Abstract
Spatial organisation of T cell receptor (TCR) and its coreceptor CD8 on the surface of live naïve and Ag-experienced CD8+ T cells was resolved by fluorescence lifetime cross-correlation microscopy. We found that exposure of naïve CD8+ T cells to antigen (Ag) causes formation of [TCR, CD8] functional ensembles on the cell surface which correlated with significantly enhanced sensitivity of these cells. In contrast, TCR and CD8 are randomly distributed on the surface of naïve cells. Our model suggests that close proximity of TCR and CD8 can increase Ag sensitivity of T cells by significant accelerating the TCR–peptide-major histocompatibility complex (pMHC) binding rate and stabilisation of this complex. We suggest that the proximity of these primary signalling molecules contributes to the mechanism of functional avidity maturation of CD8+ T cells by switching them from a low to high sensitivity mode.
Abbreviations: TCR, T cell receptor; NSOM, near-field scanning optical microscopy; EM, electron microscopy; mAb, monoclonal antibody; Ag, antigen; CTL, cytotoxic T cell lymphocyte; LN, lymph node; FCS, fetal calf serum; APC, antigen presenting cell
Keywords: T cell receptor signalling, CD8 coreceptor, Antigen recognition, Functional avidity maturation, T cell sensitivity, Fluorescence lifetime cross-correlation microscopy
1. Introduction
CD8+ T cells can substantially increase in sensitivity in the course of immune response by the mechanism of functional avidity maturation [1], [2], [3] to provide effective defence from re-infection [4]. The increase in sensitivity of T cells in contrast with B cells is not caused by the mechanism of somatic hypermutation [5]. Functional avidity maturation was explained in part by predominant expansion of clones with highest sensitivity to pMHC. However, cells expressing the same TCR can also increase sensitivity and become less dependent on CD28 co-stimulatory signal [6]. No correlation of T cell sensitivity with expression levels of the primary activation molecules (TCR and CD8) or adhesion molecules (LFA-1, CD2, CD43 and CD49d) was found [1]. All these findings suggested that that functional avidity maturation is caused by permanent changes in the signal transduction machinery regulating intermolecular events which follow the TCR–pMHC interaction.
A critical role of Lck association with CD8 has been demonstrated [7], [8], [9], [10], [11]. It was found that Lck mediated phosphorylation of immunotyrosine-based activation motifs of the cytoplasmatic tails of the CD3 is necessary for T cell signalling [12]. An important role for the alpha-chain connecting peptide motif in mediating TCR-CD8 cooperation was also shown [13]
It is noteworthy that in vitro stimulation of naïve T cells substantially increases the spatial coordination of the CD8 coreceptor and Lck which leads to more efficient organisation of the TCR signalling machinery [14]. In addition, co-capping CD8/TCR experiments suggested that optimal colocalisation of TCR and CD8 controls functional avidity of T cells [15].
Our earlier real-time data on tetramer association with T cell clones and hybridomas [16], [17] led us to suggest a kinetic promotion mechanism of CD8 cooperation which requires a colocalisation of TCR and CD8 on the cell surface which was named the proximity model. We also found that CD8 can significantly stabilise the formed TCR–pMHC complex. Disruption of rafts in the cellular membrane significantly slowed down the rate of tetramer association with T cells suggesting that the functional TCR and CD8 complexes reside in the cholesterol-rich domains [16]. Other studies investigating the involvement of CD8 in fine tuning of T cell sensitivity came to similar conclusions: namely that CD8 can influence both the TCR–pMHC association and dissociation rate and improve the binding efficiency of T cells particularly those with low affinity TCR and the CD8 involvement in Ag recognition recruits the TCR–pMHC complex to membrane microdomains where TCR mediated signalling steps proceed significantly faster [18], [19].
Using near-field scanning optical microscopy (NSOM) the physical distribution of the TCR and CD8 on the surface of antibody stimulated Rhesus monkey peripheral blood T cells was examined and showed that TCR and CD8 were organised in nanoclusters on the cell surface. In contrast, in the same study, unstimulated primary cells had a random distribution of TCR and CD8 as single molecules or small clusters of 2–4 molecules [20]. Furthermore, electron microscopy (EM) data showed that Ag-stimulated and memory T cells have more and larger TCR oligomers on the cell surface than their naïve counterparts [21]. These data indicate that the distribution of TCR and CD8 varies between naïve and antigen experienced cells although both above results were obtained in fixed cells. Nanoscale proximity of TCR and CD8 was also reported on the surface of unfixed low dose Ag-stimulated cells by fluorescence resonance energy transfer between CD8 labelled with a fluorescence donor and TCR labelled with a fluorescence acceptor by the interaction with a cognate pMHC ligand [22] however this interaction may bring about spatial coordination TCR and CD8.
Measurement of TCR and CD8 organisation in live cells represents a considerable experimental challenge. Here we employed fluorescence lifetime cross-correlation microscopy to measure the proximity of TCR and CD8 molecules in naïve and Ag-experienced CD8+ T cells. In contrast with NSOM or EM, this method does not require intensive labelling of the proteins which can disturb their organisation and allows the measurements at virtually non-invasive level of optical excitation in live cells. Our results show nearly total colocalisation of TCR with CD8 on the surface of Ag-experienced T cells but virtually their random distribution on the surface of naïve cells. Based on these results we discuss a model which can explain, at least in part, the different sensitivity of naïve and Ag-experienced cells and suggest that the proximity of these primary signalling molecules contributes to the mechanism of functional avidity maturation of CD8+ T cells by switching them from a low to high sensitivity mode.
