- | --ERIQE-- | --SPRD--- | ---FIDS-- | --AGTET-- | --GTEVF-- | --FCFKS-- |
| Rabbitf | --GPVFT-- | --ERIQE-- | --SPRD--- | ---FIDS-- | --AGTET-- | --GTEVF-- | --FRFKS-- |
NP_001164520.1 each represents the protein sequence of the corresponding homologue
In vitro protein expression and catalytic activity of novel variants
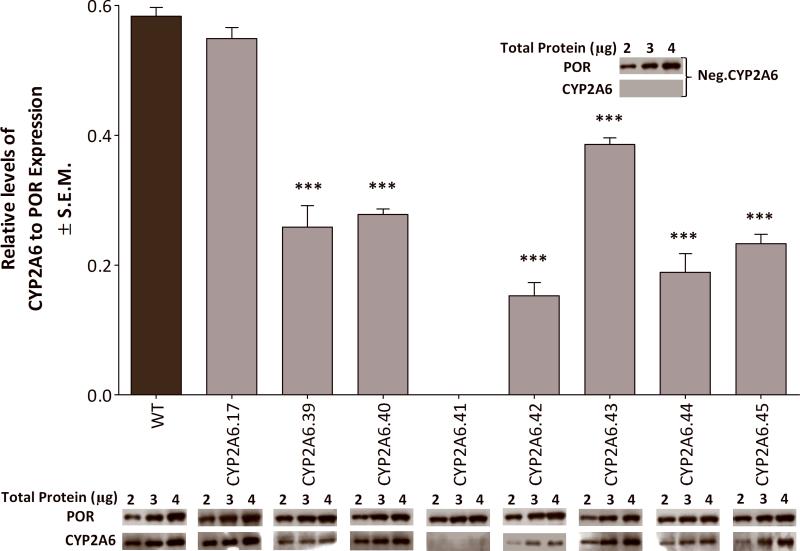
In order to characterize the potential functional impact of these variants, in vitro expression level and catalytic activity of CYP2A6 variant protein was assessed. Expression levels were determined as a ratio of CYP2A6 to POR from the bi-cistronic construct (Fig. 2); POR expression acted as an internal expression control as it was unchanged between variant constructs (Supplementary. Fig. 1). Deviation from the wildtype CYP2A6 to POR construct's ratio indicated altered CYP2A6 expression. There was an appreciable reduction in CYP2A6 protein expression among the novel variants compared to wildtype CYP2A6 (Fig. 2). There was undetectable levels of CYP2A6.41 (R265Q) protein, similar to the expression of the CYP2A6 negative construct; both had normal levels of POR expression.
FIGURE 2.
Variants had significantly lower CYP2A6 expression when compared to wildtype (normalized to POR expression). Protein expression levels of CYP2A6 and POR were determined by immunoblotting. The negative construct (Neg.CYP2A6) did not have detectable levels of CYP2A6 nor did variant R265Q, while both expressed comparable amounts of POR. ***P<0.001 Expression was determined from the averaged ratios of CYP2A6 to POR (of each variant construct), which were both in linear range, from two independent experiments. The western blots represent a visual comparison between the variants and wildtype at the same amount of total loaded protein and may not be in the linear range for all protein levels shown.
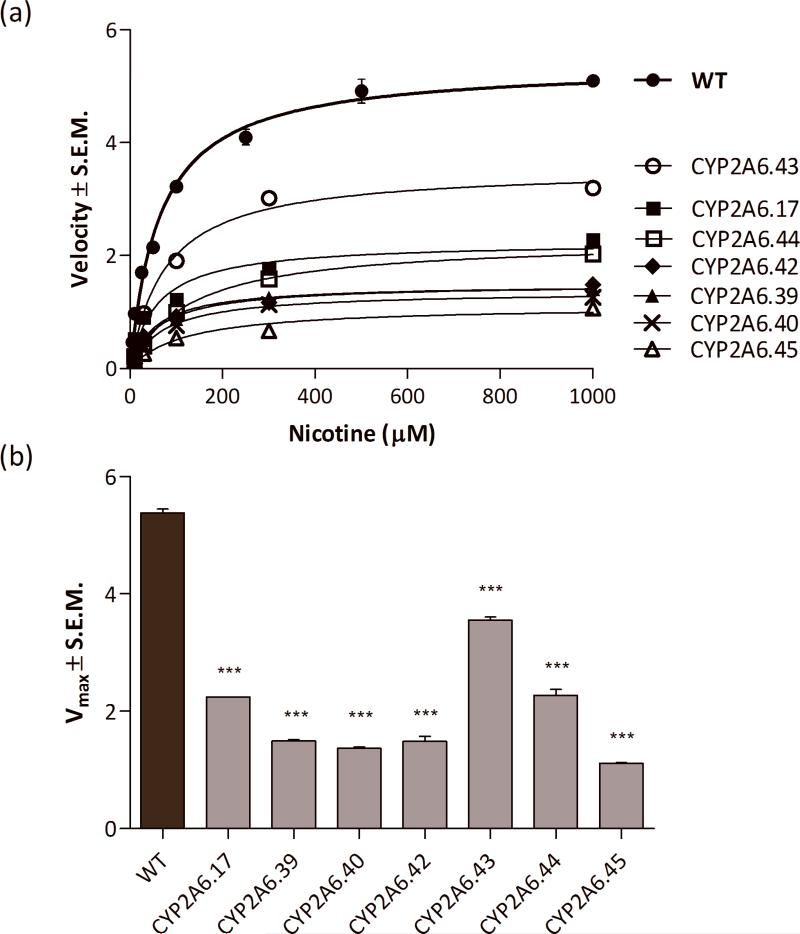
Catalytic efficiency of each variant CYP2A6 was determined by assessing cotinine formation at equivalent expression levels for each variant construct (Fig. 3; Table 2). All variants exhibited a significant reduction in velocity and turnover when compared to the wildtype (Table 2). Catalytic efficiency was also assessed at equal amounts of CYP2A6 (10 pmol), although this meant using substantially more protein (and therefore elevated levels of POR) due to the lower CYP2A6 expression (Supplementary Table 4).
