SUMMARY
The accelerated-cell-death11 (acd11) mutant of Arabidopsis provides a genetic model for studying immune response activation and localized cellular suicide that halts pathogen spread during infection in plants. Here, we elucidate ACD11 structure/function and show that acd11 disruption dramatically alters the in vivo balance of sphingolipid mediators that regulate eukaryotic programmed cell death. In acd11 mutants, normally low ceramide-1-phosphate (C1P) levels become elevated, but the relatively abundant cell death inducer, phytoceramide, rises acutely. ACD11 exhibits selective intermembrane transfer of C1P and phyto-C1P. Crystal structures establish C1P binding via a surface-localized, phosphate headgroup recognition center connected to an interior hydrophobic pocket that adaptively ensheaths lipid chains via a cleft-like gating mechanism. Point mutation mapping confirms functional involvement of binding-site residues. A π-helix (π-bulge) near the lipid-binding cleft distinguishes apo-ACD11 from other GLTP-folds. The global two-layer, α-helically-dominated, ‘sandwich’ topology displaying C1P-selective binding identifies ACD11 as the plant prototype of a new GLTP-fold subfamily.
INTRODUCTION
Sphingolipids and their metabolites, i.e. ceramide (Cer), ceramide-1-phosphate (C1P), and the long chain bases (LCB), sphingosine and sphingosine-1-phosphate (S1P), are bioactive lipids that function as messenger signals and mediators of eukaryotic processes such as cell growth, development, embryogenesis, senescence, inflammation, and programmed cell death (PCD) (Fyrst and Saba, 2010; Hannun and Obeid, 2008; Michaelson, 2010). The dynamic balance between Cer (sphingoid base amide-linked to a fatty acyl chain) and its phosphorylated derivative, C1P, critically regulates PCD in plants and animals (Berkey et al., 2012; Chen, 2009; Pata et al., 2010; Reape and McCabe, 2008).
In plants, PCD occurs during development, during disease symptoms associated with virulent infections, and during the hypersensitive response (HR) induced by avirulent stress effectors (Lam, 2004). Hallmarks of HR are local accumulation of reactive oxygen species, nitric oxide, and the phytohormone, salicylic acid. By inducing localized cell death triggered when resistance proteins recognize specific pathogen-derived molecules, HR potentiates defensive resistance. Mutants exhibiting accelerated cell death (acd) phenotypes in the absence of pathogen effectors also provide insights into HR-like PCD and defense activation. One HR mimic is the acd5 mutant, which lacks Cer kinase activity and accumulates Cers, triggering PCD (Liang et al., 2003). C1P addition partially abrogates the PCD-inducing effects of elevated Cer in acd5. In acd11 null mutant, HR-related PCD and defense genes are constitutively activated in salicylic acid-dependent fashion. The acd11 gene encodes ACD11, a lipid transfer protein able to moderately accelerate the intermembrane transfer of sphingosine and sphingomyelin, but not Cer or glycosylceramides (Brodersen et al., 2002; Petersen et al., 2008).
Structural homology modeling predicts ACD11 forms a GLTP-fold and is a glycolipid transfer protein (GLTP) superfamily member (Airenne et al., 2006; Brown and Mattjus, 2007; Petersen et al., 2008). Yet, ACD11 is unable to transfer glycolipids (Brodersen et al., 2002), consistent with the lack of essential residues needed for glycosphingolipid (GSL) sugar headgroup binding (Petersen et al., 2008). In mammalian GLTPs and HET-C2 fungal GLTP, X-ray structures reveal the molecular details of how glycolipids are recognized and bound by a conserved residue cluster (Asp, Asn, Lys, His, Trp) that form a hydrogen bond network with the GSL sugar-amide region, thus explaining the selectivity and transfer proficiency for various GSLs (Airenne et al., 2006; Kenoth et al., 2011; Kenoth et al., 2010; Malinina et al., 2006; Malinina et al., 2004; Samygina et al., 2011). Currently lacking for ACD11 is establishment of its preferred sphingolipid ligand as well as direct evidence for its functional involvement in the regulation of plant sphingolipid metabolism.
Herein, we investigated ACD11 structure and lipid transfer specificity and discovered high selectivity for C1P and phyto-C1P, but not related plant sphingolipids, i.e. glucosyl-ceramides (GlcCer), Cer, glycosylinositolphosphoceramides (GIPC), and sphingoid long chain bases (LCB). X-ray structures establish ACD11 global architecture to be a GLTP-fold and reveal the molecular basis for selective recognition of C1P. Point mutation functional analyses support structural mapping showing a cationic residue cluster mediating the selective binding of the C1P phosphate headgroup in a surface-located recognition cavity. An intra-helical distortion, i.e. π-helix (π-bulge) uniquely distinguishes ACD11 from other known GLTP-folds including the recently discovered human ceramide-1-phosphate transfer protein (CPTP) (Simanshu et al., 2013). The π-bulge involves key residues of the C1P recognition center that regulates access and encapsulation of the lipid hydrocarbon chains to an adjoining hydrophobic pocket. In Arabidopsis acd11 null mutant, normally low C1P levels are elevated, while relatively abundant phytoceramides (phyto-Cer) rise acutely, consistent with shifts in the dynamic balance and distributions of these two sphingolipids playing a key role in plant PCD regulation.
