Summary
Opioids represent widely prescribed and abused medications, although their signal transduction mechanisms are not well understood. Here we present the 1.8Å high-resolution crystal structure of the human δ-opioid receptor (δ-OR), revealing the presence and fundamental role of a sodium ion mediating allosteric control of receptor functional selectivity and constitutive activity. The distinctive δ-OR sodium ion site architecture is centrally located in a polar interaction network in the 7-transmembrane bundle core, with the sodium ion stabilizing a reduced agonist affinity state, and thereby modulating signal transduction. Site-directed mutagenesis and functional studies reveal that changing the allosteric sodium site residue Asn131 to alanine or valine augments constitutive arrestin-ergic signaling. Asp95Ala, Asn310Ala, and Asn314Ala mutations transform classical δ-opioid antagonists like naltrindole into potent β-arrestin-biased agonists. The data establish the molecular basis for allosteric sodium ion control in opioid signaling, revealing that sodium-coordinating residues act as “efficacy-switches” at a prototypic G protein-coupled receptor.
Keywords: human opioid receptor, sodium regulation, allostery, functional selectivity, GPCR signaling, constitutive activity, arrestin
Main text
The three classical opioid receptors (µ, κ and δ-OR) and the related nociceptin/orphanin FQ peptide receptor (NOP) are G protein-coupled receptors (GPCRs) essential for regulating nociception, mood and awareness1. These opioid GPCRs are activated by endogenous peptides (endorphins, enkephalins, dynorphins, N/OFQ), natural alkaloids (opiates), and an expanding number of small molecule agonists through interactions with the orthosteric site located in the extracellular portion of the seven transmembrane (7TM) bundle. Despite the progress made in understanding GPCR activation2, the underlying molecular mechanisms and structural features responsible for many processes including signal transduction, allosteric modulation, functional selectivity, and constitutive activity remain elusive 3,4.
Insights from 1.8 Å δ-OR structure
Pioneering studies initiated in 1973 on opioid receptors revealed that physiological concentrations of sodium alter opiate ligand binding and signaling, albeit by unknown mechanisms5,6. To address the molecular basis for the striking allosteric effect of sodium on opioid receptor function, we crystallized the human δ-OR (residues 36–338) with an N-terminal BRIL fusion protein [BRIL-δOR(ΔN/ΔC)] and determined the crystal structure in complex with the subtype-selective ligand naltrindole7 at 1.8 Å resolution (Fig. 1 and Table S1; Materials and Methods). Importantly, the high resolution BRIL-δOR(ΔN/ΔC)-naltrindole structure contains the wild type (WT) protein sequence, including an intact intracellular loop 3 (ICL3) providing the opportunity to study an opioid receptor that closely resembles a near native conformational state.
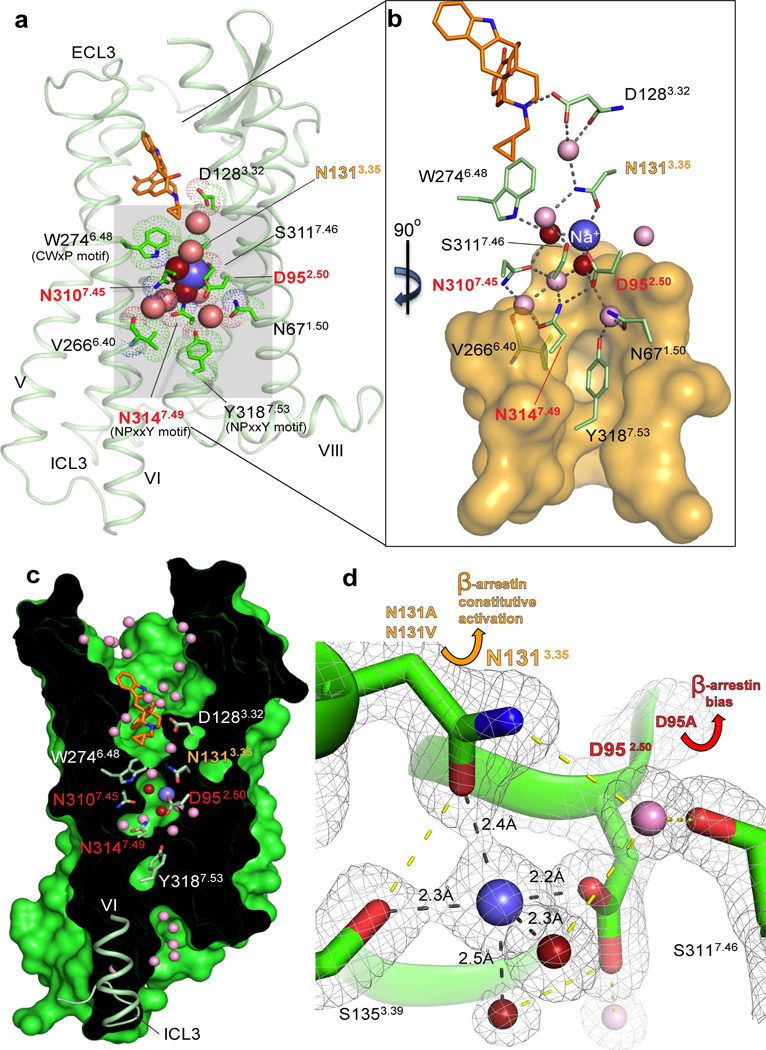
Fig. 1. Interactions in the 7TM core of BRIL-δOR(ΔN/ΔC)-naltrindole.
(a) BRIL-δOR(ΔN/ΔC)-naltrindole structure (light green, BRIL fusion is omitted) and residues in the allosteric sodium site (green sticks). Sodium is shown as a blue sphere; red and pink spheres are waters in the first and second coordination shells, respectively. Naltrindole is shown as orange sticks. (b) Interactions in the sodium site. Orange transparent surface depicts hydrophobic residues below the allosteric sodium site. Hydrogen bonds are shown as grey dotted lines. (c) ‘Sliced’ surface representation of BRIL-δOR(ΔN/ΔC)-naltrindole showing the continuous pathway connectivity between the orthosteric and allosteric sodium site. Receptor surface and interior are colored green and black, respectively. (d) 2mFo- DFcelectron density map (grey mesh) contoured at 2 σ around residues, waters and sodium in the allosteric site. Hydrogen bonds in the first sodium ion coordination shell are shown as black dotted lines; all other hydrogen bonds are shown as yellow dotted lines. Yellow arrow indicates the increased β-arrestin constitutive activity of Asn1313.35Ala and Asn1313.35Val mutants. Red arrow indicates the potent β-arrestin biased activation and concomitant abrogation of Gi signaling in response to the ligand naltrindole in the Asp952.50Ala mutant. The “efficacy switch” residues that resulted in β-arrestin bias when mutated are highlighted in orange (Asn1313.35) and red (Asp952.50, Asn3107.45 and Asn3147.49) in panels a-d.
