Abstract
The pocket proteins pRB, p107, and p130 have established roles in regulating the cell cycle through the control of E2F activity. The pocket proteins regulate differentiation of a number of tissues in both cell cycle-dependent and –independent manners. Prior studies showed that mutation of p107 and p130 in the mouse leads to defects in cartilage development during endochondral ossification, the process by which long bones form. Despite evidence of a role for pRB in osteoblast differentiation, it is unknown whether it functions during cartilage development. Here, we show that mutation of Rb in the early mesenchyme of p107-mutant mice results in severe cartilage defects in the growth plates of long bones. This is attributable to inappropriate chondrocyte proliferation that persists after birth and leads to the formation of enchondromas in the growth plates as early as 8 weeks of age. Genetic crosses show that development of these tumorigenic lesions is E2f3 dependent. These results reveal an overlapping role for pRB and p107 in cartilage development, endochondral ossification and enchondroma formation that reflects their coordination of cell cycle exit at appropriate developmental stages.
Keywords: pRB, p107, Prx1-Cre, cartilage, chondrocytes, enchondroma
INTRODUCTION
The retinoblastoma protein, pRB, is a key tumor suppressor. Inheritance of a mutant copy of the human retinoblastoma gene, RB-1, leads to early onset retinoblastoma and increases the risk of osteosarcoma by 500-fold (1). In addition, RB-1 is somatically mutated in 30% of human tumors and many other tumors carry mutations that deregulate pRB’s upstream regulators and thereby disrupt pRB function (2). The retinoblastoma gene (Rb) also acts as a tumor suppressor in mice, although Rb heterozygotes develop pituitary and thyroid tumors and not retinoblastoma (3–5). Analyses of mouse models showed that pRB is essential for normal development. Germline Rb−/− embryos die in mid-gestation with a variety of abnormalities (3–5). Some of these defects result indirectly from placental deficiency (6). The presence of a wild-type placenta extends the lifespan of Rb deficient embryos to birth, but these still display ectopic proliferation in the lens, CNS and PNS and also defective erythrocyte maturation and myogenesis (7–11). Many, but not all, of these Rb mutant defects reflect pRB’s role in suppressing cell proliferation through regulation of the E2F transcription factors (12–16).
pRB is a member of the pocket protein family, which includes p107 and p130. p107 and p130 also regulate E2F and cellular proliferation (17), and they can act as tumor suppressors in certain Rb-mutant mouse tissues. For example, chimeras generated with Rb−/−;p107−/− or Rb−/−;p130−/− ES cells develop retinoblastoma and Rb+/−;p107−/− chimeras develop osteosarcomas and other tumor types (18). The three pocket proteins also play important, overlapping roles in normal development. Mutation of p107 in the context of Rb mutation exacerbates many of the Rb mutant phenotypes and also elicits novel defects (19–21). Combined germline loss of p107 and p130 causes shorter long bones and reduced endochondral ossification, due to inappropriate proliferation of chondrocytes in the long bone epiphyses (22, 23). Interestingly, the activating E2Fs have been independently associated with normal growth plate development; E2F1 overexpression inhibits hypertrophic chondrocyte differentiation resulting in shorter long bones (24), and the combined deletion of E2f1 and E2f3a yields disorganized growth plates and abnormal chondrocytes (25, 26).
Rb mutation has been shown to impair bone differentiation in vivo and in vitro (27–30), however its role in cartilage and bone growth plate development has not been explored. Here we show that combined mutation of Rb and p107 in the early mesenchyme causes a defect in cartilage development that yields severe long bone abnormalities and the development of enchondromas, a benign cartilage lesion.
RESULTS
Mesenchymal deficiency of Rb and p107 causes decreased viability and growth plate abnormalities
To determine whether pRB and p107 function together in the growing skeleton, we used the Prx1-Cre transgene (31) to conditionally mutate Rb in early mesenchymal cells of the developing limb bud in the absence or presence of p107 mutation. By intercrossing Rb+/c;p107+/−;Prx1-Cre and Rbc/c;p107−/− mice, we generated the following test genotypes: Rb+/c;p107+/−;Prx1-Cre (herein called control), Rb+/c;p107−/−;Prx1-Cre (p107 KO), Rbc/c;p107+/−;Prx1-Cre (Rb KO), and Rbc/c;p107−/−;Prx1-Cre (DKO). We found that the DKOs were clearly under-represented at 3 weeks of age (Table 1) and thus screened for the presence of these animals at 0, 1, 2, and 3 days after birth and also at embryonic day (e) 15.5, 17.5, and 18.5. The DKOs arose at the expected Mendelian frequency before birth, but were under-represented at all perinatal stages (Table 1; data not shown). Thus, deletion of Rb and p107 in the mesenchyme of the developing limb bud reduces perinatal viability.
Table 1.
Viability of progeny from Rb+/c;p107+/−;Prx1-Cre x Rbc/c;p107−/− Cross
| Genotype | at 3 weeks | at e18.5 |
|---|---|---|
|
| ||
| ratio obs/exp (# obs)1 | ratio obs/exp (# obs)2 | |
| Rb+/c p107+/− Cre+ | 1.4 (75) | 1.1 (15) |
| Rb+/c p107−/− Cre+ | 0.7 (34) | 0.9 (12) |
| Rbc/c p107+/− Cre+ | 1.3 (70) | 1.1 (15) |
| Rbc/c p107−/− Cre+ | 0.2 (10) | 0.9 (13) |
| Rb+/c p107+/− | 1.5 (77) | 0.9 (12) |
| Rb+/c p107−/− | 0.7 (38) | 0.9 (13) |
| Rbc/c p107+/− | 1.5 (77) | 1.4 (20) |
| Rbc/c p107−/− | 0.8 (41) | 0.9 (13) |
53 mice expected for each genotype; 422 progeny counted
14 mice expected for each genotype; 113 progeny counted
To determine the cause of lethality, we screened e17.5 DKO embryos from 5 litters for mesenchymal defects. We began by staining embryos with Alizarin Red and Alcian Blue to detect calcified bone and cartilage, respectively. The cranial and other skull bones, which develop through a cartilage-independent process called intramembraneous ossification, showed no morphological abnormalities (data not shown). In contrast, we found striking defects in the DKO limbs that were absent from control and Rb KO littermates and present in a much more subtle form in a subset of p107 KO littermates. Overall limb length, and also the length of ossified regions, was reduced in DKO compared to control embryos, and the cartilaginous regions were widened (Fig. 1A). The DKO sternebrae and xiphoid processes were also under-ossified and abnormally wide (Fig. 1B). Notably, the affected bones all form through endochondral ossification in which formation of a cartilage template precedes bone deposition and mineralization. This, and the lack of any defect in intramembraneous ossification, implicates an underlying abnormality in cartilage and not bone formation. This occurs even though Prx1-Cre is expressed in a wide array of adult mesenchymal tissues including chondrocyte and osteoblast lineages (Supplementary Fig. S1) and we see efficient loss of pRb expression in Rb KO osteoblasts (Supplementary Fig. S2).