RESULTS
ACD11 Forms a GLTP-fold with a Helical π-bulge in its Lipid Headgroup Recognition Center
To experimentally establish if ACD11 forms a GLTP-fold, we crystallized wild-type (wt) protein and determined its structure (1.8 Å) (Tables S1 and S2). ACD11 adopts the two-layer, all α-helical ‘sandwich’ motif characteristic of the GLTP-fold (Figure 1A). Nonetheless, there are differences compared to the human GLTP-fold prototype (Figure S1). At the N-terminus, ACD11 has an extra α-helix (designated αN) that is lacking in human GLTP and is ~35% shorter in human CPTP. Key residues involved in lipid headgroup recognition in ACD11 (cyan) differ in GLTP (Figure 1B; pink) but not in CPTP (Figure 1C; beige) except for conserved Asp60 and His143, residues needed for Cer interaction in all GLTP-folds (Figure S1A; red). The ACD11 C-terminal region does not directly contribute to formation of the headgroup recognition cavity as occurs in GLTP (Figure 1B, red arrow) but resembles the HET-C2 fungal GLTP-fold, which terminates similarly with a Trp residue (Kenoth et al., 2010). In ACD11, C-terminal Trp206 positioning is stabilized by cation-pi interaction with Arg92, but no similar interaction occurs in GLTP or CPTP, which end with Val209 and Pro214, respectively. A relatively small, compact cavity for lipid headgroup binding exists in ACD11 and CPTP, a consequence of the α3–4 loop projecting out and over in hood-like fashion (Figure 1B, blue arrow; 1C). The nearby surface region is highly basic (Figures 1D and 1F) compared to its more neutral counterpart in human GLTP (Figure 1E). A noteworthy and novel structural feature of apo-ACD11 is the π-helix (π-bulge) in helix α2 near Asp60 resulting in close proximity to His143 via a 2.9 Å salt bridge (Figures 1G and 1J). In all other known apo-GLTP-folds including CPTP, no π-bulge occurs and the analogous Asp and His residues remain further apart (Figure 1H and 1I).
Figure 1. Crystal Structure of Arabidopsis ACD11, Comparison with Human GLTP and CPTP Structures, and π-bulge in ACD11 Lipid-headgroup Binding Pocket.
(A) GLTP-fold of apo-ACD11 (ribbon) showing arrangement of α-helices (cyan) and 310-helices (lavender).
(B) Structural superposition of ACD11 (cyan) and human GLTP (pink) comparing side-chains (stick representation) involved in lipid headgroup recognition. Blue and red arrows show insertion loop in ACD11 (lavender highlights) and human GLTP (red highlights), respectively, near the lipid headgroup binding pocket.
(C) Structural superposition of ACD11 (cyan) and human CPTP (tan) comparing side-chains (stick representation) involved in phosphate headgroup recognition.
(D) ACD11 surface electrostatics showing positively-charged region around the lipid headgroup binding cavity; blue, positive charge.
(E) Surface electrostatics of human GLTP bound to 18:1 LacCer (stick representation) showing larger, neutral (white) cavity for binding lipid sugar headgroup.
(F) Surface electrostatics of human CPTP bound to C1P (stick representation).
(G) π-bulge centered on Asp60 promotes salt-bridge formation between Asp60 (α2 helix) and His143 (α5–6 loop).
(H) In human apo-GLTP, there is no π-bulge centered on Asp48 and interaction with His140 occurs via water bridging.
(I) In human CPTP, there is no π-bulge centered on Asp56 and no water mediating interaction with His150.
(J) Stereo view of π-bulge in ACD11 α2 helix. Hydrogen bond types i→i-4 and i→i-5 are shown as black and pink dashed lines, respectively. Interaction between Asp60 and neighboring His143, Lys64 and a water molecule are shown as green dashed lines.
See also Figures S1, S7 and Tables S1–S3.
Crystal Structure of ACD11 in Complex with Lysosphingomyelin
The first tests of ACD11 transfer of glycosphingolipid and related metabolites (Brodersen et al., 2002) preceded crystal structure determination of the human GLTP-fold and mapping of the glycolipid binding site (Malinina et al., 2004). GLTP and ACD11 superpositioning (Figure 1B) reveals a positively-charged residue triad (K64, R99, R103) in ACD11 replacing N52, L92, and W96 in GLTP. This explains the lack of glycolipid transfer by ACD11 and limited transfer of SM, which has a phosphocholine headgroup (Petersen et al., 2008). Thus, initial trials focused on co-crystallization of wtACD11 complexed with SM and lysoSM (Figure 2A). Only the latter lipid yielded a crystal complex enabling 2.4 Å resolution (Figures 2B–2D and Table S1). The expected lipid-headgroup recognition cavity is occupied by a sulfate ion from crystallization solution. Also adsorbed nearby on the protein surface is the sphingoid chain of lysoSM. Notably, the choline headgroup moiety projects outward and away from the protein surface (Figures 2B–D). One phosphate oxygen undergoes hydrogen bonding with the amide nitrogen of Gly144, while the sphingoid base amine hydrogen bonds with Asp60 (Figure 2D). The π-bulge centered at Asp60 (α2 helix) persists in the ACD11/lysoSM complex. At the crystal packing interface of the asymmetric unit, an additional lysoSM molecule is observed (Figure S2A).
Figure 2. Crystal Structure of ACD11 in Complex with Lysosphingomyelin and ACD11 Lipid Transfer Specificity.
(A) Structure of lysosphingomyelin (lysoSM)
(B) Crystal structure of ACD11/lysoSM complex showing ACD11 (ribbon) bound to lysoSM and sulphate (space-filling).
(C) Surface electrostatics of ACD11 in complex with lysoSM.
(D) Lipid headgroup recognition centre residues (lavender) interacting with lysoSM (blue, ball-and-stick) and sulphate. Dashed lines show hydrogen bonds.
(E) Lipid transfer in vitro by Förster resonance energy transfer.
(F) Quantification of initial lipid transfer rates in (E).
(G) Competition against AV-C1P transfer by nonfluorescent lipids. 18:1-C1P competes strongly; lysoSM, moderately; S1P, weakly; and IPC, nearly nil (see Figures S3C–F for kinetic traces)
See also Figures S2, S3 and Tables S1 and S3.
