Abstract
The role of HIV-1-specific antibody responses in HIV disease progression is complex and would benefit from analysis techniques that examine clusterings of responses. Protein microarray platforms facilitate the simultaneous evaluation of numerous protein-specific antibody responses, though excessive data are cumbersome in analyses. Principal components analysis (PCA) reduces data dimensionality by generating fewer composite variables that maximally account for variance in a dataset. To identify clusters of antibody responses involved in disease control, we investigated the association of HIV-1-specific antibody responses by protein microarray, and assessed their association with disease progression using PCA in a nested cohort design. Associations observed among collections of antibody responses paralleled protein-specific responses. At baseline, greater antibody responses to the transmembrane glycoprotein (TM) and reverse transcriptase (RT) were associated with higher viral loads, while responses to the surface glycoprotein (SU), capsid (CA), matrix (MA), and integrase (IN) proteins were associated with lower viral loads. Over 12 months greater antibody responses were associated with smaller decreases in CD4 count (CA, MA, IN), and reduced likelihood of disease progression (CA, IN). PCA and protein microarray analyses highlighted a collection of HIV-specific antibody responses that together were associated with reduced disease progression, and may not have been identified by examining individual antibody responses. This technique may be useful to explore multifaceted host–disease interactions, such as HIV coinfections.
Introduction
HIV-1-specific antibodies may be important for long-term control of HIV-1 progression, as well as contribute to protection from transmission.1–3 During the course of HIV-1 infection, diverse combinations of antibody responses to specific HIV-1 antigens are produced, with variable intensity and duration.4 For example, anti-Env IgG is produced and maintained throughout disease, while anti-Gag IgG appears to decrease as HIV-1 disease progresses, independent of changes in HIV-1 plasma RNA.5 Although antibody responses to select HIV-1 antigens have been investigated in relation to disease progression, sample numbers and definitions of disease progression vary.5,6 In addition, there has been limited opportunity to investigate the role of combinations of antibody responses on HIV-1 disease progression.
Profiling HIV-1-specific binding antibodies using protein microarray technology may give more comprehensive insight into the role of humoral immune profiles in disease progression.7 Microarray analyses generate many variables of interest, which may be interpreted with various statistical techniques depending on the goal of the analysis.8 One use of studies profiling humoral immune responses is to identify attributes that categorize individuals by disease status.9 Consideration of all available immunologic variables as a whole, rather than a select few, may be more illustrative of what is occurring in the host, and highlight relationships between variables of interest. Principal components analysis (PCA) is a useful tool to reduce multivariate responses into fewer composite variables that account for most of the variance in a dataset.10 Previous HIV studies have used PCA to distinguish disease states based on profiling large numbers of variables related to immunity, as well as behavioral surveys.11,12 Exploring HIV-1-specific humoral immune profiles with PCA and protein microarrays may be a useful way to examine changing immune responses in complex systems, such as chronic HIV-1 infection.
Within a nested cohort study, we assessed the feasibility of utilizing protein microarray and PCA to explore HIV-1-specific antibody responses during disease progression. Using PCA, we identified relationships within humoral responses to HIV-specific antigens, in the form of shared variability. Finally, we investigated the association between these HIV-1-specific antibody responses and more traditional markers of HIV-1 disease progression, including concurrent and subsequent changes in CD4 count and plasma HIV-1 viral load.
Materials and Methods
Study design
A nested cross-sectional analysis was conducted on 100 stored samples from a large randomized controlled trial evaluating the effect of empiric deworming on markers of HIV-1 disease progression in Kenya.13 Plasma samples were collected between February 2009 and July 2010. All individuals provided written informed consent to participate in the study. The trial was independently approved by the IRB at the University of Washington and the Ethical Review Board of the Kenya Medical Research Institute. The parent trial was registered as NCT00507221 at http://clinicaltrials.gov. The parent study is now complete and significant differences between deworming treatment arms were not found for any HIV endpoints examined.13
Population
Study participants were enrolled from three sites in Kenya (Kisii Provincial Hospital, Kisumu District Hospital, and Kilifi District Hospital) who were HIV-1 infected, older than 18, were not pregnant, did not meet criteria for antiretroviral therapy (ART) initiation based on Kenyan Ministry of Health guidelines, had not used ART in the past, and were willing and able to give informed consent. From this population, participants were excluded who had started ART prior to their 12 month visit, did not have a 12 month visit by July 2010, were not from the Kisii or Kisumu study sites, had an abnormal clinical finding at the month 12 visit, took deworming medicine outside of the study, or stopped taking the study medicine before the 12 month visit. From the remaining 329 eligible participants, 100 patients were randomly selected using computer-generated random sampling; 25 participants were selected from each study site and treatment arm. Of the 100 samples selected, seven were unreadable by microarray and one participant was later found to have started ART before the 12 month visit so was not included in the analyses.