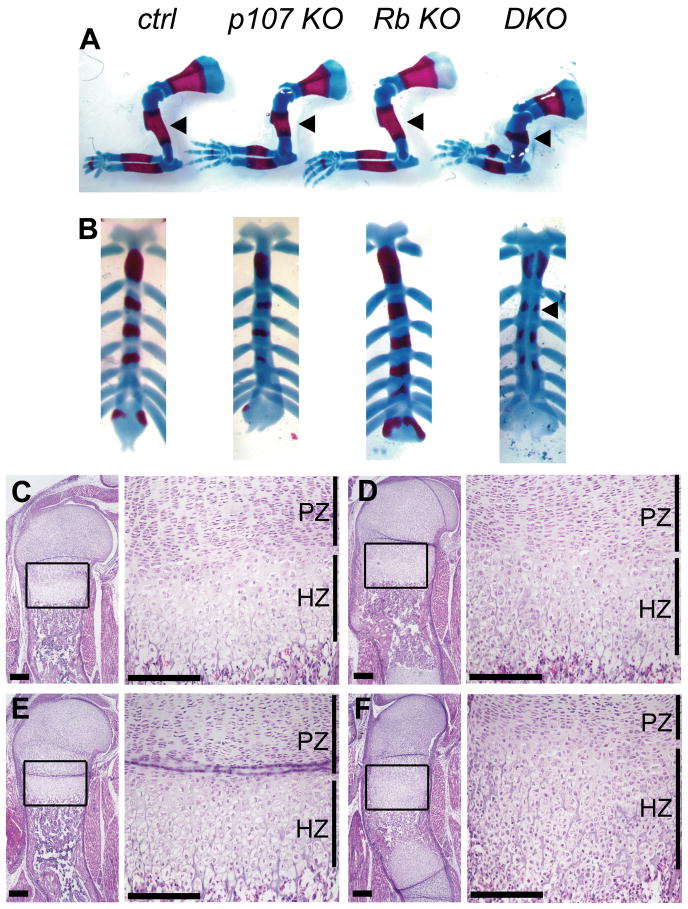
Figure 1. DKO embryos display long bone and growth plate defects.
(A and B) Embryonic skeletons from five e17.5 litters were stained with Alizarin Red and Alcian Blue to mark the bone and cartilage, respectively. Representative front limbs (A) and sternums (B) of DKO animals displayed morphological and ossification defects compared to control littermates. (C–F) Representative hematoxylin & eosin (H&E) stained sections of e17.5 proximal humoral epiphyses from control (ctrl) (C), p107 KO (D), Rb KO (E) and DKO (F) littermates. DKO epiphyses were morphologically abnormal with larger proliferative zones (PZ) and hypertrophic zones (HZ) compared to control littermates in four litters. Original magnification: panels on left, 4X; right, 20X. Boxes represent areas magnified in adjacent panel. Bar = 200μm.
To assess chondrocyte differentiation, we examined histological sections from e17.5 long bones. At this timepoint, the growth plates contain three different chondrocyte zones that represent distinct differentiation states; resting chondrocytes reside at the epiphysis or “head” of the bone, followed by proliferating chondrocytes which appear oblong and form stacks, and then terminally differentiated hypertrophic chondrocytes which are large cells comprised mostly of cytoplasm and bearing electron-lucent nuclei. The nascent bone collar develops around these hypertrophic chondrocytes. H&E staining confirmed the presence of bone defects, and particularly chondrocyte defects, in the long bones of e17.5 DKOs. The femurs (data not shown) and humerii of DKO limbs were shorter, wider, and abnormally shaped, compared to those of control littermates (Fig. 1C–F). Moreover, the proliferative (PZ) and hypertrophic zones (HZ) of chondrocytes were both greatly expanded and highly disorganized in DKO limbs. Despite the abnormal shape and organization of the DKO hypertrophic zone, H&E and Alizarin Red staining of humoral sections showed that bone collar formation (Fig. 1F), and ossification (Supplemental Fig. S3) occurred appropriately. Skeletal muscle surrounding the long bones was also histologically normal (data not shown) despite efficient deletion of Rb. Thus, we conclude that the combined deletion of Rb and p107 in mesenchymal precursors specifically disrupts chondrocyte differentiation, and thus endochondral ossification.
Embryonic long bone defects are due to inappropriate chondrocyte proliferation
Given the known roles of pRB and p107 in cell cycle control, we wished to determine whether the DKO chondrocyte defect was associated with inappropriate proliferation. Thus, we injected pregnant females with BrdU and 2 hours later collected e17.5 and e18.5 em bryos. The percentage of BrdU-positive chondrocytes in the proliferative zone of long bone growth plates was significantly higher in DKO versus other littermates at both e17.5 (Fig. 2A) and e18.5 (data not shown). Moreover, many DKOs had a sizable percentage of BrdU-positive cells within the terminally differentiated hypertrophic zone, which never occurred in the other genotypes (Fig. 2B). Thus, we conclude that loss of pRB and p107 causes ectopic proliferation of the chondrocytes and disrupts the normal coupling between cell cycle exit and terminal differentiation.