2. Materials and methods
2.1. Cells, antibodies and chemicals
Unless otherwise stated, all chemicals were purchased from Sigma–Aldrich (Haverhill, UK). Naïve T cells isolated from lymph nodes (LN) of 2–4 months old RAG1−/− F5 transgenic mice [23] which recognise an influenza virus specific peptide in the context of H-2Db, were cultured in IMDM supplemented with 5% FCS, l-glutamine, 100 U/mL penicillin and streptomycin and 50 μM β-mercaptoethanol. Ag-experienced cells were produced by in vitro stimulation of naïve F5 T cells with 100 nM NP68 peptide (ASNENMDAM) for 3 days, and then rested in 5 μg/mL IL-2 for 4–10 days. Fab fragment preparation [24] was as follows: 1 mg of CD8α-specific (YTS 169.4) mAbs [25] was digested into Fab with immobilised papain (Thermo Fisher Scientific, Loughborough, UK) in digesting buffer (50 mM PBS, 0.01 M EDTA, 0.1 M cystein, papain, mAb/enzyme ratio 100:1). The Fab preparation was confirmed to form a single band of 50 kD on SDS-PAGE. A relative affinity of Fab was measured by FACS in a competition labelling assay with intact Fluorescein-labelled YTS 169.4 mAbs. Quantum dots (Qdot) Qdot655 and Qdot800 Streptavidin conjugates were purchased from Invitrogen (Paisley, UK). Biotinylated TCRβ (H57-597) and CD8α-specific (YTS156.7.7) mAbs were purchased from BioLegend (London, UK).
2.2. FACS measurements
Naïve cells from LNs or Ag-experienced cell cultures were washed once and resuspended in IMDM medium supplemented with 5% FCS, l-glutamine, 100 U/mL penicillin and streptomycin and 50 μM β-mercaptoethanol. Triplicate cultures at 5 × 105 cells per well were added in 50 μL to 96-well plates containing media supplemented with NP68 and with or without CD8α-specific Fab fragments and incubated for 3 h at 37°/5%CO2. Stimulated cells were stained with CD8-PerCP (53–6.7), TCRβ-FITC (H57-597) and CD69-FITC (H1.2F3) specific mAbs (eBioscience, Hatfield, UK). A minimum of 30,000 events was collected using an LSRII (BD Biosciences, Oxford, UK), and data were analysed with FlowJo software (Tree Star, Olten, Switzerland).
2.3. Proximity measurements by fluorescence lifetime cross-correlation microscopy
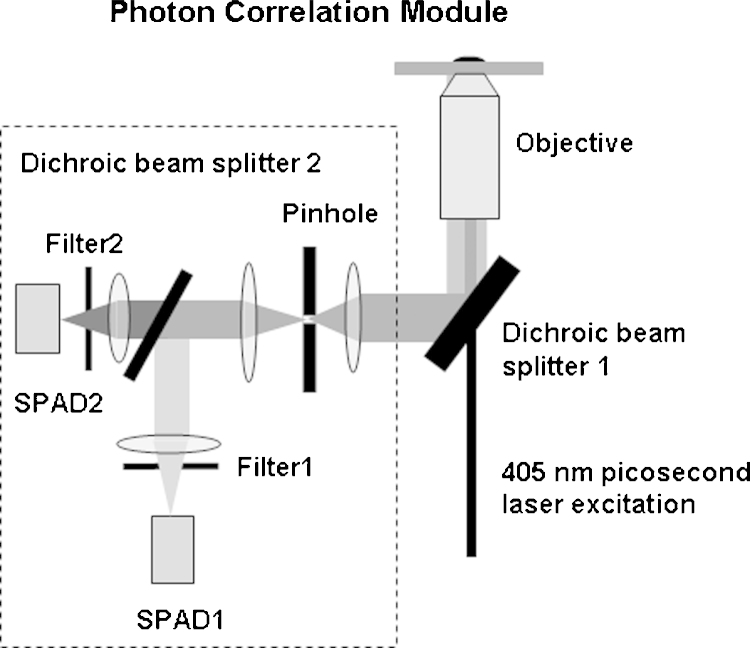
For proximity correlation measurements cells were reacted with 1 nM of Qdot655-CD8β and Qdot800-TCRβ specific mAb conjugates in 50 mM PBS 2% BSA, 0.1% NaN3 pH 7.8 buffer at room temperature for 20 min. The conjugates were prepared in 50 mM Borate buffer, pH 8 by titration of streptavidin-coated Qdots with small fractions of biotinylated antibody to 60% of the total Qdot concentration to get less than 1 quantum dot: antibody stoichiometry. 3× washed cell samples were measured in the photon correlator module (Fig. 1) attached to an Eclipse T2000 inverted fluorescence microscope (Nikon, Edinburgh, UK) through the C-mount port. Excitation light beam of a single mode fibre-coupled 405 nm picosecond laser (EPL405, Edinburgh Instruments, Livingston, UK) operated at 20 MHz was directed to the microscope through the back port and Dichroic beam splitter 1 (FF510-DiO1-25-36, Semrock, Laser 2000, Ringstead, UK) and focused on a sample by Fluor 100× Oil Immersion Objective, N.A. 1.3 (Nikon, Edinburgh, UK). Sample fluorescence separated from the excitation light by Dichroic beam splitter 1 was directed to the Photon Correlation Module, where it was filtered by 50 μm Pinhole and split into two channels by Dichroic beam splitter 2 (FF740-Di01-25-36 Semrock, Laser 2000, Ringstead, UK) to be detected by two Single-Photon Avalanche Diodes (SPAD, SPMC-AQR-13, PerkinElmer, Cambridge, UK) trough interference filters 655 nm (FF01-655/15-25 Semrock, Laser 2000, Ringstead, UK) and 800 nm (FF01-800/12-25 Semrock, Laser 2000, Ringstead, UK). Detector signals and laser reference pulses were measured by DPC-230 photon correlator operated in a Time-Tagged Time Correlated Single Photon Counting mode (Becker & Hickl, Berlin, Germany). Exported to Matlab (MathWorks, Natick, MA) data were gated in the 3–15 ns lifetime range to discriminate fluorescence background and calculate auto-and cross-correlation functions.
