Abstract
Two strains of anaerobic, Gram-stain-negative bacilli isolated from the human oral cavity (D033B-12-2T and D080A-01) were subjected to a comprehensive range of phenotypic and genotypic tests and were found to be distinct from any previously described species. 16S rRNA gene sequence analysis revealed that the strains were related most closely to the type strain of Prevotella marshii (93.5 % sequence identity). The novel strains were saccharolytic and produced acetic acid and succinic acid as end products of fermentation. The principal cellular long-chain fatty acids were C16 : 0, iso-C14 : 0, C14 : 0, anteiso-C15 : 0, iso-C16 : 0 and C16 : 0 3-OH. The G+C content of the DNA of strain D033B-12-2T was 44 mol%. Strains D033B-12-2T and D080A-01 are considered to represent a single novel species of the genus Prevotella, for which the name Prevotella saccharolytica sp. nov. is proposed. The type strain is D033B-12-2T ( = DSM 22473T = CCUG 57944T).
Members of the genus Prevotella are frequently isolated from the human oral cavity in oral and dental infections and in health. Strains D033B-12-2T and D080A-01 were among the collection of W. E. C. Moore and L. V. Holdeman Moore, formerly of the Virginia Polytechnic Institute, and had been categorized as belonging to Bacteroides group D33 (Moore et al., 1985). Preliminary screening based on partial 16S rRNA gene sequence analysis suggested that the strains belonged to the same taxon of the genus Prevotella but were distinct from recognized species. Strain D033B-12-2T was isolated from a 9-mm-deep periodontal pocket in an individual with juvenile periodontitis and strain D080A-01 from supragingival plaque in a periodontally healthy subject.
The strains were grown at 37 °C on fastidious anaerobe agar (FAA; LabM) supplemented with 5 % horse blood, under anaerobic conditions (80 % N2, 10 % H2, 10 % CO2) in an anaerobic workstation (Don Whitley Scientific). Colonial morphologies were determined by using a dissecting microscope after 4 days incubation. Cellular morphology was recorded after Gram-staining of smears prepared from 2-day-old FAA plate cultures. Hanging-drop preparations of 18 h cultures of peptone/yeast extract/glucose (PYG) broth (Holdeman et al., 1977) were examined by phase-contrast microscopy to investigate cellular motility. The range and optimum temperature for growth were determined after 48 h incubation in pre-reduced PYG broth that had been dispensed into pre-reduced, anaerobically sterilized (PRAS) tubes in an anaerobic workstation (Don Whitley Scientific). The range and optimum pH for growth were determined in peptone/yeast extract (PY) broth (Holdeman et al., 1977) incubated at 35 °C for 48 h with the initial pH adjusted by adding HCl (0.2 M) or Na2CO3 (10 %, w/v) to the PY broth.
Biochemical and physiological tests were performed by using standard methods (Jousimies-Somer et al., 2002). Fermentation tests were performed by using PRAS sugars prepared in-house in an anaerobic workstation (Holdeman et al., 1977). Susceptibility to special-potency antibiotic discs, vancomycin (5 µg), kanamycin (1 mg) and colistin (10 µg), was determined on FAA (Jousimies-Somer et al., 2002). Bacterial strains were grown in PY broth (Holdeman et al., 1977) with and without glucose, and short-chain volatile and non-volatile fatty acids produced as metabolic end products were extracted by standard methods and were analysed by GC (Holdeman et al., 1977). Enzyme profiles were generated with the Rapid ID 32A anaerobe identification kit (bioMérieux), according to the manufacturer’s instructions, by using bacteria harvested from Columbia agar plates (LabM) supplemented with 5 % horse blood in triplicate. The G+C content of the DNA of strain D033B-12-2T was determined by HPLC as described by Wade et al. (1999).
Analysis of cellular fatty acids was carried out by the Identification Service of the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany. Fatty acid methyl esters (FAMEs) were obtained from 50 mg (dry weight) cells by saponification, methylation and extraction by using minor modifications of the methods described by Kuykendall et al. (1988) and Miller (1982). The FAME mixtures were separated by using a Sherlock Microbial Identification System (MIS) (MIDI, Microbial ID), which consisted of an Agilent model 6890N gas chromatograph fitted with a 5 % phenyl-methyl silicone capillary column (0.2 mm×25 m), a flame-ionization detector, Agilent model 7683A automatic sampler and HP computer with MIDI database (Hewlett Packard Co.). Peaks were automatically integrated, and fatty acids were identified and their percentages calculated via the MIS Standard Software (Microbial ID). GC parameters were as follows: carrier gas, ultra-high-purity hydrogen; column head pressure, 60 kPa; injection volume, 2 µl; column split ratio, 100 : 1; septum purge, 5 ml min−1; column temperature, 170–270 °C at 5 °C min−1; injection port temperature, 240 °C; and detector temperature, 300 °C.
