Abstract
The 90 kDa heat-shock protein (Hsp90) and other cochaperones allow for proper folding of nascent or misfolded polypeptides. Cancer cells exploit these chaperones by maintaining the stability of mutated and misfolded oncoproteins and allowing them to evade proteosomal degradation. Inhibiting Hsp90 is an attractive strategy for cancer therapy, as the concomitant degradation of multiple oncoproteins may lead to effective anti-neoplastic agents. Unfortunately, early clinical trials have been disappointing with N-terminal Hsp90 inhibitors, as it is unclear whether the problems that plague current Hsp90 inhibitors in clinical trials are related to on-target or off-target activity. One approach to overcome these pitfalls is to identify structurally diverse scaffolds that improve Hsp90 inhibitory activity in the cancer cell milieu. Utilizing a panel of cancer cell lines that express luciferase, we have designed an in-cell Hsp90-dependent luciferase refolding assay. The assay was optimized using previously identified Hsp90 inhibitors and experimental novobiocin analogues against prostate, colon, and lung cancer cell lines. This assay exhibits good interplate precision (% CV), a signal-to-noise ratio (S/N) of ≥7, and an approximate Z-factor ranging from 0.5 to 0.7. Novobiocin analogues that revealed activity in this assay were examined via western blot experiments for client protein degradation, a hallmark of Hsp90 inhibition. Subsequently, a pilot screen was conducted using the Prestwick library, and two compounds, biperiden and ethoxyquin, revealed significant activity. Here, we report the development of an in-cell Hsp90-dependent luciferase refolding assay that is amenable across cancer cell lines for the screening of inhibitors in their specific milieu.
Introduction
For more than a decade, the 90 kDa heat-shock protein (Hsp90) has represented one of the most promising biological targets identified for the treatment of cancer. As a molecular chaperone, Hsp90 is responsible for folding many of the proteins that are directly associated with malignant progression; hence, inhibition of the Hsp90 protein folding machinery results in a combinatorial attack on numerous pathways. Furthermore, studies have revealed that Hsp90 inhibitors accumulate in tumor cells more efficiently than in normal tissue, leading to a differential selectivity of ∼200-fold.1 Currently, there are 20 Hsp90-targeted drugs in Phase I/II clinical trials, all of which target the Hsp90 N-terminal ATP-binding site.2–5 However, a major pitfall of N-terminal inhibitors is that they induce a pro-survival heat-shock response (HSR), leading to the induction of Hsp27, Hsp70, and Hsp90 at the same concentration that leads to client protein degradation and making both scheduling and dosing of these drugs extremely difficult. Thus, the induction of heat-shock proteins by N-terminal inhibitors in the tumor creates a vicious cycle in which patients require higher doses and increased frequency that ultimately pushes the patient toward the maximum tolerated dose and toxicity. At present, early clinical trials of N-terminal inhibitors have shown efficacy in only tumors that have highly sensitive and defined oncoprotein drivers, such as HER2, EML4-ALK, and c-KIT, or have inherent sensitivity toward proteotoxic stress, such as the case for multiple myelomas. It is unknown whether the lack of broad spectrum activity of N-terminal Hsp90 inhibitors against different cancers is the result of insufficient client protein depletion or a side effect of the cell-protective HSR.
While all Hsp90 inhibitors in clinical trials are directed to the N-terminal ATP-binding domain, efforts are underway to design inhibitors of Hsp90 at the C-terminal domain. Initially, the development of potent inhibitors of the C-terminal ATP-binding site of Hsp90 was based largely on the novobiocin scaffold identified by Marcu and colleagues.6–14 Previously, we have shown that targeting the C-terminal ATP-binding site results in chaperone impairment, demonstrated by client protein degradation, without compensatory induction of the HSR.9,15 The resulting drug-response profiles are often cytotoxic in nature, supporting the hypothesis that C-terminal inhibitors have the potential to overcome some of the problems associated with N-terminal inhibitors.9,15 While there are no clinical trials using C-terminal inhibitors, there is strong preclinical in vitro data and limited in vivo data demonstrating the potency of these compounds. As a result, there remains a tremendous effort in both the pharmaceutical industry and in academia to develop inhibitors of Hsp90 that can be readily modified to improve solubility and potency, diminish toxicity, and prevent induction of the pro-survival HSR. One approach to accomplish this goal is to design a high-throughput screening (HTS) assay that is specific for Hsp90 chaperone complexes within the cancer cell milieu and to identify novel scaffolds which manifest superior tumor selectivity relative to normal tissue.9
Numerous assays have been developed using HTS strategies for the identification of Hsp90 inhibitors.16 For example, several approaches have used full-length or truncated recombinant protein in biochemical assays to determine Hsp90 inhibition using different detection methods, while others have used luciferase renaturation assays. The utilization of reporter enzymes, such as luciferase and β-galactosidase, for the study of heat shock and related stress was first suggested by Nguyen and colleagues.17 In this study, the authors demonstrated that both enzymes can be rapidly inactivated within a cell during hyperthermia or exposure of the cells to ethanol. In subsequent studies, these authors demonstrated that the thermal inactivation of luciferase is a reversible process that can be minimized by pretreatment of the cells with compounds known to stabilize protein structures such D2O and glycerol.18,19 Importantly, they observed that thermally inactivated luciferase can be recovered in the absence of protein synthesis, which led to the hypothesis that enzyme recovery was an active process mediated by the heat-shock proteins.
Luciferase-based, chaperone-mediated protein renaturation was first described by Schumacher and colleagues.20 This seminal article showed that firefly luciferase could be reversibly denatured and subsequent activity regained via ATP-dependent refolding of luciferase by recombinant Hsp90 and Hsp70 or in cell-free rabbit reticulocyte lysate. Others have described the use of luciferase as a reporter of chaperone activity in Arabidopsis and in the rat myoblast cell, H9c2.21–23 Assays based on rabbit reticulocyte lysates have been successfully used to biochemically characterize the refolding kinetics of the Hsp70/Hsp90 system as well as a screening tool to identify compounds that inhibit Hsp90 activity.24 While the rabbit reticulocyte assay is quite sensitive and robust, questions remain as to the physiological relevance of the active chaperone complexes in this system, as it represents a species more related to normal tissue rather than disease.
Over the last decade, there has been considerable effort put forth to develop specific Hsp90 inhibitors toward various cancers. The current belief is that cancer cell survival is dependent on Hsp90 chaperone activity to maintain and fold many oncogenic proteins which drive tumor progression. Indeed, there is evidence which suggests that Hsp90 is mainly present in multiprotein complexes within the cancer cell, which appear to have different inhibitor binding properties than the Hsp90 homodimer present in normal tissue.9,25,26 Furthermore, since Hsp90 is present in multiprotein complexes, one might predict that unique Hsp90-binding pockets or conformational epitopes are influenced by the interaction of these proteins within the chaperone complex. Therefore, screening inhibitors against Hsp90 complexes as they exist within their physiologically relevant cancer cell niche may prove advantageous compared with cell-free systems, and may result in the identification of novel scaffolds that possess improved selectivity to cancer cells. We have developed a functional cell-based Hsp90-dependent luciferase refolding assay that is easily adaptable to a number of specific cancer cell lines and potentially patient-derived tumor cell lines. Here, we report the development and validation of this functional cell-based bioassay for the screening of Hsp90 inhibitors.