Figure 2. Chondrocyte proliferation is significantly increased in DKO embryonic growth plates.
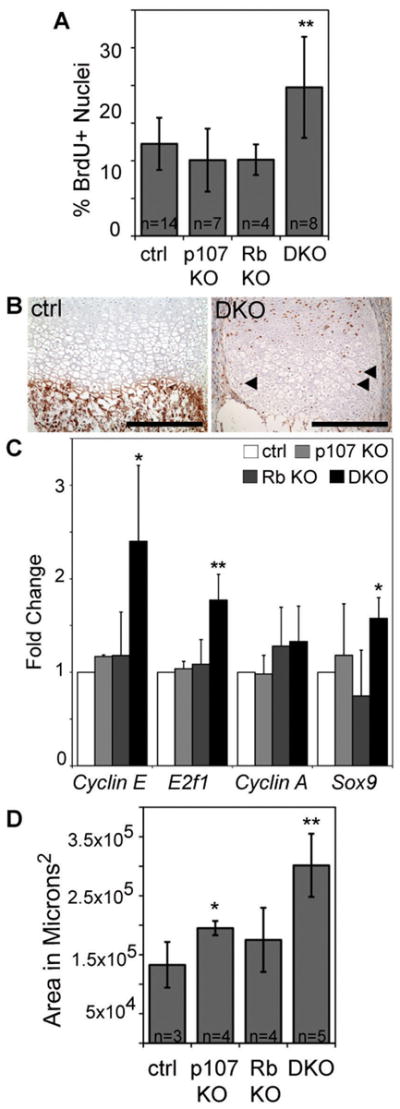
(A) Pregnant females were injected with BrdU and the e17.5 embryos harvested two hours later (litters from 5 pregnant females were analyzed). At least 2 humoral sections/mouse were stained with anti-BrdU antibodies, and BrdU incorporation was quantified in the proliferative zone chondrocytes (**p<0.01). (B) Hypertrophic terminally differentiated zone chondrocytes in DKOs stained positive for BrdU; Bar = 200μm. (C) qRT-PCR was performed on chondrocytes isolated from e18.5 sternums from three litters to determine the relative expression of E2F-target genes E2f1, Cyclin E, and Cyclin A, and the differentiation specific gene Sox9. Error bars indicate biological triplicates, *p<0.05; **p<0.01 relative to normalized control (ctrl). (D) The area of the epiphysis containing hypertrophic chondrocytes was quantified from H&E stained slides based on morphology. *p<0.05, **p<0.01
To further probe this deregulation, we conducted qRT-PCR analysis of mRNA harvested from e18.5 primary chondrocytes. Consistent with the increased proliferation of growth plate chondrocytes, we observed a significant increase in the mRNA levels of the E2F target genes E2f1 and Cyclin E, in DKO versus control littermates (Fig. 2C). Cyclin A mRNA was also increased, although not significantly (Fig. 2C). This strongly suggests that derepression of E2F target genes and consequent ectopic proliferation contributes to the chondrocyte and cartilage defects of the embryonic DKO growth plates. We also screened these mRNA samples for expression levels of chondrocyte differentiation regulators. Ihh and PTHrP showed a high degree of variability between litters, precluding conclusions about their potential contribution (data not shown). In contrast, we found that Sox9 mRNA, an early marker of chondrocyte differentiation, was significantly upregulated in the DKO chondrocytes (Fig. 2C). This upregulation could result from the increased levels of proliferative chondrocytes in the DKO growth plates (Fig. 2A) and/or a block in differentiation. To assess the latter possibility, we stained embryonic limb sections for collagen X, a marker of terminally differentiated hypertrophic chondrocytes (32). Collagen X was expressed exclusively in cells with hypertrophic chondrocyte morphology in both the control and DKO embryos (data not shown). The only difference was that the hypertrophic zone was greatly expanded in the DKOs (Fig. 2D). These data suggest that the growth plate abnormalities of DKO long bones and sternum is caused by increased cell cycling rates of the chondrocytes during the proliferative stage and an impaired ability to exit the cell cycle at hypertrophic stage, rather than a specific defect in the differentiation program.
Surviving DKO animals develop growth plate enchondromas by 8 weeks of age
Despite their skeletal abnormalities, a small number of DKOs survived to adulthood. Since most DKOs die between e18.5 and P1 (Table 1), we presume that these survivors represent animals with a weaker phenotype, likely because of variations in genetic background. To determine whether these adult DKOs develop other pathologies, we established an aging cohort of at least 10 control, p107 KO, Rb KO and DKO animals. We found that the surviving DKOs had a significantly shorter life span than all other genotypes (Table 2). This lethality was primarily due to the development of abdominal masses around 6 months of age that were gender specific. Female DKOs developed leiomyosarcomas, a tumor that derives from the smooth muscle of the uterus, a mesenchymal tissue (33). In contrast, the males developed preputial gland adenomas. PCR analysis confirmed that Cre-mediated recombination of Rb had occurred in these tumors (data not shown) despite their epithelial origin.
Table 2.
Disease Phenotypes of Aging Cohort
| Genotype (all Prx-Cre+) | Rb+/c;p107+/− | Rb+/c;p107−/− | Rbc/c;p107+/− | Rbc/c;p107−/− |
|---|---|---|---|---|
| Avg. Age at Euth. in days1 | 531 | 543 | 508 | 265 |
| # animals | 10 | 9 | 17 | 10 |
| Chondrones/Enchondromas2 | 0 | 0 | 7 | 10 |
| Cystic Uterus | 2 | 3 | 4 | 0 |
| Leiomyosarcoma (leiomyoma) | 0 | 0 | 1(1) | 2 |
| Mammary Fibroadenoma | 0 | 0 | 2 | 0 |
| Hibernoma | 0 | 0 | 1 | 0 |
| Preputial Adenoma | 0 | 0 | 3 | 4 |
| C-cell (metastatic) | 0 | 0 | 2(2) | 3(2) |
| Histiocytic Sarcoma | 4 | 2 | 1 | 0 |
| B-cell lymphoma | 2 | 0 | 4 | 0 |
Student’s T-test demonstrates p < 0.0001 for age of euthanasia of DKO versus control.