ACD11 is a Ceramide-1-phosphate Transfer Protein
Because plants contain no SM and do not produce this sphingolipid, SM transfer by ACD11 was surprising, suggesting that SM serves as a substitute analog for the plant lipid preferred in vivo (Petersen et al., 2008). Also, as noted earlier, the lipid headgroup binding cavity is relatively small, compact, and hood-like (Figure 1B, blue arrow), an arrangement expected to poorly accommodate the bulky SM phosphocholine headgroup. With that in mind, wild-type ACD11 was analyzed for intermembrane transfer of other sphingolipids and phosphoglycerides. A Förster resonance energy transfer approach involving probe lipids with acyl-linked anthrylvinyl fluorophore (AV) enabled testing of lipids with phosphate headgroups, i.e. AV-phosphatidic acid (AV-PA) and AV-ceramide-1-phosphate (AV-C1P), and controls, i.e. AV-galactosylceramide (AV-GalCer), AV-SM and AV-Cer. ACD11 robustly transferred AV-C1P (sphingoid-based) but not AV-PA (glycerol-based) (Figures 2E and 2F). Notably, ACD11 also transferred AV-phyto-C1P (Figures 2E and 2F), as expected by modeling of phyto-C1P docking in the ACD11 binding site. The sphingoid chains of “phyto” sphingolipid derivatives that predominate in plants lack the 4,5 trans double bond but contain a 4-hydroxy group (Markham et al., 2006; Markham et al., 2013). The AV-C1P and AV-phyto-C1P transfer rates depended on protein concentration, required acceptor membranes (Figures 3B, S3A, and S3B), and proceeded at ~4.5 C1P and ~5.6 phyto-C1P molecules/min/protein. Replacement of phosphate with sugar (AV-GalCer) prevented transfer by ACD11, but not by GLTP. AV-SM transfer by ACD11 was very slow (Figure 2E). The lack of AV-Cer transfer suggested a requirement for phosphate in the head-group for functionality. This was confirmed by competition assays AV-C1P transfer by lipids containing natural hydrocarbon chains (Figure 2G). Only nonfluorescent C1P competed strongly against AV-C1P (Figure S3C). IPC, S1P, lysoSM, and N-hexyl(6:0)-SM (not shown) exerted differing weak competition (Figures 2G and S3D–F). PA and lysoPA minimally slowed the initial AV-C1P transfer rate and were not effective competitors (Figure S3G).
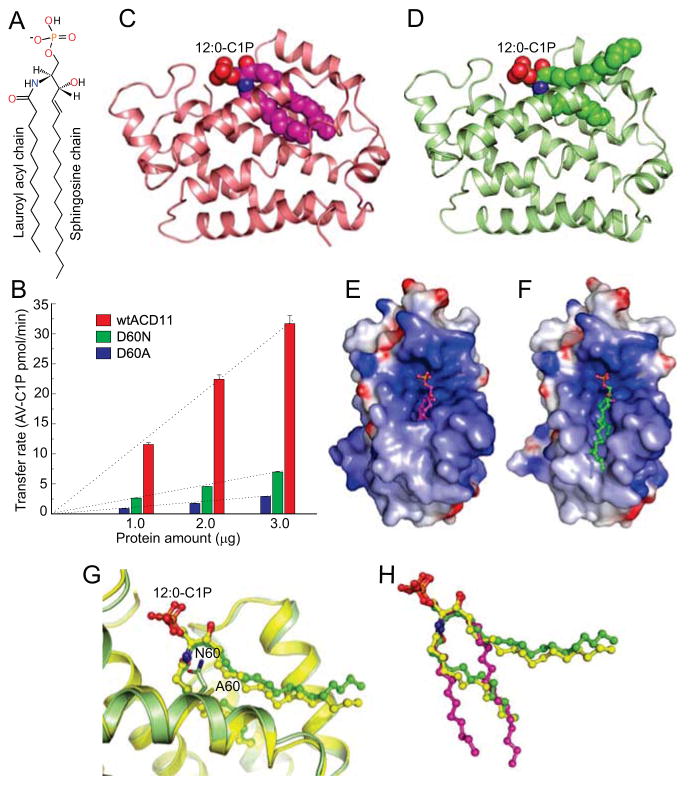
Figure 3. Crystal Structures of D60N/A-ACD11 in Complex with 12:0-C1P and Their C1P Transfer Activities.
(A) Structure of 12:0-C1P.
(B) C1P initial transfer rates by wt-ACD11 (red), D60N-ACD11 (green), and D60A-ACD11 (blue) using 3 μg each.
(C) D60N-ACD11 (ribbon) in complex with 12:0-C1P showing the acyl and sphingosine chains both buried in the hydrophobic pocket (sphingosine-in mode; space-filling) in one molecule of the crystal asymmetric unit.
(D) D60N-ACD11 (ribbon) in complex with 12:0-C1P (space-filling) with the acyl chain buried in hydrophobic pocket and the sphingosine chain adsorbing to the protein surface (sphingosine-out mode) in the second molecule of the crystal asymmetric unit.
(E) Surface electrostatics of D60N-ACD11 with bound 12:0-C1P (sphingosine-in mode; ball-and-stick).
(F) Surface electrostatics of D60N-ACD11 with bound 12:0-C1P (sphingosine-out mode).
(G) Structural superposition of C1P headgroup binding pocket of D60N-ACD11 (green) and D60A-ACD11 (yellow) in complex with 12:0-C1P showing disappearance of π-bulge in α2 helix.
(H) Structural comparison of 12:0-C1P binding to D60N-ACD11 (sphingosine-in, magenta and sphingosine-out, green) and to D60A-ACD11 (sphingosine-out, yellow).
See also Figures S4, S7 and Tables S1 and S3.