Materials and Methods
Cell Culture
A549 lung, HCT116 colon, and PC3-MM2 prostate cancer cell lines were obtained from ATCC (A549 and HCT116, Manassas, VA) and M.D. Anderson Cancer Center (PC3-MM2, Houston, TX). A549 and HCT116 cells were cultured in F-12K (ATCC) and McCoy's 5A (Sigma-Aldrich, St. Louis, MO), respectively, supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (100 IU/mL/100 mg/mL), and PC3-MM2 cells were cultured in minimum essential medium (MEM) Eagle media (Sigma-Aldrich) supplemented with 10% FBS, penicillin/streptomycin (100 IU/mL/100 mg/mL), MEM vitamins, and MEM nonessential amino acids. All cells were maintained at 37°C with 5% CO2. Freeze-down stocks of the original characterized cell lines were cryopreserved in liquid nitrogen. All experiments were performed using cells with <20 passages and <3 months in continuous culture.
Rabbit Reticulocyte Lysate Luciferase Refolding Assay
Rabbit reticulocyte assay and luciferase direct binding experiments were performed as previously described.24 Rabbit reticulocyte lysate (1:2, lysis of one volume of packed cells in two volumes of deionized water) was purchased from Green Hectares (Oregon, WI). Firefly luciferase (L-9506), luciferin, molecular biology grade acetylated bovine serum albumin, ATP, Coenzyme A, and novobiocin were purchased from Sigma-Aldrich. Luciferase activity was measured on a VICTOR III luminometer set for 0.1 s per well integration.
Cancer Cell-Based Luciferase Refolding Assay
Intracellular denaturation and refolding of luciferase
To determine the optimal time to thermally denature intracellular luciferase, initial time course experiments were conducted at 50°C in two cancer cell lines (A549 and PC3-MM2). Cells were collected by trypsinization, washed with PBS, briefly centrifuged, and suspended in prewarmed (50°C) complete media. Aliquots of cells were analyzed for cell viability and luciferase activity for ∼14 min. In subsequent experiments, A549, PC3-MM2, and HCT116 cells were heat denatured at 50°C for 6 min, which was determined to be the optimal time to thermally denature luciferase. For studies examining the effect of de novo luciferase synthesis on the measurement of luciferase refolding, cells were treated for 2 h before and during the assay incubation time with 20 μg/mL of a eukaryotic protein-translation inhibitor, cyclohexamide.
Single-point luciferase refolding screen
Luciferase refolding assay was performed in cells that were stably transduced with a lentivirus-carrying Luc2/mCherry gene (obtained from Dr. Andrew L. Kung, Dana Farber Institute).27 Briefly, cell pellets were collected from 80%–90% confluent flasks and suspended in prewarmed media (50°C) for ∼6 min. The time and temperature were sufficient to heat-denature endogenous luciferase to less than 2% of its original activity without affecting the viability of cells (Fig. 1). Cells were plated at a density of 30,000 cells per well in a 384-well white plate in the presence of inhibitors at a final concentration of 25 μM. The plates were then incubated for 60 min at 37°C to allow for luciferase refolding. After 1 h, the luciferase activity was measured by addition of a luciferin substrate solution and read on a VICTOR III luminometer set for 0.1 s per well integration.
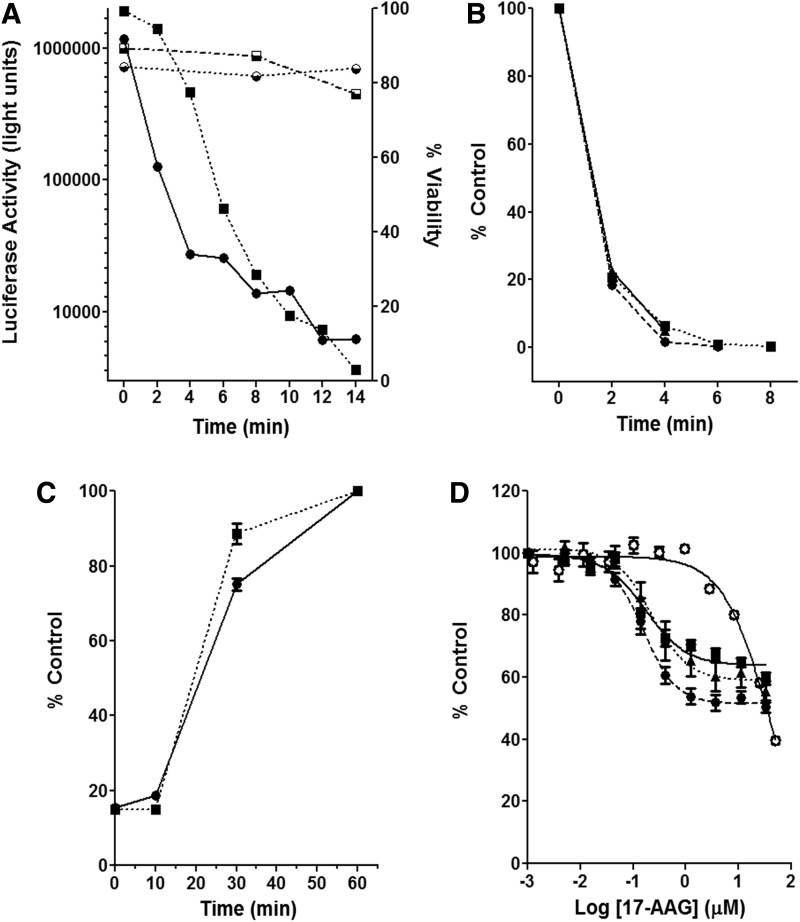
Fig. 1.
Development of a cancer cell-based luciferase refolding assay. (A) Cell-based thermal denaturation of luciferase activity (■, A549; ●, PC3-MM2) and cell viability (⬓, A549; ◒, PC3-MM2) measured concurrently over time. (B) Determination of the minimal amount of time required to thermally denature luciferase in A549 (■), PC3-MM2 (●), and HCT116 (▲) cancer cells as a percentage of its control over time. (C) Renaturation time of thermally denatured luciferase in PC3-MM2 cells pretreated in the presence (■) or absence (●) of cycloheximide. These results are the mean±SEM from two independent experiments (n=2). (D) Inhibition of luciferase refolding in A549 (■), PC3-MM2 (●), and HCT116 (▲) cancer cells and rabbit reticulocyte lysate (○) in dose response with 17-AAG. Luciferase activity is reported as a percentage of its control activity for each drug concentration. These results are the mean±SEM from three independent experiments performed in duplicate (n=3).
Dose-response cell-based luciferase refolding assay
In dose-response experiments, cancer cells transfected with Luc2/mCherry lentivirus were collected from confluent flasks and heat-denatured in prewarmed media as previously described. Cells were plated at a density of 50,000 cells/well in a white 96-well plate in the presence of inhibitor compounds. Refolding was carried out at 37°C for 1 h, and luciferase activity was measured by the addition of luciferin substrate. Direct inhibition of luciferase was analyzed for each compound as previously described.24 Raw data were plotted and normalized to control using a nonlinear regression and sigmoidal dose-response curves (GraphPad Prism).
Prestwick library high-throughput screen
The Prestwick Chemical Library® was purchased from Prestwick Chemical (Washington, DC). This library of 1,200 diverse FDA approved drugs is supplied at a concentration of 10 mM in DMSO. Dilution plates were made from original stock plates by diluting the library with saline to 100 μM. Assay plates were prepared by the addition of 10 μL of 100 μM drug to each well followed by the addition of 50,000 cells in 30 μL heated to thermally denature luciferase as described earlier. After incubation for 1 h at 37°C, 40 μL luciferase substrate was added to each well and read on a Perkin Elmer Envision 2101 multilabel reader. Data are shown as percent inhibition from a single screen of the Prestwick library.