The 1.8 Å structure of the human δ-OR is similar to the 3.4 Å Mus musculus δ-OR structure fused to T4 lysozyme8 at the ICL3 site (r.m.s. deviation of 0.91 Å over all structurally characterized Cα atoms) with the distinction that the atomic details of regions crucial for receptor activity are revealed. These include: (1) a fully resolved ICL3 adopting a ‘closed’ inactive state conformation (Fig. 2); (2) a detailed molecular characterization of the orthosteric site with water-mediated ligand-receptor interactions (Fig. S1a); (3) a distinct conformation of the human third extracellular loop (ECL3) (Fig. S2); and importantly (4) a high-resolution characterization of the allosteric sodium site, water molecules and a comprehensive network of hydrogen bond interactions inside the 7TM core (Fig. 1, Fig. S3 and Fig. S4).
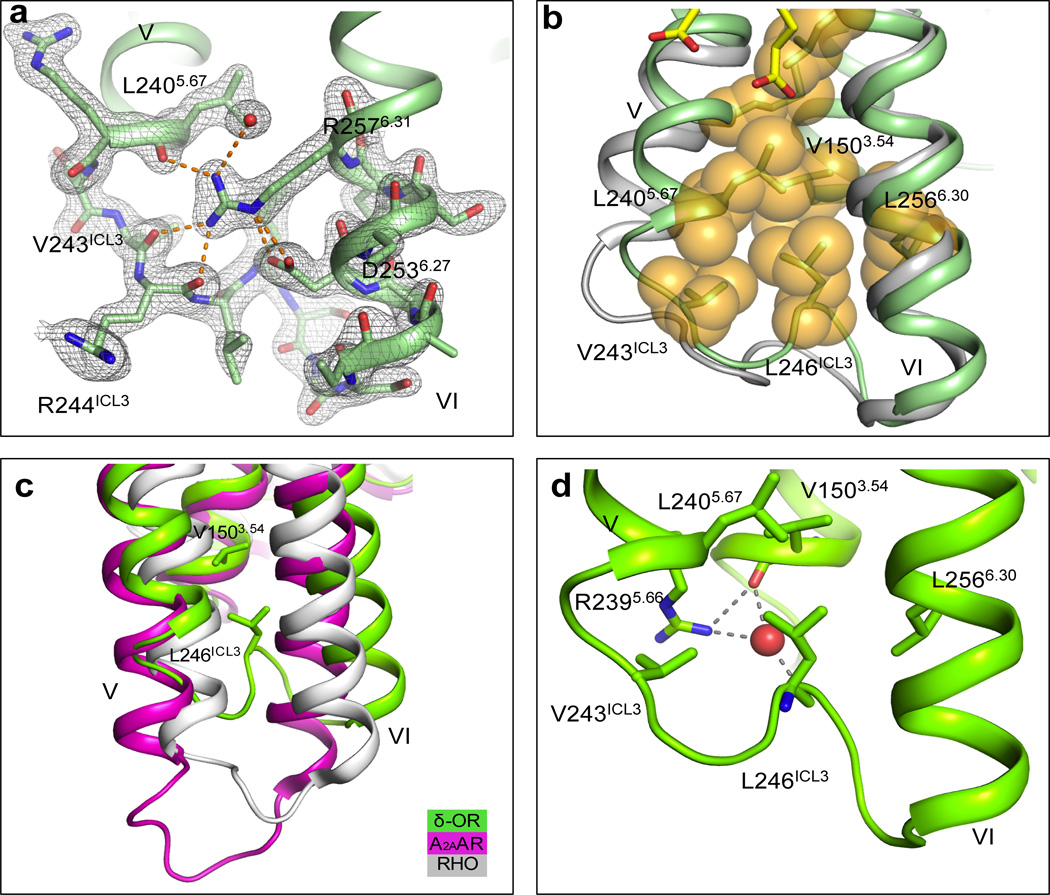
Fig. 2. Structure of the human δ-OR ICL3.
(a) 2mFo- DFcelectron density map (grey mesh) of BRIL-δOR(ΔN/ΔC)-naltrindole ICL3 contoured at 1σ. Polar and ionic interactions of Arg2576.31 shown by orange dashed lines (b) ICL3 loop comparison between BRIL-δOR(ΔN/ΔC)-naltrindole (green) and NOP12 (light grey) structures. Hydrophobic residues in the ICL3 hydrophobic cluster are shown as orange spheres and OLA molecules are represented by yellow sticks. (c) ICL3 comparison of BRIL-δOR(ΔN/ΔC)-naltrindole (green) with A2AAR (magenta; PDB ID 3PWH) and Rhodopsin (white; PDB ID 3CAP). (d) Details of the hydrogen bonds between Arg2395.66, Leu246ICL3, Val1503.54 and water molecule (red sphere) shown by grey dashed lines
All ICL3 residues are well resolved in the BRIL-δOR(ΔN/ΔC)-naltrindole structure. The side chain guanidinium group of Arg2576.31 (superscripts indicate residue numbering using the Ballesteros–Weinstein nomenclature9) appears to play a key role in stabilizing ICL3 by forming an extensive hydrogen bonding network with the main chain carbonyls of Leu2405.67, Arg244ICL3 and Val243ICL3, and a salt bridge with the carboxylate group of Asp2536.27 (Fig. 2a). The Leu246ICL3 and Val243ICL3 side chains insert back in the helical bundle and form a hydrophobic cluster with Val1503.54, Leu2405.67 and Leu2566.30 (Fig. 2b). The loop also interacts with helix III via a water mediated hydrogen bond network between the main chain carbonyl groups of Leu246ICL3 and Val1503.54, and the side chain of Arg2395.66 (Fig. 2d). These atomic details suggest a stable ‘closed’ conformation of ICL3 in the inactive δ-OR, which tethers the intracellular ends of helices V and VI. While it contrasts with the more ‘exposed’ ICL3 conformations in the thermostabilized A2A adenosine receptor (A2AAR; PDB ID 3PWH)10 and rhodopsin (PDB ID 3CAP)11 (Fig. 2c), the ICL3 in δ-OR is similar to that observed in the lower resolution NOP structure (PDB ID 4EA3)12 (Fig. 2b). A high sequence conservation of ICL3 in all four ORs, which signal primarily via Gαi/o-proteins suggests that ICL3 can adopt a similar ‘closed’ conformation in inactive states of all opioid receptor subtypes. The closed conformation of ICL3 may play a role in stabilizing the inactive state in opioid receptors, and thus compensate for the lack of a stabilizing “ionic lock” in these receptors, which have a hydrophobic Leu6.30 instead of the usual Glu6.30 side chain that is required for an “ionic lock”.