Crystal Structure of D60N/A-ACD11 with Bound Ceramide-1-phosphate
Due to the high transfer specificity for C1P, extensive co-crystallization trials were initiated for wt-ACD11 and C1P, but no positive outcome ensued. To achieve success, a point mutation strategy was used to weaken the Asp60-His143 salt-bridge associated with the π-bulge. We focused on Asp60 because mutation of the analogous Asp (D48V) in human GLTP is reasonably well tolerated (Samygina et al., 2011). Asp60 was mutated to residues expected to weaken (Asn) or eliminate (Ala) salt bridging with H143. The D60N-ACD11 mutant maintained ~25–30% activity, while D60A-ACD11 was ~10–15% active compared to wt-ACD11 (Figure 3B).
Both the D60N and D60A mutants yielded crystal complexes with N-dodecanoyl-C1P (12:0-C1P) (Figure 3; Tables S1 and S3) but not with other lipids (e.g. S1P, sphingosine, SM, lysoSM, or PA). In the D60N-ACD11/12:0-C1P crystal complex, the asymmetric unit consists of two ACD11 molecules containing 12:0-C1P bound in two ways (Figures 3C–3F). In one case, both the sphingosine and lauroyl acyl chain of 12:0-C1P are encapsulated in the hydrophobic pocket (Figures 3C and 3E). In the other case, only the lauroyl acyl chain is inserted into the hydrophobic pocket, while the sphingosine chain remains outside the pocket (Figures 3D and 3F). With D60A-ACD11/12:0-C1P complex, a different crystal form was observed involving one protein molecule with bound 12:0-C1P (Figure 3G; Tables S1 and S3).However, the overall structure resembled the sphingosine-out binding mode displayed by D60N-ACD11/12:0-C1P complex (Figures 3G and 3H). Similar sphingosine-out conformers have been observed in human GLTP complexed with glycosphingolipids (Malinina et al., 2006; Samygina et al., 2013; Samygina et al., 2011) and in human CPTP complexed with C1P (Simanshu et al., 2013). Similar positioning of different C1P species occurs in the hydrophobic pockets of ACD11 and CPTP except for the obvious differences in the bending angle of the outwardly projecting sphingoid chain in the sphingosine-out binding mode (Figure S4). The bending of sphingosine occurs immediately distal to the 4–5 trans double bond where carbon-carbon single bonds exist and torsional rotation is unrestrained, providing conformational optimization for packing at the crystal contact faces of ACD11 and CPTP.
Recognition of C1P by ACD11
It is noteworthy that the π-bulge in apo-ACD11 (helix α2) disappears upon binding of C1P in both D60N- and D60A-ACD11, presumably reflecting C1P-induced conformational changes related to portal opening and entry of one or both C1P hydrocarbon chains into the hydrophobic pocket (Figures 3G, 4A and 4B). In both 12:0-C1P conformer complexes with D60N- and D60A-ACD11, the C1P phosphate group is anchored to positively-charged residues on the protein surface via interaction of the three phosphate oxygen atoms with Lys64, Arg99 and Arg103 (Figure 4C). Their functional importance is illustrated by severe reductions in C1P transfer for the K64A, R99E, R99A, and R103A point mutants (Figure 4F). The C1P amide moiety hydrogen bonds with His143 and Asn60. The net effect is disruption of the stabilizing salt-bridge and elimination of the π-bulge characteristic of apo-ACD11.
Figure 4. π-helix Transition to α-helix Induced by C1P Binding in D60N-ACD11 and Mapping of C1P Binding Site.
(A) Stereo representation of α2 helix in the D60N-ACD11/12:0-C1P crystal complex showing absence of π-bulge.
(B) Stereo view of structural superposition of α2 helix observed in apo-ACD11 (cyan) forming a π-bulge and in D60N-ACD11 (salmon) in complex with 12:0-C1P structure without a π-bulge. For clarity, only side chains atoms of Asp60 and His143 are shown.
(C) 12:0-C1P phosphate and amide interactions with D60N-ACD11 residues in the headgroup binding cavity. Hydrogen bonds are shown by dashed lines.
(D) ACD11 residues forming the hydrophobic pocket that accommodates the C1P acyl and sphingosine chains.
(E) Localized conformational changes in apo-ACD11 and D60N-ACD11. Structural superposition of apo-ACD11 and D60N-ACD11/12:0-C1P complex showing residues undergoing large conformational changes during accommodation of C1P hydrocarbon chains. Bound C1P atoms (ball-and-stick) are colored magenta, red, and blue for carbon, oxygen, and nitrogen, respectively. apo-ACD11 and D60N-ACD11 side chains are colored cyan and salmon, respectively.
(F) C1P intervesicular transfer by ACD11 point mutants measured using Förster resonance energy transfer.
The hydrophobic pocket that accommodates the sphingosine and acyl chains of C1P is formed by a cluster of nonpolar residues, i.e. Leu, Val, Ile, Phe, Met, Ala and Tyr, that line the two layers of the α-helices in ACD11 (Figure 4D). Insertion and encapsulation of the 12:0-C1P hydrocarbon chains results in disappearance of the intra-helical π-bulge. This π-helix to α-helix structural transition involves large conformational changes for the side-chains of several residues, i.e. Phe47, Phe54, Phe56 and Leu50 (Figure 4E), which move towards the protein surface. This effectively expands the hydrophobic pocket and creates space to accommodate the hydrocarbon chains of 12:0-C1P. Introduction of polarity into the hydrophobic pocket by point mutation (F47Q-ACD11) leads to diminished activity (Figure 4F) affirming the importance of the hydrophobic environment. In the ‘sphingosine-out’ structures, the nonpolar amino acids between helices α5 and α6 interact with the sphingoid chain via hydrophobic and van der Waals interactions enabling adsorption to the protein surface when encapsulation by the hydrophobic pocket does not occur.