Data collection
HIV-1-specific antibody responses were measured from samples collected at the 12 month study visit. Primary endpoints of the parent study were time to CD4 count <350 cells/mm3 and a composite endpoint of first occurrence of (1) CD4 count <350 cells/mm3, (2) first reported use of ART, or (3) nontraumatic death. No deaths occurred among the participants evaluated. Demographic measures were collected at enrollment, while changes in health were evaluated at follow-up visits. CD4 count was measured at months 0, 6, 12, 18, and 24 and viral load (log10 plasma RNA) was measured at months 0, 12, and 24 to monitor HIV-1 disease. In this subanalysis, we consider 12 months as baseline and 24 months as endpoint.
Microarray construction
The HIV-1 microarray was constructed by amplifying and expressing the open reading frames encoding 16 HIV-1 proteins and 13 gene fragments from each of the five major subtypes (A1, A2, B, C, D) or clades of HIV-1. Subsequently, the expressed proteins were printed onto nitrocellulose-coated slides. A total of 143 HIV-1 proteins or protein fragments from five virus clades and all nine viral genes were printed on the arrays (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/aid).
Full-length infectious molecular clones of HIV-1 were used as templates for polymerase chain reaction (PCR) to create each protein or peptide-encoding gene or gene fragment. The templates used were HIV-1 clones 92UG037, subtype A1; 94CY017, subtype A2; JR-CSF, subtype B; MJ4, subtype C; and 94UG114, subtype D.14–16 All templates were obtained from the AIDS Research and Reference Reagent Program except JR-CSF (from Dr. Irvin Chen, UCLA).17–19 Primers were designed to contain 20 base pairs of homology to each ORF being amplified and 33 base pairs of homology to the plasmid vector, pXT7. The standard PCR cycle was 95°C for 5 min followed by 39 cycles of 95°C for 20 s, 50°C for 30 s, and 72°C for 30 s for every 500 base pairs of product desired, followed by a final extension of 72°C for 10 min. Amplification success was determined by PCR product visualization following agarose gel electrophoresis.
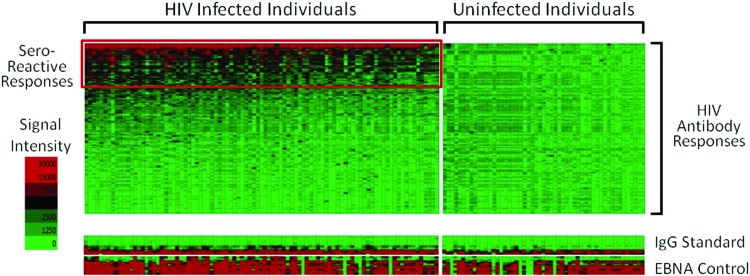
PCR products were inserted into the expression vector by in vivo recombination as previously described.20 Genes and gene fragments encoding HIV-1 proteins and protein fragments in pXT7 were expressed in an Escherichia coli-based cell-free coupled in vitro transcription-translation (IVTT; Roche) reaction, solubilized with Tween-20 and printed on nitrocellulose-coated FAST slides (GE-Schleicher and Schuell) using an Omni Grid 100 microarray printer. Microarrays were probed using human sera or control antibodies as previously described.20 Microarrays were scanned using a ScanArray 4000 machine. QuantArray software was used to quantify the intensity of the spots on the chip. The signal intensity of the “No DNA” controls were averaged and used to subtract background reactivity from the unmanipulated raw data. One-third of the spotted proteins had signals greater than the average of “no DNA” control reactions plus 2.5 times the standard deviation, and were considered seroreactive. HIV-uninfected controls were evaluated using the same batch of microarray slides to demonstrate specificity in antibody reactivity (Fig. 1).
FIG. 1.