In the orthosteric pocket, the BRIL-δOR(ΔN/ΔC)-naltrindole structure reveals an extensive network of water-mediated interactions of the morphinan group of naltrindole, including interactions with residues in helix V and ECL2 (Fig. S1a). For the ECL3 region, the key selectivity determinant for classical peptide binding to opioid receptors13, we observe that the side chain of Arg291ECL3 constrains a distinct loop conformation between helices VI and VII through hydrogen bonding networks with the main-chain carbonyl groups of Val287ECL3 and Trp2846.58, positioning the latter for a π–π interaction with naltrindole (Fig. S2). This conformation of ECL3 is quite different from the one observed for the lower resolution Mus musculus δ-OR structure8, which has an asparagine side chain instead of the Asp290ECL3 seen in the human δ-OR. These high resolution details of the binding pocket and ligand interactions in the human δ-OR orthosteric site provide an excellent framework for designing new δ-OR ligands14 and allosteric modulators15 with improved selectivity and functional profiles.
Unique features of the δ-OR sodium site
Evidence for the presence of a sodium ion in the allosteric site is similar to that observed in the high resolution A2AAR structure (PDB ID 4EIY)16, including: (1) electron density showing coordination of the proposed sodium position by five oxygen atoms; (2) short distances observed between the ion and coordinating oxygens (~2.4 Å); and (3) calculations of ion valence (Table S2). The cavity harboring the allosteric sodium is formed by the side chains of sixteen residues, fifteen of which are highly conserved in class A GPCRs (Fig. 1 and Fig. S3). Remarkably, the structure of BRIL-δOR(ΔN/ΔC)-naltrindole revealed that in addition to the highly conserved Asp952.50 and Ser1353.39 side chains16, the sodium ion is directly coordinated by a non-conserved Asn1313.35 side chain. While Asn3.35 is conserved among opioid receptors (Fig. S3b), the majority (~70%) of class A GPCRs has a hydrophobic residue in this position, and in the high-resolution A2AAR structure (PDB ID 4EIY) the side chain of Leu873.35 is pointing towards the lipidic membrane16. In contrast, in the BRIL-δOR(ΔN/ΔC)-naltrindole structure the Asn1313.35 side chain points into the sodium pocket, placing its Od1 and Nd2 atoms between the ion and the orthosteric pocket (Fig. 1). These exact atom positions are occupied by two water molecules in the allosteric sodium site of the A2AAR structure (Fig. S3a). In addition to the key role of Asn131’s side chain Od1 atom in sodium coordination, the Nd2 atom is hydrogen bonded to both side chain Od1 and main chain carbonyl atoms of Asp1283.32 via a water molecule (Fig. 1); the latter residue occupies a central position deep in the orthosteric site and establishes a salt bridge with the nitrogen group of naltrindole. These interactions between the sodium ion, Asn1313.35 and Asp1283.32 establish an apparent axis of connectivity between orthosteric and allosteric regions on the receptor characterized in the inactive state. Altogether, the δ-OR’s allosteric sodium is coordinated by five oxygen atoms, from Asp952.50, Ser1353.39 and Asn1313.35 side chains and two structurally conserved water molecules, which comprises the first coordination shell for the sodium ion (Fig. 1, Figs. S3 and S4).
The second coordination shell of the sodium ion in the allosteric site is formed by three residues (Trp2746.48, Asn3107.45and Asn3147.49) and two additional water molecules in contact with waters in the first shell (Fig. 1, Figs. S3 and S4). These conserved residues of the sodium pocket belong to two of the most well-known Class A functional motifs: CW6.48xP in helix VI and N7.49PxxY in helix VII (Fig. 1a), which play a critical role in GPCR activation processes17. As a whole, the cluster comprising the sodium ion and eight water molecules mediates extensive intra-helical hydrogen bond networks between helices I, II, III, VI and VII in the core of the receptor, when it is stabilized in an inactive state conformation. In contrast, the activated agonist bound structure of A2AAR reveals a sodium site that is collapsed by an inward movement of helix VII18, suggesting that rearrangements in this conserved sodium pocket play a major role in the activation of class A GPCRs2.
Functional characterization of δ-OR
To correlate the structural data obtained using BRIL-δOR(ΔN/ΔC) with the WT δ-OR, we performed radioligand binding assays with opioid agonists and antagonists with WT δ-OR and BRIL-δOR(ΔN/ΔC) constructs expressed in HEK293 and Sf9 (Spodoptera frugiperda) cells, respectively. We found that the BRIL-δOR(ΔN/ΔC) and WT δ-OR displayed similar ligand binding affinities (Table S3) and, when both were expressed in HEK293 cells, displayed similar functional coupling to Gαi-mediated signaling (Figs. S5a-b). Consistent with classical studies performed on ORs in situ5,19,20, physiological concentrations of NaCl (140 mM) reduced the affinity of the δ-OR peptide agonist DADLE (which is structurally related to endogenous peptide agonists) at both WT δ-OR expressed in HEK293 cells and BRIL-δOR(ΔN/ΔC) expressed in Sf9 cells, while having minimal effects on antagonist binding affinity (Table S3). To confirm the specificity of the sodium site, we also examined the effects of other monovalent cations on the binding of the 3H-DADLE in saturation binding assays. Among several monovalent cations tested, only sodium at physiological concentrations reduced 3H-DADLE binding (Fig. S6).