Crystal Structure of D60A-ACD11 in Complex with N-acetyl-C1P
To define C1P features that contribute to the π-helix-to-α-helix transition needed for C1P chain insertion into the hydrophobic pocket, D60A-ACD11 was co-crystallized with bound N-acetyl(2:0)-C1P (Figures 5A–5E; Tables S1 and S3). As expected, the phosphate headgroup hydrogen bonds with Lys64, Arg99 and Arg103, but the bidentate hydrogen bonding of Arg99 observed with 12:0-C1P (Figure 4C) is reduced to a single hydrogen bond (Figure 5C). The acetyl chain amide group is unable to hydrogen bond with Ala60 and fails to hydrogen bond with His143 (Figure 5C) leaving the acetyl group on the surface, turned away from Ala60 and outside the hydrophobic pocket. The sphingosine chain also remains outside the hydrophobic pocket (Figures 5B and 5E) adsorbed between helices α5 and α6 on the protein surface. In this altered ‘binding state’, the π-bulge persists suggesting that transitioning of π-helix to α-helix is enhanced when the C1P acyl chain is long enough to enter the hydrophobic pocket (Figure 5D). From the functional standpoint, N-acetyl-C1P competes poorly against AV-C1P transfer by ACD11 as also is the case for S1P (Figure 5F). By contrast, N-acetyl-C1P competes moderately well against CPTP-mediated AV-C1P transfer (Simanshu et al., 2013), and the structure of the CPTP/N-acetyl-C1P complex shows no missing hydrogen bond interactions with the C1P phosphate-amide region, proper engagement of the N-acetyl group in the binding cleft, and encapsulation of the sphingoid chain in the hydrophobic pocket. The differences in position and conformation of bound N-acetyl-C1P molecules in ACD11 and CPTP are shown in Figure S5.
Figure 5. Crystal Structure of D60A-ACD11 in Complex with N-acetyl-C1P.
(A) Structure of 2:0-C1P
(B) Crystal structure of D60A-ACD11 (ribbon representation) in complex with 2:0-C1P (space filling) in sphingosine-out conformation.
(C) Inverted (flipped) orientation of lipid amide-acetyl group in sphingosine-out binding mode of 2:0-C1P complexed with D60A-ACD11.
(D) Enlarged view of α2 helix showing π-bulge in the superposed structures of apo-ACD11 and D60A-ACD11 in complex with 2:0-C1P.
(E) Surface electrostatics of D60A-ACD11/2:0-C1P complex (stick representation).
(F) Competition against ACD11-mediated AV-C1P transfer by 2:0-C1P.
See also Figure S5 and Tables S1 and S3.
Perturbations of Sphingolipid Levels in Arabidopsis acd11 Mutants
To elucidate whether ACD11 involvement in Arabidopsis PCD manifests itself by altering sphingolipid metabolism, sphingolipid levels were profiled in dying leaves of homozygous acd11-1 mutants. An overall accumulation of total sphingolipids (Figure 6A) including long-chain bases (LCBs) (Figure S6A) is evident in acd11 compared to the Landsberg erecta (Ler) wild-type background, with total free ceramides showing the greatest elevation. In plants, the dominant ceramide species (>90%) are phyto-Cers (Markham et al., 2006; Markham et al., 2013) which reportedly are more potent inducers of PCD than Cer (Hwang et al., 2001). To verify and differentiate between effects caused by spontaneous cell death and reduced growth in acd11, we monitored sphingolipid levels upon PCD induction in acd11/NahG plants at 0, 12, 24, 72, and 120 hours after treatment with benzo(1,2,3)thia-diazole-7-carbothioic acid (BTH), a salicylic acid (SA) analog. Introduction of the bacterial transgene NahG into the acd11 background removes endogenous SA needed for development of the cell death phenotype. When acd11/NahG plants are then treated with BTH, cell death is fully reinstated. By 72 and 120 hours after BTH treatment (Figure 6B), a large increase in total Cer is evident compared to Ler and NahG controls, as well as a minor rise in 2-hydroxyceramide, which may reflect free Cer hydroxylation or increased sphingolipid turnover. In contrast, levels of GlcCer and GIPC remain largely unaltered (Figure 6B), suggesting that their increase in acd11 (Figure 6A) is probably due to reduced growth or the dwarf phenotype. Changes in levels of LCB(P), i.e. sphingoid-1-phosphates, also are insignificant in acd11 (Figure S6B), and thus, may take more time or require stronger inductive conditions (e.g. higher BTH/SA levels) to develop. C1P, which occurs at extremely low levels at normal growth temperature, was not detected. The observed perturbations of sphingolipid levels reveal Cer accumulation during development of acd11 cell death, suggesting that ACD11 mediates Cer synthesis in a SA-dependent manner.
Figure 6. Constitutive and Inducible Alterations in Sphingolipid Content of acd11 Mutants Grown at Normal Temperature.
(A) Sphingolipids in leaves of dying acd11-1 plants compared to the Ler wild-type control. Data are shown as four sphingolipid classes including phyto- and nonphyto- species (ceramides, Cer; hydroxyceramides, hCer; glucosylceramides, GlcCer; and glycosylinositolphosphorylceramides, GIPC). Data represent the mean ± SD (n=3) and significant differences from the control are indicated by asteriks (**P<0.01, *P<0.05) based on Students’s t test. dw, dry weight.
(B) Sphingolipid contents in leaves of acd11-1/NahG plants 0, 12, 24, 72, and 120 h after BTH treatment in comparison with Ler NahG and wild-type controls. Plants were grown for five weeks prior to BTH treatment. Sphingolipid contents are shown as in (A). Data represent the mean ± SE (n=3), and letters indicate statistically different groups using one-way ANOVA with groupings by Tukey’s HSD using 95% confidence interval.
See also Figure S6.