Z-factor estimates
The reliability of the assay was estimated by calculating a Z-factor in which the maximal signal was defined as the percent inhibition of luciferase refolding in cells treated with 25 μM of compound 174 relative to the vehicle control (0.125% DMSO), and the minimum signal was defined as the background expressed by the vehicle control. The formula used to calculate the Z-factor was 1 − 3(SDp − SDc)/|Xp − Xc|, where SD and X are the standard deviation and average, respectively, of the maximal signal (p) and the minimum signal (c). Signal-to-noise ratio (S/N) was calculated using the following formula: S/N=(Xp − Xc)/√(SDp+SDc). Z-factor and S/N were determined in both 96-well and 384-well formats in which half the plate was dosed with 25 μM of compound 174 and half was dosed with vehicle alone.
Direct Inhibition of Luciferase Enzyme
Compounds that showed significant inhibition of luciferase refolding in either the rabbit reticulocyte or cell-based assay were tested for direct luciferase enzyme inhibition in a previously described cell-free assay.24
Sulforhodamine B Assay
Sulforhodamine B (SRB) assays were conducted as described by the Developmental Therapeutics Program at NCI (dtp.cancer.gov/branches/btb/ivclsp.html). Briefly, 17-AAG, novobiocin, 174, and 401 were run in ten concentration dose-response experiments against A549, PC3-MM2, and HCT116 cancer cell lines. Percentage growth was calculated for each drug concentration using the absorbance raw data, and percentage of growth inhibition was calculated using the formula [(Ti−Tz)/(C−Tz)]×100% (where Tz=time zero, C=control growth, and Ti=test growth in the presence of drug at the five concentration levels). Data were plotted on the y-axis as 100% to −100% growth, where 0 equals the starting cell number and negative numbers indicate cell death. Results were plotted on a sigmoidal dose-response curve using nonlinear regression (GraphPad Prism).
Western Blot
PC3-MM2, A549, and HCT116 cells were seeded at a density of 1.5×106 in T75 flasks. After 24 h, the t0 flask was harvested and cells were counted for cell viability using a Vi-Cell (Beckman Coulter, Inc., Brea, CA). Remaining flasks were dosed with drugs as indicated. Total cells were collected after 24 h, cell viability was determined, and SDS-PAGE lysates were prepared in RIPA (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, and 0.5% sodium deoxycholate) with protease and phosphatase inhibitor cocktail (Sigma-Aldrich). Cells were lysed by three freezing and thawing cycles using liquid nitrogen and a 37°C water bath. Total protein concentration was determined using DC Protein Assay (Bio-Rad Laboratories, Hercules, CA), and a total of 20 μg of cell lysates were used for western blot.
Results
Intracellular Refolding of Thermally Denatured Luciferase
A critical component of this bioassay requires thermal denaturation of intracellular luciferase without decreasing the viability of the cells. Luciferase activity was observed to decrease by >95% after thermal denaturation with minimal effects on cell viability across cell lines (Fig. 1A). Subsequently, it was determined that the minimal amount of time required to thermally denature luciferase in A549, PC3-MM2, and HCT116 cell lines was 6 min such that >95% was inactive, but still capable of being refolded by Hsp90 (Fig. 1B). Optimizing the minimal time to denature luciferase is important, as excessive thermal denaturation of luciferase is irreversible and renders this assay ineffective. These results demonstrate the feasibility of thermally denaturing intracellular luciferase without significantly affecting cell viability.
To determine whether de novo luciferase synthesis had any effect on the measurement of luciferase refolding in this assay, PC3-MM2 cells were pretreated with cycloheximide to prevent translation of luciferase. In control cancer cells, refolded luciferase activity increased 27-fold from baseline denatured levels. Conversely, cancer cells pretreated with cycloheximide revealed a significant decrease in the initial luciferase counts; 723,000 counts per second (cps)/105 cells as compared with ∼107 cps/105 cells, without cycloheximide pretreatment. These data indicate that a significant portion of luciferase was inactivated or degraded during cycloheximide pretreatment before heat denaturation. Since the initial counts were lower with cyclohexamide pretreatment, only 3 min were required to heat denature luciferase and decrease activity to >99% (2,500 cps/105 cells). Consequently, cells were incubated in media containing cycloheximide to allow for refolding. Luciferase activity under these conditions increased 13-fold from denatured levels or 4% of the original basal levels. Normalizing the data to relative percent control, such that 100% is the luciferase activity regained after 60 min, revealed a minimal effect on newly translated luciferase in this assay (Fig. 1C).
After the optimization of luciferase denaturation and refolding, 17-AAG, a known N-terminal Hsp90 inhibitor, was tested for inhibition of luciferase refolding in the rabbit reticulocyte assay as well as in the PC3-MM2, HCT116, and A549 cell-based assays (Fig. 1D). In both the cell-based and the rabbit reticulocyte luciferase refolding assays, 17-AAG prevented the Hsp90-dependent refolding of luciferase. The extent of inhibition, 40%–60% at 25 μM, was similar in the rabbit reticulocyte assay compared with the cell-based assay. EC50 values in the cell-based assay typically have a range between 50 and 200 nM; whereas the rabbit reticulocyte assay typically yields EC50 values in the range of 0.5–1 μM. These data demonstrate the feasibility of testing Hsp90 inhibitors in the cancer cell-based assay, which is comparable to the previously used rabbit reticulocyte assay. However, to further validate this assay, it was important to test a diverse panel of previously established Hsp90 inhibitors, as well as a panel of experimental compounds in both the cell-based and rabbit reticulocyte assays.
Activity of Established Hsp90 Inhibitors in the Cell-Based Assay
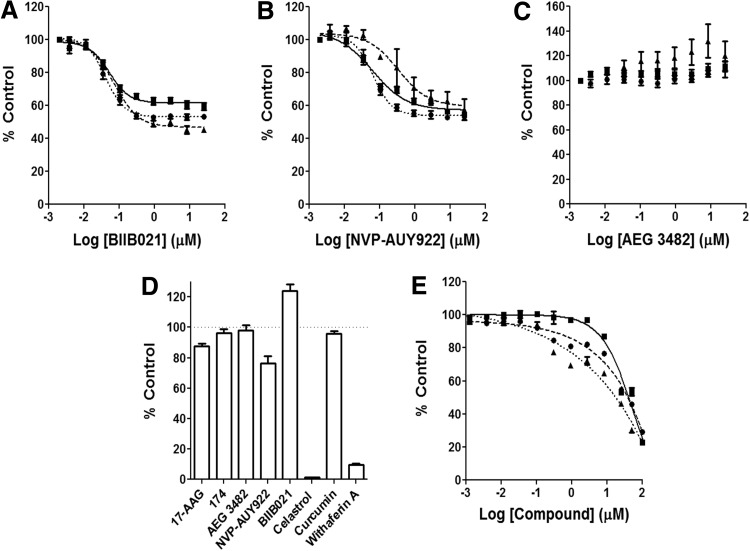
In order to further confirm the validity of the cell-based refolding assay, several known and commercially available Hsp90 inhibitors were tested in the cell-based assay. BIIB021 is an orally available, fully synthetic, highly characterized, and potent N-terminal Hsp90 inhibitor with an EC50 in the low nanomolar range against a wide variety of cell lines.28,29 BIIB021 demonstrated a dose-dependent inhibition of luciferase refolding across all three cell lines (A549, HCT116, and PC3-MM2, Fig. 2A) with an apparent EC50 in the range of 40–70 nM. Another N-terminal Hsp90 inhibitor, NVP-AUY922, was also tested in these cells lines. NVP-AUY922, being a novel resorcinylic isoxazole amide, is structurally unrelated to either 17-AAG or BIIB021 and has shown activity in a range of assays, cell lines (EC50 range of 2–40 nM), and xenograft models.30 In the cell-based assay, NVP-AUY922 showed a dose-dependent inhibition of luciferase refolding in the three cell lines, with lesser activity in the HCT116 colon cell line (Fig. 2B). The EC50 value was ∼60 nM in the PC3-MM2 and A549 cell lines and 500 nM in the HCT116 cell line. Interestingly, of all N-terminal Hsp90 inhibitors tested in this assay, 17-AAG, BIIB021, and NVP-AUY922 revealed their maximum percent inhibition at the highest concentrations to be ∼50%. As an additional control for the validity of the proposed cell-based assay, AEG 3482 was tested in the cell-based assay. AEG 3482 is known to bind to Hsp90 and disrupt Hsf-1, facilitating the induction of Hsp70 while retaining the chaperone activity of Hsp90.31 As one would predict, AEG 3482 showed no activity in the cell lines tested (Fig. 2C), and, in fact, a dose-dependent increase of chaperone activity was observed in the colon cell line HCT116. Other known Hsp90 inhibitors not considered N-terminal inhibitors were also tested and consisted of celastrol, curcumin, and withaferin A. Both celastrol and withaferin A were found to manifest direct inhibition of the luciferase enzyme and, therefore, could not be characterized further using the cell-based assay (Fig. 2D). Curcumin exhibited weak dose-dependent inhibition of luciferase refolding across all three cell lines tested (data not shown).