FIGURE 3.
(a) Measuring cotinine formation illustrated the differing effects of amino acid substitution on enzymatic function. (b) All CYP2A6 variants showed significantly lower maximum velocities when compared to wildtype. Groups were analyzed using a One-way analysis of variance with Bonferroni tests used for post-hoc analysis from three independent experiments ***P<0.001.
Table 2.
Kinetic parameters of wildtype and variant CYP2A6
| Km (μM) | Vmax (pmol/min/pmol POR)a | Vmax/Km (nl/min/pmol POR)a | |
|---|---|---|---|
| WT | 64 ± 6 | 5.4 ± 0.1 | 84 ± 2 |
| CYP2A6.17 | 57 ± 10 | 2.2 ± 0.1*** | 39 ± 2*** |
| CYP2A6.39 | 62 ± 4 | 1.5 ± 0.1*** | 24 ± 2*** |
| CYP2A6.40 | 72 ± 6 | 1.4 ± 0.1*** | 19 ± 1*** |
| CYP2A6.42 | 55 ± 10 | 1.5 ± 0.1*** | 27 ± 5*** |
| CYP2A6.43 | 76 ± 7 | 3.6 ± 0.1*** | 47 ± 5*** |
| CYP2A6.44 | 117 ± 13*** | 2.3 ± 0.1*** | 18 ± 2*** |
| CYP2A6.45 | 110 ± 18*** | 1.1 ± 0.1*** | 9 ± 1*** |
POR was used as an internal expression control among the variants.
Parameters are measured ± S.E.M. from three independent experiments
P<0.001
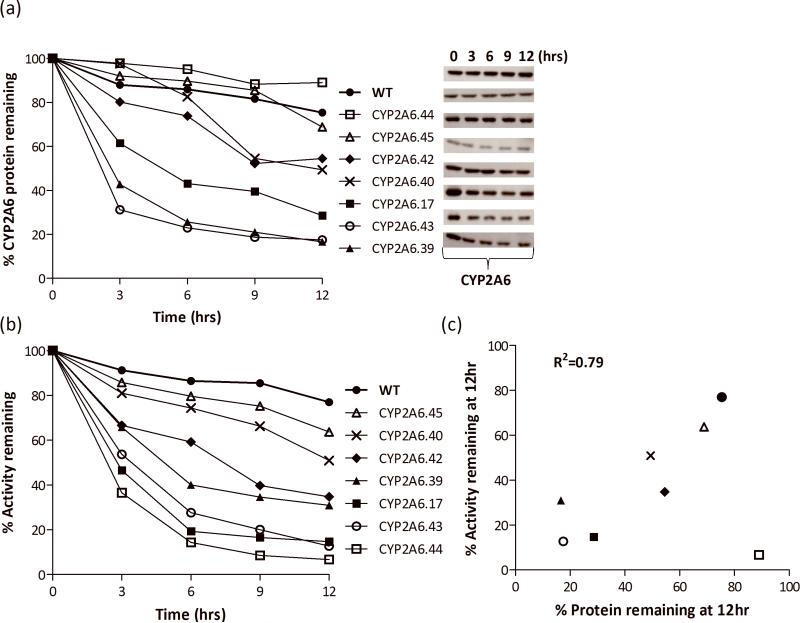
Thermal stability of novel variants assessed in vitro
To determine the impact of the variants on the stability of CYP2A6 protein structure, a thermal stability experiment was conducted from which levels of remaining protein and activity over time were assessed. The variants showed a greater decrease in protein level over time when compared to wildtype with the exception of CYP2A6.44 and CYP2A6.45 which were similar to the wildtype CYP2A6 (Fig. 4a). Variant impact on enzymatic activity also resulted in a number of variants showing a greater decrease over time when compared to wildtype (Fig. 4b). Variants differed in their degree of impact on protein stability from high to low in both measureable parameters. A correlation between remaining protein and activity was seen across each variant with the exception of CYP2A6.44 where activity declined sharply over time while protein level remained fairly constant (Fig. 4c, R2=0.79 excluding outlier, R2=0.14 with outlier).