Microarray heat map of all HIV-specific antibody responses, with seroreactive responses boxed in red. Each column represents a patient and each row represents an antigen-specific antibody response. HIV-uninfected controls, on the right, were evaluated using the same batch of microarray slides showing no reactivity to the HIV-specific antigens. Color images available online at www.liebertpub.com/aid
Statistical analysis
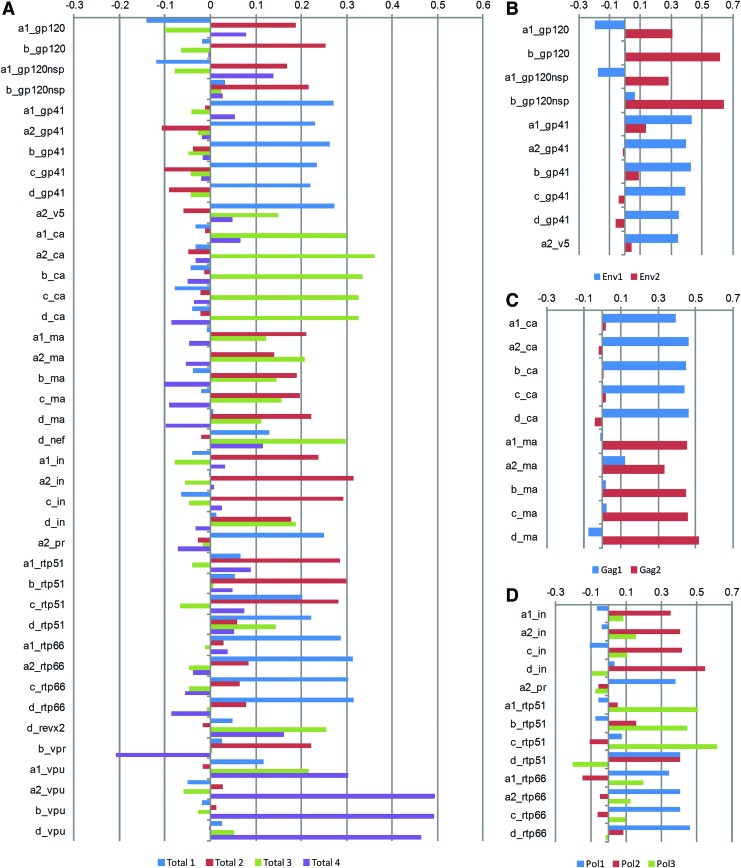
PCA was used to reduce the dimensionality of the HIV-1-specific antibody responses and explore relationships between the variables in the dataset.10 All 40 seroreactive antigens were used to generate the total antigen principal components, which determine grouping based on the variance of the antibody responses rather than a prespecified grouping. Utilizing the hierarchical structure within the HIV-1 antigens, gene-specific PCA was also performed. All seroreactive Env, Gag, or Pol antibody responses were used to generate gene-specific components. Components were retained if at least three variables loaded, and the eigenvalue was greater than 2 or the proportion of variance explained was >5%.10 All retained components were transformed using orthogonal (varimax) rotation to ease interpretation of the components. Rotation causes antigens to load predominantly to one component by maximally aligning the principle component axes with the projected points in the coordinate space.10 Variables with large positive and negative loadings are most influential in the component, and therefore were used to describe the component. Loadings are similar to the weight each antibody response contributes to the components. Contributions of antibody responses to the retained components after PCA for total (A), and gene-specific analyses: Env (B), Gag (C), and Pol (D), are illustrated in Fig. 2. The y-axis represents the loadings of each antibody response to the retained components after rotation.
FIG. 2.
Graphic representation of the contributions of each antibody response to the retained components after principle components analysis for total (A) and gene-specific analyses: Env (B), Gag (C), and Pol (D). The y-axis represents the loadings of each antibody response to the retained components after orthogonal rotation. Loadings are similar to the weight each antibody response provides to the components. Rotation causes antigens to load predominately to one component by maximally aligning the principle component axes with the projected points in the coordinate space. Color images available online at www.liebertpub.com/aid
Linear regression with robust standard errors was used to assess the association between HIV-1-specific antibody responses (using total and gene-specific components) and the following: CD4 count at baseline, viral load at baseline, change in CD4 count per year adjusted for baseline CD4 count, and change in viral load per year adjusted for baseline viral load. Logistic regression was used to assess the effects of HIV-1-specific antibody responses (using total and gene-specific components) on disease progression using CD4 count <350 cells/mm3 or ART initiation as the disease progression endpoint. We also evaluated whether deworming treatment modified any associations examined with HIV-specific antibody responses. All interaction terms assessing this effect modification were not statistically significant. Statistical analyses were performed using Stata 11.2 (StataCorp).
Results
Among the 92 individuals evaluated and analyzed, 76.1% were female and the mean age was 33.6 years. Most participants were married (56.5%), had some education, and were relatively immunocompetent with a mean CD4 count and viral load of 515.8 cells/mm3 and 4.01 log10 copies/ml, respectively. The study ended prior to four participants completing their 24 month study visit, making 18 months their final study visit. Nine of 92 participants began ART between baseline and endpoint. Evaluating change in CD4 and viral load prior to ART initiation from baseline to endpoint, the mean change in CD4 count was −108.2 cells/mm3 per year and the viral load was 0.02 log10 plasma RNA copies/ml per year. Among ART-naive participants, at baseline and endpoint there were 23 and 25 patients with CD4 <350, respectively. Using a composite indicator of disease progression (CD4 <350 or ART use), at baseline and endpoint there were 23 and 34 participants with progressive HIV-1 disease, respectively. Of the 88 individuals observed at 24 months, 25 had a CD4 count <350, two had started ART, and seven had started ART as well as had a CD4 count <350 (Table 1).