Fig. 1.
Schematics of the photon correlation module. The laser light is focused by objective with high numerical aperture to the diffraction limit on the cell surface. Fluorescence from the confocal volume separated from the excitation by Dichroic beam splitter 1 and filtered by 50 μm Pinhole is split into two channels by Dichroic beam splitter 2 and focused onto SPAD1 and SPAD2 detectors through interference filters 1 and 2.
Autocorrelation function was calculated as
| (1) |
where n(t) and n(t + τ) are 0 or 1, Np is a total number of photons and N is a number of time-intervals (laser periods) in the fluctuation measurement. Cross-correlation between the first and second channels was calculated as
| (2) |
where Np1 and Np2 total numbers of photons in channels 1 and 2.
Relative concentration of a spatially coordinated components were calculated as
| (3) |
3. Results
3.1. Naïve and antigen-experienced T-cells express similar levels of TCR and CD8
The aim of this study was to compare the spatial organisation of the TCR and CD8 in live naïve versus Ag-experienced CD8+ T cells and compare it with T cell sensitivity. Cells were obtained from a TCR transgenic mice in order to keep the TCR and CD8 constant. Naïve T cells were obtained directly from LN of a transgenic F5 Rag1−/− mice and Ag-experienced cells were produced by in vitro stimulation of naïve F5 T cells with NP68 peptide and then further cultured in presence of IL-2.
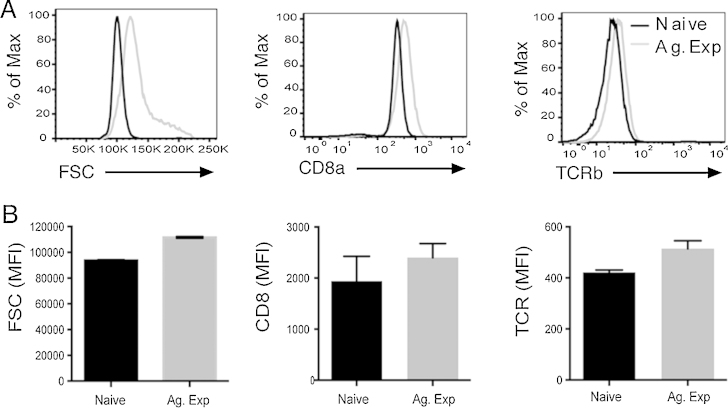
First we compared expression levels of TCR and CD8 in naïve and Ag-experienced cells. Flow cytometry results of the cells reacted with labelled CD8 and TCR-specific mAbs show that Ag-experienced cells expressed slightly higher levels of these molecules than naïve cells. However, Ag-experienced cells were also larger as evaluated from the forward scatter signal (Fig. 2). Hence, the naïve and Ag experienced cells showed similar average densities of TCR and CD8.
Fig. 2.
Relative expression of CD8 and TCR on naïve and Ag experienced cells. Ag-experienced CD8+ T cells were generated by stimulation of splenocytes cells from Rag1−/–F5 mice with 100 nM NP68 for 3 days, rested in 5 μg/mL IL-2 for 4 days, and then compared to naïve LN cells. (A) CD8+ T cells were assessed by flow cytometry to determine cell size (FSC) and CD8α and TCRβ expression. (B) Graphs show the mean values of FSC and CD8α and TCRβ associated fluorescence (MFI). Values are the mean of triplicate samples ± SD. All data are representative of at least two independent experiments.
3.2. Involvement of CD8 in Ag recognition strongly affects T cell sensitivity
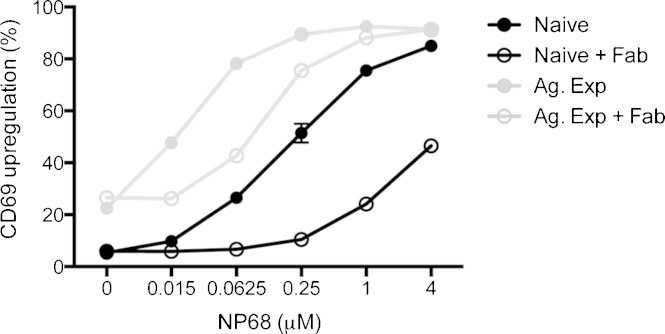
Then we studied sensitivity of naïve and Ag-experienced cells to Ag. CD69 is a convenient marker of T cell activation as its expression rapidly upregulates upon triggering of the TCR [26]. We employed this marker to compare the sensitivity of Ag-experienced and naïve F5 T cells (Fig. 3). As shown previously [27], [28] Ag-experienced F5 T cells are more sensitive to stimulation than naïve F5 T cells. Half-maximal upregulation of CD69 in naïve cells (black closed circles) occurred at ∼16-fold higher peptide concentration than in Ag-experienced cells (gray closed circles), i.e. Ag-experienced cells are ∼16-fold more sensitive than naïve cells. To examine the extent to which CD8 binding to pMHC influences the sensitivity of F5 T cells we blocked the CD8-MHC interaction by Fab fragments (100 μg/mL) of a CD8α-specific mAb (YTS 169.4) before pulsing the cells with NP68 peptide. The Fab concentration required to block the CD8-MHC interaction was measured in a competition binding assay with intact YTS 169.4 antibody (Supplementary Fig. S1). The abrogation of the CD8-pMHC interaction brought about an ∼8-fold reduction in sensitivity of Ag-experienced cells compared to a ∼20-fold reduction in naïve cells showing that CD8 strongly affects sensitivity to antigenic stimulation in the both cell types.