Sodium modulates δ-OR ligand binding
Although the phenomenon of allosteric modulation of GPCR ligand affinity by sodium ions has been previously described for a number of class A GPCRs (e.g. opioid, adrenergic, adenosine and dopamine receptors)21–24, the nature of this allosteric effect, as well as sodium’s affinity for the allosteric site is unknown. To quantify sodium’s affinity at its allosteric binding site and to clarify the nature of the apparent negative cooperativity with respect to the peptide agonist DADLE, we performed a series of radioligand binding assays in varying NaCl concentrations and then analyzed the results using a standard allosteric model25. Our studies revealed that sodium had essentially the same affinity and produced a similar degree of negative cooperativity at both WT δ-OR expressed in HEK293 cells and the BRIL-δOR(ΔN/ΔC) expressed in Sf9 cells, although the negative cooperativity with DADLE was slightly decreased in the crystallized construct (Fig. 3a, Figs. S5c-d, and Table 1). These findings confirm that the crystallized construct maintains essentially the same capacity for sodium-dependent allosteric regulation as the WT δ-OR. Moreover, by revealing the relatively high sodium affinity to δ-OR (KB = 13.3 mM), our studies demonstrate that at physiological sodium concentrations (140 mM) the sodium site is likely to be saturated.
Fig. 3. Effect of sodium site mutations on allosteric sodium effect and β-arrestin constitutive activity.
The effects of graded doses of sodium on DADLE affinity on (a) WT and (b) sodium site mutant D95A were determined, results normalized and then pooled for analysis in Prism using the allosteric model with fitted parameters summarized in Table 1. (c) The basal activity of N131A and N131V is compared to WT receptor over a range of DNA dosages (0.5 – 5.0 µg per 15-cm dish) and shows that receptors were expressed at comparable levels (WT 226 – 758 fmol/mg; N131A 77 – 553 fmol/mg; N131V 176 – 715 fmol/mg). Basal activity of N131A and N131V exceeded the activity achieved with a saturating concentration of DADLE (10 µM) at WT δ-OR (open square). Results represent average ± SEM from a minimum of 64 replicates from a representative assay; error bars are smaller than the corresponding symbols. (d) The sodium site mutations N131A and N131V responded to DADLE (10 µM) with a modest degree of stimulation (*p<0.01 (t-test) vs. no drug addition). Receptors were transfected at a low level in these experiments, allowing the detection of activation by a saturating concentration of DADLE.
Table 1. Allosteric parameters for sodium at BRIL-δOR(ΔN/ΔC) and WT δ-OR.
Radioligand binding assays (Figs. 3a and 3b, Figs. S5c and S5d) were analyzed by the allosteric model (see 25 for details). Here α defines the effect of allosteric ligand (in this case, sodium) on orthosteric ligand (in this case, DADLE); an α>1 indicates a positive effect thereby increasing binding affinity, while an α<1 indicates a negative effect thereby reducing binding affinity. For the D95A, D95N and N131A mutations no allosteric effect of sodium was observed (see Fig. 3b for representative data with the D95A mutant).
| Na+ pKB ± SEM |
Na+ KB (mM) |
pα ± SEM | α | Hill Coefficient | |
|---|---|---|---|---|---|
| δ-OR WT (HEK293) | 1.88 ± 0.10 | 13.3 | 1.93 ± 0.34 | 0.012 | 0.56 ± 0.16 |
| D95A (HEK293) | NAE | NAE | NAE | NAE | NAE |
| D95N (HEK293) | NAE | NAE | NAE | NAE | NAE |
| N131A (HEK293) | NAE | NAE | NAE | NAE | NAE |
| N131V (HEK293) | 1.11 ± 0.15 | 77 | 1.61 ± 0.86 | 0.025 | 0.61 ± 0.02 |
| BRIL-δOR(ΔN/ΔC) (Sf9) | 1.79 ± 0.11 | 15.9 | 0.86 ± 0.05 | 0.138 | 0.79 ± 0.02 |
NAE=No Allosteric Effect of sodium observed
Asn131 regulates β-arrestin bias
Given the intimate relationships between residues coordinating allosteric sodium and motifs implicated in GPCR activation (e.g. Asn3147.49 and Tyr3187.53 of the NP7.50xxY motif; Fig. 1)4, we predicted that mutating selected sodium site residues could modulate δ-OR functionality. Accordingly, we performed functional studies on selected sodium site mutants (Figs. 3 and 4, Table 1, Tables S4-S6 and Figs. S7 and S8) and discovered that mutating the sodium anchoring δ-OR residue Asn1313.35 into alanine or valine dramatically enhanced constitutive activity for the β-arrestin pathway (Fig. 3c-d, Fig. S8). Remarkably, the β-arrestin constitutive activity of Asn131Ala or Asn131Val mutants exceeded the activation levels of WT δ-OR achieved with a saturating concentration of the agonist DADLE (Fig. 3c), whereas Gαi protein basal activity remained unaffected (Fig. S7). The Asn1313.35 mutants increased receptor β-arrestin constitutive activity as well as DADLE binding affinity, as compared with WT δ-OR, albeit DADLE efficacy for the β-arrestin pathway was greatly reduced (Tables S4-S6). Importantly, while the Asn1313.35Ala mutation abolished the ‘sodium effect’, the receptor carrying the Asn1313.35Val mutation retained sodium ion binding, although with lower affinity compared to WT receptor (Table 1).
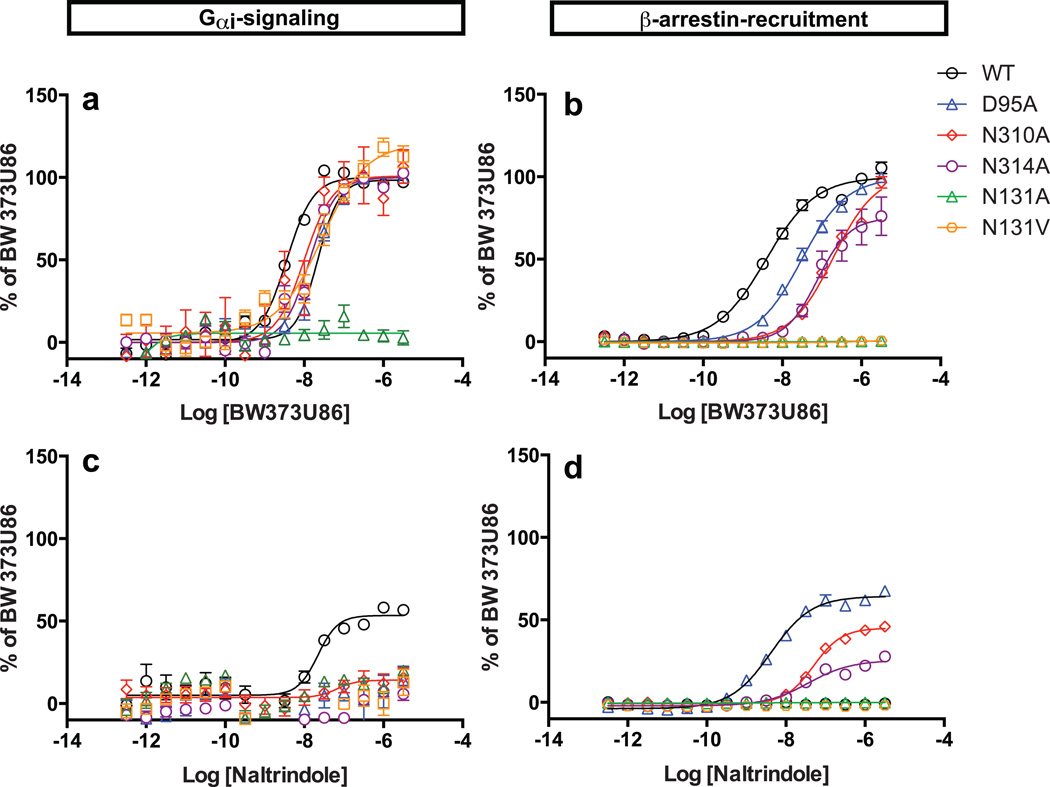
Fig. 4. Sodium-coordinating residues form an efficacy switch regulating biased signaling.