Table 1.
Patient Characteristics at Enrollment (n=92)
| Characteristics at enrollment | N (%) or mean (SD) |
|---|---|
| Female | 70 (76.1%) |
| Age at enrollment | 33.6 (8.2) |
| Deworming treatment arm | 46 (50.0%) |
| Clinic location | |
| Kisii | 44 (47.8%) |
| Kisumu | 48 (52.2%) |
| Health measures among ART naive | Month 12 (n=92) | Month 24 (n=79) |
|---|---|---|
| BMI (kg/m2) | 22.6 (3.8) | 22.7 (4.4)a |
| CD4 count (cells/mm3) | 515.8 (232.6) | 438.6 (176.7) |
| Viral load (log10 copies/ml) | 4.01 (0.97) | 3.94 (0.95) |
| Change in CD4 per year, 12–24M [n=89]b | −108.2 (169.3) | |
| Change in log10 viral load per year, 12–24M [n=79]b | 0.02 (0.56) | |
| CD4 count <350 | 23 | 25 |
| Disease progression (CD4 count <350 or ART initiation) | 23 | 34c |
Missing=3.
Slope is change in CD4 or viral load before antiretroviral treatment (ART) initiation, for those who started ART.
25 CD4 <350, 2 ART use, 7 ART use plus CD4 <350 (n=88).
We used PCA to identify shared variance among the 40 seroreactive HIV-1 antibody responses. In Fig. 2, the contribution of each antibody response to the retained components is graphically demonstrated. The first four components of the total PCA accounted for 72% of the total variance of the 40 seroreactive antigens. After orthogonal rotation, total components 1–4 accounted for 23%, 21%, 19%, and 9% of the total variance, respectively. Five HIV-1 viral clades were included in the protein microarray to capture the full range of HIV-specific antibody responses circulating in sub-Saharan Africa. However, antigen-specific antibody responses were similar across clades, and differences in clade-specific responses were not observed between individuals. The infecting HIV-1 clade of the participants was not known.
Evaluating Env, Gag, and Pol-specific antibody responses using gene-specific PCA, two Env, two Gag, and three Pol-specific components were retained, accounting for 79%, 87%, and 77% of the total gene-specific variance, respectively. After rotation, each retained gene-specific component contained mostly one protein-specific antibody response, simplifying the interpretation of each component (Fig. 2). Env1 (gp41: TM) and Env2 (gp120: SU) accounted for 56% and 23% of the total variance, respectively. Gag1 (p24: CA) and Gag2 (p17: MA) accounted for 47% and 40% of the total variance, respectively. Finally, Pol1 (p66; RT), Pol2 (IN), and Pol3 (p51: RT) accounted for 34%, 23%, and 19% of the total variance, respectively. HIV-uninfected samples were also identically evaluated by protein microarray and demonstrated no seroreactivity to HIV-specific antigens, demonstrating the specificity of the HIV-specific antibody responses (Fig. 1).
We compared HIV-1-specific antibody responses, represented by PCA components, at baseline among ART-naive individuals with more traditional markers of HIV-1 disease. Total and gene-specific components were significantly associated with higher or lower viral load at baseline, indicated by the sign of the coefficient (Table 2). Total1 (RT p66, TM, V5, PR, p=0.023), Env1 (TM, p=0.007), and Pol1 (RT p66, p=0.028) were associated with greater viral load while Total2 (IN, RT p51, SU, MA, p=0.001), Total3 (CA, Nef, Rev, p=0.018), Env2 (SU, p=0.002), Gag1 (CA, p=0.028), Gag2 (MA, p=0.004), and Pol2 (IN, p=0.006) were associated with lower viral loads. No total or gene-specific components were significantly associated with CD4 count at baseline, though the components associated with higher viral loads had coefficients suggesting a relationship with lower CD4 counts.
Table 2.