FIGURE 4.
Thermal stability of wildtype and variant CYP2A6 protein was assessed at 37°C over time. (a) CYP2A6 protein levels were measured by immunoblotting, assessed at each time point and plotted relative to 0hr. The rate of protein loss among variants differed from high to moderate to low. The levels of protein were assessed from one experimental procedure. (b) CYP2A6 activities are shown as percent remaining relative to 0hr. The degree of impact on activity varied over time from high to low. The activity assessment was run in duplicate. (c) There is a significant correlation between % activity and % protein remaining of variants at 12hr with the exclusion of the outlier (R2 value excludes outlier; P=0.01).
Novel variants associated with slow nicotine metabolizers in vivo
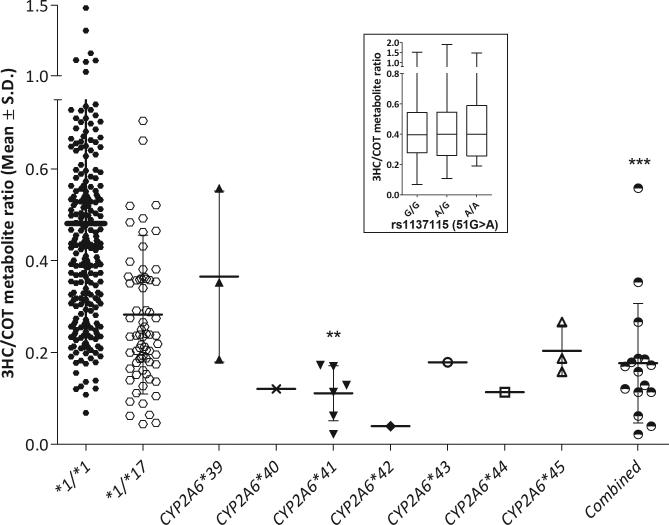
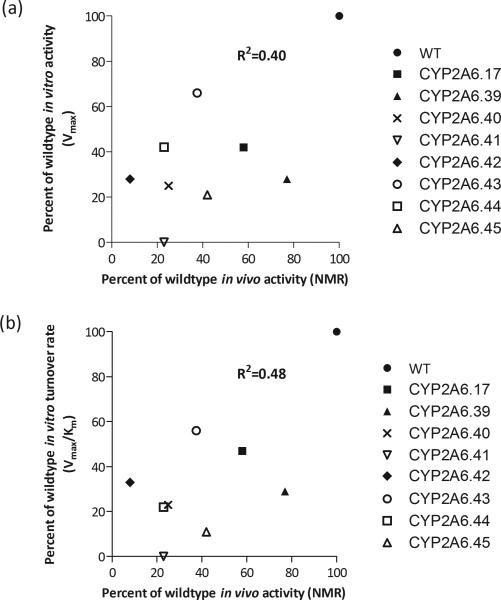
A population of 534 African American treatment seekers was genotyped to determine variant allelic frequency. Genotype frequencies were as high as 1.2% in the case of 3515G>A along with some variants found in 0.6% of the sample. There was a collective frequency of 3.2%. Four variants were identified in a single individual (Table 3). Allele frequencies ranged from 0.1 to 0.6% (Table 3). The majority of individuals who were found to have a novel variant had a 3HC/COT ratio lower than the group mean of individuals without any known genetic variation (Fig. 5). The collective group mean of individuals with a novel variant was also significantly lower (Fig. 5) suggesting these variants are associated with slow nicotine metabolizers in vivo. There was a positive correlation found between in vivo and in vitro CYP2A6 activity (assessed by correlation between in vivo NMR with in vitro Vmax or turnover rate, Vmax/Km) (Fig. 6a; R2=0.40; Fig. 6b; R2=0.48, respectively).
Table 3.