Mutation of the sodium-anchoring residues Asp95, Asn310 and Asn314 promotes efficacy switching of the cyclopentene-containing antagonist naltrindole into a potent β-arrestin-biased agonist. Normalized concentration-responses of δ-OR-mediated Gαi signaling induced by (a) BW373U86 and (c) naltrindole, and δ-OR mediated β-arrestin recruitment with (b) BW373U86 and (d) naltrindole were quantified as in Methods. Results represent average ± SEM of four independent experiments each in quadruplicate and are presented as % of activation by BW373U86. Receptors were all transfected with 15 µg DNA revealing weak partial agonist activity of naltrindole at Gαi signaling as previously described29,30.
The key differences in sodium ion binding affinity between the Asn1313.35 mutants provided us a model system to further clarify the role of sodium on canonical Gαi-protein mediated signaling. Remarkably, the Asn1313.35Ala mutant was inactive while the Asn1313.35Val mutant, which retains a significant ‘sodium effect’, maintained Gαi-activity though with reduced agonist potency compared with the WT δ-OR (Fig. 4a, Table S4). These data indicate that a complete disruption of the interactions between Asn1313.35 and the sodium ion can induce high levels of constitutive activity at non-canonical β-arrestin signaling, while simultaneously abolishing canonical G-protein signaling, essentially inducing an ‘efficacy switch’ from the Gαi-protein pathway to an β-arrestin pathway. These results reveal that the non-conserved residue Asn1313.35 plays an essential role in controlling both δ-OR functional selectivity and constitutive activity, likely via its structural role in coordinating the allosteric sodium. We also compared basal Gαi-protein activity for the sodium site δ-OR mutants (Fig. S7) and found that only the Asn3107.45Ala mutant displayed enhanced constitutive Gαi-protein activity as indicated by lower basal pertusis-toxin sensitive activity.
Sodium-dependent opioid receptor pharmacology
We next examined Asp952.50, another key sodium site residue, and found that mutation of this residue into either alanine or asparagine abolished the ‘sodium effect’ in radioligand binding assays (Fig. 3b and Table 1). The small molecule agonist BW373U86 has been previously described as a selective orthosteric δ-OR agonist for which binding to the receptor is minimally affected by sodium ions26. Consequently, BW373U86 displays a low to moderate reduction in G-protein and β-arrestin signaling at sodium-anchored residue mutants compared to WT (Fig. 4a-b). On the other hand, the peptide agonist DADLE (which is structurally related to endogenous peptide agonists) has a diminished potency for β-arrestin recruitment at the δ-OR mutant Asp952.50Ala and is inactive at both the Asn3107.45Ala and the Asn3147.49Ala mutants, while showing low to moderate reduction in G protein agonist efficacy (Table S4). Importantly, we discovered that classical δ-OR ligands containing a cyclopentene functional group (naltrindole, naltriben, and BNTX), which display no apparent agonist activity at the β-arrestin pathway with the WT δ-OR, gained potent β-arrestin-biased agonist activity at the Asp952.50Ala sodium site mutant (Fig. 4d and Table S4). A similar transformative effect is observed when the sodium site residues Asn3107.45and Asn3147.49 were mutated to alanine (Fig. 4d and Table S4). The conversion of these antagonists/weak partial agonists into β-arrestin biased-agonists by mutations in the allosteric sodium site, together with the effects of Asn1313.35 mutants described above uncovered what we characterize as ‘efficacy switches’ within δ-OR. These efficacy switches are apparently distinct from those previously reported at which only G protein signaling was enhanced27,28.
The allosteric sodium binding pocket described here in atomic details is potentially an attractive drug discovery target. While the binding of small molecule allosteric modulators in this highly conserved site is unlikely to have desired subtype selectivity, extension of selective orthosteric ligands into the sodium cavity may lead to bitopic compounds with new pharmacological properties, e.g. inverse agonism or strong functional bias. The detailed crystal structure may also help to identify the binding site and shed light on the mode of action for the other types of PAM compounds, such as those recently reported by Burford et al.15 .
Our results reveal a profound and essential role for allosteric sodium anchoring residues at specifying GPCR signal transduction and pharmacology. Mutation of sodium anchoring residues within the allosteric site selectively modulates not only agonist binding, but also dramatically changes GPCR functional activity by augmenting β-arrestin constitutive activity and introducing new patterns of biased signaling. In particular, these findings highlight the surprisingly essential role for sodium-coordinating amino acids as “efficacy-switches” for GPCR signaling.
Methods
Cloning, expression and purification
The WT human δ-OR gene (OPRD1; UniProt accession P41143) was synthesized by DNA2.0 with codon optimization for expression in Spodoptera frugiperda (Sf9), and then cloned into a modified pFastBac1 vector (Invitrogen) containing an expression cassette with a haemagglutinin signal sequence followed by a Flag tag, a 10× His tag and a TEV protease recognition site at the N-terminus. Thirty-four amino acids were deleted from the C-terminus (residues 339–372), and 35 residues of the N-terminus (residues 1–35) of δ-OR were replaced with the thermostabilized apocytochrome b562RIL from Escherichia coli (M7W, H102I and R106L) (BRIL)31 protein using splicing by overlap extension PCR32. A Pro37Ser mutation was introduced in the N-terminus of δ-OR to facilitate crystallization. Recombinant baculoviruses were generated using the Bac-to-Bac system (Invitrogen) and were used to infect Sf9 insect cells at a density of 2 × 106 cells ml−1 at a multiplicity of infection of 5 as previously described33. Infected cells were grown at 27 °C for 48 h before being harvested, and the cell pellets were stored at −80 °C.