Association Between HIV-1-Specific Antibody Responses and CD4 Count or log10 Viral Load
| CD4 count at 12 months [n=92]a | Log10 viral load at 12 months [n=92]a | |||||
|---|---|---|---|---|---|---|
| Coefficient | p-value | 95% CI | Coefficient | p-value | 95% CI | |
| Total antigens 1 (RT p66, TM, V5, PR) | −9.861 | 0.229 | −26.055, 6.331 | 0.078 | 0.023 | 0.011, 0.145 |
| Total antigens 2 (IN, RT p51, SU, MA) | 11.073 | 0.364 | −13.052, 35.198 | −0.102 | 0.001 | −0.164, −0.040 |
| Total antigens 3 (CA, nef, rev) | 14.312 | 0.149 | −5.221, 33.844 | −0.087 | 0.018 | −0.159, −0.015 |
| Total antigens 4 (vpu, vpr) | −12.155 | 0.324 | −36.500, 12.191 | −0.053 | 0.289 | −0.151, 0.045 |
| Env antigens 1 (TM, V5) | −12.474 | 0.242 | −33.523, 8.574 | 0.117 | 0.007 | 0.032, 0.201 |
| Env antigens 2 (SU) | 19.456 | 0.438 | −30.149, 69.062 | −0.181 | 0.002 | −0.296, −0.067 |
| Gag antigens 1 (CA) | 22.490 | 0.080 | −2.717, 47.697 | −0.104 | 0.028 | −0.197, −0.011 |
| Gag antigens 2 (MA) | 12.599 | 0.508 | −25.023, 50.221 | −0.140 | 0.004 | −0.233, −0.046 |
| Pol antigens 1 (RT p66) | −15.221 | 0.174 | −37.278, 6.836 | 0.105 | 0.028 | 0.011, 0.199 |
| Pol antigens 2 (IN) | 16.892 | 0.267 | −13.122, 46.905 | −0.148 | 0.006 | −0.253, −0.043 |
| Pol antigens 3 (RT p51) | 16.069 | 0.377 | −19.882, 52.021 | −0.075 | 0.184 | −0.187, 0.037 |
| Change in CD4 count per year after 12 months (slope) [n=89]b | Change in Log10 viral load per year after 12 months (slope) [n=79]b | |||||
|---|---|---|---|---|---|---|
| Total antigens 1 (RT p66, TM, V5, PR) | −1.241 | 0.786 | −10.314, 7.832 | 0.016 | 0.473 | −0.029, 0.061 |
| Total antigens 2 (IN, RT p51, SU, MA) | 8.148 | 0.185 | −3.986, 20.282 | −0.011 | 0.574 | −0.051, 0.029 |
| Total antigens 3 (CA, nef, rev) | 11.347 | 0.012 | 2.562, 20.131 | −0.001 | 0.965 | −0.051, 0.049 |
| Total antigens 4 (vpu, vpr) | 3.863 | 0.628 | −11.929, 19.656 | 0.035 | 0.316 | −0.035, 0.106 |
| Env antigens 1 (TM, V5) | −2.930 | 0.633 | −15.077, 9.218 | 0.027 | 0.328 | −0.028, 0.082 |
| Env antigens 2 (SU) | 13.674 | 0.226 | −8.632, 35.979 | −0.014 | 0.694 | −0.083, 0.056 |
| Gag antigens 1 (CA) | 12.139 | 0.034 | 0.924, 23.354 | −0.013 | 0.653 | −0.072, 0.046 |
| Gag antigens 2 (MA) | 15.722 | 0.037 | 0.940, 30.505 | −0.007 | 0.810 | −0.067, 0.053 |
| Pol antigens 1 (RT p66) | −0.593 | 0.920 | −12.365, 11.179 | 0.011 | 0.724 | −0.053, 0.075 |
| Pol antigens 2 (IN) | 14.004 | 0.063 | −0.759, 28.767 | −0.006 | 0.860 | −0.078, 0.065 |
| Pol antigens 3 (RT p51) | 0.322 | 0.979 | −23.540, 24.185 | −0.003 | 0.945 | −0.088, 0.082 |
Linear regression with robust standard errors, excluding measures after ART initiation.
Linear regression with robust standard errors, excluding measures after ART initiation, adjusted for measure at 12 months.
Significant associations are in bold for easier identification.
After baseline, Total3 (CA, Nef, Rev, p=0.012), Gag1 (CA, p=0.034), Gag2 (MA, p=0.037), and Pol2 (IN, p=0.063) were significantly associated with less decrease in CD4 counts from baseline to endpoint, adjusted for baseline CD4 measures and excluding time after ART among the subset who initiated ART. Additionally, when also adjusted for baseline viral load, the association between change in CD4 count and the following components was largely unchanged: Total3 (CA, Nef, Rev, p=0.024), Gag1 (CA, p=0.066), and Gag2 (MA, p=0.084). In contrast, no total or gene-specific components were associated with change in viral load, comparing individuals with similar baseline viral load measures and no ART use.
