SUMMARY
A handful of tumor-derived cell lines form the mainstay of cancer therapeutic development, yielding drugs with impact typically measured as months to disease progression. To develop more effective breast cancer therapeutics and more readily understand their clinical impact, we constructed a functional metabolic portrait of 46 independently-derived breast cell lines. Our analysis of glutamine uptake and dependence identified a subset of triple negative samples that are glutamine auxotrophs. Ambient glutamine indirectly supports environmental cystine acquisition via the xCT antiporter, which is expressed on 1/3 of triple negative tumors in vivo. xCT inhibition with the clinically approved anti-inflammatory Sulfasalazine decreases tumor growth revealing a therapeutic target in breast tumors of poorest prognosis, and a lead compound for rapid, effective drug development.
INTRODUCTION
Current cancer therapeutic development methodologies are expensive, slow, and unable to predict early on, the palette and prevalence of responses tumors can mount when challenged with a prototypic drug. Breast cancer is a challenging example. Large heterogeneity exists within and between well-established subtypes and drug responses (reviewed in Weigelt and Reis-Filho, 2009). Three distinct nomenclatures group breast tumors based on: morphological criteria (e.g. ductal, lobular, invasive, in situ); expression of the estrogen receptor (ER), progesterone receptor (PR), and Her2 receptor tyrosine kinase (Her2); or molecular phenotype, derived from comprehensive mRNA similarities (e.g. luminal, basal). About 1/4 of breast tumors are "triple negative" (ER− / PR− / Her2− ; TNBC), and usually basal molecular phenotype. They are aggressive, with poorest prognosis, high mitotic index, and intrinsic DNA damage repair defects (reviewed in Weigelt and Reis-Filho, 2009; Alli et al., 2009). A subset termed claudin low, and related metaplastic tumors have rapid disease courses, stem cell features, and chemotherapy-resistant characteristics (Hennessy et al., 2009, Prat et al., 2010). No TNBC-targeted therapeutic exists, and patient prognosis is grim.
Many tumors increase uptake and reliance on environmental nutrients such as glucose, glutamine (reviewed in Souba, W. W., 1993; Gatenby and Gillies, 2004; DeBerardinis, et al., 2008), cystine, and asparagine (Iglehart, et al., 1977; Asselin, et al., 1989). Seminal work in tumor series of increasing proliferation rate and de-differentiation (Erlich ascites, Knox et al., 1970; Morris hepatoma, Linder-Horowitz et al., 1969; Nb2 lymphoma, Gout et al., 1997) correlated these features with malignant progression, fostering drug development efforts focused on specific nutrients. But resulting nutrient mimetics were systemically toxic (reviewed in Souba, W. W., 1993), and inexplicable variability among increasing numbers of tumor isolates eventually discouraged these endeavors. Only leukemia dependence on asparagine was successfully pursued to a molecular understanding and effective drug (Asparaginase, reviewed in Narta, et al., 2007).
The xCT cystine/glutamate antiporter is the major means of increasing cystine uptake and the rate limiting step for glutathione (GSH) synthesis in fibroblasts, rat hepatocytes, and Nb2 lymphoma (Bannai, S., and Tateishi, N.,1986; Gout et al., 1997). Dual roles in reactive oxygen species (ROS) neutralization and detoxification of xenobiotics such as chemotherapeutics make GSH an appealing drug target. But inhibitors of glutathione synthesis failed clinical trials due to toxicities related to systemic GSH depletion (reviewed in Hamilton and Batist, 2004). xCT may provide a target for cell-specific GSH depletion. Drug screens identified off target effects of the anti inflammatory pro drug Sulfasalazine (SASP) as an xCT inhibitor (Gout, P.W., et al., 2001). SASP, glutamate, monosodium glutamate, and chemical inhibitors of xCT reduce GSH, increase ROS, potentiate chemotherapeutic effects, and attenuate growth in a handful of tumor-derived cell lines in vitro and xenografts (reviewed in: Lo et al. 2008a). SASP is labile and insoluble under physiological conditions, limiting anti-xCT use to preclinical experiments. Other effects ascribed to SASP include the on-target anti inflammatory activity of an SASP metabolite, NFkB inhibition, and direct interaction with GSH in cell free extracts.
Molecular explanations for glutamine reliance remain elusive although the phenomenon is well described (Coles and Johnstone, 1962; Kovacevic and Morris, 1972; Reitzer et al., 1979; DeBerardinis et al., 2007; Yuneva et al., 2007; Wise et al., 2008). Glutamine provides carbon and nitrogen for independent metabolic events, either directly, (e.g. nucleotide and protein synthesis), via the de-amidated product glutamate (e.g. polysaccharide synthesis, membrane antiporter activities), or via further glutamate deamination to 2-ketoglutarate (2-kg; e.g. respiratory/TCA cycle substrates; reviewed in Deberardinis and Cheng 2010). Abundant serum levels maintained by skeletal muscle reserves allow most cells to be glutamine consumers, although they may also be capable of synthesis (reviewed in Curthoys and Watford, 1995; Kovacevic and McGivan 1983). Glutaminase (GLS; GLS) and glutamine synthase (GS; GLUL) modulate intracellular glutamine/glutamate levels; GLS deamidates glutamine, producing glutamate and ammonia, while GS synthesizes glutamine from these products. Reciprocal expression precludes futile substrate cycling (Curthoys and Watford, 1995; Kovacevic and McGivan 1983). GLS expression levels were correlated with proliferation rate, respiratory glutamine use, and environmental glutamine reliance (Knox et al., 1970; Linder-Horowitz et al., 1969; reviewed in Wise et al., 2008). Thus GLS is a commonlyproposed biomarker of glutamine-dependence and therapeutic target (Lobo et al., 2000; Lora et al., 2004; Wang et al., 2010, Yuneva et al. 2007; van den Heuvel et al., 2012), and GLUL a marker of glutamine independence (Collins et al., 1998; Kung et al., 2011).
Resurgence of interest in nutrient reliance followed realizations that oncogenes can direct nutrient uptake and dependence (reviewed in Wise and Thompson 2010). But as in other preclinical discoveries, findings in exemplar cell lines do not identify appropriate patient populations or estimate their sizes. For example, the metabolic effects of oncogenic Myc vary substantially between tissue types (Yuneva et al., 2012), making a simple cause-effect relationship between oncogenic Myc and glutamine dependence unlikely. Nor does restricting expectations to tumors of one tissue type improve the probable efficacy; in singular glioma lines oncogenic Myc confers glutamine reliance (Wise et al., 2008), but studies of multiple glioma lines report little glutamine dependence (Dranoff et al., 1985). Also, naturally-arising tumors without dominant oncogenic driver may overcome nutrient scarcity by switching nutrient sources (Zielke et al., 1978), attenuating cell cycle progression (Jones et al., 2005), or other activities reported in non-transformed cells. Finally, nutritional requirements of tumors versus proliferating normal cells, rather than quiescent tissues have seldom been reported (exceptions: Iglehart et al., 1977; Jelluma et al., 2006), but are a critical part of the therapeutic development puzzle. Here we present analyses of metabolic activities implied by microarray data, and identification of therapeutic targets enriched in basal and claudin low TNBC.
RESULTS
Expression profiles of culture adapted, proliferating non-tumorigenic cells (human mammary epithelial cell derivatives; HMECd); freshly purified normal breast epithelia; and purified tumor cells from patient pleural effusions were derived and merged with previously published expression profiles of 45 independently-derived breast carcinoma cell lines (Supplemental Methods, Neve et al., 2006). These represent all major breast cancer subtypes and common breast oncogenes in their natural genetic contexts (Figure 1A; Table S1, media, and name abbreviations). Significance analysis contrasting purified tumors and tumor-derived cell lines against purified normal breast epithelia and HMECd lines identified about 760 differentially expressed probeset IDs (>30%) encoding metabolic proteins (Figure S1A; Table S2).
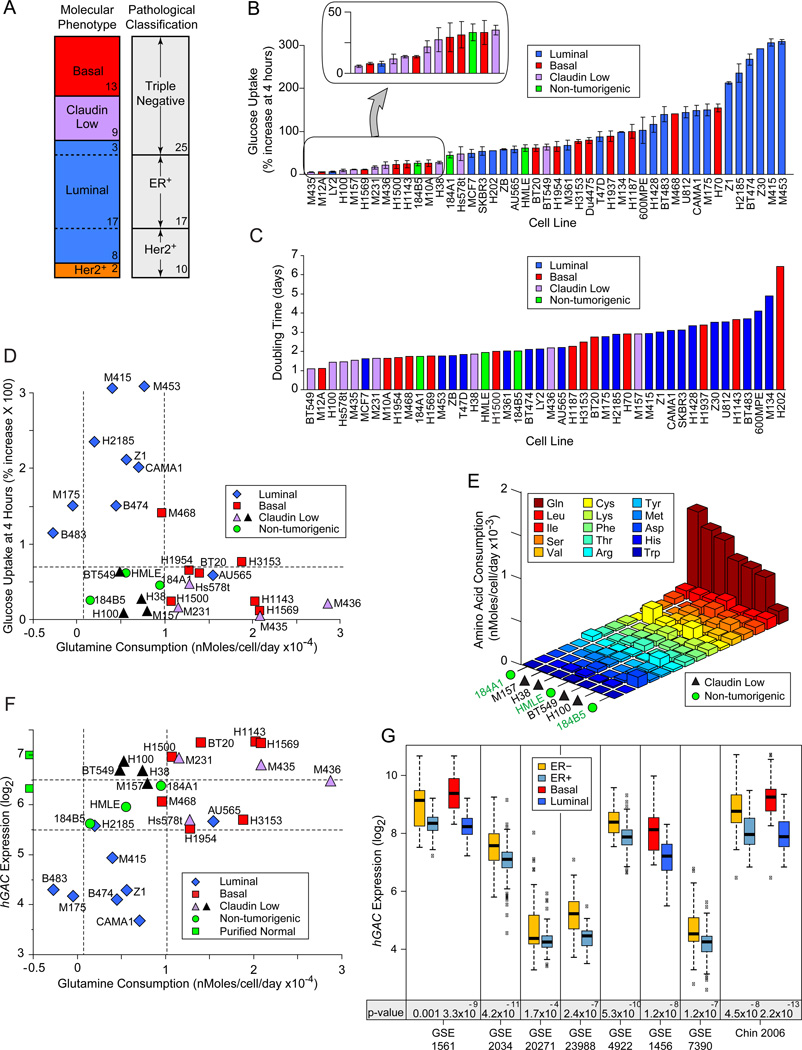
Figure 1. Nutrient Consumption in Breast-Derived Cell Lines.