Genotype and allele frequencies of variants in African Americans
| Variant Allele | % of genotyped individuals with one or two copies of a variant (N=) | % of genotyped alleles with a variant (N=) |
|---|---|---|
| CYP2A6*39 | 0.6 (3) | 0.3 (3) |
| CYP2A6*40 | 0.2 (1) | 0.1 (1) |
| CYP2A6*41 | 1.2 (6) | 0.6 (6) |
| CYP2A6*42 | 0.2 (1) | 0.1 (1) |
| CYP2A6*43 | 0.2 (1) | 0.1 (1) |
| CYP2A6*44 | 0.2 (1) | 0.1 (1) |
| CYP2A6*45 | 0.6 (3) | 0.3 (3) |
| Combined Group | 3.2 (16) | 1.6 (16) |
FIGURE 5.
Novel variants were associated with slow nicotine metabolizers in vivo as indicated by lower group means compared to CYP2A6 *1/*1 genotype group. Novel variants are shown individually along the X axis as well as together as a group (combined). Variant R265Q had three individuals identified via sequencing and variants V68M and L462P had one individual identified via sequencing (the remaining individuals within these variant groups were identified via genotyping). Association with lower activity of the novel variant groups was tested using a one-way analysis of variance with Bonferroni tests used for post-hoc analysis **P<0.01. A comparison between CYP2A6*1/*1 and the combined group was tested using an unpaired t-test ***P<0.001. Inset indicates the lack of association between 51G>A (rs1137155) among the CYP2A6*1/*1 individuals.
Figure 6.
(a) In vivo impact (assessed as differences in NMR) is correlated with the in vitro impact on Vmax, when Vmax of each variant when compared to the wildtype. (b) There is a significant correlation between in vivo and in vitro CYP2A6 activity when in vitro activity is assessed using turnover rate (Vmax/Km). P=0.04
Many of the novel variants were in linkage disequilibrium with the 51G>A variant (rs1137115, Fig. 1), which has been predicted by others to reduce CYP2A6 activity in vivo due to the possibility of creating an exonic suppressor site in exon 1 [30, 31]. To investigate if this 51G>A variant was associated with lower in vivo CYP2A6 activity in African Americans we examined 1) the prevalence of the 51G>A variant among those with, versus without, reduce/loss of function variants, 2) the association of 51G>A variant on CYP2A6 activity among those with or without variants. We found 1) a higher prevalence of the 51G>A among wildtype vs. variant alleles carriers (29.5% vs 12.7%, Chi2 P<0.0001), and 2) no association between 51G>A genotypes and CYP2A6 activity among those with any known or novel variant (P=0.32) or among the CYP2A6 wildtype individuals (P=0.86, Fig. 5 Insert), suggesting the association between the novel variants identified here, and variant alleles in general, and CYP2A6 activity was not mediated through linkage disequilibrium with 51A.
DISCUSSION
After sequencing the CYP2A6 gene of low activity individuals in a population of African American light smokers, seven novel nonsynonymous variants were identified and characterized. These variants were present at allelic frequencies of 0.1 to 0.6%, resulting in approximately 3% of this population containing at least one of these variants; this constitutes an initial step towards identification of rare variants in this highly variable gene loci. High conservation across species at the positions of the variants suggests important structural or functional regions; both the specific amino acid in each variant allele, and also the surrounding amino acid region, were highly conserved (Table 1). This high level of conservation was consistent with the predictions made by PolyPhen-2 that all changes were damaging. When expressed in vitro, each variant had a deleterious impact on the enzyme characteristics tested, protein expression, enzyme activity and/or stability, consistent with their low CYP2A6 activity in vivo.
Assessing the variant locations using the three dimensional crystal structure of CYP2A6 and bioinformatics predicted that six variants were surface alterations which would negatively impact function/stability and one, CYP2A6*43, resided internally in a substrate recognition site [32]. As both CYP2A6 and POR were translated from the same transcript, POR was used as an internal control for cDNA expression. CYP2A6 variants had widely differing levels of expression relative to the consistently expressed POR levels suggesting that some of the variants were particularly unstable. CYP2A6*40, CYP2A6*42 and CYP2A6*43 were located within helical motifs of CYP2A6 whereas CYP2A6*39, CYP2A6*44 and CYP2A6*45 were located within β-sheets. CYP2A6*41 was located within a connection loop between two α-helices and had undetectable levels of expression. CYP2A6*42, with an alteration located quite close to 3515G>A (CYP2A6*41), also had very low levels of expression suggesting that this region of CYP2A6 protein structure is important in maintaining stability. CYP2A6*39 and CYP2A6*40 had similar modest amino acid substitutions and changes in levels of expression, which was approximately half of wildtype; thus these surface alterations may have played a role in their reduced expression as interactions with the surrounding hydrophilic environment may have been altered. Expression of CYP2A6.44 and CYP2A6.45 were approximately one-third of wildtype, consistent with the more dramatic change in amino acids in these two variants. The CYP2A6*44 allele resulted in a change from a negatively to positively charged amino acid likely to have a larger impact on the proteins interaction with the environment [33]. CYP2A6*45 introduced a proline into the protein structure which may alter protein conformation due to the irregular chemical structure of this amino acid. Expression was slightly altered with CYP2A6.43 suggesting this location had less impact on initial stability/expression levels.