Fig. 3.
Comparison of sensitivity of naïve and Ag experienced cells by upregulation of CD69. Naïve and Ag. Exp. CD8+ T cells were incubated with Fab (100 μg/mL) prior to stimulation with NP68 and assessed by flow cytometry. The graph represents the percentage of cells that upregulated CD69 expression in response to NP68 measured at 3 h. Values are the mean of triplicate samples ± SD. All data are representative of one experiment.
3.3. TCR and CD8 spatial organisation
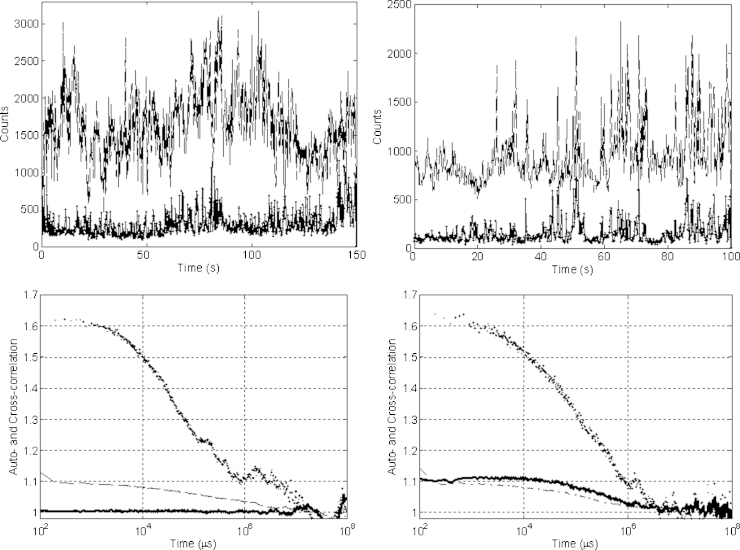
We employed fluorescence lifetime cross-correlation microscopy to investigate TCR and CD8 organisation on the surface of naïve and Ag-experienced cells expressing the same TCR but having different sensitivity. T cells were reacted with mono-functionalised Qdots with either CD8 or TCR-specific mAb, which have non-overlapping emission spectra with maxima at 655 nm or 800 nm, to get optimal density of labelled TCR and CD8 molecules on the cell surface. Live cells were imaged through a 50 μm pinhole aperture and events caused by the passage of individual labelled molecules through the confocal volume were measured. In this manner associations between TCR and CD8 could be monitored as the coincident emission of both wavelengths if both molecules passed through the confocal volume together at the moment of excitation. The upper panels in Fig. 4 show typical fluctuation traces of emission intensity of CD8-Qdot655 (dash line) and TCR-Qdot800 (dots) on the surface of naïve (left panel) and Ag-experienced (right panel) cells taken day 7 post-stimulation. The lower panels show auto-correlation functions of the QD655-CD8 (dash line) and QD800-TCR (dots) fluctuation signals and their cross-correlation function (solid line) for naïve (left panel) and Ag-experienced (right panel) cells. Similar results were observed for Ag-experienced cells between days 5–10 post-stimulation (data not shown). The cross-correlation trace (solid line) shows remarkable coincidences of TCR-Qdot800 and CD8-Qdot655 emission photons excited by the same or adjacent laser pulses in Ag-experienced cells only, indicating that, in this cell population most of the TCR molecules are colocalised with a CD8. In contrast, naïve T cells show much lower level of colocalisation of these two molecules. The amplitudes of the auto-correlation functions can be used to calculate the average number (N) of labelled molecules present in the confocal volume: N = 1/(G(0) − G(∞)), where G(0) and G(∞) are the values of the auto-correlation function in 0 and ∞ moments of time-delay. Thus, we calculate ∼10 CD8 and ∼1.4 TCR labelled molecules on the surface of both naïve and Ag-experienced cells. Fractions of TCR and CD8 correlated molecules can be calculated using Eq. (3). Averaging over 10 cells of each type gave (5 ± 5%) colocalisation for naïve cells and (90 ± 10%) for Ag-experienced cells. Because the surface density of CD8 was 7-fold higher than that of TCR the TCR/CD8 colocalisation implies that a TCR is surrounded by one or several CD8 coreceptors.
Fig. 4.
Proximity measurements of TCR and CD8. Intensity fluctuations of Qdot labelled TCR-Qdot800 (dots) and CD8-Qdot655 (dash line) specific mAbs on the surface of naïve (top left) and Ag-experienced cells (top right). Auto-correlation (TCR-Qdot800 (dots), CD8-Qdot655, (dash line)) and cross-correlation functions (solid line) for naïve (bottom left) and Ag-experienced cells (bottom right). Further examples are shown in Supplementary Figure S2 (Supplementary materials).