Icon codes in figure keys; dotted lines bracket proliferating, non-tumorigenic sample values. Icons represent mean values +/− SD. (A) Comparison of two nomenclatures used to describe samples in our study. Sample numbers of each type indicated. (B) Fluorescence increase of cell lines after 4 hour culture with 2-NBDG or 6-NBDG -labeled glucose relative to unlabeled cultures. (C) Population doubling times, calculated from standard growth curves. (D) Glucose uptake at 4 hrs (from part B) vs. glutamine consumption, derived from media depletion at 24 hours culture, normalized to cell number. (E) Amino acid consumption by 4 claudin low samples that consume little glucose or glutamine compared to HMECd samples. Values derived as in D; standard three letter amino acid codes in key. (F) Glutamine consumption at 24 hrs vs. hGAC GeneChip hybridization signal. Green squares on y-axis are the average hGAC signals from 3 CD10+, and 3 BerEP4+ purified normal breast samples. (G) Differential hGAC expression by basal vs. luminal or ER+ vs. ER− samples in 8 clinical breast tumor datasets, downloaded from NCBI GEO and Chin 2006 (Chin et al., 2006). t-test p-values below paired box plots. See also Figure S1, and Tables S1, S2, S3.
Nutrient Preference Varies Widely Among Breast Tumors
Elevated glucose consumption relative to adjacent quiescent tissue is a canonical hallmark of tumors, but whether this is due to proliferative glycolytic demands or specific oncogenic activities is unclear. In analysis of in vitro glucose uptake rates by tumorigenic versus HMECd lines, we found highest glucose consumption in luminal carcinoma-derived isolates (Figure 1B). Claudin low TNBC and about 1/3 of basal samples consume as little or less glucose than HMECd, although they preserve many aspects of the aggressive tumors from which they are derived; (e.g. similar molecular signatures (Neve et al., 2006), rapid doubling times, (Figure 1C) and extracellular matrix invasiveness (Sommers et al., 1994, Han et al., 2010)). Thus glucose consumption varies widely among breast isolates, and tumorigenic lines do not necessarily consume more glucose than non-tumorigenic proliferating cells.
Glutamine is an alternate bioenergetic substrate for many metabolic processes. In amino acid depletion analyses, we found that most basal and claudin low TNBC consume more glutamine than the luminal or HMECd samples (Figure 1D). However, at least 4 claudin low TNBC consume little glutamine (black triangles), and do not significantly increase consumption of another amino acid over that of the HMECd lines, with the possible exception of cystine (Figure 1E). Thus while nutrient preference often associates with molecular phenotype, the 4 claudin low exceptions reveal this as a generalization. Significantly enhanced nutrient consumption may not be an absolute requisite of aggressive breast tumors.
Reduced hGAC Expression Identifies Luminal Breast Carcinomas
We tested historical associations between glutamine consumption and glutaminase expression in our cells. Probeset IDs targeting most portions of GLS (p= 0.6, 0.9, 0.4), and total GLS protein levels (Figure S1B, p= 0.19) are not statistically different between tumor molecular subtypes. But a GLS carboxy terminal splice variant (hGAC) is reduced in luminal carcinomas (Figure 1F; hGAC: 221510_s_at; p= 3.1E−9; Elgadi et al., 1999; Figure S1C). hGAC but not other GLS probeset ID signals are also lower in ER+ and luminal tumors compared to ER− and basal tumors in 8/8 clinical breast tumor microarrays examined (Figure 1G; Table S3). Thus reduced hGAC expression is a strong indicator of luminal carcinoma identity (t-test luminal vs. all p= 4.1E−13). While hGAC expression and glutamine consumption modestly correlate (Figure 1F, p= 0.005), the exceptions in our panel (black icons between dotted vertical lines) reveal that hGAC and GLS probeset IDs poorly identify high glutamine consumers.
Glutamine Restriction Slows Expansion of Most Breast Cell Lines
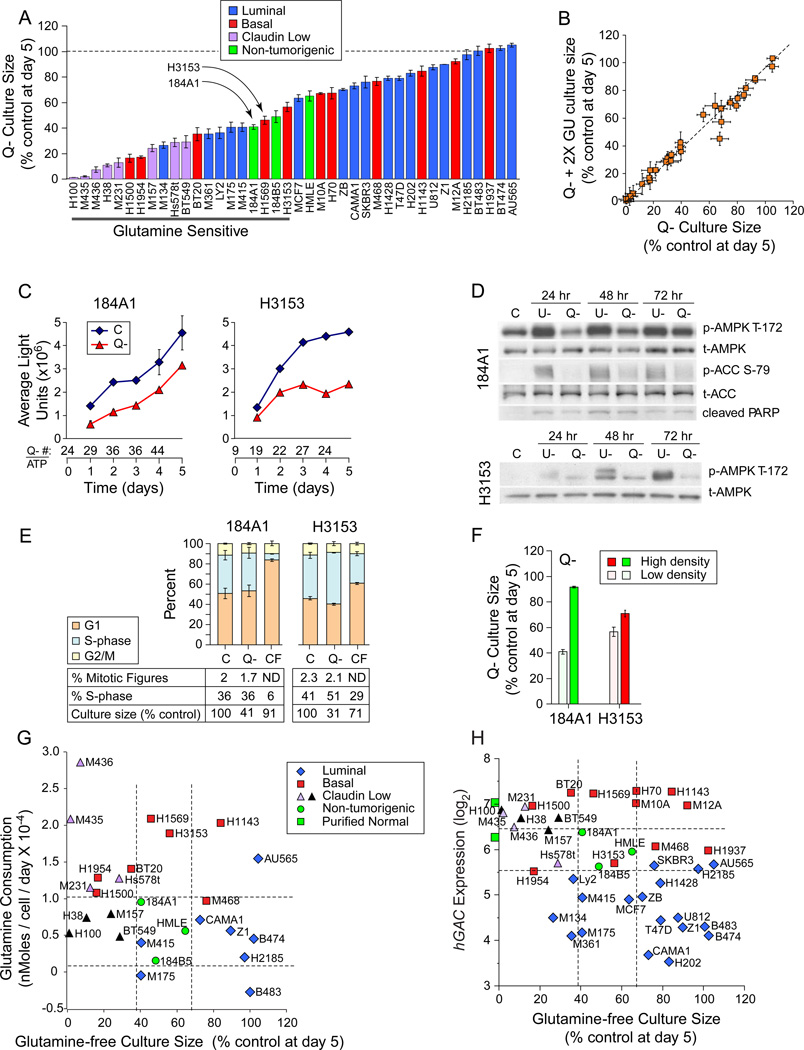
Comparison of glutamine restriction responses between tumorigenic and HMECd samples revealed that 68% of luminal and 54% of basal TNBC samples were more restriction-resistant than HMECd (Figure 2A). Restriction deficits were not rescued by increasing the glucose concentration 2 or 5 fold (Figure 2B, 2X glucose; 5X not shown), suggesting that cells do not simply switch nutrients like fibroblasts and Erlich ascites (Zielke et al., 1978; Kvamme and Svenneby, 1961), to exhaust media glucose. Growth curves of HMECd and similarly sensitive tumor lines revealed that glutamine restriction simply slows culture expansion of each cell type (Figure 2A, examples at arrows; Figure 2C; Figure S2A). Unlike glucose restriction, glutamine restriction produces little AMPK activation (T-172 phosphorylation) indicative of ATP depletion, and little ACC phosphorylation that would inhibit fatty acid synthesis (Figure 2D). Only a modest increase in cell number / ATP ratios (Figure 2C, Q- # / ATP) and little evidence of apoptosis by Annexin V staining (data not shown), PARP cleavage (Figure 2D), or nuclear morphology (Figures S2B-E) are seen. Day 5 cell cycle fractions and mitotic figure counts are also unchanged (Figure 2E). Culture confluence reduces S-phase fractions and glutamine sensitivity, indicating that proliferative drive imparts glutamine reliance (Figure 2E C vs. CF, % S-phase; Figure 2F). These data predict that normal proliferating breast progenitors and most breast tumors would survive a glutamine restricting therapeutic by simply slowing expansion rates. Glutamine-free culture sizes do not correlate with glutamine consumption (Figure 2G, p= 0.13) or hGAC expression (Figure 2H, p=0.014). We conclude that historical correlations between these parameters are not applicable to breast tumors.
Figure 2. Glutamine Restriction Slows Culture Expansion.