Rbc/c;p107+/−;Prx1-Cre mice only developed chondrones, not enchondromas.
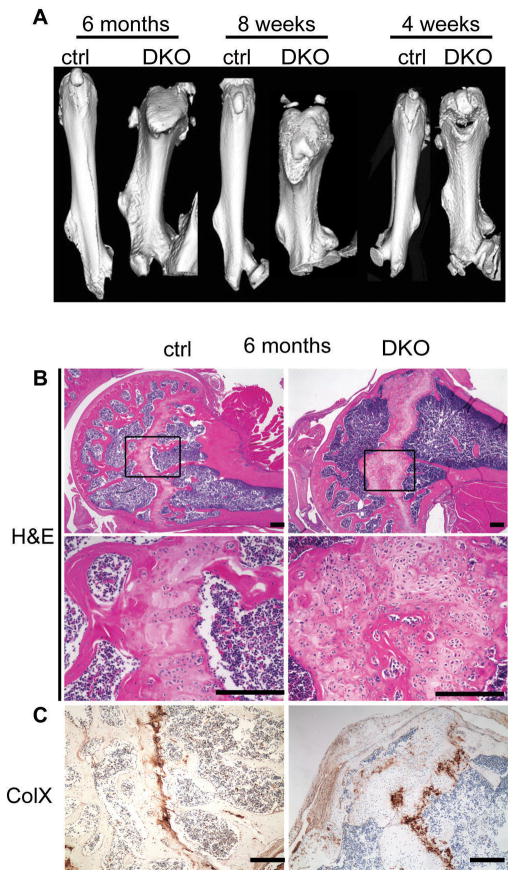
Importantly, our analysis of these ≥6 month-old DKOs showed that they possess defects in the humerii and femurs that were more pronounced than those of the embryonic DKOs. First, the long bones were both highly misshapen and markedly thicker and shorter than those of controls, as illustrated by micro-computerized tomography (μCT) of 6 month-old littermate femurs (3 animals analyzed/genotype; Fig. 3A). Second, histological analysis revealed that the adult long bone growth plates of all DKOs, and 40% of Rbc/c;p107+/−;Prx1-Cre mice, developed chondrones, an osteoarthritic-like cartilage abnormality characterized by circular clusters of chondrocytes (Table 2; Fig. 3B, data not shown). By 6 months of age, and only in the DKOs, these lesions had progressed to yield benign cartilage tumors called enchondromas (Table 2; Fig 3B). These lesions closely resembled human enchondromas (34, 35) in that they were predominantly comprised of non-hypertrophic chondrocytes with dense nuclei, but also contained smaller regions of hypertrophic chondrocytes that stained positive for collagen X expression (Fig. 3C). The enchondromas were surrounded by abnormal cartilage matrix (Fig. 3B). Finally, like their human counterparts, these tumors were largely non-proliferative as judged by analysis of the expression of Ki67, a direct E2F target, and also BrdU incorporation (Supplementary Fig. S4 and data not shown).
Figure 3. Cartilage abnormalities and enchondroma formation in 6-month-old DKO growth plates.
(A) Femurs were analyzed by μCT at 4 weeks, 8 weeks, and 6 months. DKO femurs were shorter, wider, and abnormally shaped compared to control genotypes at all timepoints. (B) Representative H&E stained humoral sections from 6-month-old mice revealed the presence of enchondromas in the growth plate of DKOs but not control (ctrl) animals. Boxes represent regions magnified in lower panel. (C) Enchondromas and the corresponding growth plate regions of control animals were stained for collagen X expression. Bar = 200μm.
The absence of proliferative cells within the enchondromas of 6 month old animals suggests that these lesions formed at earlier timepoints. In normal development, murine growth plates continue to proliferate until approximately 8 weeks of age, when mice reach sexual maturity, analogous to the development of human growth plates (36). Micro-CT analysis showed that 4 and 8 week old DKO femurs displayed severe morphological defects, including reduced bone length and expanded bone width, that were more pronounced than those of DKO embryos, and similar to those of 6 month old DKOs (Fig. 3A). Thus, we conducted careful histological analysis of 4 and 8 week timepoints. At 4 weeks, DKOs had greatly expanded cartilage regions in both the proximal epiphysis of the humerus (Supplementary Figure S5A), and the distal epiphysis of the femur (data not shown). The DKO growth plates were highly disorganized and contained chondrocytes with pleiomorphic morphologies. Moreover, the DKO articular cartilage regions were expanded with disorganized chondrocytes. We also observed a higher level of proliferation in growth plates of DKO versus control littermates, and proliferating cells were present in disorganized regions of the DKO growth plate expressing the terminal differentiation marker collagen X (Supplementary Figure S5B, C). This spectrum of bone defects was similar to, but more pronounced, than those of DKO embryos.
By 8-weeks, the growth plate and articular cartilage regions were further expanded in DKO versus control littermates (Supplementary Figure S5D). Moreover, enchondromas were detected in 3/3 DKOs analyzed at this timepoint. Regions of the DKO growth plate that retained normal architecture still contained a higher percentage of BrdU-positive cells than the littermate controls. In contrast, the enchondromas typically lacked BrdU-positive cells, suggesting that they had ceased proliferation by this timepoint (Supplementary Figure S5E). These tumors contained non-hypertrophic chondrocytes with dense nuclei, and there was little evidence of the collagen X-positive differentiated chondrocytes present at the 6 month timepoint (Supplementary Figure S5F).
Taken together, these data show that the loss of Rb and p107 in early mesenchymal cells leads to the formation of enchondromas that arise from inappropriately proliferating growth plate chondrocytes between 4 and 8 weeks of age. Some of these cells retain a poorly differentiated phenotype, while others succeed in undergoing terminal differentiation between 8 weeks and 6 months of age. The acquisition of these hypertrophic chondrocytes, and the accumulation of abnormal cartilage matrix, seems toaccount for the increased tumor size at the later timepoint.