Because cold temperature treatment of Arabidopsis induces substantial and rapid elevation of C1P and LCB(P) by a transduction process regulated by endogenous nitric oxide (Cantrel et al., 2011), the responses of C1P and related sphingolipids to reduced temperature were analyzed in the acd11 background. Figure 7A shows that acd11 loss-of-function results in 3-to 5-fold elevations in the levels of different C1P species of plants subjected to cold treatment. To our knowledge, these are the first quantitative determinations of C1P mass levels in plants, which previously were detected by radiolabeling (Cantrel et al., 2011). Also evident are moderate increases in LCBP, but not LCB (Figure 7B) and dramatic elevations of phyto-Cers (Figure 7C). Interestingly, with phyto-Cer, the palmitoyl (16:0) species and very long acyl species (22–26 carbons) are most affected; whereas, the elevations with C1P are shifted to the moderately long acyl species (16–22 carbons). These changes are consistent with a complex regulatory mechanism involving ACD11.
Figure 7. Constitutive and Inducible Alterations in Sphingolipid Content of acd11 Mutants in Response to Cold-Temperature Treatment.
(A–C) Sphingolipid contents of acd11, acd11-1/NahG, and control plants 72 h after treatment with BTH. Plants were grown for five weeks and subjected to cold (4°C) for 4 h prior to BTH treatment. A) C1P; B) LCB(P); C) PhytoCer. Data represent the mean ± SE (n=3) and significant differences from controls are indicated by asteriks (**P<0.01, *P<0.05) based on Students’s t test.
DISCUSSION
Unique Structural Aspects of the Lipid Headgroup Recognition Center and Hydrophobic Pocket of the ACD11 GLTP-fold
Despite low sequence homology of ACD11 and other GLTP homologs including human CPTP (Figure S1A), our crystallographic data establish the conserved structural homology shared by Arabidopsis ACD11, human GLTP, and human CPTP (Simanshu et al., 2013) while revealing important differences. Unlike GLTP but like CPTP, ACD11 contains a modified lipid headgroup recognition center that selectively binds C1P, an important signaling lipid linked to cell survival. Comparison of ACD11, GLTP, and CPTP with their preferred bound lipids suggests adaptation and evolutionary conservation of key residues in their GLTP-folds. The net outcome is two divergent subfamilies within the GLTP superfamily. Residues adapted to focus lipid specificity to C1P in ACD11 include Lys64 for Asn52, Arg99 for Leu92, and Arg103 for Trp96 in GLTP. The clustered Lys/Arg residues of ACD11 form a positively-charged triad that is ideally arranged for binding phosphate, explaining the inability of ACD11 to bind sugar headgroups and transfer glycolipids (Petersen et al., 2008). It is noteworthy that Arg103 occupies the same position where Trp acts as a stacking plate for the Cer-linked headgroup sugar in human GLTP, fungal HET-C2, plant GLTP1, and human FAPP2 (Kamlekar et al., 2013; West et al., 2008). Conversely, residues analogous to Asp60 and His143 of ACD11 are absolutely conserved in the lipid headgroup recognition centers of all known GLTP-folds. The ‘pincher-like’ clamping that occurs when Asp and His hydrogen bond with the Cer moiety amide nitrogen and oxygen ensure a highly conserved and oriented entry of the sphingolipid hydrocarbon chains into the hydrophobic pocket regardless of lipid headgroup composition. The X-ray data rectify earlier 3D homology modeling involving identification of key residues of the ACD11 lipid headgroup recognition center and location of the C-terminus, i.e. Trp206 (Airenne et al., 2006).
A unique feature of the apo-ACD11 GLTP-fold compared to other known GLTP-folds including CPTP is the presence of π-helix, i.e. π-bulge, in helix α2 near the entrance portal of the lipid binding cleft (Figures 1G and 1J). π-bulges exist in only 15% of known proteins but often at locations that enhance/regulate function (Cartailler and Luecke, 2004; Cooley et al., 2010). The π-bulge in apo-ACD11 brings Asp60 and His143 sufficiently close (2.9 Å) to form a salt bridge (Figure 1G), thus providing a potential regulatory mechanism for the ACD11 GLTP-fold. In other GLTP-folds, a water molecule often bridges the Asp and His residues (Figure 1H). In apo-ACD11, the Asp60-His143 salt-bridge created by the π-bulge appears to tightly seal the entry portal region of the hydrophobic pocket (Figure 1G). In D60A-ACD11, the π-bulge persists after binding 2:0-C1P but not 12:0-C1P, suggesting that salt-bridge disruption between Asp60 and His143 by itself is insufficient to induce the π-helix-to-α-helix conformational change needed for the ACD11/C1P complex to become ‘transfer viable’. In addition, the C1P acyl chain needs to be longer than only two carbons. This conclusion is supported by the structure of wt-ACD11 complexed with lysoSM, which has no acyl chain, but displays a bound conformation resembling that of 2:0-C1P in D60A-ACD11 (Figures 5 and S2C). LysoSM is tethered to the surface via its amine group interacting with Asp60, while a sulfate anion occupies the lipid headgroup (phosphate) binding pocket lending credence to the authenticity of the lysoSM binding site. Analogous behavior is observed in human GLTP/hexyl glucoside crystal complexes where the sugar headgroup occupies the glycolipid recognition center despite weak binding affinity and no measureable transfer (Malinina et al., 2006; Zhai et al., 2009). In the ACD11/lysoSM complex, occupation of the phosphate headgroup recognition center by the sulfate anion and the bulky, zwitterionic nature of the phosphocholine lipid headgroup are likely contributors to its minimal interaction and outward projection from the protein (Figure S2A). Similar conformation and surface localization are observed for the 2:0-C1P sphingosine chain in complex with D60A-ACD11 (Figure S2). Thus, despite seemingly adequate positioning on the ACD11 surface, π-bulge persistence renders the lipid binding interaction insufficient to drive robust transfer.