Compared to wildtype, and consistent with the impact on expression described above, the variants also had significant and differing impacts on protein stability over time at 37°C, ranging from high (CYP2A6.17, CYP2A6.39 and CYP2A6.43) to moderate (CYP2A6.40 and CYP2A6.42) to low (CYP2A6.44 and CYP2A6.45) impact. The enzymatic activity also dropped with time at 37°C resulting in a relatively high correlation between protein stability and remaining activity; this suggests that the protein instability resulted in a loss of activity. CYP2A6.44 exhibited a minimal loss of protein over time whereas the enzymatic activity dropped significantly suggesting that the impact of this variant was not on the structure/stability of the entire apoprotein but rather the change altered some key aspect of the enzyme such that it was less able to bind and transform nicotine to cotinine, such as the substrate, cofactor, or perhaps heme binding site.
All variants had an impact on the enzymatic activity in vitro. Decreases in Vmax were observed among the six variants assessed (CYP2A6.41 had no appreciable levels of CYP2A6 protein), while nicotine's substrate affinity (Km) was unaltered for most CYP2A6 variants (Table 2). Turnover rate (Vmax/Km) was also assessed (Table 2). Each surface variant exhibited a significant reduction in turnover rate signifying an overall reduction in enzymatic function (Table 2). Surface changes altering the positioning of internal bonding sites for substrate recognition and/or the shape of the active site could play a role in decreasing the function of the enzyme. A limitation to be considered is that these in vitro analyses were conducted in a bacterial system rather than within a mammalian cell system. A mammalian cell system has specific advantages, but some disadvantages for characterization including limited protein yields (for the numerous characterizations performed). As this and previous studies have successfully characterized nonsynonymous changes using this bacterial system, with in vitro characterization being consistent with the impact observed in vivo, we do not believe this is a significant limitation [24, 25, 34].
An association between these variants and slow metabolism in vivo was observed. The collective group of individuals with at least one novel variant had a significantly lower NMR than individuals without any known genetic variation. The lower NMR correlates with the observed in vitro metabolism suggesting that the slow metabolism may be attributed to these specific novel variants. A positive correlation was observed between both in vitro parameters, Vmax and turnover rate, and NMR with R2 values of 0.40 and 0.48, respectively (Figure 6). As some of these variants were only found in the specific proband sequenced, larger sequencing programs of phenotyped individuals will be needed to confirm the association with decreased activity in vivo, although the in vitro characterization is relatively strong evidence that these are casual changes to protein structure. A concerted effort to sequence using approaches as performed here, or those optimized for next generation sequencing, is needed to fully understand the impact of genetic variation in this enzyme on smoking behaviors, particularly cessation, as well as disease related risk and pharmacogenetic implications.
Some inferences with respect to other substrates of CYP2A6 can be made from these findings. As most of the variants characterized here altered protein expression and/or stability, they are likely to alter the enzymatic activity of all substrates, including those of clinical and toxicological significance. If the structural instability observed from this study occurs in situ in human liver, this would suggest that individuals with these variants will have elevated response to drugs inactivated by CYP2A6, and reduced response to drugs activated by CYP2A6. CYP2A6 plays a principal role in the metabolic activation of the antineoplastic prodrug tegafur to 5-fluorouracil (5-FU) which is a precursor of the active anti-cancer metabolites. Reduced tegafur metabolic activation occurs in those with decrease of function CYP2A6 alleles in vitro and in vivo which may result in decreased clinical response [35, 36]. Letrozole, another antineoplastic, is metabolically inactivated by CYP2A6; CYP2A6 slow metabolizers have reduced letrozole clearance and higher plasma levels [14, 37]. CYP2A6 slow metabolizers have longer plasma exposure to the antiretroviral substrate drug efavirenz in the presence of impaired CYP2B6, suggesting a minor role of CYP2A6 [13, 38]. Thus the reduced expression and/or stability of the variants observed in the present study could impact dosing regimens required for maximal therapeutic response. In addition to drug metabolism, CYP2A6 plays a role in influencing procarcinogen exposure and activation [39]. Slow metabolizers have decreased lung cancer risk due to a reduced likelihood of being a smoker, a reduced exposure to nitrosamines (due to the impact on lowering cigarette consumption) and a reduced activation of tobacco procarcinogen nitrosamines [7, 40]. Thus it is likely that the majority of the variants characterized here will also have reduced activity towards both nicotine and the highly structurally related nicotine-derived nitrosamine substrates resulting in a reduction in risk for lung cancer; this is important as African Americans, while generally light smokers have a disproportionately high morbidity and mortality from smoking, including lung cancer [41, 42].