Icon codes in figure keys; dotted lines bracket non-tumorigenic sample values; C, complete media; Q-, glutamine-free media; U-, glucose free media. Icons represent mean values +/−SD. (A) Day 5 culture sizes for each cell line grown in glutamine-free media, normalized to culture in control media. (B) Day 5 culture sizes in glutamine-free media with twice the normal glucose concentration (2X GU, y-axis) vs. glutamine-free media with normal glucose levels (x-axis), each normalized to culture in control media. (C-F) Glutamine deprivation responses in the non-tumorigenic exemplar 184A1 and a similarly-sensitive tumorigenic line H3153 (arrows, panel A). (C) Growth curves derived from Cell Titer Glow/ATP content analysis. Numbers below the x-axis, ratio of cell number derived from manual counting (trypan blue) / ATP values. (D) Comparison of AMPK activating phosphorylation, PARP cleavage, and ACC inhibitory phosphorylation. (E) Cell cycle distribution, mitotic figure counts, S-phase fractions, and culture sizes at day 5 culture in indicated conditions. CF, confluent cells in complete media. (F) Day 5 culture sizes differences for high density vs. low density cultures in glutamine-free media. (G) Glutamine consumption (from Figure 1D) vs. glutamine-free culture sizes from part A. (H) hGAC GeneChip hybridization signal vs. glutamine-free culture sizes from part A. Green squares on y-axis are the average hGAC hybridization signals from 3 CD10+, and 3 BerEP4+ purified normal breast samples. See also Figure S2.
Restriction Induces S-phase Stalling in a Subset of TNBC
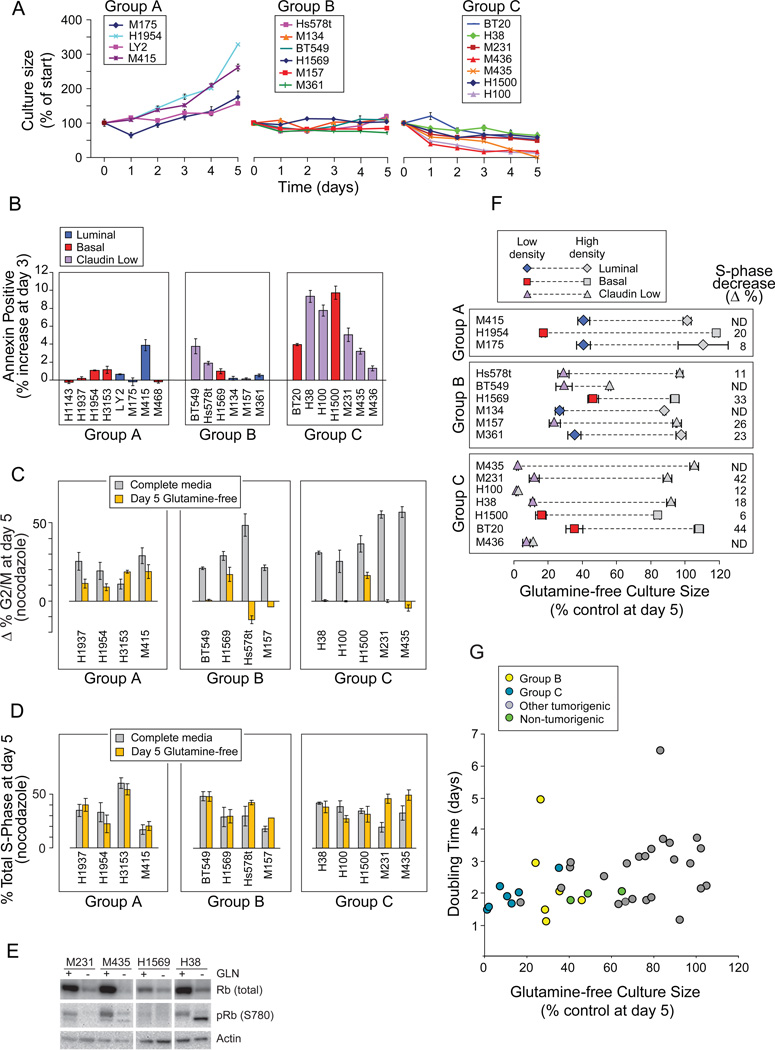
Growth curves of the most restriction-sensitive tumors (Figure 2A, “Glutamine Sensitive” underbar) revealed cultures that A) continued expansion, similar to HMECd; B) did not significantly expand; and C) decreased (Figure 3A). Groups B and C (B+C) include two ER+ (luminal) and 11 TNBC (3 basal, 8 claudin low) independently-derived samples, including the 4 claudin low cells that consume little glutamine (Figures 1D, black). Group C cells increase Annexin V reactivity (Figure 3B) and apoptotic figures (Figure S3A) on days 2–4 of restriction. Thus about 1/2 of our TNBC lines are unable to survive or expand without glutamine.
Figure 3. Glutamine Restriction Induces S-phase Stalling in a Subset of Basal TNBC.
Icon codes in figure keys. Icons represent mean values +/−SD. (A) Growth curves of “Glutamine Sensitive” carcinomas (underlined in Figure 2A) in glutamine-free media. (B) Percent increases in Annexin V reactivity of cells in glutamine-free media at day 3. Group averages; A, 0.8%,+/−1.4; B 1.2% +/−1.4; C, 5.8% +/−3.2; t-test A vs C p= 0.005. (C) Paired bars representing the change in percent of cells in G2/M (Δ% G2/M) with nocodazole treatment at day 5 in control (gray) versus glutamine-free (yellow) media, using cell cycle curve fitting software (FLOJO, Treestar). (D) Paired bars representing the percent S-phase fraction with nocodazole treatment of day 5 cultures in control (gray) vs. glutamine-free (yellow) media. (E) S phase stalling is accompanied by a decrease in total and Serine 780 phosphorylated retinoblastoma protein. (F) Culture confluence reduces S-phase fractions and glutamine sensitivity; S-phase decrease (Δ%), is the decrease in % S-phase with high density culture (gray icons) versus low density culture (colored icons); paired icons connected by dashed lines represent a single cell line. (G) Doubling time (from Figure 1C) vs. glutamine-free culture sizes (from Figure 2H). See also Figure S3.
We tested whether the few live cells in day 5 Group B+C cultures had arrested in G1, reasoning that they might survive glutamine restriction to seed tumor recurrences, discouraging development of glutamine-restricting therapeutics. In complete media, nocodazole treatment increased G2/M and late S-phase fractions of all samples tested, demonstrating transit to and activation of an intact mitotic checkpoint (Figure 3C, gray bars; late S-phase data not shown; Figure S3B). Glutamine-free cultures of group A exemplars exhibited similar G2/M increases (gray vs. yellow bars), but not Group B or C samples. The unchanged S-phase fractions rule out G1 arrest by Group B+C samples (Figure 3D, paired gray vs. yellow bars). Their cell cycle profiles are identical with or without nocodazole treatment (Figure S3C) indicating S-phase stalling, and total and phosphorylated retinoblastoma proteins are reduced similar to S-phase stalling in DNA damage responses (Figure 3E, Knudsen et al. 2000). But like glutamine-restricted Myc-transformed fibroblasts, γH2A.X phosphorylation is not increased, indicating that the intra-S-phase DNA damage checkpoint remains inactive (data not shown; Yuneva et al., 2007). Culture confluence reduces the S-phase content and glutamine sensitivity of all but two Group C samples (Figure 3F, colored vs. gray icons). However Group B+C samples are not simply the most rapidly dividing cells (Figure 3G; t-tests: all vs. C+B, p= 0.11; all basal and claudin low vs. C+B, p= 0.2), indicating that they harbor specific defects which makes them unable to surmount restriction.