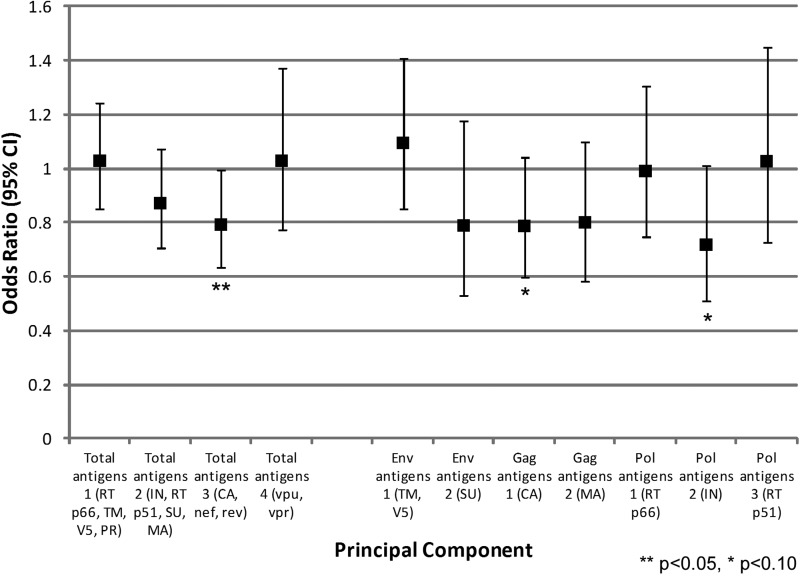
We also compared HIV-1-specific antibody responses at baseline with disease progression (CD4 <350 or ART initiation) at endpoint (Fig. 3). Total3 was significantly associated with a 21% reduced odds of disease progression (CA, Nef, Rev, p=0.043). Among gene-specific components, there was a trend for decreased odds of disease progression for both Gag1 (CA, p=0.092) and Pol2 (IN, p=0.059), corresponding to a 21% and 29% reduced odds respectively.
FIG. 3.
Odds of disease progression at 24 months by total and gene-specific components. Disease progression was determined by antiretroviral treatment (ART) use or CD4 count <350, excluding those with CD4 <350 or ART use at 12 months (n=66).
Discussion
Using principal components analysis, we were able to examine a broad array of HIV-specific antibody responses, as well as evaluate associations between complex patterns of antibody responses and disease progression. In this analysis, we clustered antibody responses at two levels. The first level considered all seroreactive HIV-specific antibody responses, and through PCA, relationships between responses were identified. From our 40 seroreactive antibody responses, we extracted four meaningful total antigen components that summarized 72% of the variance in the data. The HIV-specific antibody responses demonstrated varying relationships with disease control, suggesting not all are equally effective, or involved in controlling the HIV virus. Among total seroreactive components, Total1 (RT p66, TM, V5, PR) was associated with higher viral load, while Total2 (IN, RT p51, SU, MA) and Total3 (CA, Nef, Rev) were associated with lower viral loads at baseline. Over 12 months, Total3 was associated with smaller decreases in CD4 count and less likelihood of disease progression (CD4 <350 or ART use). No associations with change in viral load were observed. In this cohort, average viral load increased minimally over 12 months.
A challenge with this type of analysis is identifying the contributions of individual antibody responses to the associations detected. Further refining the PCA by gene specificity created seven components that happened to be protein specific. Each protein-specific component represented antibody responses from only one protein, allowing easier interpretation of the components. Additionally, the associations demonstrated between the total antibody response components and HIV disease progression were also observed using the gene-specific components. Among protein-specific antibody responses, TM and RT p66 were associated with higher baseline viral loads, while SU, CA, MA, and IN were associated with lower viral loads. Over 12 months, antibody responses to CA and MA were significantly associated with smaller decreases in CD4 count, and antibody responses to CA and IN showed a trend for an association with reduced likelihood of disease progression, though this was not significant (p<0.10).
Evaluating individual antibody responses would not have identified relationships between antibody responses, and many statistical comparisons would have been needed. These comparisons would not have been independent, as antibody responses were related by clade, gene, and protein, adding further issues with adjusting for multiple comparisons. Additionally, antibody responses within total response components exhibit similar variation, but protein-specific responses were not associated with disease progression to the same degree of significance as total response components. If antigen-specific responses were considered individually, significant associations may have been obscured. Therefore, protein microarray with PCA was a useful technique for exploring humoral immune profiles that may then encourage and guide further confirmatory investigations.
We observed differences in the relationship between individual antigens and HIV-1 disease progression similar to other studies. In studies of Thai and U.S. populations, investigators have observed significantly lower CA-specific antibody responses among rapid compared to slow progressors.21,22 Here, CA and MA-specific responses were associated with lower baseline viral load and less CD4 decline over time. We also observed that higher Gag-specific responses (CA and MA) were associated with decreased progression independent of baseline viral load. Though Gag-specific antibodies have little to no antiviral activity, they may be important in T cell helper responses.5 Additionally, over 12 months, higher CA, Nef, and Rev-specific antibody responses considered together (Total3) were significantly associated with reduced risk of disease progression. Anti-Nef and anti-CA antibody responses have exhibited parallel relationships with disease progression before.23 Efficient Nef-specific antibody-dependent cellular cytotoxicity has been observed, and along with Nef's involvement in the evasion of host adaptive immunity, there may be a role for anti-Nef antibodies.24 However, the involvement of anti-Rev antibody responses is less clear and may not be significantly associated with disease progression independent of CA and Nef.