As variant genotype frequencies ranged from 0.2% to 1.2% most would be classified as rare variants. The combined genotype frequency was 3.2% in this sample, suggesting that while individually they are rare they combine to be a potentially important contribution to variation in the population. The characterization of rare variants will be an important next step in disease genetics and pharmacogenomics versus the focus on genome wide association studies of common gene variation [43, 44]. Rare variants may play a larger role in the unidentified heritability for common diseases than previously expected. Here we sequenced just 32 alleles, using low activity phenotypes to enhance identification of new variants, and identified seven novel amino acid changing variants. Characterizing the 5’ and 3’ UTR regions and the intronic variation in these samples without amino acid alterations characterized here, will likely lead to further sources of variability; detrimental alterations outside of the 10kb gene fragment assessed in this study may also contribute to this variability. This suggests that the wide variation observed in rates of nicotine metabolism among individuals without currently identified genetic variation may be due to rare variants which alter structure, like those characterized in this study, as well as regulatory variants. Next generation sequencing analyses, once optimized for the complexities of CYP genes (which are highly homologous, have many copy number variants, hybrids and deletions), should provide further insight into both the prevalence of rare variants and their impact on nicotine metabolism and resulting impacts on variation in smoking behaviors, smoking related disorders and smoking cessation [45, 46].
In conclusion, this study identified seven novel nonsynonymous variants in a population of African American light smokers; each reduced in vitro CYP2A6 protein expression, enzymatic activity and/or stability, consistent with the associated slow metabolism phenotype. Identifying novel genetic variation establishes a more complete picture of CYP2A6 variability, improving our understanding of genetic variation in smoking behaviors and pharmacogenetic responses to CYP2A6 substrate drugs and toxins.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge the support provided by NIH grant CA091912, CIHR grants MOP86471, the Endowed Chair in Addiction for the Department of Psychiatry, Ontario Graduate Scholarship, NIH grant DA020830, CAMH and the CAMH foundation, the Canada Foundation for Innovation (#20289 and #16014) and the Ontario Ministry of Research and Innovation. We would also like to thank Dr. Bin Zhao for assistance with the metabolism studies.
Footnotes
Conflicts of Interest
Dr. Neal L. Benowitz serves as a consultant to several pharmaceutical companies that market smoking cessation medications and has been a paid expert witness in litigation against tobacco companies. Dr. Rachel F. Tyndale has participated in one-day advisory meetings for Novartis and McNeil.
REFERENCES
- 1.Benowitz NL. Nicotine Addiction. New England Journal of Medicine. 2010;362:2295–2303. doi: 10.1056/NEJMra0809890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC Current Cigarette Smoking Among Adults — United States, 2011. Morbidity and Mortality Weekly Report 2012. 61:889–894. [PubMed] [Google Scholar]
- 3.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996;24:1212–1217. [PubMed] [Google Scholar]
- 4.Ho MK, Mwenifumbo JC, Al Koudsi N, Okuyemi KS, Ahluwalia JS, Benowitz NL, et al. Association of Nicotine Metabolite Ratio and CYP2A6 Genotype With Smoking Cessation Treatment in African-American Light Smokers. Clinical Pharmacology & Therapeutics. 2009;85:635–643. doi: 10.1038/clpt.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malaiyandi V, Sellers EM, Tyndale RF. Implications of CYP2A6 Genetic Variation for Smoking Behaviors and Nicotine Dependence. Clinical Pharmacology & Therapeutics. 2005;77:145–158. doi: 10.1016/j.clpt.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14:615–626. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103:1342–1346. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lerman C, Tyndale R, Patterson F, Wileyto E, Shields P, Pinto A, et al. Nicotine metabolite ratio predicts efficacy of transdermal nicotine for smoking cessation. Clinical Pharmacology & Therapeutics. 2006;79:600–608. doi: 10.1016/j.clpt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Patterson F, Schnoll RA, Wileyto EP, Pinto A, Epstein LH, Shields PG, et al. Toward Personalized Therapy for Smoking Cessation: A Randomized Placebo-controlled Trial of Bupropion. Clinical Pharmacology & Therapeutics. 2008;84:320–325. doi: 10.1038/clpt.2008.57. [DOI] [PubMed] [Google Scholar]