Insect cell membranes were disrupted as previously described34. In brief, cell pellets were homogenized by douncing in a hypotonic buffer containing 10 mM HEPES pH 7.5, 10 mM MgCl2, 20 mM KCl and EDTA-free complete protease inhibitor cocktail tablets (Roche). Washing of the membranes was performed by repeated dounce homogenization and centrifugation in the same hypotonic buffer (once more), followed by high osmotic buffer containing 1.0 M NaCl, 10 mM HEPES pH 7.5, 10 mM MgCl2, 20 mM KCl and EDTA-free complete protease inhibitor cocktail tablets (three times). Purified membranes were resuspended in 10 mM HEPES pH 7.5, 10 mM MgCl2, 20 mM KCl and 30% (v/v) glycerol, flash frozen with liquid nitrogen, and stored at −80°C.
Prior to receptor purification membranes were thawed and washed once again in a buffer containing 1.0 M NaCl, 10 mM HEPES pH 7.5, 10 mM MgCl2, 20 mM KCl. Washed membranes were resuspended in buffer containing 50 µM naltrindole (Tocris), 2 mg/ml−1 iodoacetamide (Sigma), 500 mM NaCl, 50 mM HEPES pH 7.5, and incubated at 4 °C for 1 h before solubilization. The membranes were then solubilized in 50 mM HEPES, pH 7.5, 500 mM NaCl, 1.5% (w/v) n-dodecyl-β-D-maltopyranoside (DDM; Anatrace), 0.3% (w/v) cholesteryl hemisuccinate (CHS; Sigma) and 25 µM naltrindole for 3 h at 4 °C. The supernatant was isolated by centrifugation at 160,000 g for 45 min, and incubated in 20 mM buffered imidazole (pH 7.5), 0.7 M NaCl with 1 ml of TALON IMAC resin (Clontech) overnight at 4 °C. After binding, the resin was washed with 15 column volumes of wash buffer I (50 mM HEPES, pH 7.5, 800 mM NaCl, 10% (v/v) glycerol, 0.1% (w/v) DDM, 0.02% (w/v) CHS, 10 mM ATP, 10 mM MgCl2 and 50 µM naltrindole), followed by 10 column volumes of wash buffer II (50 mM HEPES, pH 7.5, 500 mM NaCl, 10% (v/v) glycerol, 0.02% (w/v) DDM, 0.004% (w/v) CHS, 50 mM imidazole and 50 µM naltrindole). The protein was then eluted by 5 column volumes of Elution Buffer (50 mM HEPES, pH 7.5, 500 mM NaCl, 10% (v/v) glycerol, 0.02% (w/v) DDM, 0.004% (w/v) CHS, 250 mM imidazole and 100 µM naltrindole). PD miniTrap G-25 column (GE Healthcare) was used to remove imidazole. The protein was then treated overnight with His-tagged TEV protease to cleave the N-terminal His-tag and Flag-tag. TEV protease and the cleaved N-terminal fragment were removed by TALON IMAC resin incubation for 1 h at 4 °C. Purified receptor was concentrated to 20 mg ml−1 with a 100 kDa molecular weight cut-off Vivaspin centrifuge concentrator (GE healthcare). Protein purity and monodispersity were tested by SDS–PAGE and analytical size-exclusion chromatography (aSEC). Typically, the protein purity exceeded 95%, and the aSEC profile showed a single peak, indicative of receptor monodispersity.
Crystallization
Receptor samples in complex with naltrindole were reconstituted into lipidic cubic phase (LCP) by mixing with molten lipid using a mechanical syringe mixer35. The protein-LCP mixture contained 40% (w/w) protein solution, 54% (w/w) monoolein (Sigma) and 6% (w/w) cholesterol (AvantiPolar Lipids). Crystallization trials were performed in 96-well glass sandwich plates36 (Marienfeld) by an NT8-LCP crystallization robot (Formulatrix) using 40 nl protein-laden LCP overlaid with 0.8 µl precipitant solution in each well, and sealed with a glass coverslip. Protein reconstitution in LCP and crystallization trials was carried out at room temperature (~20–23 °C). The crystallization plates were stored and imaged in an incubator/imager (RockImager 1000, Formulatrix) at 20 °C. Diffraction quality crystals of an average size of 50×10×3 µm were obtained within ~10 d in 31–34% (v/v) PEG 400, 0.095 to 0.12 M K/Na tartrate, 5% (v/v) ethylene glycol, 100 mM MES buffer at pH 6.1–6.2 and 1 mM naltrindole. Crystals were harvested directly from LCP using 50 µm MiTeGen micromounts and immediately flash frozen in liquid nitrogen.
X-ray data collection and processing
Crystallographic data were collected on the 23ID-D beamline (GM/CA CAT) of the Advanced Photon Source at the Argonne National Laboratory using a 20 µm collimated minibeam at a wavelength of 1.0330 Å and a MarMosaic 300 detector. To reduce radiation damage crystals were translated to a fresh position, if possible, or replaced after collecting 10 frames at 1 s exposure and 1.0° oscillation with an unattenuated beam. Datasets from 47 different crystals were integrated, scaled and merged together using HKL200037(Table S1).
Structure determination and refinement
Initial molecular replacement solution was obtained by PHASER38 in the CCP4 suite, using the 3.4 Å δ-OR receptor structure (PDB ID 4EJ4) with deleted T4L, and BRIL from A2AAR (PDB ID 4EIY) as independent search models. The resulting BRIL-δ-OR model was refined by manually building in the excessive 2Fo-Fc density and by repetitive cycling between COOT39, REFMAC540, and simulated annealing using PHENIX41 until convergence. Ten translation, libration and screw-rotation atomic displacement (TLS) groups were used throughout refinement. The elongated electron density tubes near the protein hydrophobic surface were modeled as oleic acids (OLA), with the exception of the few that were better fit with monooleins (OLC), the major lipid component used for crystallization. The data collection and refinement statistics are shown in Table S1.