The 16S rRNA genes of the two strains were sequenced as described by Downes et al. (2005). Sequences were assembled by using the BioEdit program (Hall, 2004) and their closest relatives were identified by blast interrogation of the GenBank database (Altschul et al., 1990). Sequences were aligned by using clustal w within the BioEdit program. Phylogenetic trees were constructed via mega version 4 (Tamura et al., 2007), by using the neighbour-joining method, from distance matrices prepared with the Jukes–Cantor correction.
The results of phenotypic tests for the two strains are summarized in the species description below and in Supplementary Table S1 in IJSEM Online. Cells of strains D033B-12-2T and D080A-01 were obligately anaerobic, non-motile, non-pigmented, Gram-negative rods that were 0.6–0.7 µm wide and 0.9–3 µm long (occasionally up to 5 µm long). After 4 days of incubation on FAA plates, colonies were 0.9–1.2 mm in diameter, circular, entire, convex, smooth, opaque and grey with an off-white centre when viewed under a plate microscope. The optimum temperature for growth was 35 °C, with good growth at 30 °C, marginal growth at 25 and 42 °C and no growth at 20 or 45 °C. The optimum pH for growth was pH 7, with reduced growth at pH 6 and no growth at pH 5 or 8.
The two strains were resistant to vancomycin, kanamycin and colistin. Growth in PY broth produced a moderately turbid suspension (2–3+ on a scale of 0–4+) and growth was enhanced by the addition of 1 % fermentable carbohydrates (3–4+). Strains D033B-12-2T and D080A-01 were saccharolytic (sugar reactions are given in the species description) and moderate amounts of acetic acid and succinic acid were produced as end products of metabolism. The two strains hydrolysed aesculin and hydrolysed gelatin weakly but other biochemical tests were negative (see species description). There was no growth on 20 % bile. The G+C content of the DNA of strain D033B-12-2T was 44 mol%.
Strains D033B-12-2T and D080A-01 gave strong positive reactions in the Rapid ID 32A panel for β-galactosidase, α-glucosidase, α-arabinosidase, N-acetyl-β-glucosaminidase, alkaline phosphatase, leucyl glycine arylamidase, alanine arylamidase and glutamyl glutamic acid arylamidase and weakly positive reactions for α-galactosidase, β-galactosidase 6-phosphate, β-glucosidase and raffinose fermentation. Reactions for mannose fermentation were variable and weak and negative reactions were obtained for the remaining 16 enzymes. These results thus corresponded to a Rapid ID 32A profile 4735/7 4402 02.
The cellular FAME profile of strain D033-12-2T is given in Supplementary Table S2. The principal cellular long-chain fatty acids were C16 : 0 (24.4 %), iso-C14 : 0 (15.7 %), C14 : 0 (11.4 %), anteiso-C15 : 0 (11.4 %), iso-C16 : 0 (9.5 %) and C16 : 0 3-OH (12.2 %), consistent with those of recognized Prevotella species analysed previously (Downes et al., 2007).
Phylogenetic analysis of the 16S rRNA gene sequences of strains D033B-12-2 and D080A-01 and tree construction via the neighbour-joining method revealed that they belonged to the genus Prevotella (Fig. 1; an extended version of this tree is presented in Supplementary Fig. S1). Maximum-parsimony analysis yielded a tree with virtually identical topology (Supplementary Fig. S2). The two strains showed 99.4 % 16S rRNA gene sequence similarity over 1452 unambiguously aligned bases, and were related most closely to the type strain of Prevotella marshii (93.5 % similarity between strain D033-12-2T and P. marshii E9.34T).
Fig. 1.
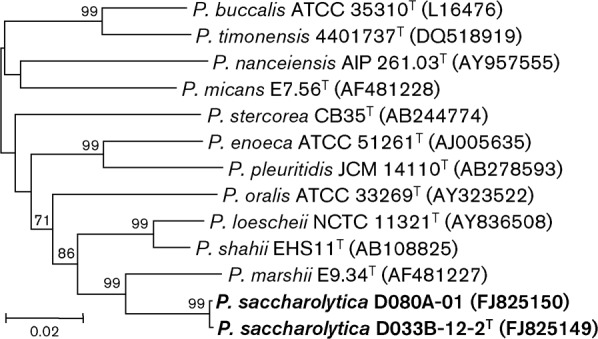
Phylogenetic tree based on 16S rRNA gene sequence comparisons over 1360 aligned bases showing the relationship between strains D033B-12-2T and D080A-01 and related Prevotella species. The tree was constructed by using the neighbour-joining method from a distance matrix constructed from aligned sequences with the Jukes–Cantor correction. The tree was rooted with Porphyromonas endodontalis ATCC 35406T (GenBank accession no. AY253728; not shown). Numbers at nodes are bootstrap percentages based on 500 replications; only values >50 % are shown. Accession numbers are given in parentheses. Bar, 0.02 substitutions per site. An extended version of this tree is presented in Supplementary Fig. S1.