- 10.Pianezza ML, Sellers EM, Tyndale RF. Nicotine metabolism defect reduces smoking. Nature. 1998;393:750. doi: 10.1038/31623. [DOI] [PubMed] [Google Scholar]
- 11.Ikeda K, Yoshisue K, Matsushima E, Nagayama S, Kobayashi K, Tyson CA, et al. Bioactivation of tegafur to 5-fluorouracil is catalyzed by cytochrome P-450 2A6 in human liver microsomes in vitro. Clin Cancer Res. 2000;6:4409–4415. [PubMed] [Google Scholar]
- 12.Murai K, Yamazaki H, Nakagawa K, Kawai R, Kamataki T. Deactivation of anti-cancer drug letrozole to a carbinol metabolite by polymorphic cytochrome P450 2A6 in human liver microsomes. Xenobiotica. 2009;39:795–802. doi: 10.3109/00498250903171395. [DOI] [PubMed] [Google Scholar]
- 13.di Iulio J, Fayet A, Arab-Alameddine M, Rotger M, Lubomirov R, Cavassini M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genomics. 2009;19:300–309. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]
- 14.Desta Z, Kreutz Y, Nguyen AT, Li L, Skaar T, Kamdem LK, et al. Plasma Letrozole Concentrations in Postmenopausal Women With Breast Cancer Are Associated With CYP2A6 Genetic Variants, Body Mass Index, and Age. Clinical Pharmacology & Therapeutics. 2011;90:693–700. doi: 10.1038/clpt.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fushiya N, Takagi I, Nishino H, Akizuki S, Ohnishi A. Genetic polymorphisms of enzymes related to oral tegafur/uracil therapeutic efficacy in patients with hepatocellular carcinoma. Anti-Cancer Drugs. 2013:1. doi: 10.1097/CAD.0b013e3283614fef. [DOI] [PubMed] [Google Scholar]
- 16.Dempsey D. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity*1. Clinical Pharmacology & Therapeutics. 2004;76:64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, et al. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996;277:1010–1015. [PubMed] [Google Scholar]
- 18.St Helen G, Novalen M, Heitjan DF, Dempsey D, Jacob P, 3rd, Aziziyeh A, et al. Reproducibility of the nicotine metabolite ratio in cigarette smokers. Cancer Epidemiol Biomarkers Prev. 2012;21:1105–1114. doi: 10.1158/1055-9965.EPI-12-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binnington MJ, Zhu AZ, Renner CC, Lanier AP, Hatsukami DK, Benowitz NL, et al. CYP2A6 and CYP2B6 genetic variation and its association with nicotine metabolism in South Western Alaska Native people. Pharmacogenet Genomics. 2012;22:429–440. doi: 10.1097/FPC.0b013e3283527c1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lea RA, Dickson S, Benowitz NL. Within-subject variation of the salivary 3HC/COT ratio in regular daily smokers: prospects for estimating CYP2A6 enzyme activity in large-scale surveys of nicotine metabolic rate. J Anal Toxicol. 2006;30:386–389. doi: 10.1093/jat/30.6.386. [DOI] [PubMed] [Google Scholar]
- 21.Swan GE, Lessov-Schlaggar CN, Bergen AW, He Y, Tyndale RF, Benowitz NL. Genetic and environmental influences on the ratio of 3′hydroxycotinine to cotinine in plasma and urine. Pharmacogenetics and Genomics. 2009;19:388–398. doi: 10.1097/FPC.0b013e32832a404f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitarque M, von Richter O, Rodriguez-Antona C, Wang J, Oscarson M, Ingelman-Sundberg M. A nicotine C-oxidase gene (CYP2A6) polymorphism important for promoter activity. Hum Mutat. 2004;23:258–266. doi: 10.1002/humu.20002. [DOI] [PubMed] [Google Scholar]
- 23.Gillam EM, Aguinaldo AM, Notley LM, Kim D, Mundkowski RG, Volkov AA, et al. Formation of indigo by recombinant mammalian cytochrome P450. Biochem Biophys Res Commun. 1999;265:469–472. doi: 10.1006/bbrc.1999.1702. [DOI] [PubMed] [Google Scholar]
- 24.Ho MK, Mwenifumbo JC, Zhao B, Gillam EM, Tyndale RF. A novel CYP2A6 allele, CYP2A6*23, impairs enzyme function in vitro and in vivo and decreases smoking in a population of Black-African descent. Pharmacogenet Genomics. 2008;18:67–75. doi: 10.1097/FPC.0b013e3282f3606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al Koudsi N, Ahluwalia JS, Lin S-K, Sellers EM, Tyndale RF. A novel CYP2A6 allele (CYP2A6*35) resulting in an amino-acid substitution (Asn438Tyr) is associated with lower CYP2A6 activity in vivo. The Pharmacogenomics Journal. 2009;9:274–282. doi: 10.1038/tpj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siu ECK, Wildenauer DB, Tyndale RF. Nicotine self-administration in mice is associated with rates of nicotine inactivation by CYP2A5. Psychopharmacology. 2006;184:401–408. doi: 10.1007/s00213-006-0306-6. [DOI] [PubMed] [Google Scholar]