4. Discussion
The molecular mechanisms behind the functional affinity maturation of Ag-experienced T cells which endows them with increased sensitivity to stimulation by pMHC are not well understood. In this study we show a different cell surface spatial organisation of TCR and CD8 between live naïve and Ag-experienced CD8+ T cells. These molecules are present as preformed [TCR, CD8] functional ensembles on the surface of Ag-experienced cells while they are predominantly randomly distributed on the surface of naïve T cells. Importantly, the surface density of TCR and CD8 were almost equal in both these cell types, so changes in molecular association were not due to alterations in relative abundance of the two proteins. The sensitivity of Ag-experienced cells was greatly reduced by reacting the cells with CD8α-specific Fab fragments known to abrogate the pMHC-CD8 interaction but to not affect expression levels of TCR or CD8 or concentrations of downstream signalling molecules. Hence, the T cell sensitivity is likely to be controlled by the proximity of CD8 and TCR which interact with pMHC at the very first steps of Ag recognition.
There is a significant difference in the interaction of TCR and CD8 with pMHC. The latter binds to the conservative α3 domain of the MHC heavy chain and to β2m whereas TCR binds to the pMHC antigenic epitope shaped by the peptide structure. The first interaction with the affinity range of 100–200 μM is characterised by a relatively fast on-rate constant (≥105 M−1 s−1) and weak temperature dependence [29], [30], [31]. This binding rate complies with the rate predicted by the “grey spheres” model (5 × 105–5 × 106 M−1 s−1) which is about an order of magnitude smaller than the diffusion controlled limit for globular proteins of this molecular mass, ∼107 M−1 s−1, due to the geometrical factor accounting for their mutual orientation. In contrast, the interaction of TCR with non-self peptide-MHC class I restricted antigen is characterised by higher affinity (1–50 μM) [30], [32] and may involve significant conformational adjustments of the TCR and pMHC binding sites [33], [34] proceeding in the millisecond range [35]. As a result this interaction has typically a slower on-rate constant (102–104 M−1 s−1) and is temperature dependent [36], [37], [38]. Namely this interaction endows the specificity to Ag whereas CD8 can just facilitate antigen recognition and is not always necessary. The binding sites of these two interactions on pMHC are spatially separated by ∼4 nM and no cooperativity was observed in binding of soluble CD8 and TCR to the same pMHC ligand [31]. Nevertheless a cooperation between TCR and CD8 expressed on the cell surface has been reported [32], [39]. Indeed, being anchored in the cell surface membrane and bound to the same pMHC ligand the CD8-MHC interaction can stabilise the TCR–pMHC complex by increasing its rebinding probability. This mechanism has been discussed in the literature [32], [40] and considered as the major mechanism of CD8 cooperation.
Another less obvious mechanism of CD8 cooperation is a possibility to affect the binding rate of the TCR–pMHC interaction. Assume that a T cell forms a stable contact interface with an APC and both TCR and CD8 can interact with the same pMHC ligand by a two-step reversible reaction mechanism which can proceed by one of the following reaction schemes starting either with the association of pMHC with CD8 (Eq. (4)) or with TCR (Eq. (5)). For both these reaction schemes the first step forward rate constants, k1 or k3, are proportional to the surface densities of pMHC and TCR or CD8, and on-rate constants of pMHC-CD8 () or pMHC–TCR () interactions respectively (Eqs. (6), (7)). For simplicity we assume that TCR and CD8 form a functional ensemble [TCR, CD8] with the minimal (1:1) stoichiometry when TCR and CD8 have equal local surface density. Hence, k1 and k3 differ only by the on-rate constants of the pMHC-CD8 or pMHC-TCR interactions. CD8 binds pMHC much faster than TCR and hence the first step operates faster in the first reaction scheme. If TCR and CD8 are colocalised at a short enough distance (≤5 nm), at which they can form a ternary complex with pMHC without involvement of lateral diffusion, their local surface density is to be ≥4 × 104 molecules/μm2. At such a high density the forward rate constants k2 or k4 of the second step will be considerably faster than the corresponding first step backward rate constant, i.e. k2 ≫ k−1 and k4 ≫ k−3. Hence, in both these cases the overall forward rate constants (Eq. (8)) will be close to the corresponding forward rate constants of the first step, i.e. in the first case to the rate of the CD8-pMHC binding () and in the second case to the rate of the TCR–pMHC binding (). Because k1 > k3, the first reaction route dominates in the formation of the pMHC·CD8·TCR ternary complex and the major fraction of pMHC will bind TCR at the rate of association of CD8 with pMHC (k1), i.e. significantly faster than it would be without the CD8 cooperation (k3).
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
A different scenario is expected if TCR and CD8 are randomly distributed on the cell surface and separated such far that the formation of TCR·pMHC·CD8 ternary complex requires lateral diffusion. In this case the time between the first (pMHC-TCR or pMHC-CD8) and the second (TCR·pMHC-CD8 or CD8·pMHC-TCR) binding steps is expected to be ∼0.014 s [16]. The reciprocal of this value gives the frequency factor of the TCR–pMHC on-rate constant ∼70 s−1. Thus, the second step on-rate constant in the first reaction scheme being a product of the frequency factor by the geometrical (∼10−1) and activation energy factors (∼10−2) will be ≤0.1 s−1, whereas the dissociation rate constant of the CD8·pMHC complex is ∼10 s−1, i.e. k−1 ≫ k2. In contrast, the off-rate constant of the first step k−3 in the second reaction scheme is 0.1 − 1 s−1 and the on-rate constant of the second step k4 is ∼1 s−1 (because the activation energy factor for the CD8·pMHC interaction is at least 10-fold larger and than that of the TCR–pMHC interaction) and hence k−3 ≤ k4. Thus, for randomly distributed TCR and CD8 the second reaction scheme (Eq. (5)) predominantly operates where pMHC binds first to TCR and then CD8 binds to this complex. Hence, in this case CD8 can only stabilise the already formed TCR–pMHC complex by binding to the same pMHC ligand and getting thereby spatially coordinated with the TCR.