Known Regulators of Glutamine Metabolism do not Identify Group B or C Cells
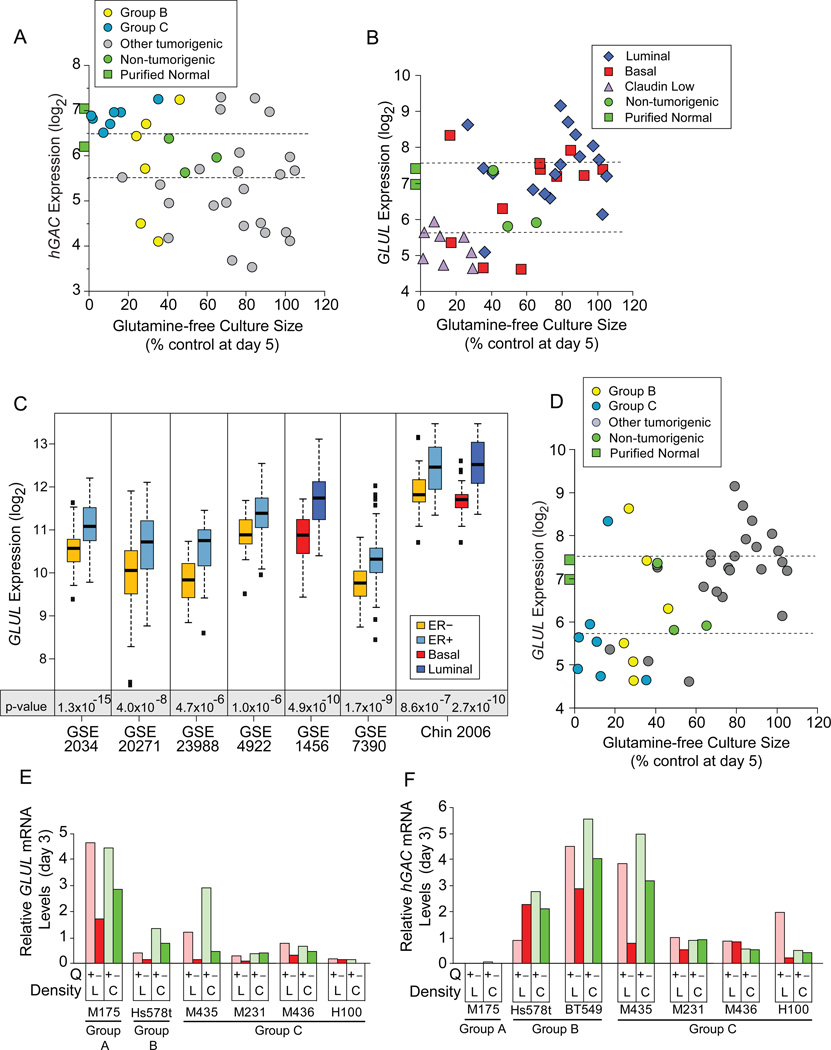
We tested proposals that hGAC and GLUL identified glutamine dependent cells. While hGAC statistically identifies Group C (p= 5.6×10−9) and Group B+C (p= 0.008) cells, only 9/13 (69%) samples expressing higher hGAC than HMECd cells are Group B or C (Figure 4A, above upper dotted line), and these levels are similar to purified normal epithelia (green squares). hGAC also poorly discerns Group C (p=0.03) or B+C (p=0.17) from all other TNBC. While GLUL levels correlate with restricted culture sizes (Figure 4B; p= 0.005 to 2.3E−4, 3 probeset IDs; Figure S4A), GLUL also more accurately discerns luminal from claudin low samples (Figure 4B; p=1.9E−5). At lower p-value GLUL discerns luminal from all basal + claudin low (p= 2.5E−5 to 1.9E−4), or ER+ from ER− (p= 4.4E−3 to 1.2E−2) cell lines, and samples in 7/8 clinical expression datasets (Figure 4C; Table S4). Most Group B+C cells express less GLUL than normal samples (Figure 4D), but low expression is not unique to Group B+C (grey circles below lower dotted line). Nor can GLUL discern Group C (p=0.20–0.67) or B+C samples (p=0.015–0.09) from other TNBC. Thus hGAC and GLUL are not strong biomarkers for Group B or C type tumors.
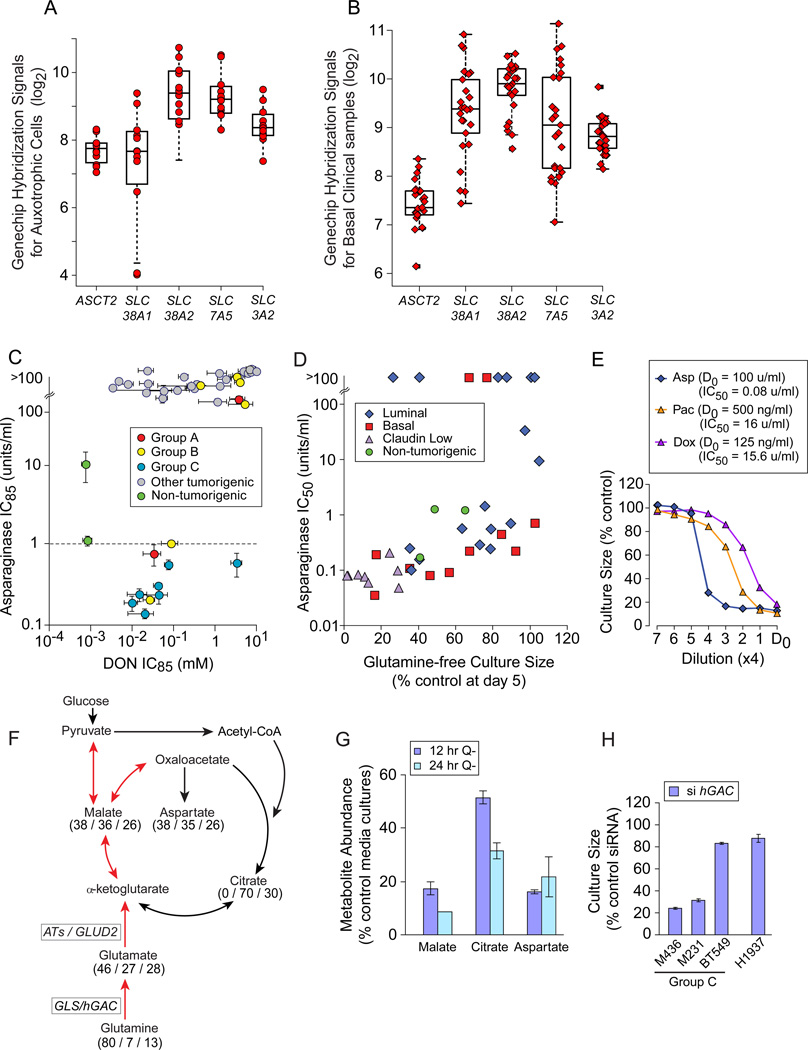
Figure 4. Common Regulators of Glutamine Metabolism do not Identify Auxotrophic Cells.
Icon codes in figure keys; green squares, average hybridization signals from 3 CD10+, and 3 BerEP4+ purified normal breast samples. Icons represent mean values+/−SD. (A) hGAC GeneChip hybridization signal (log2) vs. glutamine-free culture sizes (from Figure 2H) coded as restriction Groups B and C vs. others. (B) GLUL GeneChip hybridization signal (log2) vs. day 5 glutamine-free culture sizes, coded by molecular subtype, dotted lines bracket proliferating, non-tumorigenic sample values. (C) GLUL expression by basal vs. luminal or ER+ vs. ER- samples in 8 clinical breast tumor expression datasets, downloaded from NCBI GEO and Chin 2006 (Chin et al., 2006); t-test p-values below paired boxplots. (D) Correlation of glutamine-free culture sizes and GLUL expression as in panel B, but coded by restriction groups. (E, F) Comparison of (E) GLUL and (F) hGAC mRNA levels at day 3 in glutamine replete versus deficient media, from quantitative PCR analysis; L, low density; C, confluent; Q, glutamine. See also Figure S4 and Tables S4, S5.
Oncogenic Myc can drive glutamine uptake and dependence in exemplar fibroblasts and glioma (Wise et al., 2008; Gao et al., 2009; Yuneva et al., 2007). CMYC is enriched in our basal and claudin low cells (p=0.013). But neither CMYC, other Myc family members, or core MYC expression signature genes (Chandriani et al., 2009) are differentially expressed in Group C or group B+C cells versus other basal + claudin low cells (Figure S4B; Table S5). We found no correlation between TP53 mutational status and Group B or C membership using the IARC TP53 Database (Supplemental Methods, p=0.7; http://p53.iarc.fr/CellLines.aspx). Significance analysis contrasting gene expression in the Group C or B+C samples versus other basal and claudin low TNBC did not identify potential biomarkers or offer molecular explanations for S-phase stalling (data not shown).
Group B and C Carcinomas are Glutamine Auxotrophs
Glutamine restriction reduces hGAC and GLUL mRNA levels in low-density cultures, while producing little change in GS protein levels (Figures 4E, 4F, S4C, S4D). Confluent cultures can induce GLUL mRNA (Figure 4E, e.g. M435), suggesting one mechanism for their starvation resistance. We conclude that proliferating breast tumors cannot induce GLUL to escape glutamine restriction.
Titrations of 17 common carbon sources over 4 concentration logs were tested for rescue of an exemplar Group C carcinoma (M436) from glutamine restriction (Figure S4E). Neither these, or nitrogen sources ammonium chloride, choline chloride, or putrescine improved cultures (Figure S4F). Only glutamate (2–3 fold), oxaloacetate (2–6%), and dimethyl 2-ketoglutarate (2–10%; Figure S4G) increased ATP values slightly, but only glutamate increased viable cell numbers in multiple Group C cell lines (4–6%; Figure S4H). Combining GS substrates, glutamate and nitrogen sources, did not further improve cultures (Figure S4I). We conclude it unlikely that other commonly available nutrients can substitute for glutamine during restriction, and that the Group C+B tumors are functional glutamine auxotrophs.
Glutamine Auxotrophy Presents Therapeutic Opportunities
We tested three approaches to therapeutic development targeting auxotrophic TNBC: preventing glutamine access; inhibiting glutamine-dependent enzymes; and inhibiting activities requiring glutamine metabolites. Auxotrophs (Figure 5A) and basal carcinoma subsets in clinical microarray datasets (Figure 5B, example; Table S6) express multiple glutamine transporters. Thus we prevented glutamine access by treating cultures with the leukemia therapeutic Asparaginase, reducing asparagine and glutamine to their acidic derivatives (Figure S5A; Narta et al., 2007). This produced an apparent synthetic lethality equal to that of leukemia in all Group C and about 1/3 of Group B cells (Figure 5C, IC85 below dotted line; Asselin et al., 1989). Correlated Asparaginase sensitivities and glutamine-free culture sizes (Figure 5D, p=5.7 E−6; Asparaginase IC84 p= 9.3E−9, not shown) reveal that glutamine re-synthesis from the Asparaginase products glutamate and ammonia is uncommon. We propose that local asparaginase / glutaminase delivery would kill auxotrophic tumors without requiring selective identification and targeting of their potentially varied molecular defects. Cells relatively resistant to Paclitaxel or Doxorubicin are exquisitely Asparaginase sensitive, indicating that an asparaginase-like therapeutic could become a critical, independent alternative for drug resistant tumors (Figure 5E).
Figure 5. Glutamine Auxotrophy Presents Therapeutic Opportunities.