Also unclear are the roles of anti-IN and anti-Env antibody responses (TM and SU) in disease control. HIV-1 integrase (IN) enzyme integrates viral DNA into the host's genomic DNA, allowing the virus to establish a chronic infection. The enzyme is highly conserved and few mutations are seen among integrase inhibitor-naive patients.25 Other studies detect anti-IN antibody responses during initial infection, but relationships with disease progression have not been shown.26 Here we observed a trend for an association between greater anti-IN antibody responses and reduced disease progression, as well as less decline in CD4 count. Finally, previous studies of Env-specific IgG responses have shown mixed or no associations with disease progression.5,6 In this analysis, greater anti-TM IgG was associated with higher viral load at baseline but not with subsequent changes in CD4 count, viral load, or disease progression at follow-up. In contrast, greater anti-SU IgG was associated with lower baseline viral load, and the two responses were negatively correlated (p<0.001), demonstrating inverse relationships with HIV-1 disease.
A recent study also sought to define host immune parameters that were associated with HIV virus control using clinical and humoral measures, supporting the importance of this work.27 Select antibody binding, as well as functional activity, was evaluated among 42 individuals divided into five distinct disease progression categories. A statistical technique was employed to predict the classification of patients by disease progression utilizing the included measures. Interestingly, anti-TM responses were found to be important to define patient status. In our analysis, binding antibody responses were more thoroughly examined among 92 ART-naive adults. Disease progression was also more finely evaluated, identifying patterns among the responses that were associated with concurrent and subsequent changes in CD4 count and viral load.
In both cohorts, associations between antibody measures and disease progression were observed. Further studies to elucidate the biological mechanisms of these humoral responses in relation to timing of HIV infection and future disease progression will be useful. The associations between HIV-specific antibody responses and disease progression may not be related to antibody function and could reflect other unmeasured immune parameters. However, identifying meaningful associations between disease progression and antibody responses may provide important insight into the mechanisms involved in disease control.
The strengths of this analysis include the use of PCA to reduce a large dataset to a subset of created variables that account for most of the observed variation. This allowed the identification of patterns within the antibody responses that could be explored and refined, reducing the number of comparisons made. The association of collections of HIV-specific antibody responses with disease progression may have been missed if individual antibody responses were considered, or if only a select few were chosen for analysis. In addition, the inclusion of 92 individuals in the analysis is a larger sample size than prior studies of HIV-specific humoral responses and disease progression.5,6,21,22
There were some limitations to this analysis. First, the study included samples collected over a 12-month period, which may be a short time frame of observation to adequately measure disease progression. Additionally, time of HIV infection is unknown in this cohort. Despite this, interesting antibody-specific associations were observed. Second, only 40 of the 143 antigens printed on the slide were seroreactive. Finally, we did not examine the subclass or function of binding antibody responses or the role of conformational epitopes in this analysis. Identifying IgG subclass specificity or antiviral function would be important to further understand the mechanism behind antibody-specific responses and disease control.4
High-throughput immune profiling by protein microarray, along with PCA to reduce data dimensionality, is a feasible and straightforward approach to explore complex immune responses, such as seen in HIV infection. HIV viral load is known to be associated with HIV disease progression and may stimulate higher antibody levels. However, we did not observe that higher viral loads were driving higher HIV-specific IgG responses; rather we saw the converse association of lower viral loads with higher antibody levels for some specific antibody responses (Table 2). While we are unable to determine whether antibody class or function is involved in shaping HIV antibody responses, the associations detected in this analysis warrant further examination to understand the role of HIV-specific antibody responses in disease control. Future applications of this technique may also include profiling immune responses of HIV coinfections, as defective antibody responses are characteristic of HIV progression.
Sequence Data
The templates and GenBank accession numbers of the HIV-1 clones used are as follows: subtype A1 (clone: 92UG037, accession # U51190), subtype A2 (clone: 94CY017, accession # AF286237), subtype B (clone: JR-CSF, accession # M38429), subtype C (clone: MJ4, accession # AF321523), and subtype D (clone: 94UG114, accession # U88824).
Supplementary Material
Acknowledgments
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: p92UG037.1 (Near-Full-Length) from Drs. Beatrice Hahn and Feng Gao, and the UNAIDS Network for HIV Isolation and Characterization; p94CY017.41 from Drs. Stanley A. Trask, Feng Gao, Beatrice H. Hahn, and the Aaron Diamond AIDS Research Center; pYK-JRCSF from Dr. Irvin S.Y. Chen and Dr. Yoshio Koyanagi; HIV-1 MJ4 from Drs. Thumbi Ndung'u, Boris Renjifo, and Max Essex; and p94UG114.1.6 (Full-Length) from Drs. Beatrice Hahn and Feng Gao, and the UNAIDS Network for HIV Isolation and Characterization.