Mutation of E2f3a or E2f3b suppresses enchondroma formation in DKO animals
Our data suggest that ectopic proliferation underlies both the developmental abnormalities and transformation of the DKO growth plates. Thus, we sought to address the role of E2F in these defects, particularly E2f1 and E2f3a, as these have been shown previously to influence cartilage development (24, 25). Unfortunately, the E2f1 and p107 loci are closely linked, and our attempts to combine the E2f1 and p107 mutant alleles were unsuccessful. However, we were able to cross the E2f3a and E2f3b mutant strains into our model to yield Rbc/c;p107−/−;E2f3a−/−;Prx1-Cre (herein called TKO-a) or Rbc/c;p107−/−;E2f3b−/−;Prx1-Cre (TKO-b) mice. Because of the complexity of these crosses, and thus the rarity of the TKO animals, we focused our attention on assessing how E2f mutation affected the tumor phenotype. For these studies, we generated intermediate mutant genotypes, and conducted crosses that yielded TKO or DKO animals at reasonable frequencies (1 in 8 or 1 in 16).
Initially, we compared long bone histology at 4 weeks of age. At this timepoint, TKO-a and TKO-b growth plates looked similar to DKO growth plates; that is, they were abnormally expanded and more proliferative than controls, as shown by BrdU staining (Supplementary Fig. 6A, B). By 8 weeks of age, however, the abnormal growth plate phenotype in the TKO-a and TKO-b animals was reduced compared to DKOs, and this was concomitant with a decrease in the number of BrdU-positive chondrocytes (Supplementary Fig. 6D, E). Like the DKO at these timepoints, Collagen X staining was restricted to hypertrophic chondrocytes (Supplementary Fig. 6C, F).
We then examined 6 month old animals for the presence of enchondromas. Remarkably, these lesions were almost completely absent in the TKO-a (1/11 had chondrones) and TKO-b (1/6 had chondrones) animals, but were observed in 15/17 DKO and also 1/1 Rbc/c;p107−/−;E2f3a+/−;Prx1-Cre and 9/10 Rbc/c;p107−/−;E2f3b+/−;Prx1-Cre animals (Fig. 4A, Table 3). The chondrocytes in the TKO-a and TKO-b growth plates appeared disorganized but still expressed collagen X in comparison with growth plates of control animals by histology and collagen X staining respectively (Fig. 4A, B). These results strongly suggest that mutation of E2f3a or E2f3b ameliorates some of the proliferative defects caused by Rb and p107 loss. Enchondromas in these TKO animals either don’t form at all, or resolve themselves by 6 months of age. These results demonstrate that E2F3a and E2F3b play a key role in the formation of enchondromas resulting from pRB and p107 loss.
Figure 4. TKO-a and TKO-b animals do not develop enchondromas.
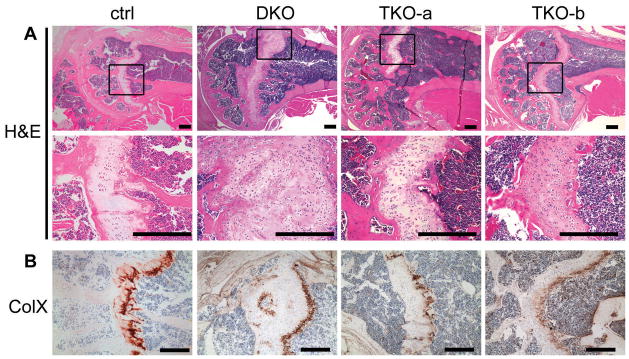
(A) Representative H&E stained humoral sections from 6-month-old TKO-a and TKO-b mice lack enchondromas compared to sections from DKO animals, but have a disorganized growth plate relative to control animals from the same litter. Boxes represent regions magnified in lower panel. (B) Growth plates were stained for collagen X expression. TKO-a: Rbc/c;p107−/−;E2f3a−/−;Prx1-Cre. TKO-b: Rbc/c;p107−/−; E2f3b−/−;Prx1-Cre. Bar = 200μm.
Table 3.
Phenotypes of DKO and TKO animals at 6 months
| Rbc/c;p107−/− Prx1-Cre1 | Rbc/c;p107−/−;E2f3a−/− Prx1-Cre | Rbc/c;p107−/−;E2f3b−/− Prx1-Cre | |
|---|---|---|---|
| # mice | 28 | 11 | 6 |
| Enchondroma | 25 | 1 | 1 |
| Leiomyosarcoma | 2 | 0 | 1 |
| Preputial Tumors | 3 | 3 | 1 |
This set is comprised of Rbc/c;p107−/−;E2f3a+/+;Prx1-Cre, Rbc/c;p107−/−;E2f3a+/−;Prx1-Cre, Rbc/c;p107−/−;E2f3b+/+;Prx1-Cre and Rbc/c;p107−/−;E2f3b+/−;Prx1-Cre littermates.
DISCUSSION
By mutating Rb in the early mesenchyme of p107−/− embryos, we have uncovered a novel role for pRB in murine cartilage development. We found that in the absence of p107, loss of pRB causes significant growth plate abnormalities during embryonic development that persist throughout adulthood. Furthermore, loss of both pRB and p107 leads to increased chondrocyte proliferation resulting in growth plate expansion, disorganization, and the development of enchondromas, a benign cartilage lesion. We show that these phenotypes are, at least in part, due to the deregulation of E2F3, and subsequent uncontrolled proliferation of growth plate chondrocytes.