Fig. 2.
Inhibition of luciferase refolding by established Hsp90 inhibitors. (A–C) Inhibition of luciferase refolding with commercially available Hsp90 inhibitors BIIB021, NVP-AUY922, and AEG 3482 in A549 (■), PC3-MM2 (●), and HCT116 (▲) cell lines using the cell-based luciferase refolding assay. The data shown represent the mean±SEM from three independent experiments performed in duplicate (n=3). (D) Direct luciferase inhibition assay of established Hsp90 inhibitors. The data shown here represent the mean±SEM from three independent experiments (n=3). (E) Inhibition of luciferase refolding in the rabbit reticulocyte assay using AEG 3482 (■), NVP-AUY922 (●), and BIIB021 (▲). The data shown represent the mean±SEM from two independent experiments performed in duplicate (n=2). Hsp90, heat-shock protein 90.
Screening of Novobiocin Analogues in the Rabbit Reticulocyte and Cell-Based Assays
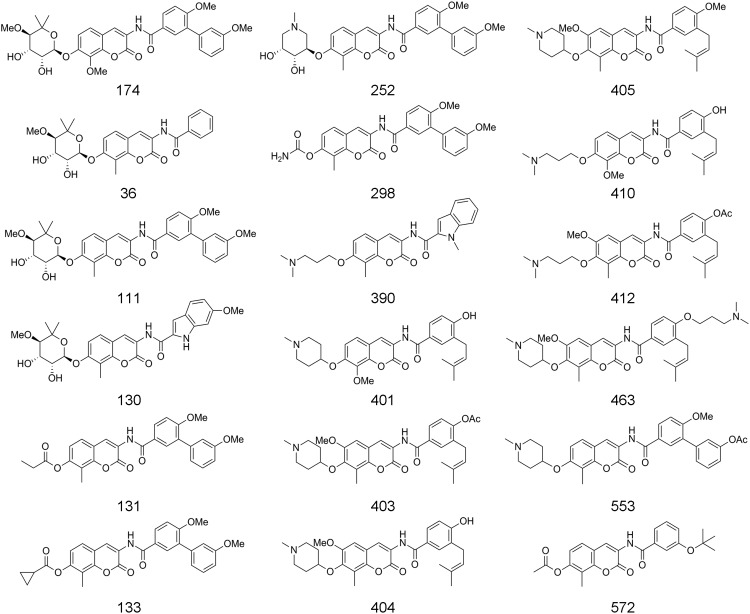
A series of novobiocin analogues (Fig. 3) was evaluated for their ability to inhibit the refolding of denatured luciferase in either a rabbit reticulocyte system or the cell-based assay. The data were normalized to percent control such that the results for both assays could be plotted for comparison (Table 1). From these data, it was apparent that the rabbit reticulocyte assay results in more active hits than the cell-based assay. For example, novobiocin analogue 390 was quite active in the reticulocyte system but showed no activity in the cell-based assay. Still other analogues such as 174, 252, 401, and 403 yielded similar results for both assays. However, from this testing, it appears that certain compounds manifest increased activity towards specific cell lines. For example, compound 412 showed significantly higher activity against A549 and HCT116 than PC3-MM2 cells. These data support the necessity of testing Hsp90 inhibitors in specific cancer cell lines to minimize the number of false positives, while maximizing the number of cancer cell-specific inhibitors. These results also demonstrate that the rabbit reticulocyte assay, while being quite sensitive to Hsp90 inhibitors, cannot serve to reliably predict cell-specific Hsp90 inhibition, thus demonstrating a significant strength of the cell-based assay.
Fig. 3.
Structures of novobiocin analogs.
Table 1.
Comparison of a Single-Point Luciferase Screen in Rabbit Reticulocyte Lysate Compared with the Cell-Based Luciferase Assay
| Compound | RRL (% inhibition) | HCT116 (% inhibition) | PC3-MM2 (% inhibition) | A549 (% inhibition) |
|---|---|---|---|---|
| DMSO | 0 | 0 | 0 | 0 |
| 17-AAG | 81±9 | 51±8 | 46±6 | 46±8 |
| 36 | 43±3 | 34±45 | −1±25 | 6±4 |
| 111 | 55±5 | 36±24 | 9±13 | −2±9 |
| 130 | 44±5 | 4±6 | −2±21 | −11±16 |
| 131 | 38±6 | −2±8 | −5±15 | −10±16 |
| 133 | 39±16 | −2±16 | −6±12 | −9±16 |
| 174 | 72±1 | 51±20 | 40±25 | 44±15 |
| 252 | 83±14 | 45±16 | 30±27 | −4±6 |
| 298 | 55±0 | −13±69 | −16±19 | −7±9 |
| 390 | 64±0 | 6±2 | 6±12 | −19±18 |
| 401 | 78±2 | 90±7 | 52±14 | 51±29 |
| 403 | 72±1 | 61±13 | 34±15 | 35±23 |
| 404 | 56±1 | 14±3 | 26±15 | −1±25 |
| 405 | 56±1 | 16±7 | 31±18 | 11±9 |
| 410 | 41±31 | 7±11 | 26±5 | 11±9 |
| 412 | 41±2 | 56±8 | 27±13 | 63±39 |
| 463 | 36±27 | 59±50 | 12±28 | 20±1 |
| 553 | 32±25 | 65±41 | 32±36 | 17±10 |
| 572 | 73±16 | 56±47 | 26±15 | 22±4 |
RRL and the cancer cell-based luciferase assay in HCT116, PC3-MM2, and A549 cells were used to test novobiocin analogues and identify potential Hsp90 inhibitors at a final concentration of 25 μM. The data displayed here represent the mean±standard deviation from two independent experiments (n=2).
Hsp90, heat-shock protein 90; RRL, rabbit reticulocyte lysate.