Icons represent mean values+/−SD. (A) GeneChip hybridization signals for 4 glutamine transporters and the common heavy chain (SLC3A2) in glutamine auxotrophic cells. (B) GeneChip hybridization signals for transporters as in panel A, for basal carcinoma subsets of clinical datasets downloaded from NCBI GEO and Chin 2006 (Chin et al., 2006), 1 example dataset shown. (C) Relative Asparaginase (y-axis), and DON (x-axis) sensitivities (IC85) of our cell panel, coded by restriction Groups B and C vs. others; dotted line, Asparaginase concentration used to kill sensitive leukemia cells in vitro (1u/ml). (D) Day 5 glutamine-free culture sizes (from Figure 2A) vs. Asparaginase sensitivity (IC50 shown). (E) Four fold drug titrations and calculated IC50s for an exemplar Group C auxotroph (M436). D0, highest drug concentration and calculated IC50s in figure key; Asp, Asparaginase; Pac, Paclitaxel; Dox, Doxorubicin. (F) TCA cycle diagram illustrating respiratory use of glutamine in red arrows. Numbers indicate mass spectroscopy determination of the percent of each metabolite that contains (all / several / no) 13C-carbons derived from culture with 13C-5-glutamine in M436. (G) Decrease in key TCA cycle metabolite pools with glutamine restriction, expressed as % control media cultures; Q-, glutamine-free media. (H) Proliferative effects of siRNA-mediated hGAC mRNA reduction in exemplar cell lines, expressed as a percent of transfection with a scrambled siRNA. See also Figure S5, Tables S6, S7.
Analysis of 13C-5-glutamine-derived metabolites in the Group C TNBC M436 (Figure 5F; Table S7) revealed that 80% of intracellular glutamine is imported (all 5 carbon positions 13C-labeled). About 40% of TCA cycle metabolites and their derivatives are directly produced from this pool, and another third (27–30%) are partially 13C labeled. Glutamine restriction depletes these pools (Figure 5G) suggesting that inhibition of glutaminase (GLS/hGAC) or aminotrasferases (ATs) might kill or slow growth of the auxotrophs. siRNA-mediated reduction of hGAC attenuated culture expansion of auxotrophs with high glutamine consumption rates (Figure 5H, M436, M231; Figure S5B), but provided little efficacy against BT549, an auxotroph that consumes little glutamine (Figure 1D), or H1937, a Group A TNBC. In comprehensive tests, treatment with a broad spectrum inhibitor of amidotransferases including GLS, (6-diazo-5-oxo-L-norleucine; DON; reviewed in Souba, W. W., 1993) placed auxotrophs among the most sensitive samples (Figure 5C). We propose that inhibitors of single DON targets should be refined for use against auxotrophic TNBC.
Glutamine is Required for ROS Control in TNBC
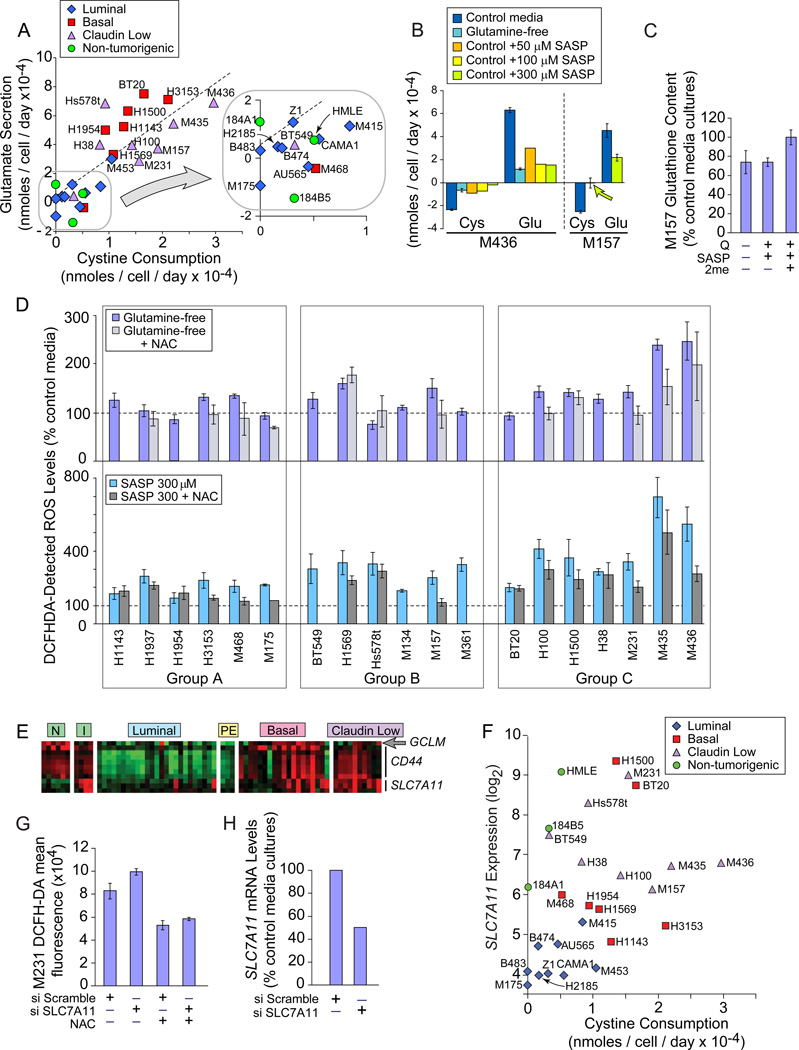
Finally, we tested the strategy of inhibiting an activity that requires glutamine metabolites. In the normal human fibroblast IMR-90, 1/3 of glutamine uptake supplies glutamate for xCT activity (Bannai and Ishii, 1988). Analysis of amino acid consumption revealed highly correlated cystine depletion and glutamate secretion in 27 of our lines (Figure 6A, p= 8.8E−11; Figure S6A), suggesting xCT activity. Glutamine restriction strongly reduced exchange (Figure 6B), modestly decreased GSH levels (Figure 6C) and increased intracellular ROS by at least 30% in 8/19 TNBC (Figure 6D, light blue). This is partially corrected by the ROS scavenger N-acetylcysteine (NAC) in 12/13 samples (light gray). NAC does not allow culture expansion (Figure S6B), thus glutamine use for ROS control is common, but other glutamine-influenced factors are also required for auxotroph proliferation.
Figure 6. Glutamine Restriction and xCT Inhibition Increase ROS.
Icon codes in figure keys; NAC, N-acetylcystine; SASP, Sulfasalazine. Icons represent mean values+/−SD. Amino acid quantitation by HPLC. (A) Change in media cystine (xaxis) and glutamate (y-axis) concentrations of cells cultured 24 hours in control media. (B) 24 hrs of glutamine restriction or SASP treatment reduces cystine / glutamate exchange by exemplar Group C auxotrophs; cys, cystine; glu, glutamate. (C) Effects of 24 hour glutamine restriction or SASP treatment on GSH content in an exemplar Group C TNBC; Q-, glutamine-free media; SASP, SASP treatment in complete media; SASP + 2me, SASP treatment in the presence of beta mercaptoethanol. (D) ROS levels in basal carcinomas assessed by DCFHDA fluorescence, normalized to control media reactivity. Light blue, 2 day cultures in glutamine-free media; Group averages; A, 112% +/− 21; B, 121% +/−31; C,162% +/−59; t-test group A vs. C p=0.07; Group A vs. B+C p= 0.074. Light gray, glutamine-free media +NAC. Teal, cultures treated 24 hours with SASP; Group averages; A, 205% +/− 43; B, 287% +/− 62; C, 405% +/− 168; t-test Group A vs. C p= 0.019; Group A vs. B+C p= 0.003. Dark gray, SASP +NAC. (E) Heatmap of genes involved in xCT function; red, increased; green, decreased; N, purified normal CD10+ and BerEp4+ breast epithelial cells; I, proliferating non-tumorigenic cell lines; PE, ER+ tumor cells purified from pleural effusions. (F) SLC7A11 GeneChip hybridization signal (log2) vs. cystine consumption, icons coded by molecular subtype. (G) ROS levels in M231, 48 hours after targeting SLC7A11 or a scrambled siRNA, in the presence or absence of N-acetylcystine (NAC). (H) SLC7A11 knockdown efficiency of siRNAs used in part G. See also Figure S6.
We directly assessed the xCT expression and function in TNBC that was implied by the glutamine restriction effects on ROS. Basal and claudin low lines overexpress the exchange-specific subunit SLC7A11 (Figure 6E, p=0.06–7.4E−4; Figure S6C), the glutamate-cysteine ligase regulatory subunit of glutathione synthase (arrow; GCLM, p= 0.011), and a membrane interacting protein CD44 p=0.005–5.9E−8; Ishimoto et al., 2011). Cystine consumption and SLC7A11 mRNA levels correlate (Figures 6F, S6D), and siRNA-mediated reduction of SLC7A11 mRNA increases intracellular ROS (Figure 6G, H). SASP treatment reduces cystine/glutamate exchange and GSH content (Figure 6B, 6C). 2-mercaptoethanol (2me) provides cystine as mixed 2me-cysteine disulfides (Ishii et al., 1981), and normalizes GSH levels, demonstrating SASP specificity for cystine and GSH production. In 18/19 TNBC, SASP increases endogenous ROS by at least 50% (Figure 6D, teal bars), which are reduced by NAC in 14/16 samples (dark gray bars). Using reagents for specific ROS species, we found that SASP increases hydroxyl radicals, in keeping with the expected effects of GSH depletion (Figure S6E; Franco et al., 2007). Thus the xCT antiporter is commonly expressed and functional in TNBC. HMECd express these genes (Figure 6E, columns “I”) but is much less active (Figure 6A, green).