- 27.Cox LS, Nollen NL, Mayo MS, Choi WS, Faseru B, Benowitz NL, et al. Bupropion for Smoking Cessation in African American Light Smokers: A Randomized Controlled Trial. JNCI Journal of the National Cancer Institute. 2012;104:290–298. doi: 10.1093/jnci/djr513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu AZX, Cox LS, Nollen N, Faseru B, Okuyemi KS, Ahluwalia JS, et al. CYP2B6 and Bupropion's Smoking-Cessation Pharmacology: The Role of Hydroxybupropion. Clinical Pharmacology & Therapeutics. 2012;92:771–777. doi: 10.1038/clpt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloom AJ, Harari O, Martinez M, Madden PA, Martin NG, Montgomery GW, et al. Use of a predictive model derived from in vivo endophenotype measurements to demonstrate associations with a complex locus, CYP2A6. Hum Mol Genet. 2012;21:3050–3062. doi: 10.1093/hmg/dds114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bloom AJ, Harari O, Martinez M, Zhang X, McDonald SA, Murphy SE, et al. A compensatory effect upon splicing results in normal function of the CYP2A6*14 allele. Pharmacogenet Genomics. 2013;23:107–116. doi: 10.1097/FPC.0b013e32835caf7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 33.Betts MJ, Russel RB. Bioinformatics for geneticists. Wiley; New York: 2003. Amino Acid Properties and Consequences of Substitutions. [Google Scholar]
- 34.Mwenifumbo JC, Al Koudsi N, Ho MK, Zhou Q, Hoffmann EB, Sellers EM, et al. Novel and established CYP2A6 alleles impair in vivo nicotine metabolism in a population of Black African descent. Human Mutation. 2008;29:679–688. doi: 10.1002/humu.20698. [DOI] [PubMed] [Google Scholar]
- 35.Daigo S, Takahashi Y, Fujieda M, Ariyoshi N, Yamazaki H, Koizumi W, et al. A novel mutant allele of the CYP2A6 gene (CYP2A6*11 ) found in a cancer patient who showed poor metabolic phenotype towards tegafur. Pharmacogenetics. 2002;12:299–306. doi: 10.1097/00008571-200206000-00005. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Bian T, Liu D, Jin T, Chen Y, Lin A, et al. Association analysis of CYP2A6 genotypes and haplotypes with 5-fluorouracil formation from tegafur in human liver microsomes. Pharmacogenomics. 2011;12:481–492. doi: 10.2217/pgs.10.202. [DOI] [PubMed] [Google Scholar]
- 37.Tanii H, Shitara Y, Horie T. Population pharmacokinetic analysis of letrozole in Japanese postmenopausal women. Eur J Clin Pharmacol. 2011;67:1017–1025. doi: 10.1007/s00228-011-1042-3. [DOI] [PubMed] [Google Scholar]
- 38.Kwara A, Lartey M, Sagoe KW, Rzek NL, Court MH. CYP2B6 (c.516G-->T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol. 2009;67:427–436. doi: 10.1111/j.1365-2125.2009.03368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossini A, de Almeida Simao T, Albano RM, Pinto LF. CYP2A6 polymorphisms and risk for tobacco-related cancers. Pharmacogenomics. 2008;9:1737–1752. doi: 10.2217/14622416.9.11.1737. [DOI] [PubMed] [Google Scholar]
- 40.Zhu AZ, Binnington MJ, Renner CC, Lanier AP, Hatsukami DK, Stepanov I, et al. Alaska Native smokers and smokeless tobacco users with slower CYP2A6 activity have lower tobacco consumption, lower tobacco-specific nitrosamine exposure and lower tobacco-specific nitrosamine bioactivation. Carcinogenesis. 2013;34:93–101. doi: 10.1093/carcin/bgs306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;354:333–342. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 42.Pinsky PF. Racial and ethnic differences in lung cancer incidence: how much is explained by differences in smoking patterns? (United States). Cancer Causes Control. 2006;17:1017–1024. doi: 10.1007/s10552-006-0038-2. [DOI] [PubMed] [Google Scholar]
- 43.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marth GT, Yu F, Indap AR, Garimella K, Gravel S, Leong WF, et al. The functional spectrum of low-frequency coding variation. Genome Biol. 2011;12:R84. doi: 10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maxmen A. Exome sequencing deciphers rare diseases. Cell. 2011;144:635–637. doi: 10.1016/j.cell.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 46.Raychaudhuri S. Mapping rare and common causal alleles for complex human diseases. Cell. 2011;147:57–69. doi: 10.1016/j.cell.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.