Our data provide strong evidence that coordination of cell cycle exit with differentiation by pRB acts as a mechanism of tumor suppression. This reinforces similar conclusions made from studies in other tissues. In the bone, Rb is required for proper osteoblast differentiation (28–30) while mutation of Rb and p53 in this tissue leads to osteosarcoma (37, 38). Additionally, Rb is required in both mature hair cells and blood vessel endothelial cells to prevent uncoupling of proliferation and differentiation, which leads to significant tissue defects (19, 39). Analogously, our work shows that Rb is also required for regulating hypertrophic differentiation in growth plate chondrocytes. Combined loss of Rb and p107 in chondrocyte precursors leads to ectopic proliferation of chondrocytes in the embryo that results in the development of benign tumors arising in the growth plate cartilage. Notably, these benign tumors completely ceased proliferation by 8 weeks of age. This explains the lack of progression to malignant disease. It also strongly suggests that the tumorigenic signal resulting from pRB/p107 loss is restrained sometime between 4 and 8 weeks of age. Normal growth plates are known to cease proliferation around this time (36), but we also note that our histological analyses detect non-proliferative tumor tissue adjacent to proliferative normal tissue. Thus, we speculate that the observed suppression of tumor proliferation reflects a combination of normal developmental switches and tumor-specific events. These could include downregulation of mitogenic signals, upregulation of p130 or the pocket protein-independent repressor E2Fs, E2F6, 7 and 8, and the activation of tumor-surveillance mechanisms such as senescence. Most importantly, we note that our tumors closely model human enchondromas, which are predominantly non-proliferative (34, 35).
Enchondroma is the most common benign neoplasm of the bone, and likely the precursor of chondrosarcoma (35). Alterations in the Rb pathway, such as mutation and LOH of RB1 or alterations in the expression of upstream pRB regulators p16, CYCLIN D1 or CDK4, are observed in human cartilage tumors (35, 40–43). This study shows that mutation of Rb and p107 in early mesenchymal precursor cells can yield enchondroma development. These results are consistent with the notion that cartilage tumors, including chondrosarcomas, may arise in a chondrocyte progenitor cell. Although the cell of origin of chondrosarcoma is uncertain, there is evidence to suggest it is a mesenchymal progenitor cell, or at least an undifferentiated chondrocyte precursor (44). Indeed, the most frequently altered pathway in human enchondroma and chondrosarcoma formation is the Hedgehog signaling pathway, which is active in undifferentiated chondrocytes and required for growth plate differentiation (36, 45, 46).
The p16-CYCLIN D/CDK4-pRB pathway is strongly associated with cell cycle control. Accordingly, our data suggest that the growth plate developmental abnormalities and the enchondroma formation stem from a failure to curtail proliferation at the appropriate stage of development. The E2F transcription factors E2F1 and E2F3 have been shown to act downstream of Rb mutation in many tissue types and their deletion rescues some tumor phenotypes caused by pocket protein loss. For example, mutation of E2f1 in Rb heterozygous mice extends lifespan and suppresses formation of pituitary and thyroid tumors (47). E2f3-mutation extends lifespan and suppresses tumorigenesis in Rb+/− mice in a similar manner (16). Prior studies have linked E2F1 and E2F3 to growth plate development in the mouse. E2f1 overexpression inhibits hypertrophic chondrocyte differentiation resulting in shorter long bones (24). Additionally, in E2f1−/−;E2f3a−/− embryos, the growth plate is disorganized and the chondrocytes are larger than normal (25, 26). Our results show that in the context of Rb and p107 mutation, E2f3a and E2f3b both act as oncogenes to promote unscheduled proliferation in the cartilage growth plate and ultimately enchondroma formation. Given the observed upregulation of E2F-responsive mRNAs in DKO growth plate chondrocytes, it seems highly likely that E2F3a and E2F3b both act to promote E2F-target genes expression in this setting. By extension of this logic, we presume that loss of either E2F3 isoform is sufficient to lower the total level of E2F activity below the threshold required to trigger unscheduled cell division.
In summary, we have developed a mouse model of enchondroma that recapitulates the key features of the human disease. This model provides a platform to study the genetic factors important for proper long bone development, as well as the events that drive enchondroma progression to chondrosarcoma, open questions that warrant further study.
MATERIALS AND METHODS
Animal Maintenance
The generation of Rbc/c mice, p107−/− mice, Prx1-Cre mice, and Rosa26-LSL-LacZ mice has been described previously (5, 20, 31, 48). All animal procedures followed protocols approved by MIT’s Committee on Animal Care. All mice were maintained on a mixed background. Gestation was dated by detection of a vaginal plug. Mice were injected with 10μl/gm body weight of 5mg/ml 5-Bromo-2′-deoxyuridine(BrdU)(Sigma) in phosphate buffered saline (PBS) two hours prior to tissue collection. Collected tissues were fixed in 10% formalin (adult) or 4% paraformaldehyde (embryonic) and embedded in paraffin. Adult tissues containing bone were decalcified in 0.46M EDTA, 2.5% Ammonium Hydroxide, pH7.2, for two weeks prior to paraffin embedding. Histological sections were cut at 5μm.
Histological Analyses
Adult tissues were stained for β-galactosidase activity as described (49). Skeletal staining of embryos was conducted as previously described (28). Alizarin Red S staining was performed by deparaffinizing and dehydrating in ethanol, rinsing briefly in distilled H2O, and staining in freshly made 2% Alizarin Red S (Sigma) in H2O (pH 4.2) for 5 minutes. Slides were dipped 20 times in acetone, 20 times in acetone:xylene (1:1), cleared in xylene and mounted. Immunohistochemistry for BrdU and collagen X were performed using mouse anti-BrdU (1:50, BD Biosciences) and mouse anti-Collagen X (1:20, Quartett clone ColX53) antibodies as described (25). HRP-conjugated secondary antibodies were provided in the Vectastain ABC Kit Elite (Vector Labs), and antibody staining was visualized with DAB (Vector Labs). Immunohistochemistry for pRB was conducted using the UltraVision LP Detection System (Lab Vision Corporation) with the primary antibody (G3-245, BD Biosciences) at a concentration of 1:100. All quantification was performed using ImageJ software (NIH). All statistical analysis was performed using the two-tailed t-test.
Micro-computerized Tomography (μCT)
Analysis of 3D bone structure was performed as described in (50). 3D image data was collected from euthanized animals using a GE Healthcare Micro-CT machine and analyzed using the Microview 2.2 software from GE Healthcare.