Summarising the reaction mechanism we note that the formation of [TCR, CD8] functional ensembles can significantly accelerate the binding of pMHC and TCR and switch thereby a T cell to a high sensitivity mode. Importantly, the model predicts that all pMHC ligands irrespective of peptide structure will bind TCR with nearly the same rate close to the rate of the CD8-pMHC association. This high sensitivity mode is more important for slow binding pMHC ligands and less so for those which bind TCR with rate constants comparable with that of the CD8-pMHC interaction, i.e. T cell clones can be more or less CD8-dependent.
In respect to a pMHC ligand a [TCR, CD8] functional ensemble can be considered as a bivalent receptor. However, in contrast with B cell receptor affinities of its binding sites are not equal. A question arises why the evolution developed the mechanism of non-constitutive association of TCR and CD8 to enhance the TCR–pMHC interaction instead of including CD8 into the TCR complex or by increasing TCR affinity? It is apparently because the proximity mechanism allows fine tuning of T cell sensitivity by the possibility to regulate the fraction of functional [TCR, CD8] ensembles and optimise the strength of immune response. Any structural changes in the binding site of a clonally preselected TCR may extend the TCR repertoire and increase the risk of autoimmunity. In contrast, the proximity mechanism does not affect the T cell repertoire but only kinetically promotes the binding of pMHC to TCR by giving an equal chance to any pMHC ligand to produce a transient complex with a TCR with a similar rate constant independently of its structure. At the same time the possibility that a stable TCR–pMHC complex will be formed is strictly determined by the structure of the pMHC antigenic epitope.
Affinity of the CD8-pMHC interaction should be significantly lower than that of a TCR-cognate pMHC interaction to maintain peptide Ag specificity of the TCR–pMHC interaction [41], [42]. This is consistent with our model which suggests that the major role of this interaction is not to stabilise the TCR–pMHC complex but to provide the highest robustness of Ag screening on the surface of APC by stopping every pMHC ligand in front of a TCR and giving to it an equal chance to produce a stable TCR–pMHC complex with a lifetime longer than a threshold of Ag recognition. To provide the highest robustness of the Ag screening CD8 should bind pMHC as quickly as possible and the on-rate constant ≥105 M−1 s−1 is apparently the maximal rate which can be achieved for binding of these two globular proteins. Thus, lifetime of this complex should be long enough to facilitate the formation of a TCR·pMHC complex which in some cases involves millisecond conformational adjustments [35] but on the other hand it should not be too long to avoid increasing the lifetime of the CD8·pMHC·TCR ternary complexes with non-cognate ligands which comprise the major fraction of the pMHC population on the surface of APCs and to not largely reduce in the number of non-engaged TCRs available for Ag recognition. Thus, the delicate balance between CD8-pMHC and TCR–pMHC affinities and the proximity of TCR and CD8 are key factors providing the highest robustness to Ag recognition without compromising its specificity. Lifetime of the CD8-pMHC complex in the range of 50–100 ms is apparently a reasonable trade-off for the above conditions which gives the 100–200 μM range of CD8-pMHC affinity.
Our data show that blocking the CD8-pMHC interaction significantly reduces sensitivity of Ag-experiences cells in accord with the prediction of the kinetic model. However the sensitivity did not decrease to the level of sensitivity of naïve cells. This suggests that together with the spatial CD8-TCR coordination there are other mechanisms affecting sensitivity of T cells. We know that TCRs can form nanoclusters on the surface of Ag-experienced cells [20], [21] and this differs them from naïve cells. Assembling of TCR into nanoclusters increases their local density and consequently the probability of pMHC rebinding, which can also contribute to the sensitivity of Ag-experienced cells.
Previously we showed that CD8-pMHC interaction plays an important role in binding of Qdot-based pMHC nano-sensors to Ag-experienced T cells: the nano-sensors can bind to T cells even when all MHC molecules are “loaded” with a non-cognate peptide but do not bind when the CD8-MHC interaction was abrogated by a mutation in the α3 domain of the MHC heavy chain [43]. This means that CD8 also plays a role of adhesion molecule in the formation of T cell–APC contacts. We suggest that the random distribution of TCR on the surface of naïve cells makes this role even more important for naïve cells whereas in Ag-experienced cells the blockage of the CD8-MHC interaction can be partially compensated by a higher rebinding probability of pMHC to a TCR nanocluster. Therefore, naïve cells greatly loose their capacity to form stable contacts with APCs without the CD8-MHC interaction and their sensitivity dramatically decreases. In addition, the abrogation of CD8-MHC interaction can also reduce lifetime of TCR–pMHC complex which is likely to be me more important for naïve cells.
In conclusion we note that the spatial organisation of the primary signalling molecules is formed in naïve CD8+ T cells by the interaction with Ag and remains over the lifespan of these cells. The proximity of TCR and CD8 increases the rate of Ag recognition and increases thereby the sensitivity of T cells. Detailed molecular mechanisms controlling TCR and CD8 organisation and the switching of T cells from a low to high sensitivity mode are yet to be elucidated.