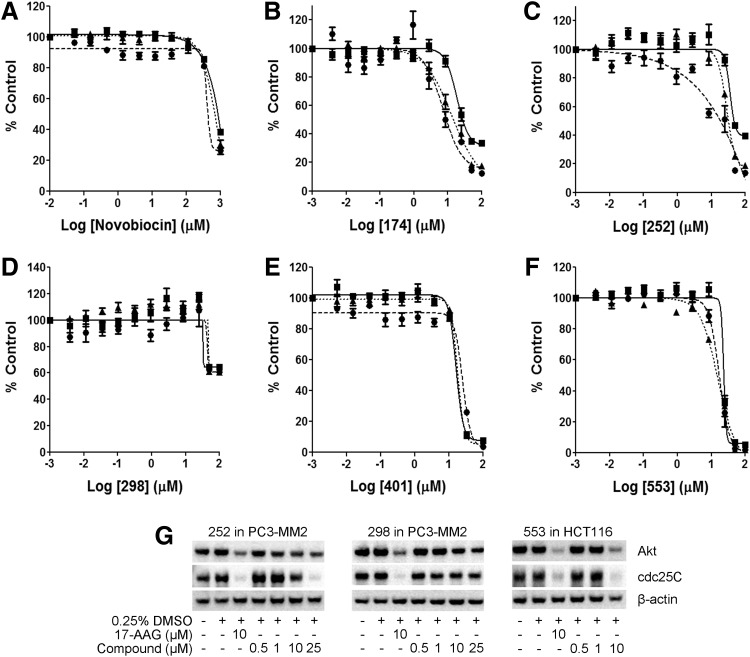
Compounds exhibiting diverse activities, 174, 252, 298, 401, and 553, were selected for full-dose-response studies against these cell lines along with novobiocin (Fig. 4). These compounds showed results that were consistent with those observed in single-dose studies. Compound 174 was most active against PC3-MM2 and HCT116 cells, and to a lesser extent in the A549 cell line (Fig. 4B) with EC50 values in the low micromolar range. Alternatively, compound 298 only revealed activity at the two highest doses (Fig. 4D) despite being very active in the rabbit reticulocyte assay (Table 1). Similarly, compound 252 was active in the rabbit reticulocyte assay and HCT116 and PC3-MM2 cell lines but showed no activity in the A549 cell line (Fig. 4C), which is in agreement with the single-dose study. Furthermore, compounds 401 and 553 demonstrated a dose-dependent inhibition of luciferase refolding in all three cell lines evaluated (Fig. 4E, F). In follow-up studies, compounds 252 and 298 were tested in PC3-MM2 cells and 553 in HCT116 cells for their ability to induce client protein degradation of AKT and cdc25C. We have previously demonstrated that AKT is sensitive to C-terminal Hsp90 inhibition by 174 and recently discovered that cdc25C is also very sensitive to degradation on Hsp90 inhibition.9 Consistent with the cell-based assay, compounds 252 and 553 showed a dose-dependent degradation of both proteins; whereas compound 298, being significantly less active in the cell-based assay, showed minimal decrease in protein levels (Fig. 4G).
Fig. 4.
Cell-based luciferase refolding assay in cancer cell lines. (A–F) Inhibition of luciferase refolding with novobiocin, 174, 252, 298, 401, and 553 in A549 (■), PC3-MM2 (●), and HCT116 (▲) cancer cell lines using the luciferase refolding assay. The data presented are the mean±SEM of three independent experiments performed in duplicate (n=3). (G) Degradation of client protein AKT and cdc25C in PC3-MM2 cells treated with 252 or 298 and HCT116 cells treated with 553. These results are representative of three independent experiments (n=3).
Novobiocin Analogue 401 Induces Cytotoxicity Compared with the N-Terminal Inhibitor 17-AAG
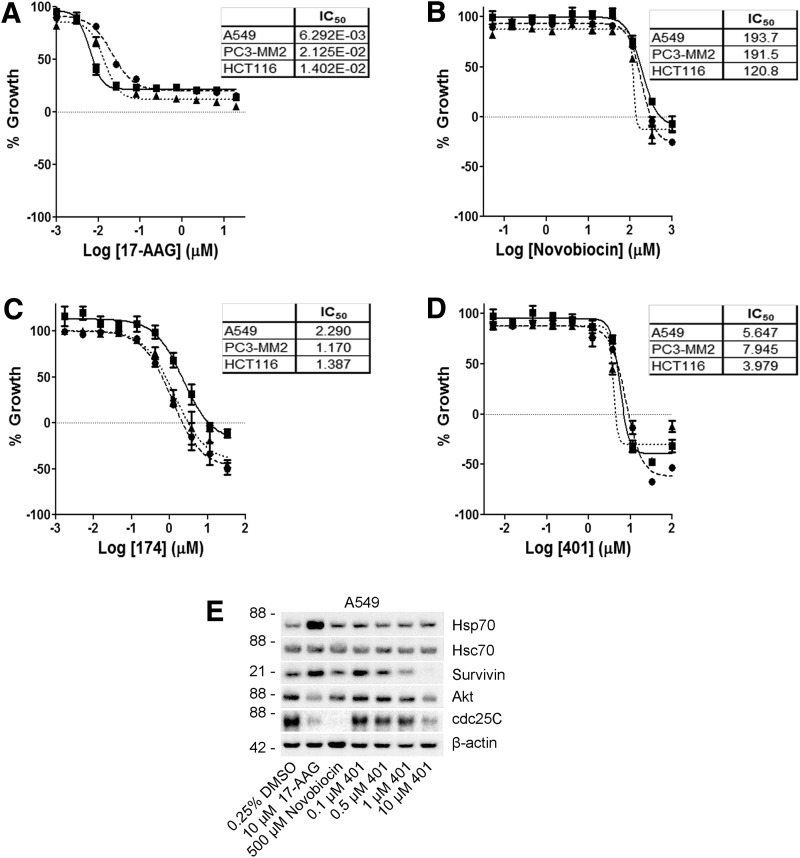
One of the major pitfalls of N-terminal Hsp90 inhibitors is that they are largely cytostatic against many cell lines, presumably as a consequence of the pro-survival HSR. For example, when the N-terminal inhibitor 17-AAG was tested in A549, PC3-MM2, and HCT116 cells using the SRB assay, the results consistently showed a cytostatic profile (Fig. 5A). Our lead C-terminal Hsp90 inhibitor 174 appears to be cytotoxic against PC3-MM2 and HCT116 cells, while A549 cells were less sensitive, which is again consistent with the luciferase refolding inhibition (Fig. 5C). Thus, based on the results from the cell-based luciferase refolding assay, we predicted that 401, a putative C-terminal inhibitor that exhibits >50% activity in the single-point screen, would manifest improved activity across these cell lines as compared with 174 (see Table 1). Indeed, this compound manifests significant cytotoxicity against all three cell lines (Fig. 5D) and resulted in the dose-dependent degradation of the client proteins AKT, survivin, and cdc25C without increasing the heat-shock proteins that are usually associated with N-terminal Hsp90 inhibitors, such as 17-AAG (Fig. 5E). The parent compound novobiocin revealed significantly less antiproliferative activity and exhibited the same client protein degradation profile as compound 401, although at lower potency (Fig. 5B, E).
Fig. 5.
Antiproliferative effects of 17-AAG, novobiocin, 174, and 401 in a panel of cancer cell lines. (A–D) Antiproliferative effects of 17-AAG, novobiocin, 174, and 401 in A549 (■), PC3-MM2 (●), and HCT116 (▲) cells. These data are the mean±SEM from three independent experiments performed in duplicate (n=3) and are represented as percent growth, where 100 to 0 is the antiproliferative response and 0 to -100 is the cytotoxic response. (E) Western blots for client protein degradation of survivin, AKT, and cdc25C as well as the induction of heat-shock response in A549 cells after treatment with 401 along with positive (17-AAG and novobiocin) and negative (vehicle, 0.25% DMSO) controls. These results are representative of three independent experiments (n=3).