δ-OR β-arrestin recruitment assay
Assays were performed using modifications of the original Tango assay42 as described previously43,44. Briefly, δ-OR constructs were codon optimized for mammalian expression and synthesized by Blue Heron Biotech (Bothell WA), and inserted into the modular vector system (described in 42) with all constructs confirmed by automated dsDNA sequencing. HTLA cells (kindly provided by Dr. Richard Axel) were transfected by the calcium phosphate precipitation method. The next day, the cells were plated in DMEM supplemented with 1–2% dialyzed FBS into Poly-L-Lys (PLL) coated 384-well white clear bottom cell culture plates at a density of 15,000 cells per well in a total volume of 50 µl. In the following day, ligand solutions were prepared in filtered assay buffer (20 mM HEPES, 1× Hanks’ balanced salt solution (HBSS), pH 7.40) at 3.5× and added to cells (20 µl per well) for overnight incubation (16–20 h). On the next day, media and drug solutions were removed and 20 µl per well of Bright-Glo reagent (Promega, 1:20 dilution in assay buffer) was added. Plates were incubated for 20 min at room temperature in the dark before being counting using a TriLux luminescence reader (1 s/well). To determine basal and constitutive activity of WT δ-OR and N131A or N131V mutant δ-ORs, HTLA cells were transfected as above at 0.5 – 5 µg DNA per 15-cm dish. An aliquot of the transfected cells was plated and incubated for overnight without agonist stimulation, and luminescence was determined as above. The remainders of the transfected cells were subjected to membrane preparation and binding assays to estimate receptor expression levels with 3H-naltrindole (see below). Data were subjected to non-linear least-squares regression analysis using the sigmoidal dose-response function provided in GraphPad Prism 5.0 or 6.0. Data of four independent experiments (N=4) performed in quadruplicate are presented as Relative Luminescence Unit (Fig. 3) or as % of BW373U86 (Fig. 4).
cAMP assays
HEK293T cells were co-transfected with human δ-OR or various mutants along with a split-luciferase-based cAMP biosensor (GloSensor; Promega) and assays performed in a manner similar to prior studies with Gαi-coupled receptors 45. Transfected cells were plated into Poly-L-Lys coated 384-well white clear bottom cell culture plates with DMEM + 1% dialyzed FBS at a density of 15,000 cells per 40 µl per well for overnight. On the day of assay, cells were removed of culture medium and received 20 µl/well assay buffer (20 mM HEPES, 1× HBSS, pH 7.40) followed by addition of 10 µl of 3× drug solutions for 15 min at room temperature. To measure agonist activity for Gαi coupled receptors, 10 µl of luciferin (4 mM final concentration) supplemented with Isoproterenol (400 nM final concentration were added to activate Gs via endogenous β2-adrenergic receptors) and luminescence intensity was quantified 15 min later. Data were normalized compared to the agonist BW373U86 and regressed using the sigmoidal dose-response function. Data of four independent experiments (N = 4) conducted in quadruplicate are presented as % of BW373U86 (Fig. 4). Analyses were performed using the software GraphPad Prism.
Radioligand binding assays
3H-naltrindole binding assays were performed using Sf9 membrane fractions expressing the crystallization construct BRIL-δOR(ΔN/ΔC) or HEK293 T membrane preparations transiently expressing WT or mutant δ-OR receptors. HEK293 T cells were transfected to make membranes and binding assays were set up in 96-well plates as previously described46. To examine the effects of sodium on ligand binding at δ-OR receptors, all binding assays were conducted in the δ-OR binding buffer (50 mM Tris HCl, 2 mM EDTA, pH 7.40) in the absence or presence of NaCl at designed concentrations. Saturation binding assays with 0.2 – 30 nM 3H-naltrindole in δ-OR binding buffer were performed to the determine equilibrium dissociation constant (Kd); while 10 µM naltrindole was used to define nonspecific binding. To quantify the allosteric effects of sodium ions, test ligands (e.g. DADLE) in a series of 11 concentrations (ranging from 0.1 nM – 300 µM) were incubated with a fixed concentration of 3H-naltrindole in the absence and presence of increasing concentrations of sodium chloride (0.3 mM – 1 M). Reactions (either saturation or competition binding) were incubated for 2 h at room temperature in the dark, and terminated by rapid vacuum filtration onto chilled 0.3% PEI-soaked GF/A filters followed by three quick washes with cold washing buffer (50 mM Tris HCl, pH 7.40) and quantified as previously described46. Results (with or without normalization) were analyzed using GraphPad Prism using one-site, two-site, or allosteric IC50 shift models where indicated.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Common Fund grant P50 GM073197 for technology development (V.C. and R.C.S.), PSI:Biology grant U54 GM094618 for biological studies and structure production (target GPCR-39) (V.K., V.C. and R.C.S.); R01 DA017204 and the NIMH Psychoactive Drug Screening Program (P.G. X-P.H and B.L.R.) and the Michael Hooker Chair for Protein Therapeutics and Translational Proteomics to B.L.R. We thank J. Velasquez for help with molecular biology, T. Trinh and M. Chu for help with baculovirus expression, G.W. Han for help with structure analysis and quality control review, E. Abola for help with sodium site analysis, A. Walker for assistance with manuscript preparation and J. Smith, R. Fischetti and N. Sanishvili for assistance in development and use of the minibeam and beamtime at beamline 23-ID at the Advanced Photon Source, which is supported by National Cancer Institute grant Y1-CO-1020 and National Institute of General Medical Sciences grant Y1-GM-1104.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
G.F. designed, optimized and purified δ-OR receptor constructs for structural studies, crystallized the receptor in LCP, collected and processed diffraction data, determined the structure, analyzed the data and wrote the paper. P.M.G. performed mutagenesis and signaling studies, analyzed the data and wrote the paper. X.-P.H. performed ligand binding and signaling studies, analyzed the data and wrote the paper. V.K. analyzed the data and wrote the paper. A.A.T. design and cloned initial δ-OR constructs. V.C. analyzed the data and wrote the paper. B.L.R. supervised the pharmacology and mutagenesis studies, analyzed the data and wrote the paper. R.C.S. was responsible for the overall project strategy and management, analyzed the data and wrote the paper.
The coordinates and the structure factors have been deposited in the Protein Data Bank under accession code 4N6H. Reprints and permissions information is available at www.nature.com/reprints.
The authors state there are no conflicting financial statements.