This publication was made possible by NIH Grant TL1 RR 025016, 5U01AI078213-05, and by the University of Washington Center for AIDS Research, Scientific Program on HIV and coinfections.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hessell AJ, et al. : Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med 2009;15:951–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haynes BF, et al. : Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 2012;366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang KH, et al. : B-cell depletion reveals a role for antibodies in the control of chronic HIV-1 infection. Nat Commun 2010;1:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomaras GD. and Haynes BF: HIV-1-specific antibody responses during acute and chronic HIV-1 infection. Curr Opin HIV AIDS 2009;4:373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley JM, et al. : Differential regulation of the antibody responses to Gag and Env proteins of human immunodeficiency virus type 1. J Virol 1997;71:2799–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loomis-Price LD, et al. : Correlation between humoral responses to human immunodeficiency virus type 1 envelope and disease progression in early-stage infection. J Infect Dis 1998;178:1306–1316 [DOI] [PubMed] [Google Scholar]
- 7.Vigil A, Davies DH, and Felgner PL: Defining the humoral immune response to infectious agents using high-density protein microarrays. Future Microbiol 2010;5:241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JR, et al. : Statistical methods for analyzing immunosignatures. BMC Bioinformatics 2011;12:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundaresh S, et al. : Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics 2006;22:1760–1766 [DOI] [PubMed] [Google Scholar]
- 10.Jolliffe IT: Principal Component Analysis. 2nd ed. Springer, New York, 2002 [Google Scholar]
- 11.Loke P, et al. : Correlating cellular and molecular signatures of mucosal immunity that distinguish HIV controllers from noncontrollers. Blood 2010;116(13):2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang JT, et al. : A population-based study of Kaposi sarcoma-associated herpesvirus seropositivity in Uganda using principal components analysis. Infect Agent Cancer 2013;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walson J, et al. : Empiric deworming to delay HIV disease progression in adults with HIV who are ineligible for initiation of antiretroviral treatment (the HEAT study): A multi-site, randomised trial. Lancet Infect Dis 2012;12:925–932 [DOI] [PubMed] [Google Scholar]
- 14.Gao F, et al. : A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol 1998;72:5680–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao F, et al. : Evidence of two distinct subsubtypes within the HIV-1 subtype A radiation. AIDS Res Hum Retroviruses 2001;17:675–688 [DOI] [PubMed] [Google Scholar]
- 16.Ndung'u T, Renjifo B, and Essex M: Construction and analysis of an infectious human immunodeficiency virus type 1 subtype C molecular clone. J Virol 2001;75:4964–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haltiner M, Kempe T, and Tjian R: A novel strategy for constructing clustered point mutations. Nucleic Acids Res 1985;13:1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cann AJ, et al. : Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J Virol 1990;64:4735–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koyanagi Y, et al. : Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 1987;236:819–822 [DOI] [PubMed] [Google Scholar]
- 20.Davies DH, et al. : Profiling the humoral immune response to infection by using proteome microarrays: High-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci USA 2005;102:547–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee K, et al. : IgG subclass profiles in infected HIV type 1 controllers and chronic progressors and in uninfected recipients of Env vaccines. AIDS Res Hum Retroviruses 2010;26:445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuenchitra T, et al. : Longitudinal study of humoral immune responses in HIV type 1 subtype CRF01_AE (E)-infected Thai patients with different rates of disease progression. AIDS Res Hum Retroviruses 2003;19:293–305 [DOI] [PubMed] [Google Scholar]
- 23.Chen YM, et al. : Decreasing levels of anti-Nef antibody correlate with increasing HIV type 1 viral loads and AIDS disease progression. AIDS Res Hum Retroviruses 1999;15:43–50 [DOI] [PubMed] [Google Scholar]
- 24.Yamada T, et al. : Antibody-dependent cellular cytotoxicity via humoral immune epitope of Nef protein expressed on cell surface. J Immunol 2004;172:2401–2406 [DOI] [PubMed] [Google Scholar]
- 25.Ceccherini-Silberstein F, et al. : Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev 2009;11:17–29 [PubMed] [Google Scholar]
- 26.Barin F, et al. : Development and validation of an immunoassay for identification of recent human immunodeficiency virus type 1 infections and its use on dried serum spots. J Clin Microbiol 2005;43:4441–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brombin C, et al. : A nonparametric procedure for defining a new humoral immunologic profile in a pilot study on HIV infected patients. PLoS One 2013;8(3):e58768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.