Miniaturization of the Cell-Based Luciferase Refolding Assay to 384-Well Format
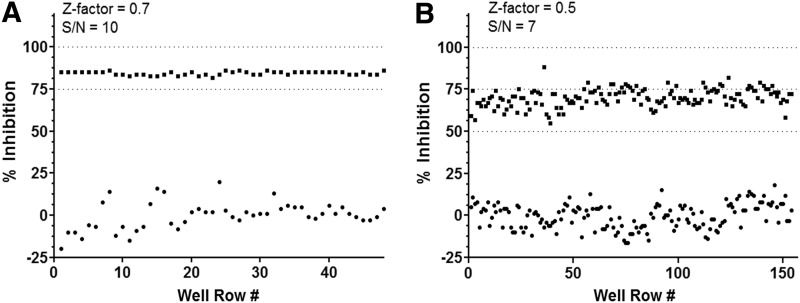
The refolding assay was further optimized and miniaturized to a 384-well format, as described in Table 2. Results for the 96-well and 384-well formats yielded a Z-factor of 0.7 (Fig. 6A) and 0.5 (Fig. 6B), respectively, as well as S/N of ≥7 and serve to highlight the feasibility of this assay for HTS.
Table 2.
HTS Assay Protocol
| Step | Parameter | 96-well plate | 384-well plate | Description |
|---|---|---|---|---|
| 1 | Plate cells | 50 μL | 12.5 μL | 50,000 cells/well for 96-well plate; 12,500 cells/well for 384-well plate |
| 2 | Controls | 50 μL | 12.5 μL | DMSO, 17-AAG, 174 |
| 3 | Library compounds | 50 μL | 12.5 μL | Dilution range 1.2 nM–25 μM |
| 4 | Incubation time | 1 h | 1 h | 37°C, 5% CO2 |
| 5 | Reporter reagent | 100 μL | 50 μL | Luciferin substrate |
| 6 | Assay readout | CCD imager, luminescent mode |
Step Notes
1. Prepare drug plates in DMSO by serial dilution of 20 mM drug stocks (dilution range 100 nM–2 mM). Final volume is 30 μL for 96-well plate or 20 μL for 384-well plate.
2. Dilute control and test compounds from drug plates in media to make dilution plates. Final volume is 200 μL for 96-well plate or 80 μL for 384-well plate.
3. Add drug in media from dilution plate to 96-well plate (Costar 96-well white opaque plate) or 384-well plate (Corning 384-well black plate with clear bottom) at the volume indicated earlier.
4. Add heat-treated (50°C) cells to each well of cell plate at the volume and concentration indicated earlier.
5. Incubate plate at 37°C for 1 h.
6. Add reporter reagent (75 mM tricine, pH 7.8, 24 mM MgSO4, 0.3 mM EDTA, 2 mM DTT, 0.313 mM luciferin, 0.64 mM coenzyme A, 0.66 mM ATP, 150 mM KCl, 10% Triton X-100, 20% glycerol, and 3.5% DMSO) at the volume indicated earlier. Shake plate for 2 min.
7. Read luciferase activity (relative light units, Rel. LU) across plate.
Fig. 6.
Optimization of the cancer cell-based high-throughput screening (HTS) assay in 96-well and 384-well formats. (A) The Z-factor was first established in 96-well format using PC3-MM2 cells treated with 25 μM 174 (■) or vehicle control (●, 0.125% DMSO). The resulting 96-well-format Z-factor was 0.7, with a signal-to-noise ratio (S/N) of 10. (B) After miniaturizing the cancer cell-based assay to a 384-well format, the Z-factor and S/N observed were 0.5 and 7, respectively.
Screening a Small-Molecule Library for Hsp90 Inhibition
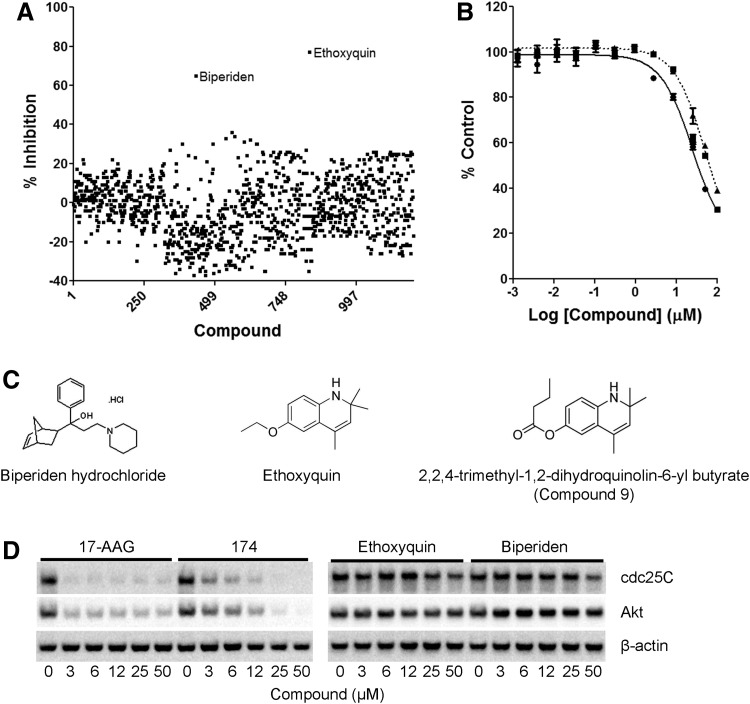
As a proof of concept, the cell-based assay was used to screen the Prestwick small-molecule library for compounds that inhibit Hsp90 activity as measured by luciferase refolding in PC3-MM2 cells. This library represents the highest degree of diverse small molecules that possess “drug-like properties.” From this screen, two structurally diverse compounds, ethoxyquin and biperiden showed significant inhibition (>50%) of luciferase refolding (Fig. 7A). Control experiments to rule out direct inhibition of the luciferase enzyme were negative (data not shown). Neither ethoxyquin nor biperiden has been previously reported to inhibit Hsp90 chaperone activity. Subsequently, these compounds were evaluated in the rabbit reticulocyte assay, and both revealed a dose-dependent inhibition of luciferase refolding (Fig. 7B). Interestingly, an HTS screen by Galam et al. using the rabbit reticulocyte assay reported a compound, designated as compound 9, that contains a similar structure to ethoxyquin, but only differing in the side chain (Fig. 7C).24 PC3-MM2 cells treated with ethoxyquin or biperiden resulted in a dose-dependent decrease of the highly sensitive Hsp90 client protein cdc25C and, to a lesser extent, AKT (Fig. 7D).32,33 Overall, these data suggest that these compounds represent novel Hsp90 inhibitor scaffolds and may prove useful for future structure–function analysis.
Fig. 7.
HTS of a 1,200 compound library in the cancer cell-based assay utilizing PC3-MM2 cells. (A) One thousand two hundred compounds from the Prestwick library were screened using a single-point cell-based luciferase refolding drug screen that resulted in two potential hits, ethoxyquin and biperiden. (B) Inhibition of luciferase refolding with ethoxyquin (■), biperiden (▲), and 17-AAG (●) in the rabbit reticulocyte assay. These data are the mean±SEM from two independent experiments performed in duplicate (n=2). (C) The molecular structures of ethoxyquin and biperiden, along with a previous scaffold (compound 9), identified in a rabbit reticulocyte screen by Galam et al.24 (D) Western blots for client protein degradation of cdc25C and AKT in PC3-MM2 cells treated with 17-AAG, 174, ethoxyquin, and biperiden. These results are representative of three independent experiments (n=3).