The strains studied here constitute a homogeneous group that is clearly distinct from any recognized species. Strains D033B-12-2 and D080A-01 are thus considered to represent a novel species of the genus Prevotella, for which we propose the name Prevotella saccharolytica sp. nov. Phenotypic characteristics that distinguish P. saccharolytica from related Prevotella species are shown in Table 1.
Table 1. Differential phenotypic characteristics between strains D033B-12-2T and D080A-01 and related Prevotella species.
Taxa: 1, strains D033B-12-2T and D080A-01 (data from this study); 2, P. oralis (Shah & Collins, 1990); 3, P. micans (Downes et al., 2009); 4, P. buccalis; 5, P. loescheii (both from Shah & Collins, 1990); 6, P. nanceiensis (Alauzet et al., 2007); 7, P. shahii (Sakamoto et al., 2004); 8, P. stercorea (Hayashi et al., 2007); 9, P. enoeca (Moore et al., 1994); 10, P. pleuritidis (Sakamoto et al., 2007); 11, P. timonensis (Glazunova et al., 2007); 12, P. marshii (Downes et al., 2005). +, Positive; −, negative; w, weakly positive; v, variable.
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| Pigment* | − | − | + | − | + | − | + | − | − | − | − | − |
| Fermentation of: | ||||||||||||
| Arabinose | + | − | − | − | − | − | − | − | − | − | − | − |
| Cellobiose | + | + | + | + | + | v | − | − | − | − | − | − |
| Lactose | + | + | + | + | + | + | + | + | + | + | + | − |
| Mannose | + | + | + | + | + | + | + | + | v | + | − | v |
| Raffinose | + | + | + | + | + | + | + | + | − | − | − | − |
| Salicin | + | + | + | − | − | − | − | − | − | − | − | − |
| Sucrose | + | + | + | + | + | + | + | + | − | − | − | − |
| Indole | − | − | + | − | − | − | − | − | − | − | − | − |
| Aesculin | + | + | − | + | + | + | − | − | v | − | − | − |
| Gelatin | w | v | + | − | + | − | + | − | + | + | + | + |
*Pigmentation on blood agar may take up to 14 days and varies from tan to brown to black depending on the species.
Description of Prevotella saccharolytica sp. nov.
Prevotella saccharolytica (sac.cha.ro.ly′ti.ca. Gr. n. saccharon sugar; N.L. fem. adj. lytica from Gr. fem. adj. lutikê able to loosen; N.L. fem. adj. saccharolytica saccharolytic, breaking down multiple sugars).
The description is based on two strains isolated from the human oral cavity. Cells are obligately anaerobic, non-motile, non-pigmented, Gram-negative bacilli (0.6–0.7×0.9–5 µm). After 4 days of incubation on FAA plates, colonies are 0.9–1.2 mm in diameter, circular, entire, convex, smooth, opaque and grey with an off-white centre. The optimum temperature and pH for growth are 35 °C and pH 7. Growth in broth media produces a moderate turbidity that is enhanced by the addition of fermentable carbohydrates. Cells are saccharolytic and are able to ferment arabinose, cellobiose, fructose, glucose, lactose, maltose, mannose, raffinose, salicin and sucrose, but not mannitol, melezitose, sorbitol or trehalose. Moderate amounts of acetic acid and succinic acid are produced as end products of metabolism. Hydrolyses aesculin and gelatin (weakly) but not arginine or urea. Indole and catalase are not produced and nitrate is not reduced. The Rapid ID 32A enzyme profile is 4735/7 4402 02. There is no growth in 20 % bile. The principal cellular long-chain fatty acids are C16 : 0, iso-C14 : 0, C14 : 0, anteiso-C15 : 0, iso-C16 : 0 and C16 : 0 3-OH. The species is Human Oral Taxon 781 in the Human Oral Microbiome Database (http://www.homd.org). The G+C content of the DNA of the type strain is 44 mol%.
The type strain, D033B-12-2T ( = DSM 22473T = CCUG 57944T), was isolated from dental plaque in the human oral cavity. D080A-01, also isolated from dental plaque, is a second strain of the species.