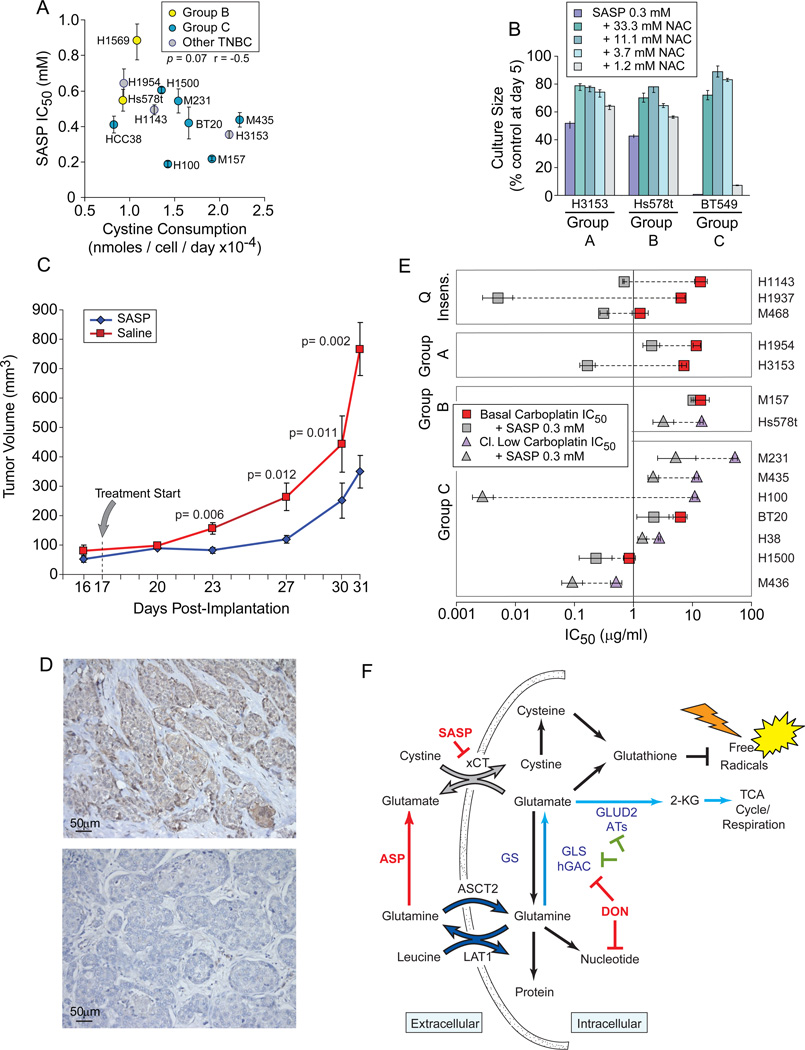
SASP Treatment Attenuates Tumor Growth
In proliferation assays we found that SASP treatment dramatically reduces TNBC culture sizes with IC50 values modestly correlate to cystine uptake in complete media (Figure 7A, p= 0.07). Cultures are significantly restored by NAC (Figure 7B), and not effected by the active anti-inflammatory fragment of SASP, 5-Aminosalicylic acid (5-ASA; Figure S7A). Thus inhibition of cystine acquisition, not anti-inflammatory activity, is responsible for TNBC proliferative sensitivity. This concentration range (0.2 to 0.7 mM) is not far from serum concentrations measured in bowel inflammatory patients (0.18 mM; Guastavino et al., 1988). Under normoxic conditions (5% O2) SASP sensitivity increased 2–3 fold (Figure S7B), and SASP significantly slowed growth of an auxotrophic line xenograft (Figure 7C), indicating that xCT activity is also critical for growth in vivo. Accordingly, 8/20 anonymous TNBC clinical specimens strongly express xCT (Figure 7D; Figures S7C-K). TNBC can be treated with Carboplatin, and Carboplatin + SASP reduces the Carboplatin IC50 in 13/14 of our TNBC samples (Figure 7E). We propose that SASP be derived for clinical use.
Figure 7. SASP Attenuates Proliferation In vitro and In vivo.
Icon codes in figure keys. Icons represent mean values+/−SD. (A) Cystine consumption in complete media derived from HPLC analysis (x-axis) versus SASP sensitivity (IC50). (B) NAC treatment rescues SASP-induced culture size defects in an exemplar cell from each restriction group A-C. (C) SASP treatment attenuates xenograft growth; Group average tumor volumes separation p-values noted on graph. (D) Examples of xCT expression in exemplar human TNBC tumor sections; nuclei, blue; xCT-specific HRP signal, brown Upper positive, lower negative. (E) SASP reduces the Carboplatin IC50 of most basal TNBC; paired icons connected by dashed lines represent a single TNBC; colored icons, Carboplatin IC50; gray icons, IC50 of Carboplatin plus 300 µM SASP; Q insens., basal TNBC with less glutamine-sensitivity than non-tumorigenic cells. (F) Summary of discussed glutamine catabolic activities. Red, compounds tested in this manuscript; green, activities with potential therapeutic inhibitory importance in glutamineavid TNBC; dark blue, LAT1 glutamine/leucine antiporter and ASCT2, system ASC glutamine transporter; gray, xCT, the glutamate/cystine antiporter; light blue, glutamine anaplerosis path; SASP, Sulfasalazine; ASP, Asparaginase; DON, 6-diazo-5-oxo-Lnorleucine; ATs, various aminotransferases; GS, glutamine synthase; GLS, glutaminase; hGAC, carboxy-terminal splice variant of glutaminase. Not all uses of intracellular glutamine or DON targets are illustrated. See also Figure S7.
DISCUSSION
Rodent breast tumors (Erlich Ascites series) were instrumental in the definition of central metabolic pathways and tumor-specific aberrations, but work in human breast tumors is largely limited to aspects of fatty acid metabolism (see Menendez and Lupu, 2007). Reports of other metabolic features are less frequent and use only one or a few samples, producing biased conclusions due, for example, to the mistaken use claudin low lines to represent typical TNBC; misidentification of GeneChip probeset IDs that are hGAC splice variant specific as reporting total GLS mRNA; the lack of gene expression comparisons between tumors and normal breast epithelia; and the inability to directly compare non-tumorigenic breast derivative and tumorigenic behaviors (examples see Collins et al., 1998; Kung et al., 2011; Simpson, et al., 2011). Our data clarify these misunderstandings and provide a detailed nutrient utilization portrait of a comprehensive organ site-specific tumor collection, contrasting gene expression and functional assays to define common nutrient utilization patterns and responses to drugs that leverage associated metabolic activities.
Historically-Associated Metabolic Features Vary Substantially in Breast
Doubling time, glutamine consumption, and glutamine reliance are historically correlated. Among this triad, only doubling time and glutamine-free culture sizes modestly correlate across our large sample collection (Figure 3G, p=0.002). But the glutamine auxotrophs are not simply the most rapidly dividing samples. Nor are they the largest glutamine consumers; 4/8 auxotrophic, claudin low samples consume no more glutamine than proliferating non-tumorigenic cells (Figure 1D, black triangles: BT549, H38, H100, M157). Glutamine can be cycled by import via ASCT2 and export in exchange for essential amino acids via the LAT1/4f2hc antiporter (SLC7A5/SLC3A2, Figure 7F, dark blue arrows; Nicklin et al., 2009). Our 4 low glutamine consumers express all antiporter components (Figure 5A), and glutamate cycling would not alter their measured ambient glutamine levels. Thus these cells may require glutamine to fuel both this exchange activity and the xCT antiporter, and use relatively less glutamine for respiration. This may explain the relative proliferative resistance to siRNA-mediated reduction of hGAC seen in BT549 (Figure 5H). Differences between glutamine cycling and catabolism may also partially explain historically variable correlations between glutamine uptake and glutamine reliance in other tumors.
Conversely, auxotrophs of high glutamine consumption (Figure 1D, M436) that require glutamine as a major respiratory fuel (Figures 5F, 5G; Figure 7F, light blue arrows) and for glutamate/cystine exchange (Figure 6B, 7F, gray arrows), may be more susceptible to inhibition of hGAC (Figure 5H, M436, M231) and subordinate glutamate-dependent activities such as aminotransferases (ATs; Thornburg et al., 2008). Thus within a single tumor cell line, multiple critical requirements for glutamine may exist and provide multiple therapeutic targets, either individually or in combination. We hypothesize that xCT inhibition may be further potentiated by limiting glutamate availability (Figure 7F).