Acknowledgments
Funding for this project was provided by Royal Society Industry Fellowship and R2 research grant (DMG), MRC project grant G1100116 (JGB&RZ) and Wellcome Trust grant No. 096669/Z/11/Z (RZ). We thank Dr. Anna Gakamsky for help with data evaluation and Dr. Kaija Allikmets for excellent assistance with T cell culture.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.imlet.2013.11.005.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Slifka M.K., Whitton J.L. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat Immunol. 2001;2:711–717. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]
- 2.Walker L.J., Sewell A.K., Klenerman P. T cell sensitivity and the outcome of viral infection. Clin Exp Immunol. 2010;159:245–255. doi: 10.1111/j.1365-2249.2009.04047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Essen M.R., Kongsbak M., Geisler C. Mechanisms behind functional avidity maturation in T cells. Clin Dev Immunol. 2012 doi: 10.1155/2012/163453. 8 pp. [Article ID 163453] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho B.K., Wang C., Sugawa S., Eisen H.N. Functional differences between memory and naive CD8 T cells. Proc Natl Acad Sci U S A. 1999;96:2976–2981. doi: 10.1073/pnas.96.6.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peled J.U., Kuang F.L., Iglesias-Ussel M.D., Roa S. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 6.Flynn K., Mullbacher A. Memory alloreactive cytotoxic T cells do not require costimulation for activation in vitro. Immunol Cell Biol. 1996;74:413–420. doi: 10.1038/icb.1996.71. [DOI] [PubMed] [Google Scholar]
- 7.Veillette A., Bookman M.A., Horak E.M., Bolen J.B. The CD4 and CD8 T cell surface antigens are associated with the internal membrane tyrosine-protein kinase p56lck. Cell. 1988;55:301–308. doi: 10.1016/0092-8674(88)90053-0. [DOI] [PubMed] [Google Scholar]
- 8.Molina T.J., Kishihara K., Siderovski D.P., van Ewijk W. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357:161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- 9.Barber E.K., Dasgupta J.D., Schlossman S.F., Trevillyan J.M. The CD4 and CD8 antigens are coupled to a protein-tyrosine kinase (p56lck) that phosphorylates the CD3 complex. Proc Natl Acad Sci U S A. 1989;86:3277–3281. doi: 10.1073/pnas.86.9.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovatt M., Filby A., Parravicini V., Werlen G. Lck regulates the threshold of activation in primary T cells, while both Lck and Fyn contribute to the magnitude of the extracellular signal-related kinase response. Mol Cell Biol. 2006;26:8655–8665. doi: 10.1128/MCB.00168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmond R.J., Filby A., Qureshi I., Caserta S. T-cell receptor proximal signaling via the Src-family kinases, Lck and Fyn, influences T-cell activation, differentiation, and tolerance. Immunol Rev. 2009;228:9–22. doi: 10.1111/j.1600-065X.2008.00745.x. [DOI] [PubMed] [Google Scholar]
- 12.Palacios E.H., Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23:7990–8000. doi: 10.1038/sj.onc.1208074. [DOI] [PubMed] [Google Scholar]
- 13.Naeher D., Luescher I.F., Palmer E. A role for the alpha-chain connecting peptide motif in mediating TCR-CD8 cooperation. J Immunol. 2002;169:2964–2970. doi: 10.4049/jimmunol.169.6.2964. [DOI] [PubMed] [Google Scholar]
- 14.Bachmann M.F., Gallimore A., Linkert S., Cerundolo V. Developmental regulation of Lck targeting to the CD8 coreceptor controls signaling in naive and memory T cells. J Exp Med. 1999;189:1521–1530. doi: 10.1084/jem.189.10.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cawthon A.G., Alexander-Miller M.A. Optimal colocalization of TCR and CD8 as a novel mechanism for the control of functional avidity. J Immunol. 2002;169:3492–3498. doi: 10.4049/jimmunol.169.7.3492. [DOI] [PubMed] [Google Scholar]
- 16.Gakamsky D.M., Luescher I.F., Pramanik A., Kopito R.B. CD8 kinetically promotes ligand binding to the T-cell antigen receptor. Biophys J. 2005;89:2121–2133. doi: 10.1529/biophysj.105.061671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pecht I., Gakamsky D.M. Spatial coordination of CD8 and TCR molecules controls antigen recognition by CD8+ T-cells. FEBS Lett. 2005;579:3336–3341. doi: 10.1016/j.febslet.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Laugel B., Van Den Berg H.A., Gostick E., Cole D.K. Different T cell receptor affinity thresholds and CD8 coreceptor dependence govern cytotoxic T lymphocyte activation and tetramer binding properties. J Biol Chem. 2007;282:23799–23810. doi: 10.1074/jbc.M700976200. [DOI] [PubMed] [Google Scholar]
- 19.van den Berg H.A., Wooldridge L., Laugel B., Sewell A.K. Coreceptor CD8-driven modulation of T cell antigen receptor specificity. J Theor Biol. 2007;249:395–408. doi: 10.1016/j.jtbi.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong L., Zeng G., Lu X., Wang R.C. NSOM/QD-based direct visualization of CD3-induced and CD28-enhanced nanospatial coclustering of TCR and coreceptor in nanodomains in T cell activation. PLoS ONE. 2009;4:e5945. doi: 10.1371/journal.pone.0005945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar R., Ferez M., Swamy M., Arechaga I. Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity. 2011;35:375–387. doi: 10.1016/j.immuni.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Perica K., Bieler J.G., Edidin M., Schneck J. Modulation of MHC binding by lateral association of TCR and coreceptor. Biophys J. 2012;103:1890–1898. doi: 10.1016/j.bpj.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mamalaki C., Elliott J., Norton T., Yannoutsos N. Positive and negative selection in transgenic mice expressing a T-cell receptor specific for influenza nucleoprotein and endogenous superantigen. Dev Immunol. 1993;3:159–174. doi: 10.1155/1993/98015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rousseaux J., Rousseaux-Prevost R., Bazin H. Optimal conditions for the preparation of Fab and F(ab′)2 fragments from monoclonal IgG of different rat IgG subclasses. J Immunol Methods. 1983;64:141–146. doi: 10.1016/0022-1759(83)90392-7. [DOI] [PubMed] [Google Scholar]