For ACD11 to become fully ‘transfer viable’, uptake of the sphingolipid acyl chain into the hydrophobic pocket and repositioning of specific residues appear to be required. At the molecular level, π-bulge formation at Asp60 results in the Phe56 nonpolar phenyl ring projecting into the hydrophobic pocket to function as a ‘portal gate’ that swings open during lipid acyl chain uptake (Figure 4E). Phe54 orients into the hydrophobic pocket providing conformational stability to apo-ACD11 in the absence of a lipid acyl chain. When C1P contains a sufficiently long acyl chain (e.g. 12:0-C1P), the acyl chain enters deep into the hydrophobic pocket, as shown for D60N-ACD11 and D60A-ACD11. A ‘peristaltic-like shift’ of Ala57 to occupy the position of Phe56 as well as Phe54 being pushed outwards accompanies transformation from π-bulge to α-helix, enabling hydrophobic pocket formation/expansion sufficient to accommodate either one or both hydrocarbon chains of Cer (Figures 4D and 4E). The key role played by Phe56 of helix α2 in functioning as a ‘portal gate’ represents a fundamental difference between the ACD11 GLTP-fold and human GLTP-fold, which uses an ‘oppositely-located’ Phe (Phe148 of helix α6) as the ‘portal gate’ that swings open during hydrocarbon chain insertion (Malinina et al., 2004; Samygina et al., 2011). The global folding topology of ACD11 and conformational adaptability of its flexible, single-cavity, hydrophobic pocket contrast with Cer transfer protein, which uses an α/β fold built around an incomplete U-shaped β-barrel to bind Cer via a START domain lipid cavity (Kudo et al., 2008) (see Supplemental Discussion).
ACD11 Modulates Arabidopsis PCD by Intermediary Regulation of Sphingolipid Levels
The hypersensitive response (HR) in plants generates localized cell death to minimize the spread of pathogens. HR-like PCD also is exhibited by the recessive Arabidopsis acd11 mutant. Despite the known ties between ACD11 and HR-like PCD (Brodersen et al., 2002), determination of the molecular structure and lipid specificity of ACD11 remained unclear until now. Establishment of ACD11 architecture as a C1P-selective GLTP-fold capable of binding/transferring either C1P or phyto-C1P at similar rates provides insights into how this GLTP superfamily member impacts PCD-related processes regulated by key sphingolipid metabolites. While fungal GLTP (HET-C2) and human FAPP2 (C-terminal GLTP-like domain) have both been implicated in PCD-related processes (Fedorova et al., 2005; Paoletti and Clave, 2007; Tritz et al., 2009), no sphingolipid analyses were performed upon in vivo depletion of these glycosylceramide-selective GLTP-folds.
Compared to mammals, plant membranes show fundamental differences in sphingolipid content including large contributions by GlcCer, GIPC, and modified sphingoid chains (e.g. phyto-derivatives) as well as a lack of sphingomyelin and gangliosides (Markham et al., 2006; Markham et al., 2013; Pata et al., 2010). Adjustment to major lipid content during cold acclimation is well established in plants. Recent studies also show rapid elevations of low level LCB(P)s and C1P (Cantrel et al., 2011). The established signaling functions of such lipids make them leading candidates as early signals during cold acclimation. Because LCB(P)s and C1P content is low under both normal and cold-stressed growth conditions, radiolabeling was initially needed for detection (Cantrel et al., 2011). Determination of LCB(P) and C1P derivative mass levels has been challenging, a situation exacerbated by a dearth of authentic standards (Markham and Jaworski, 2007). This has been especially true for C1P derivatives, which had not been mass quantified in plants until the present study.
Our finding that acd11 deficiency not only alters C1P levels, but also acutely elevates phyto-Cer levels (and LCBP to a lesser extent) establishes a functional link between acd11 expression and sphingolipid metabolic regulation in plants where the dynamic balance between Cer and C1P appears to be critical for regulating PCD (Chen, 2009; Liang et al., 2003; Pata et al., 2010; Reape and McCabe, 2008). While elevated C1P levels induced by acd11 disruption in Arabidopsis also are observed upon RNAi-induced depletion of the ACD11 ortholog CPTP in human cells, it is noteworthy that the dramatic elevations in phyto-Cer levels in acd11 mutants are not duplicated in the Cer levels of CPTP-depleted human cells (Simanshu et al., 2013). This suggests some differing aspects of ACD11 involvement in the regulation of sphingolipid metabolism in plants. Elucidating the mechanistic details and associated kinetics of this involvement will first require detailed analyses of plant sphingolipid metabolic pathways, related regulatory signaling pathways, and changes triggered during cold acclimation.
What is known is that the acd5 mutant lacks Cer kinase (CerK) activity, accumulates Cers, and exhibits PCD (Liang et al., 2003). ACD11 may act in concert with ACD5 (CerK) to maintain the balance of Cer and C1P levels, thus controlling HR-associated PCD. In this context, the loss of IPC synthase activity (erh1 mutant) also results in total Cer accumulation, and both erh1 and acd5 exhibit enhanced HR-associated cell death triggered by the RPW8 resistance protein (Wang et al., 2008). However, Cer accumulation and cell death in acd11, acd5, and erh1 are dependent on the phytohormone, salicylic acid (SA). This suggests that perturbations in sphingolipid metabolism, such as occur in acd11, may regulate SA levels or signaling during R gene mediated HR. Plant R proteins confer recognition of pathogen avirulence proteins and trigger effective innate immune responses (e.g. HR). A genetic screen for suppressors of acd11 cell death (laz mutants) identified the R gene LAZ5. Thus, the absence of ACD11 in acd11 leads to inappropriate HR activation by LAZ5 (Palma et al., 2010). Since sphingolipids are important in both microbial pathogenesis and host defense (Heung et al., 2006), LAZ5 may ‘guard’ ACD11 function(s) in the sphingolipid metabolism that are targeted by pathogen effectors. Also, transgenic expression of human wt-GLTP and D48V-GLTP (Petersen et al., 2008) suppresses acd11 cell death raising the possibility that the C1P binding/transfer activity of ACD11 is partially dispensable for PCD suppression. This could implicate LAZ5 as a response amplifier that triggers the HR when the local distribution and balance between phyto-Cer and C1P is disturbed, thereby intensifying the response through SA accumulation. Pathogen effector-induced modification or loss-of-function of ACD11 could interfere with normal sphingolipid distribution and trigger a defense response strong enough to deter microbial colonization. Since sphingolipid bases are up-regulated early during an infection or HR, it is probable that sphingolipids are signaling mediators, and not the de facto cell death inducers via membrane perturbations (Mackey et al., 2003; Peer et al., 2010). Testing this hypothesis will require research to clarify the interplay between cellular sphingolipid metabolism and basal immunity in plants.