Historically-Proposed Genetic Indicators of Glutamine Reliance do not Define Auxotrophy
With the resurgent interest in tumor metabolism, metabolic genes such as GLS and GLUL have been re-asserted as potential therapeutic targets and biomarkers, but we find that therapeutic relevance is not so easily defined. Gene expression may suggest metabolic behaviors that are more likely active in specific tumor groups, such as a statistical association of hGAC with high glutamine consumption in basal and claudin low versus luminal tumors (Figures 1F). However neither hGAC nor GLUL defines high glutamine consumption or identifies the true auxotrophic Group B and/or C cells with appropriate sensitivity to be considered independent clinical biomarkers. We also find that responses to interruption of ongoing metabolic activities can vary substantially due to unknown cell-intrinsic factors. For example individual tumors can respond to glutamine restriction by slowing culture expansion or stalling in S-phase and dying (Figure 3A, B, C). Molecular explanation(s) for S-phase stalling remain unclear, and may be due to tumor defects far removed from direct glutamine interaction.
xCT is a Compelling Therapeutic Target for Triple Negative Tumors
Inhibitor and RNAi studies reveal xCT induction as the dominant means of increasing cystine acquisition for GSH synthesis (reviewed in Lo et al. 2008a). Thus xCT may be a target for cell-specific GSH depletion, since SASP and other xCT inhibitors can slow growth of exemplar cell lines in xenograft without significant effects on other organs, and can cooperate with chemotherapeutics such as Cisplatin (Okuno, et al., 2003), Geldanamycin (Huang et al., 2005), Doxorubicin (Narang, et al., 2007) and Gemcitabine (Lo, et al., 2008b). However the SASP structure is labile, designed to be cleaved by enteric bacteria to release an active anti-inflammatory fragment. It is also insoluble in aqueous solutions and not optimized for the fortuitous interaction with xCT. Thus while direct clinical applications to TNBC are unrealistic, SASP is a strong lead compound for development of xCT inhibitory therapeutics. Our studies reveal that SLC7A11 expression, cystine/glutamate exchange activity (Figures 6E, 6A, 7D), and deleterious proliferative effects of xCT inhibition are common in basal and claudin low carcinomas (Figure 7A, C). CD44 and the claudin low gene expression signature associate with breast cancer stem-cell phenotypes (Hennessy et al., 2009, Prat et al., 2010), implying that SASP-derived therapeutics may target breast tumor stem cells. This is reminiscent of CD44 and xCT dependent ROS regulation in gastric tumor progenitors (Ishimoto 2011). Thus we have identified a compelling therapeutic target commonly expressed by breast tumors of poorest prognosis, and a lead compound for rapid, effective drug development.
EXPERIMENTAL PROCEDURES
Dataset Preparation
Cell Culture
Tumorigenic cell lines were adapted to RPMI or DMEM + 5% Fetal Bovine Serum (GIBCO 11875, 11965; Table S2). Culture expansion assays performed at least 3 times in triplicate, in 96-well format. Relative cell number determined (Cell Titer Glow, Promega), verified by microscopy. Averages reported +/−SD. (See Supplemental Experimental Procedures, and below).
siRNA Effects
Transfections (Oligofectamine, Life Technologies 12252-011) used SLC7A11 (SilencerSelect Validated s24291, Life Technologies), hGAC (Dharmacon custom synthesis, sense: GGAAAGUCUGGGAGAGAAAUU, antisense: UUUCUCUCCCAGACUUUCCUU), or nonspecific siRNAs (sc-37007, Santa Cruz Biotechnology; SN-1002, Bioneer), in triplicate sub-confluent 6-well or 96-well plates and the Life Technology protocol. RNA and ROS quantitation at day 2.
Glucose Uptake
Cultures in DMEM/low glucose (GIBCO 11885), RPMI, or HMEC (Medium 171, Cascade Biologics), were treated +/− 2-NBDG or 6-NBDG glucose (30µM, Molecular Probes N13195, N23106) for 0–8 hrs, harvested, external fluorescence quenched (0.4% trypan blue), 30,000 cells analyzed in triplicate FACSCalibur (Becton Dickinson) or C6 Flow Cytometer (Accuri). Average mean fluorescence values at 4 hours normalized to unstained controls are reported +/−SD.
ROS Detection
Cells were incubated 15 min. with 10 µM 2',7'-dichlorofluorescein diacetate (DCFH-DA, Sigma D6883), or 1 hour with 5µM 3’-(p-hydroxyphenyl) fluorescein (HPF, Molecular Probes H36004), harvested, and 30,000 cells analyzed in triplicate by FACS, normalized to unstained controls. Average mean fluorescence values reported +/−SD.
GSH Quantitation
Used the ApoGSH Glutathione Detection Kit (BioVision) per manufacturer's instructions in triplicate. 2-me at 60µM. Average values reported +/−SD.
Amino Acid Analysis
Supernatants from 24 hr sub-confluent duplicate or triplicate cultures and cell-free media were analyzed using standard HPLC techniques (Biochemical Genetics Laboratory, Stanford University; UC Davis Genome Center, UC Davis). Values subtracted from media controls, normalized to cell number. Average values reported +/−SD.
Metabolite Analysis
Cells were cultured in the presence of 13C-5 labeled glutamine (Cambridge Isotope) for 0–12 hrs, PBS washed, frozen (dry ice), lyophilized, pellets weighed, and homogenized in cold 60% acetonitrile (40 µl per 1mg of protein with acetonitrile washed glass beads. Samples incubated 30 min/−80°C, supernatants collected, pellets washed (60% acetonitrile). Pooled supernatants lyophilized, re-suspended in 200 µl H20, and 40 µl mixed with 30 µl of 40% TCA and 50 µl of 0.1 mM noreleucine (Sigma; internal standard). Lyophilized samples were silylated (50 µl acetonitrile: MTBSTFA (N-methyl-N- [tert-butyl-dimethylsilyltrifluoroacetamide, Regis Chemical, Morton Grove, IL, v/v 1:1), sonicated 3 hours, incubated overnight. Analysis used a PolarisQ GC-ion trap mass spectrometer (ThermoFinnigan, Austin, TX) as previously described (Yuneva et al., 2012). Metabolites identified and quantitated using XCalibur software (ThermoFinnigan). Results normalized to dry pellet weight, and noreleucine standard. Average values reported +/−SD.
Immunohistochemistry
Cell lines and anonymous, de-identified tumor sections (UCSF/SPORE Tissue Core, collected under UCSF Internal Review Board approval) with high and low SLC7A11 values (Chin, et al., 2006), were stained to correlate anti xCT reactivity (Novus Biologicals NB300-318) with SLC7A11 mRNA levels (data not shown). Detection used citrate antigen retrieval, ABC Kit (Vector Labs), and the Novus antibody-specific protocol. Commercial tissue arrays of anonymous breast tumor and normal sections with known ER/PR/Her2 status were purchased and analyzed (Biomax, Inc).
Bioinformatics
Association of GLS and GLUL expression with ER status and molecular subtype was determined in clinical microarrays available via http://www.ncbi.nlm.nih.gov/gds. Data were preprocessed (RMA algorithm in R), analyzed with Matlab R2011a and R version 2.12.0 for MacOS X. Pearson correlations reported, class distinctions by Student's t-test.
Xenografts
The claudin low auxotroph M231 was implanted into mammary fatpad 4 in 14 6-week old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) female mice. Animals were randomized at day 16 and injected IP twice daily with 250 µl Saline or 50mM SASP in 0.1N NaOH pH 7.5 from days 17–31(Guan, et al., 2009). Tumor volume measured twice weekly, average values reported +/−SD. Experiments performed following UCSF Institutional Animal Care and Use Committee approval, in accordance with institutional and national guidelines.
Supplementary Material
SIGNIFICANCE.
There is a strong disconnect between compound efficacy in tumor cell lines used for cancer drug development versus clinical response rates, slowing the production of effective therapeutics. This would be improved if the frequency and types of tumor responses to a particular perturbation could be identified early in the process. While drug target expression is currently used to identify applicable clinical populations, these patient cohorts contain both responders and non-responders. Here we use functional analyses in 47 independent breast-derived cell lines to measure metabolic responses related to perturbations in glutamine metabolism, and estimate their frequency. We identify a therapeutic target in a severely underserved population of breast cancer patients, and a lead compound for rapid, durable therapeutic development.
HIGHLIGHTS.
Most breast tumors survive glutamine restriction, limiting its therapeutic use
A subset of triple negative breast tumors are true glutamine auxotrophs
Conventional biomarkers of glutamine reliance do not identify true auxotrophs
Triple negative tumors require cystine import via the xCT transporter
ACKNOWLEDGMENTS
The UCSF/Gladstone Genome Core: microarray dataset generation help. T. Cowan, Stanford Biochemical Genetics Laboratory: HPLC amino acid analysis. R. Higashi, CREAM, University of Louisville, Kentucky: mass spectrometry. UCSF Preclinical Therapeutics Core: xenografts. D. Albertson, T. Tlsty, J. Korkola, and W. Kinlaw for valuable manuscript critiques. LT support: National Institutes of Health, National Cancer Institute, Bay Area Breast Cancer SPORE (P50 CA 58207); the Durra Family Fund; and the Mount Zion Health Fund, a Supporting Foundation of the Jewish Community Federation (4/01/12).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Number
NCBI GEO GSE48984
SUPPLEMENTAL INFORMATION
Supplemental information includes 7 Figures, 7 Tables, Supplemental Experimental Procedures, and References.
We declare no conflicts of interest.