Acknowledgements
This research was supported by grants from the Guy’s and St Thomas’ Charity (ref. R050724) and the US National Institutes of Health (DE015847 and DE016937). The authors acknowledge financial support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London.
Abbreviations:
- FAME
fatty acid methyl ester
- PRAS
pre-reduced, anaerobically sterilized
Footnotes
Neighbour-joining and maximum-parsimony phylogenetic trees based on 16S rRNA gene sequence comparison and tables detailing phenotypic characteristics and FAMEs of the novel strains are available as supplementary material with the online version of this paper.
References
- Alauzet C., Mory F., Carlier J. P., Marchandin H., Jumas-Bilak E., Lozniewski A. (2007). Prevotella nanceiensis sp. nov., isolated from human clinical samples. Int J Syst Evol Microbiol 57, 2216–2220 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410 [DOI] [PubMed] [Google Scholar]
- Downes J., Sutcliffe I., Tanner A. C., Wade W. G. (2005). Prevotella marshii sp. nov. and Prevotella baroniae sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 55, 1551–1555 [DOI] [PubMed] [Google Scholar]
- Downes J., Sutcliffe I. C., Booth V., Wade W. G. (2007). Prevotella maculosa sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 57, 2936–2939 [DOI] [PubMed] [Google Scholar]
- Downes J., Liu M., Kononen E., Wade W. G. (2009). Prevotella micans sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 59, 771–774 [DOI] [PubMed] [Google Scholar]
- Glazunova O. O., Launay T., Raoult D., Roux V. (2007). Prevotella timonensis sp. nov., isolated from a human breast abscess. Int J Syst Evol Microbiol 57, 883–886 [DOI] [PubMed] [Google Scholar]
- Hall T. (2004). BioEdit. Biological sequence alignment editor for Win95/98/NT/2K/XP. Carlsbad, CA: Ibis Biosciences [Google Scholar]
- Hayashi H., Shibata K., Sakamoto M., Tomita S., Benno Y. (2007). Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 57, 941–946 [DOI] [PubMed] [Google Scholar]
- Holdeman L. V. H., Cato E. P., Moore W. E. C. (1977). Anaerobe Laboratory Manual, 4th edn Blacksburg, VA: Virginia Polytechnic Institute and State University [Google Scholar]
- Jousimies-Somer H., Summanen P., Citron D. M., Baron E. J., Wexler H. M., Finegold S. M. (2002). Wadsworth Anaerobic Bacteriology Manual, 6th edn Belmont, CA: Star Publishing [Google Scholar]
- Kuykendall L. D., Roy M. A., O’Neill J. J., Devine T. E. (1988). Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradorhizobium japonicum. Int J Syst Bacteriol 38, 358–361 [Google Scholar]
- Miller L. T. (1982). Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol 16, 584–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W. E. C., Holdeman L. V., Cato E. P., Smibert R. M., Burmeister J. A., Palcanis K. G., Ranney R. R. (1985). Comparative bacteriology of juvenile periodontitis. Infect Immun 48, 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. V., Johnson J. L., Moore W. E. (1994). Descriptions of Prevotella tannerae sp. nov. and Prevotella enoeca sp. nov. from the human gingival crevice and emendation of the description of Prevotella zoogleoformans. Int J Syst Bacteriol 44, 599–602 [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Suzuki M., Huang Y., Umeda M., Ishikawa I., Benno Y. (2004). Prevotella shahii sp. nov. and Prevotella salivae sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 54, 877–883 [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Ohkusu K., Masaki T., Kako H., Ezaki T., Benno Y. (2007). Prevotella pleuritidis sp. nov., isolated from pleural fluid. Int J Syst Evol Microbiol 57, 1725–1728 [DOI] [PubMed] [Google Scholar]
- Shah H. N., Collins D. M. (1990). Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int J Syst Bacteriol 40, 205–208 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). mega4: molecular evolutionary genetics analysis (mega) software version 4.0. Mol Biol Evol 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- Wade W. G., Downes J., Dymock D., Hiom S. J., Weightman A. J., Dewhirst F. E., Paster B. J., Tzellas N., Coleman B. (1999). The family Coriobacteriaceae: reclassification of Eubacterium exiguum (Poco et al. 1996) and Peptostreptococcus heliotrinreducens (Lanigan 1976) as Slackia exigua gen. nov., comb. nov. and Slackia heliotrinireducens gen. nov., comb. nov., and Eubacterium lentum (Prevot 1938) as Eggerthella lenta gen. nov., comb. nov. Int J Syst Bacteriol 49, 595–600 [DOI] [PubMed] [Google Scholar]