Isolation of Primary Chondrocytes
Chondrocytes were isolated by modification of a prior procedure (51). e18.5 embryos were euthanized and sternums removed and rinsed in 1% FBS/PBS. Sternums were subjected to a series of digestions at 37°C with vigorous rocking: 2mg/ml pronase (Roche) in PBS for 30 min., then 3mg/ml Collagenase D (Roche) in 10% FBS in DME for 1hr, then fresh 3mg/ml Collagenase D for an additional 4–6 hours until the cartilage regions were released from the ossified nodules. The cell preparation was filtered through a 70μm cell strainer, washed and collected for mRNA analysis.
Quantitative real-time PCR
RNA was isolated from embryonic sternum chondrocytes using the Qiagen RNeasy kit. First-strand cDNA synthesis was performed using 0.5–1μg RNA and Superscript III Reverse Transcriptase (Invitrogen). Quantitative RT-PCR with 50–100ng cDNA was performed using SYBR Green (Applied Biosystems) using Ubiquitin as an internal control. Reactions were run on the ABI Prism 7000 Sequence Detection System and analyzed using the 7000 SDS software. Primers are listed in Supplementary Table 1.
Supplementary Material
Acknowledgments
Rbc/c, p107−/−, and Rosa26-LSL-LacZ transgenic mice were kindly provided by Tyler Jacks (MIT). We are grateful to Seth Berman for initial mouse colony generation, and Julie West for immuno-histochemistry assistance, as well as all members of the Lees lab for helpful discussions throughout this work. We thank MIT Swanson Biotechnology Center members Eliza Vasile, for assistance with μCT data collection and analysis, and Alicia Caron, for histological sectioning and H&E staining. We also thank Roderick Bronson for histopathological analyses and advice. This work was supported by a grant to J.A.L from the National Cancer Institute (CA121921), a Ludwig Graduate Fellowship to A.S.L. J.A.L. is a Ludwig Scholar.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Gurney JG, Severson RK, Davis S, Robison LL. Incidence of cancer in children in the United States. Sex-, race-, and 1-year age-specific rates by histologic type. Cancer. 1995 Apr 15;75(8):2186–2195. doi: 10.1002/1097-0142(19950415)75:8<2186::aid-cncr2820750825>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995 May 5;81(3):323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 3.Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, et al. Requirement for a functional Rb-1 gene in murine development. Nature. 1992 Sep 24;359(6393):328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 4.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359(6393):295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 5.Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, et al. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992 Sep 24;359(6393):288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 6.Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003 Feb 27;421(6926):942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- 7.Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996 Dec 1;10(23):3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 8.MacPherson D, Sage J, Crowley D, Trumpp A, Bronson RT, Jacks T. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol Cell Biol. 2003 Feb;23(3):1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipinski MM, Macleod KF, Williams BO, Mullaney TL, Crowley D, Jacks T. Cell-autonomous and non-cell-autonomous functions of the Rb tumor suppressor in developing central nervous system. EMBO J. 2001 Jul 2;20(13):3402–3413. doi: 10.1093/emboj/20.13.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson KL, Vanderluit JL, Hebert JM, McIntosh WC, Tibbo E, MacLaurin JG, et al. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 2002 Jul 1;21(13):3337–3346. doi: 10.1093/emboj/cdf338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bruin A, Wu L, Saavedra HI, Wilson P, Yang Y, Rosol TJ, et al. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci U S A. 2003 May 27;100(11):6546–6551. doi: 10.1073/pnas.1031853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirlam A, Spike BT, Macleod KF. Deregulated E2f-2 underlies cell cycle and maturation defects in retinoblastoma null erythroblasts. Mol Cell Biol. 2007 Dec;27(24):8713–8728. doi: 10.1128/MCB.01118-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saavedra HI, Wu L, de Bruin A, Timmers C, Rosol TJ, Weinstein M, et al. Specificity of E2F1, E2F2, and E2F3 in mediating phenotypes induced by loss of Rb. Cell Growth Differ. 2002 May;13(5):215–225. [PubMed] [Google Scholar]
- 14.Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998 Sep;2(3):293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 15.Wenzel PL, Wu L, de Bruin A, Chong JL, Chen WY, Dureska G, et al. Rb is critical in a mammalian tissue stem cell population. Genes Dev. 2007 Jan 1;21(1):85–97. doi: 10.1101/gad.1485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziebold U, Reza T, Caron A, Lees JA. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 2001 Feb 15;15(4):386–391. doi: 10.1101/gad.858801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002 Jan;3(1):11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 18.Robanus-Maandag E, Dekker M, van der Valk M, Carrozza ML, Jeanny JC, Dannenberg JH, et al. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 1998 Jun 1;12(11):1599–1609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman SD, West JC, Danielian PS, Caron AM, Stone JR, Lees JA. Mutation of p107 exacerbates the consequences of Rb loss in embryonic tissues and causes cardiac and blood vessel defects. Proc Natl Acad Sci U S A. 2009 Sep 1;106(35):14932–14936. doi: 10.1073/pnas.0902408106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003 Jul 10;424(6945):223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz S, Santos M, Segrelles C, Leis H, Jorcano JL, Berns A, et al. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development. 2004 Jun;131(11):2737–2748. doi: 10.1242/dev.01148. [DOI] [PubMed] [Google Scholar]
- 22.Cobrinik D, Lee MH, Hannon G, Mulligan G, Bronson RT, Dyson N, et al. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996 Jul 1;10(13):1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 23.Rossi F, MacLean HE, Yuan W, Francis RO, Semenova E, Lin CS, et al. p107 and p130 Coordinately regulate proliferation, Cbfa1 expression, and hypertrophic differentiation during endochondral bone development. Dev Biol. 2002 Jul 15;247(2):271–285. doi: 10.1006/dbio.2002.0691. [DOI] [PubMed] [Google Scholar]