REFERENCES
- Alli E, Sharma VB, Sunderesakumar P, Ford JM. Defective repair of oxidative dna damage in triple-negative breast cancer confers sensitivity to inhibition of poly(ADP-ribose) polymerase. Cancer Res. 2009;69:3589–3596. doi: 10.1158/0008-5472.CAN-08-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin BL, Ryan D, Frantz CN, Bernal SD, Leavitt P, Sallan SE, Cohen HJ. In vitro and in vivo killing of acute lymphoblastic leukemia cells by L-asparaginase. Cancer Res. 1989;49:4363–4368. [PubMed] [Google Scholar]
- Bannai S, Ishii T. A novel function of glutamine in cell culture: utilization of glutamine for the uptake of cystine in human fibroblasts. J Cell Physiol. 1988;137:360–366. doi: 10.1002/jcp.1041370221. [DOI] [PubMed] [Google Scholar]
- Bannai S, Tateishi N. Role of membrane transport in metabolism and function of glutathione in mammals. J Membr Biol. 1986;89:1–8. doi: 10.1007/BF01870891. [DOI] [PubMed] [Google Scholar]
- Chandriani S, Frengen E, Cowling VH, Pendergrass SA, Perou CM, Whitfield ML, Cole MD. A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS One. 2009;4:e6693. doi: 10.1371/journal.pone.0006693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Coles NW, Johnstone RM. Glutamine metabolism in Ehrlich ascitescarcinoma cells. Biochem J. 1962;83:284–291. doi: 10.1042/bj0830284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CL, Wasa M, Souba WW, Abcouwer SF. Determinants of glutamine dependence and utilization by normal and tumor-derived breast cell lines. J Cell Physiol. 1998;176:166–178. doi: 10.1002/(SICI)1097-4652(199807)176:1<166::AID-JCP18>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Dranoff G, Elion GB, Friedman HS, Campbell GL, Bigner DD. Influence of glutamine on the growth of human glioma and medulloblastoma in culture. Cancer Res. 1985;45:4077–4081. [PubMed] [Google Scholar]
- Elgadi KM, Meguid RA, Qian M, Souba WW, Abcouwer SF. Cloning and analysis of unique human glutaminase isoforms generated by tissuespecific alternative splicing. Physiol Genomics. 1999;1:51–62. doi: 10.1152/physiolgenomics.1999.1.2.51. [DOI] [PubMed] [Google Scholar]
- Franco R, Panayiotidis MI, Cidlowski JA. Glutathione depletion is necessary for apoptosis in lymphoid cells independent of reactive oxygen species formation. J Biol Chem. 2007;282:30452–30465. doi: 10.1074/jbc.M703091200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Gout PW, Kang YJ, Buckley DJ, Bruchovsky N, Buckley AR. Increased cystine uptake capability associated with malignant progression of Nb2 lymphoma cells. Leukemia. 1997;11:1329–1337. doi: 10.1038/sj.leu.2400739. [DOI] [PubMed] [Google Scholar]
- Gout PW, Buckley AR, Simms CR, Bruchovsky N. Sulfasalazine, a potent suppressor of lymphoma growth by inhibition of the x(c)- cystine transporter: a new action for an old drug. Leukemia. 2001;15:1633–1640. doi: 10.1038/sj.leu.2402238. [DOI] [PubMed] [Google Scholar]
- Guan J, Lo M, Dockery P, Mahon S, Karp CM, Buckley AR, Lam S, Gout PW, Wang YZ. The xc- cystine/glutamate antiporter as a potential therapeutic target for small-cell lung cancer: use of sulfasalazine. Cancer Chemother Pharmacol. 2009;64:463–472. doi: 10.1007/s00280-008-0894-4. [DOI] [PubMed] [Google Scholar]
- Guastavino E, Litwin NH, Heffes Nahmod L, Licastro R. Ulcerative colitis in children. Levels of salicylazosulfapyridine and sulfapyridine during treatment. Acta Gastroenterol Latinoam. 1988;18:107–113. [PubMed] [Google Scholar]
- Hamilton D, Batist G. Glutathione analogues in cancer treatment. Curr Oncol Rep. 2004;6:116–122. doi: 10.1007/s11912-004-0023-4. [DOI] [PubMed] [Google Scholar]
- Han J, Chang H, Giricz O, Lee GY, Baehner FL, Gray JW, Bissell MJ, Kenny PA, Parvin B. Molecular predictors of 3D morphogenesis by breast cancer cell lines in 3D culture. PLoS Comput Biol. 2010;6:e1000684. doi: 10.1371/journal.pcbi.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-tomesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zunyan D, Barbacioru C, Sadee W. Cystine-Glutamate Transporter SLC7A11 in Cancer Chemosensitivity and Chemoresistance. Cancer Res. 2005;65:7446–7454. doi: 10.1158/0008-5472.CAN-04-4267. [DOI] [PubMed] [Google Scholar]
- Iglehart JK, York RM, Modest AP, Lazarus H, Livingston DM. Cystine requirement of continuous human lymphoid cell lines of normal and leukemic origin. J Biol Chem. 1977;252:7184–7191. [PubMed] [Google Scholar]
- Ishii T, Hishinuma I, Bannai S, Sugita Y. Mechanism of growth promotion of mouse lymphoma L1210 cells in vitro by feeder layer or 2-mercaptoethanol. J Cell Physiol. 1981;107:283–293. doi: 10.1002/jcp.1041070215. [DOI] [PubMed] [Google Scholar]
- Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011;19:387–400. doi: 10.1016/j.ccr.2011.01.038. [DOI] [PubMed] [Google Scholar]
- Jelluma N, Yang X, Stokoe D, Evan GI, Dansen TB, Haas-Kogan DA. Glucose withdrawal induces oxidative stress followed by apoptosis in glioblastoma cells but not in normal human astrocytes. Mol Cancer Res. 2006;4:319–330. doi: 10.1158/1541-7786.MCR-05-0061. [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Knox WE, Linder M, Friedell GH. A series of transplantable rat mammary tumors with graded differentiation, growth rate, and glutaminase content. Cancer Res. 1970;30:283–287. [PubMed] [Google Scholar]
- Knudsen KE, Booth D, Naderi S, Sever-Chroneos Z, Fribourg AF, Hunton IC, Feramisco JR, Wang JY, Knudsen ES. RB-dependent S-phase response to DNA damage. Mol Cell Biol. 2000;20:7751–7763. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic Z, Morris HP. The role of glutamine in the oxidative metabolism of malignant cells. Cancer Res. 1972;32:326–333. [PubMed] [Google Scholar]
- Kovacevic Z, McGivan JD. Mitochondrial metabolism of glutamine and glutamate and its physiological significance. Physiol. Rev. 1983;63:547–605. doi: 10.1152/physrev.1983.63.2.547. [DOI] [PubMed] [Google Scholar]
- Kung HN, Marks JR, Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet. 2011;7:e1002229. doi: 10.1371/journal.pgen.1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvamme E, Svenneby G. The effect of glucose on glutamine utilization by Ehrlich ascites tumor cells. Cancer Res. 1961;21:92–98. [PubMed] [Google Scholar]
- Linder-Horowitz M, Knox WE, Morris HP. Glutaminase activities and growth rates of rat hepatomas. Cancer Res. 1969;29:1195–1199. [PubMed] [Google Scholar]
- Lo M, Wang YZ, Gout PW. The x(c)− cystine/glutamate antiporter: a potential target for therapy of cancer and other diseases. J Cell Physiol. 2008a;215:593–602. doi: 10.1002/jcp.21366. [DOI] [PubMed] [Google Scholar]
- Lo M, Ling V, Wang YZ, Gout PW. The xc− cystine/glutamate antiporter: a mediator of pancreatic cancer growth with a role in drug resistance. Br J Cancer. 2008b;99:464–472. doi: 10.1038/sj.bjc.6604485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo C, Ruiz-Bellido MA, Aledo JC, Marquez J, Nunez De Castro I, Alonso FJ. Inhibition of glutaminase expression by antisense mRNA decreases growth and tumourigenicity of tumour cells. Biochem J. 2000;348(Pt 2):257–261. [PMC free article] [PubMed] [Google Scholar]
- Lora J, Alonso FJ, Segura JA, Lobo C, Marquez J, Mates JM. Antisense glutaminase inhibition decreases glutathione antioxidant capacity and increases apoptosis in Ehrlich ascitic tumour cells. Eur J Biochem. 2004;271:4298–4306. doi: 10.1111/j.1432-1033.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Narang VS, Pauletti GM, Gout PW, Buckley DJ, Buckley AR. Sulfasalazine-induced reduction of Glutathione levels in breast cancer cells: enhancement of growth-inhibitory activity of doxorubicin. Chemotherapy. 2007;53:210–217. doi: 10.1159/000100812. [DOI] [PubMed] [Google Scholar]
- Narta UK, Kanwar SS, Azmi W. Pharmacological and clinical evaluation of L-asparaginase in the treatment of leukemia. Crit Rev Oncol Hematol. 2007;61:208–221. doi: 10.1016/j.critrevonc.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno S, Sato H, Kuriyama-Matsumura K, Tamba M, Wang H, Sohda S, Hamada H, Yoshikawa H, Kondo T, Bannai S. Role of cystine transport in intracellular glutathione level and cisplatin resistance in human ovarian cancer cell lines. Br J Cancer. 2003;88:951–956. doi: 10.1038/sj.bjc.6600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- Simpson NE, Tryndyak VP, Beland FA, Pogribny IP. An in vitro investigation of metabolically sensitive biomarkers in breast cancer progression. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1871-x. Online: 10.1007/s10549-011-1871-x. [DOI] [PubMed] [Google Scholar]
- Sommers CL, Byers SW, Thompson EW, Torri JA, Gelmann EP. Differentiation state and invasiveness of human breast cancer cell lines. Breast Cancer Res Treat. 1994;31:325–335. doi: 10.1007/BF00666165. [DOI] [PubMed] [Google Scholar]
- Souba WW. Glutamine and cancer. Ann Surg. 1993;218:715–728. doi: 10.1097/00000658-199312000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg JM, Nelson KK, Clem BF, Lane AN, Arumugam S, Simmons A, Eaton JW, Telang S, Chesney J. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008;10:R84. doi: 10.1186/bcr2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel AP, Jing J, Wooster RF, Bachman KE. Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol Ther. 2012;13:1185–1194. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, Cerione RA. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke HR, Ozand PT, Tildon JT, Sevdalian DA, Cornblath M. Reciprocal regulation of glucose and glutamine utilization by cultured human diploid fibroblasts. J Cell Physiol. 1978;95:41–48. doi: 10.1002/jcp.1040950106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.