References
- 1.Pasternak GW. Opioids and their receptors: Are we there yet? Neuropharmacology. 2013 doi: 10.1016/j.neuropharm.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katritch V, Cherezov V, Stevens RC. Structure-function of the g protein-coupled receptor superfamily. Annual review of pharmacology and toxicology. 2013;53:531–556. doi: 10.1146/annurev-pharmtox-032112-135923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wootten D, Christopoulos A, Sexton PM. Emerging paradigms in GPCR allostery: implications for drug discovery. Nature reviews. Drug discovery. 2013;12:630–644. doi: 10.1038/nrd4052. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pert CB, Pasternak G, Snyder SH. Opiate agonists and antagonists discriminated by receptor binding in brain. Science. 1973;182:1359–1361. doi: 10.1126/science.182.4119.1359. [DOI] [PubMed] [Google Scholar]
- 6.Cooper DM, Londos C, Gill DL, Rodbell M. Opiate receptor-mediated inhibition of adenylate cyclase in rat striatal plasma membranes. Journal of neurochemistry. 1982;38:1164–1167. doi: 10.1111/j.1471-4159.1982.tb05365.x. [DOI] [PubMed] [Google Scholar]
- 7.Portoghese PS, Sultana M, Nagase H, Takemori AE. Application of the message-address concept in the design of highly potent and selective non-peptide delta opioid receptor antagonists. Journal of medicinal chemistry. 1988;31:281–282. doi: 10.1021/jm00397a001. [DOI] [PubMed] [Google Scholar]
- 8.Granier S, et al. Structure of the delta-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 10.Dore AS, et al. Structure of the Adenosine A(2A) Receptor in Complex with ZM241385 and the Xanthines XAC and Caffeine. Structure. 2011;19:1283–1293. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 12.Thompson AA, et al. Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic. Nature. 2012;485:395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonner G, Meng F, Akil H. Selectivity of mu-opioid receptor determined by interfacial residues near third extracellular loop. European journal of pharmacology. 2000;403:37–44. doi: 10.1016/s0014-2999(00)00578-1. [DOI] [PubMed] [Google Scholar]
- 14.Filizola M, Devi LA. Grand opening of structure-guided design for novel opioids. Trends in pharmacological sciences. 2013;34:6–12. doi: 10.1016/j.tips.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burford NT, et al. Discovery of positive allosteric modulators and silent allosteric modulators of the mu-opioid receptor. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10830–10835. doi: 10.1073/pnas.1300393110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Audet M, Bouvier M. Restructuring g-protein- coupled receptor activation. Cell. 2012;151:14–23. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Xu F, et al. Structure of an Agonist-Bound Human A2A Adenosine Receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selley DE, Cao CC, Liu Q, Childers SR. Effects of sodium on agonist efficacy for G-protein activation in mu-opioid receptor-transfected CHO cells and rat thalamus. British journal of pharmacology. 2000;130:987–996. doi: 10.1038/sj.bjp.0703382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yabaluri N, Medzihradsky F. Regulation of mu-opioid receptor in neural cells by extracellular sodium. Journal of neurochemistry. 1997;68:1053–1061. doi: 10.1046/j.1471-4159.1997.68031053.x. [DOI] [PubMed] [Google Scholar]
- 21.Horstman DA, et al. An aspartate conserved among G-protein receptors confers allosteric regulation of alpha 2-adrenergic receptors by sodium. The Journal of biological chemistry. 1990;265:21590–21595. [PubMed] [Google Scholar]
- 22.Costa T, Lang J, Gless C, Herz A. Spontaneous association between opioid receptors and GTP-binding regulatory proteins in native membranes: specific regulation by antagonists and sodium ions. Molecular pharmacology. 1990;37:383–394. [PubMed] [Google Scholar]
- 23.Gao ZG, Ijzerman AP. Allosteric modulation of A(2A) adenosine receptors by amiloride analogues and sodium ions. Biochemical pharmacology. 2000;60:669–676. doi: 10.1016/s0006-2952(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 24.Neve KA. Regulation of dopamine D2 receptors by sodium and pH. Molecular pharmacology. 1991;39:570–578. [PubMed] [Google Scholar]
- 25.Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacological reviews. 2002;54:323–374. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- 26.Childers SR, Fleming LM, Selley DE, McNutt RW, Chang KJ. BW373U86: a nonpeptidic delta-opioid agonist with novel receptor-G protein-mediated actions in rat brain membranes and neuroblastoma cells. Molecular pharmacology. 1993;44:827–834. [PubMed] [Google Scholar]
- 27.Holst B, et al. Identification of an efficacy switch region in the ghrelin receptor responsible for interchange between agonism and inverse agonism. The Journal of biological chemistry. 2007;282:15799–15811. doi: 10.1074/jbc.M609796200. [DOI] [PubMed] [Google Scholar]
- 28.Steen A, et al. Biased and constitutive signaling in the CC-chemokine receptor CCR5 by manipulating the interface between transmembrane helices 6 and 7. The Journal of biological chemistry. 2013;288:12511–12521. doi: 10.1074/jbc.M112.449587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szekeres PG, Traynor JR. Delta opioid modulation of the binding of guanosine-5'-O-(3-[35S]thio)triphosphate to NG108-15 cell membranes: characterization of agonist and inverse agonist effects. The Journal of pharmacology and experimental therapeutics. 1997;283:1276–1284. [PubMed] [Google Scholar]
- 30.Liu JG, Prather PL. Chronic agonist treatment converts antagonists into inverse agonists at delta-opioid receptors. The Journal of pharmacology and experimental therapeutics. 2002;302:1070–1079. doi: 10.1124/jpet.102.035964. [DOI] [PubMed] [Google Scholar]
- 31.Chu R, et al. Redesign of a four-helix bundle protein by phage display coupled with proteolysis and structural characterization by NMR and X-ray crystallography. Journal of molecular biology. 2002;323:253–262. doi: 10.1016/s0022-2836(02)00884-7. [DOI] [PubMed] [Google Scholar]
- 32.Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nature protocols. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- 33.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wacker D, et al. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. Journal of the American Chemical Society. 2010;132:11443–11445. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nature protocols. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta crystallographica. Section D, Biological crystallography. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnea G, et al. The genetic design of signaling cascades to record receptor activation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:64–69. doi: 10.1073/pnas.0710487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen JA, et al. Discovery of beta-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18488–18493. doi: 10.1073/pnas.1104807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlsson J, et al. Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nature chemical biology. 2011;7:769–778. doi: 10.1038/nchembio.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wacker D, et al. Structural features for functional selectivity at serotonin receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Besnard J, et al. Automated design of ligands to polypharmacological profiles. Nature. 2012;492:215–220. doi: 10.1038/nature11691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.