- 24.Scheijen B, Bronk M, van der Meer T, Bernards R. Constitutive E2F1 overexpression delays endochondral bone formation by inhibiting chondrocyte differentiation. Mol Cell Biol. 2003 May;23(10):3656–3668. doi: 10.1128/MCB.23.10.3656-3668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danielian PS, Friesenhahn LB, Faust AM, West JC, Caron AM, Bronson RT, et al. E2f3a and E2f3b make overlapping but different contributions to total E2f3 activity. Oncogene. 2008 Nov 20;27(51):6561–6570. doi: 10.1038/onc.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai SY, Opavsky R, Sharma N, Wu L, Naidu S, Nolan E, et al. Mouse development with a single E2F activator. Nature. 2008 Aug 28;454(7208):1137–1141. doi: 10.1038/nature07066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, Lees JA. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010 Aug 4; doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman SD, Yuan TL, Miller ES, Lee EY, Caron A, Lees JA. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res. 2008 Sep;6(9):1440–1451. doi: 10.1158/1541-7786.MCR-08-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez GM, Kong E, Sabbagh Y, Brown NE, Lee JS, Demay MB, et al. Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1−/− mice. Proc Natl Acad Sci U S A. 2008 Nov 25;105(47):18402–18407. doi: 10.1073/pnas.0805925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001 Aug;8(2):303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 31.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002 Jun;33(2):77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Q, Keller B, Zhou G, Napierala D, Chen Y, Zabel B, et al. Localization of the cis-enhancer element for mouse type X collagen expression in hypertrophic chondrocytes in vivo. J Bone Miner Res. 2009 Jun;24(6):1022–1032. doi: 10.1359/JBMR.081249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Politi K, Szabolcs M, Fisher P, Kljuic A, Ludwig T, Efstratiadis A. A mouse model of uterine leiomyosarcoma. Am J Pathol. 2004 Jan;164(1):325–336. doi: 10.1016/S0002-9440(10)63122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller MD. Review of orthopaedics. 4. Saunders; Philadelphia, Pa: 2004. p. xv.p. 704. [Google Scholar]
- 35.Bovee JV, Hogendoorn PC, Wunder JS, Alman BA. Cartilage tumours and bone development: molecular pathology and possible therapeutic targets. Nat Rev Cancer. 2010 Jul;10(7):481–488. doi: 10.1038/nrc2869. [DOI] [PubMed] [Google Scholar]
- 36.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003 May 15;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 37.Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A. 2008 Aug 19;105(33):11851–11856. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008 Jun 15;22(12):1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sage C, Huang M, Vollrath MA, Brown MC, Hinds PW, Corey DP, et al. Essential role of retinoblastoma protein in mammalian hair cell development and hearing. Proc Natl Acad Sci U S A. 2006 May 9;103(19):7345–7350. doi: 10.1073/pnas.0510631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asp J, Inerot S, Block JA, Lindahl A. Alterations in the regulatory pathway involving p16, pRb and cdk4 in human chondrosarcoma. J Orthop Res. 2001 Jan;19(1):149–154. doi: 10.1016/S0736-0266(00)00022-X. [DOI] [PubMed] [Google Scholar]
- 41.Asp J, Sangiorgi L, Inerot SE, Lindahl A, Molendini L, Benassi MS, et al. Changes of the p16 gene but not the p53 gene in human chondrosarcoma tissues. Int J Cancer. 2000 Mar 15;85(6):782–786. doi: 10.1002/(sici)1097-0215(20000315)85:6<782::aid-ijc7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi T, Toguchida J, Wadayama B, Kanoe H, Nakayama T, Ishizaki K, et al. Loss of heterozygosity and tumor suppressor gene mutations in chondrosarcomas. Anticancer Res. 1996 Jul-Aug;16(4A):2009–2015. [PubMed] [Google Scholar]
- 43.Ropke M, Boltze C, Meyer B, Neumann HW, Roessner A, Schneider-Stock R. Rb-loss is associated with high malignancy in chondrosarcoma. Oncol Rep. 2006 Jan;15(1):89–95. doi: 10.3892/or.15.1.89. [DOI] [PubMed] [Google Scholar]
- 44.Boeuf S, Kunz P, Hennig T, Lehner B, Hogendoorn P, Bovee J, et al. A chondrogenic gene expression signature in mesenchymal stem cells is a classifier of conventional central chondrosarcoma. J Pathol. 2008 Oct;216(2):158–166. doi: 10.1002/path.2389. [DOI] [PubMed] [Google Scholar]
- 45.Tiet TD, Hopyan S, Nadesan P, Gokgoz N, Poon R, Lin AC, et al. Constitutive hedgehog signaling in chondrosarcoma up-regulates tumor cell proliferation. Am J Pathol. 2006 Jan;168(1):321–330. doi: 10.2353/ajpath.2006.050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schrage YM, Hameetman L, Szuhai K, Cleton-Jansen AM, Taminiau AH, Hogendoorn PC, et al. Aberrant heparan sulfate proteoglycan localization, despite normal exostosin, in central chondrosarcoma. Am J Pathol. 2009 Mar;174(3):979–988. doi: 10.2353/ajpath.2009.080623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamasaki L, Bronson R, Williams BO, Dyson NJ, Harlow E, Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−)mice. Nat Genet. 1998 Apr;18(4):360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 48.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999 Jan;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 49.Parisi T, Yuan TL, Faust AM, Caron AM, Bronson R, Lees JA. Selective requirements for E2f3 in the development and tumorigenicity of Rb-deficient chimeric tissues. Mol Cell Biol. 2007 Mar;27(6):2283–2293. doi: 10.1128/MCB.01854-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glatt V, Canalis E, Stadmeyer L, Bouxsein ML. Age-related changes in trabecular architecture differ in female and male C57BL/6J mice. J Bone Miner Res. 2007 Aug;22(8):1197–1207. doi: 10.1359/jbmr.070507. [DOI] [PubMed] [Google Scholar]
- 51.Lefebvre V, Garofalo S, Zhou G, Metsaranta M, Vuorio E, De Crombrugghe B. Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol. 1994 Aug;14(4):329–335. doi: 10.1016/0945-053x(94)90199-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.