Discussion
In the present study, we extend the use of endogenously expressed luciferase as a reporter for chaperone activity and use this approach to detect and screen for Hsp90 inhibitors against specific cancer cells. To date, no study has been reported showing the utility of a cell-based luciferase refolding assay that is specifically designed for the detection and screening of Hsp90 inhibitors as anti-cancer agents. Earlier studies have used a similar approach using a cell-based system to monitor the Hsp90 protein folding machinery; however, that approach utilized aliquots of cell extracts to examine Hsp90 inhibition, which is not amenable to HTS.34,35 Furthermore, lysing of the cells to prepare the cell extract has the potential to physically disrupt the Hsp90 heteroprotein complexes, leading to a loss of physiological relevance. In contrast, this approach is novel, as Hsp90 inhibition can be measured in intact cancer cells without disruption of the native complexes. In addition, this assay has been miniaturized and can be used for HTS. By screening the small library of novobiocin analogues, this cell-based assay can be adapted to identify inhibitors against a variety of cancer cells, allowing for the rapid screening of compound libraries against specific cancer types. Importantly, both the N-terminal and C-terminal domains of Hsp90 participate independently in binding denatured luciferase and support the rationale of using this assay for screening to identify both N-terminal and C-terminal Hsp90 inhibitors.36 In validating this assay with previously established Hsp90 inhibitors of different classes and structures, it was observed that the Hsp90 inhibitors are more potent in the cell-based assay, while the rabbit reticulocyte assay has the potential for false positives. For example, BIIB021, NVP-AUY922, and AEG 3482 had significant activity in the rabbit reticulocyte assay, while only BIIB021 and NVP-AUY922 demonstrate potent activity in the cell-based assay, indicating that AEG 3482 would be false positive for Hsp90 in human cancer cells. Furthermore, all three of these compounds exhibit a lower potency in the rabbit reticulocyte assay compared with the cell-based assay (Fig. 2E), indicating that the cell-based assay is more sensitive for identifying Hsp90 inhibitors. Moreover, it appears that N-terminal inhibitors can only achieve a maximal inhibition of 50% in the cell-based assay despite increasing concentrations of inhibitors, while in the rabbit reticulocyte assay they are capable of greater than 50% inhibition with percent inhibition being dose dependent. These data indicate that the rabbit reticulocyte assay misrepresents the efficacy of N-terminal inhibitors with regard to the inhibition of Hsp90 in human cancer cells. Novobiocin analogues screened in the cell-based assay revealed varying degrees of activity, and several compounds were found to be inactive across cell lines. Conversely, when these compounds were tested in the rabbit reticulocyte assay, every compound exhibited inhibitory activity. An explanation of the differences in activities between assays with the known Hsp90 inhibitors could be due to species differences (human versus rabbit) or differences in the Hsp90 heteroprotein complexes. Hsp90 complexes in the rabbit reticulocyte assay can be considered similar to those present in normal tissue, while complexes found in cancer cells have been reported to be significantly different.9,25,26 In these and other studies, it was found that novobiocin and many of the novobiocin analogs differ from N-terminal Hsp90 inhibitors because they do not induce an HSR (Hsp27 and Hsp70).9,15 Here, we describe another major difference between these two classes of Hsp90 inhibitors. In the cell-based luciferase assay, N-terminal Hsp90 inhibitors can only achieve a maximal inhibition of 50%, while C-terminal Hsp90 inhibitors are capable of 100% inhibition in a dose-dependent manner. To date, there are no reports of an Hsp90-independent luciferase refolding mechanism in cells. Thus, we believe the differences between N-terminal and C-terminal Hsp90 inhibitors in this assay to be Hsp90 dependent and a result of inhibition at the different termini. However, the precise mechanism leading to these differences remains unclear and is the focus of future studies. A benefit of this assay is that one could use the different inhibitory profiles to screen for N-terminal and C-terminal Hsp90 inhibitors based on the percent inhibition.
One should carefully consider the contributions of de novo luciferase synthesis as a confounding element for any cell-based luciferase refolding assay. In this study, pretreatment with cycloheximide was used to inhibit protein synthesis, and minimal contribution was observed from de novo synthesis. This finding is in complete agreement with previous studies that estimated the synthesis rate of luciferase to be 2.6% per hour relative to the basal amount of luciferase already present within a cell.22,23
While the described cell-based assay appears to be sensitive and sufficiently robust for the detection of Hsp90 inhibition, we recognize some limitations of the assay. For example, since this is a cell-based assay, any compounds that are not able to cross the cell membrane or compounds rapidly effluxed out of the cell will be misrepresented in this assay. Small compound efflux is mediated by ATP-binding cassette (ABC) transporters that form a class of membrane-associated proteins thought to be involved in the resistance of tumors to chemically unrelated anti-cancer drugs, termed multidrug resistance. Therefore, potential Hsp90 inhibitors that are substrates for ABC transporters may be missed or their potency may be underestimated with a cell-based assay. Moreover, confounding limitations for this assay may also arise from compounds that are cytotoxic or deplete ATP levels within the cell during the initial hour incubation step, resulting in false-positive hits. However, despite these potential limitations, this cell-based assay, when used in combination with secondary confirmatory Hsp90 inhibitor screens such as western blot for client protein degradation or Hsp70 induction, has the possibility to yield highly selective Hsp90 inhibitors to specific cancers.
Future studies plan to test the use of conformationally destabilized luciferase mutants described elsewhere.37 The rationale for these studies is that by increasing the sensitivity of luciferase to thermal denaturation or by increasing its dependence on Hsp90 chaperone activity, greater inhibition may be observed in this assay.
Abbreviations
- ABC
ATP-binding cassette
- cps
counts per second
- FBS
fetal bovine serum
- Hsp90
heat-shock protein 90
- HSR
heat-shock response
- HTS
high-throughput screening
- RRL
rabbit reticulocyte lysate
- S/N
signal-to-noise ratio
- SRB
sulforhodamine B
Acknowledgments
The authors thank Andrew L. Kung at the Dana Farber Cancer Institute (Boston, MA) for providing the FUW-Luc-mCherry-puro construct.
Disclosure Statement
The authors have no conflicts of interest with any institutional or commercial affiliations regarding the publication of this article.