- 25.Cobbold S.P., Jayasuriya A., Nash A., Prospero T.D. Therapy with monoclonal antibodies by elimination of T-cell subsets in vivo. Nature. 1984;312:548–551. doi: 10.1038/312548a0. [DOI] [PubMed] [Google Scholar]
- 26.Arva E., Andersson B. Kinetics of cytokine release and expression of lymphocyte cell-surface activation markers after in vitro stimulation of human peripheral blood mononuclear cells with Streptococcus pneumoniae. Scand J Immunol. 1999;49:237–243. doi: 10.1046/j.1365-3083.1999.00470.x. [DOI] [PubMed] [Google Scholar]
- 27.Pihlgren M., Arpin C., Walzer T., Tomkowiak M. Memory CD44(int) CD8 T cells show increased proliferative responses and IFN-gamma production following antigenic challenge in vitro. Int Immunol. 1999;11:699–706. doi: 10.1093/intimm/11.5.699. [DOI] [PubMed] [Google Scholar]
- 28.Borger J.G., Filby A., Zamoyska R. Differential polarization of C-terminal Src kinase between naive and antigen-experienced CD8+ T cells. J Immunol. 2013;190:3089–3099. doi: 10.4049/jimmunol.1202408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao G.F., Willcox B.E., Wyer J.R., Boulter J.M. Classical and nonclassical class I major histocompatibility complex molecules exhibit subtle conformational differences that affect binding to CD8alphaalpha. J Biol Chem. 2000;275:15232–15238. doi: 10.1074/jbc.275.20.15232. [DOI] [PubMed] [Google Scholar]
- 30.van der Merwe P.A., Davis S.J. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- 31.Wyer J.R., Willcox B.E., Gao G.F., Gerth U.C. T cell receptor and coreceptor CD8 alphaalpha bind peptide-MHC independently and with distinct kinetics. Immunity. 1999;10:219–225. doi: 10.1016/s1074-7613(00)80022-9. [DOI] [PubMed] [Google Scholar]
- 32.Gao G.F., Rao Z., Bell J.I. Molecular coordination of alphabeta T-cell receptors and coreceptors CD8 and CD4 in their recognition of peptide-MHC ligands 5. Trends Immunol. 2002;23:408–413. doi: 10.1016/s1471-4906(02)02282-2. [DOI] [PubMed] [Google Scholar]
- 33.Reiser J.B., Gregoire C., Darnault C., Mosser T. A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex 52. Immunity. 2002;16:345–354. doi: 10.1016/s1074-7613(02)00288-1. [DOI] [PubMed] [Google Scholar]
- 34.Reiser J.B., Darnault C., Gregoire C., Mosser T. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat Immunol. 2003;4:241–247. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- 35.Gakamsky D.M., Lewitzki E., Grell E., Saulquin X. Kinetic evidence for a ligand-binding-induced conformational transition in the T cell receptor. Proc Natl Acad Sci U S A. 2007;104:16639–16644. doi: 10.1073/pnas.0707061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gakamsky D.M., Luescher I.F., Pecht I. T cell receptor-ligand interactions: a conformational preequilibrium or an induced fit. Proc Natl Acad Sci U S A. 2004;101:9063–9066. doi: 10.1073/pnas.0402840101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis-Harrison R.L., Armstrong K.M., Baker B.M. Two different T cell receptors use different thermodynamic strategies to recognize the same peptide/MHC ligand. J Mol Biol. 2005;346:533–550. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 38.Armstrong K.M., Insaidoo F.K., Baker B.M. Thermodynamics of T-cell receptor-peptide/MHC interactions: progress and opportunities. J Mol Recognit. 2008;21:275–287. doi: 10.1002/jmr.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wooldridge L., Van Den Berg H.A., Glick M., Gostick E. Interaction between the CD8 coreceptor and major histocompatibility complex class I stabilizes T cell receptor-antigen complexes at the cell surface. J Biol Chem. 2005;280:27491–27501. doi: 10.1074/jbc.M500555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniels M.A., Jameson S.C. Critical role for CD8 in T cell receptor binding and activation by peptide/major histocompatibility complex multimers. J Exp Med. 2000;191:335–346. doi: 10.1084/jem.191.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melenhorst J.J., Scheinberg P., Chattopadhyay P.K., Lissina A. Detection of low avidity CD8(+) T cell populations with coreceptor-enhanced peptide-major histocompatibility complex class I tetramers. J Immunol Methods. 2008;338:31–39. doi: 10.1016/j.jim.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wooldridge L., Clement M., Lissina A., Edwards E.S. MHC class I molecules with Superenhanced CD8 binding properties bypass the requirement for cognate TCR recognition and nonspecifically activate CTLs. J Immunol. 2010;184:3357–3366. doi: 10.4049/jimmunol.0902398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anikeeva N., Gakamsky D., Scholler J., Sykulev Y. Evidence that the density of self peptide-MHC ligands regulates T-cell receptor signaling. PLoS ONE. 2012;7:e41466. doi: 10.1371/journal.pone.0041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.