References
- 1.Chiosis G. Huezo H. Rosen N, et al. 17AAG: low target binding affinity and potent cell activity—finding an explanation. Mol Cancer Ther. 2003;2:123–129. [PubMed] [Google Scholar]
- 2.Travers J. Sharp S. Workman P. HSP90 inhibition: two-pronged exploitation of cancer dependencies. Drug Discov Today. 2012;17:242–252. doi: 10.1016/j.drudis.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Neckers L. Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kummar S. Gutierrez ME. Gardner ER, et al. Phase I trial of 17-dimethylaminoethylamino-17-demethoxygeldanamycin (17-DMAG), a heat shock protein inhibitor, administered twice weekly in patients with advanced malignancies. Eur J Cancer. 2010;46:340–347. doi: 10.1016/j.ejca.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YS. Alarcon SV. Lee S, et al. Update on Hsp90 inhibitors in clinical trial. Curr Top Med Chem. 2009;9:1479–1492. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burlison JA. Avila C. Vielhauer G, et al. Development of novobiocin analogues that manifest anti-proliferative activity against several cancer cell lines. J Org Chem. 2008;73:2130–2137. doi: 10.1021/jo702191a. [DOI] [PubMed] [Google Scholar]
- 7.Burlison JA. Blagg BS. Synthesis and evaluation of coumermycin A1 analogues that inhibit the Hsp90 protein folding machinery. Org Lett. 2006;8:4855–4858. doi: 10.1021/ol061918j. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly AC. Mays JR. Burlison JA, et al. The design, synthesis, and evaluation of coumarin ring derivatives of the novobiocin scaffold that exhibit antiproliferative activity. J Org Chem. 2008;73:8901–8920. doi: 10.1021/jo801312r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eskew JD. Sadikot T. Morales P, et al. Development and characterization of a novel C-terminal inhibitor of Hsp90 in androgen dependent and independent prostate cancer cells. BMC Cancer. 2011;11:468–484. doi: 10.1186/1471-2407-11-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu XM. Shen G. Neckers L, et al. Hsp90 inhibitors identified from a library of novobiocin analogues. J Am Chem Soc. 2005;127:12778–12779. doi: 10.1021/ja0535864. [DOI] [PubMed] [Google Scholar]
- 11.Zhao H. Brandt GE. Galam L. Matts RL. Blagg BS. Identification and initial SAR of silybin: an Hsp90 inhibitor. Bioorg Med Chem Lett. 2011;21:2659–2664. doi: 10.1016/j.bmcl.2010.12.088. [DOI] [PubMed] [Google Scholar]
- 12.Kusuma BR. Duerfeldt AS. Blagg BS. Synthesis and biological evaluation of arylated novobiocin analogs as Hsp90 inhibitors. Bioorg Med Chem Lett. 2011;21:7170–7174. doi: 10.1016/j.bmcl.2011.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kusuma BR. Peterson LB. Zhao H, et al. Targeting the heat shock protein 90 dimer with dimeric inhibitors. J Med Chem. 2011;54:6234–6253. doi: 10.1021/jm200553w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcu MG. Chadli A. Bouhouche I. Catelli M. Neckers LM. The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP-binding domain in the carboxyl terminus of the chaperone. J Biol Chem. 2000;275:37181–37186. doi: 10.1074/jbc.M003701200. [DOI] [PubMed] [Google Scholar]
- 15.Matthews SB. Vielhauer GA. Manthe CA, et al. Characterization of a novel novobiocin analogue as a putative C-terminal inhibitor of heat shock protein 90 in prostate cancer cells. Prostate. 2010;70:27–36. doi: 10.1002/pros.21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matts RL. Manjarrez JR. Assays for identification of Hsp90 inhibitors and biochemical methods for discriminating their mechanism of action. Curr Top Med Chem. 2009;9:1462–1478. doi: 10.2174/156802609789895692. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen VT. Morange M. Bensaude O. Protein denaturation during heat shock and related stress. Escherichia coli beta-galactosidase and Photinus pyralis luciferase inactivation in mouse cells. J Biol Chem. 1989;264:10487–10492. [PubMed] [Google Scholar]
- 18.Pinto M. Morange M. Bensaude O. Denaturation of proteins during heat shock. In vivo recovery of solubility and activity of reporter enzymes. J Biol Chem. 1991;266:13941–13946. [PubMed] [Google Scholar]
- 19.Dubois MF. Hovanessian AG. Bensaude O. Heat-shock-induced denaturation of proteins. Characterization of the insolubilization of the interferon-induced p68 kinase. J Biol Chem. 1991;266:9707–9711. [PubMed] [Google Scholar]
- 20.Schumacher RJ. Hurst R. Sullivan WP, et al. ATP-dependent chaperoning activity of reticulocyte lysate. J Biol Chem. 1994;269:9493–9499. [PubMed] [Google Scholar]
- 21.Forreiter C. Kirschner M. Nover L. Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo. Plant Cell. 1997;9:2171–2181. doi: 10.1105/tpc.9.12.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Souren JE. Wiegant FA. van Hof P. van Aken JM. van Wijk R. The effect of temperature and protein synthesis on the renaturation of firefly luciferase in intact H9c2 cells. Cell Mol Life Sci. 1999;55:1473–1481. doi: 10.1007/s000180050386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souren JE. Wiegant FA. Van Wijk R. The role of hsp70 in protection and repair of luciferase activity in vivo; experimental data and mathematical modelling. Cell Mol Life Sci. 1999;55:799–811. doi: 10.1007/s000180050333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galam L. Hadden MK. Ma Z, et al. High-throughput assay for the identification of Hsp90 inhibitors based on Hsp90-dependent refolding of firefly luciferase. Bioorg Med Chem. 2007;15:1939–1946. doi: 10.1016/j.bmc.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamal A. Thao L. Sensintaffar J, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 26.Moulick K. Ahn JH. Zong H, et al. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol. 2011;7:818–826. doi: 10.1038/nchembio.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimbrel EA. Davis TN. Bradner JE. Kung AL. In vivo pharmacodynamic imaging of proteasome inhibition. Mol Imaging. 2009;8:140–147. [PubMed] [Google Scholar]
- 28.Kasibhatla SR. Hong K. Biamonte MA, et al. Rationally designed high-affinity 2-amino-6-halopurine heat shock protein 90 inhibitors that exhibit potent antitumor activity. J Med Chem. 2007;50:2767–2778. doi: 10.1021/jm050752+. [DOI] [PubMed] [Google Scholar]
- 29.Lundgren K. Zhang H. Brekken J, et al. BIIB021, an orally available, fully synthetic small-molecule inhibitor of the heat shock protein Hsp90. Mol Can Ther. 2009;8:921–929. doi: 10.1158/1535-7163.MCT-08-0758. [DOI] [PubMed] [Google Scholar]
- 30.Eccles SA. Massey A. Raynaud FI, et al. NVP-AUY922: a novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 2008;68:2850–2860. doi: 10.1158/0008-5472.CAN-07-5256. [DOI] [PubMed] [Google Scholar]
- 31.Salehi AH. Morris SJ. Ho WC, et al. AEG3482 is an antiapoptotic compound that inhibits Jun kinase activity and cell death through induced expression of heat shock protein 70. Chem Biol. 2006;13:213–223. doi: 10.1016/j.chembiol.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Samadi AK. Zhang X. Mukerji R, et al. A novel C-terminal HSP90 inhibitor KU135 induces apoptosis and cell cycle arrest in melanoma cells. Cancer Lett. 2011;312:158–167. doi: 10.1016/j.canlet.2011.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senju M. Sueoka N. Sato A, et al. Hsp90 inhibitors cause G2/M arrest associated with the reduction of Cdc25C and Cdc2 in lung cancer cell lines. J Cancer Res Clin Oncol. 2006;132:150–158. doi: 10.1007/s00432-005-0047-7. [DOI] [PubMed] [Google Scholar]
- 34.Hernandez A. Lopez-Lluch G. Bernal JA. Navas P. Pintor-Toro JA. Dicoumarol down-regulates human PTTG1/Securin mRNA expression through inhibition of Hsp90. Mol Can Ther. 2008;7:474–482. doi: 10.1158/1535-7163.MCT-07-0457. [DOI] [PubMed] [Google Scholar]
- 35.Schneider C. Sepp-Lorenzino L. Nimmesgern E, et al. Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc Natl Acad Sci USA. 1996;93:14536–14541. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minami M. Nakamura M. Emori Y. Minami Y. Both the N- and C-terminal chaperone sites of Hsp90 participate in protein refolding. Eur J Biochem. 2001;268:2520–2524. doi: 10.1046/j.1432-1327.2001.02145.x. [DOI] [PubMed] [Google Scholar]
- 37.Gupta R. Kasturi P. Bracher A, et al. Firefly luciferase mutants as sensors of proteome stress. Nat Methods. 2011;8:879–884. doi: 10.1038/nmeth.1697. [DOI] [PubMed] [Google Scholar]