It’s also possible that loss of ACD11 as a selective carrier blocks C1P exit from the Golgi (or ER) resulting in organelle stress, as occurs in human cells depleted of the ACD11 ortholog, CPTP (Simanshu et al., 2013), and leading to local accumulation of Cer that alters membrane component organization. As ACD11 may be indirectly guarded (Palma et al., 2010), triggering of HR cell death may not rely directly on the absence of ACD11 in the acd11 mutant. In mammals, nonmicrobial ‘danger signals’ instigate obesity-induced inflammation via NLRP3 which senses increasing Cer and induces apoptosis (Vandanmagsar et al., 2011), providing a potential clue as to how the HR might be induced in acd11 by LAZ5 via detection of the accumulation of specific sphingolipid species. Future localization studies on ACD11 and LAZ5 to evaluate possible co-restriction in specific organelles could provide more insights.
EXPERIMENTAL PROCEDURES
Cloning, Expression and Purification
Arabidopsis acd11 ORF (NCBI NP_181016.1) expression in BL21(DE3) pLysS cells using pET-SUMO vector (Invitrogen) enabled Ni2+-NTA affinity chromatography purification of ACD11 N-terminally-tagged with His6-SUMO (see Supplemental Experimental Procedures). Pure proteins were either used for crystallization immediately or flash frozen in liquid N2 and stored at −80 °C. ACD11 mutants were generated by PCR-based overlap extension and confirmed by DNA sequencing. Expression/purification was the same as for wt-ACD11.
Crystallization
A Mosquito crystallization robot (Molecular Dimensions) was used for initial co-crystallization screening of wt-ACD11 and the D60N and D60A mutants with lysoSM and C1P species (see Supplemental Experimental Procedures). Positive hits were optimized using hanging drop vapor diffusion method by varying pH and concentration of individual components (Table S3). For data collection, crystals were flash frozen (100 K) in crystallization condition containing 25% (v/v) ethylene glycol. Diffraction datasets were collected on 24-ID-C and 24-ID-E beamlines at the Advanced Photon Source and X29A beamline at National Synchrotron Light Source. Collected datasets were integrated and scaled using the HKL2000 suite (Otwinowski and Minor, 1997). All crystals have different packing interactions leading to different unit cell dimensions and space groups (Table S1).
Structure Determination and Refinement
Ab initio phasing was obtained by soaking apo-ACD11 crystals in 1 mM ethyl mercuric phosphate for 24 h and collecting data (2.45 Å resolution) using a Rigaku RU-H3R X-ray generator equipped with a RAXIS-HTC detector (Table 2). ACD11 structure was determined by the SIRAS method using Hg isomorphous as well as anomalous scattering data, with the 8 Hg sites located and refined for phasing using SHARP (Vonrhein et al., 2007). Apo-ACD11 crystals belonged to space group P212121 and contained four protein molecules per asymmetric unit. The structures of ACD11-sphingolipid complexes were solved by molecular replacement (MOLREP program) using apo-ACD11 (Hg derivative) structure as the search model (Vagin and Teplyakov, 1997) (see Supplemental Experimental Procedures).
ACD11 Intermembrane Lipid Transfer Activity
Förster resonance energy transfer provided kinetic insights into lipid transfer by ACD11. Donor POPC vesicles, containing 1 mole% AV-lipid acylated with (11E)-12-(9-anthryl)-11-dodecenoate and 1.5 mole% 1-acyl-2-[9-(3-perylenoyl)-nonanoyl]-3-sn-glycero-3-phosphocholine [Per-PC] were prepared by rapid ethanol injection (Mattjus et al., 1999). In competition assays, donor vesicles contained 1.5 mol% AV-C1P (Boldyrev et al., 2013) as well as 0.5, 1.0 or 2.0 mole% competitor lipids (Samygina et al., 2011). Both fluorescent lipids were present initially only in donor vesicles where minimal AV emission occurs upon excitation (370 nm) because of energy transfer to Per-PC. ACD11 addition results in an exponential increase in AV emission intensity as the protein transports AV-C1P from donor vesicles (creating separation from the ‘nontransferable’ Per-PC) and delivers to the 10-fold excess POPC acceptor vesicles. The time-dependent increase in AV emission at 425 nm, relative to baseline fluorescence in the absence of ACD11, yields the AV-C1P transfer kinetics (see Supplemental Experimental Procedures).
Plant Material and Sphingolipid Analyses
Arabidopsis acd11 (acd11-1), acd11/NahG and NahG plants in Landsberg erecta (Ler) background have been described (Brodersen et al., 2005). For sphingolipid analyses, plants were grown in soil under short days (8 h light/16 h dark) in chambers at 150 mE/m2s, 21°C and 70% relative humidity. Ler wild type and acd11-1 mutants were grown untreated for 4 weeks before sampling. acd11-1/NahG together with NahG and Ler plants were grown for five weeks prior to spraying with the SA analog BTH (100 μM) and sampling after 0, 12, 24, 72, and 120 hours. Leaf material was harvested from three biological replicates for each genotype and time point. Sphingolipid analysis was performed by mass spectrometry (Bielawski et al., 2009; Markham and Jaworski, 2007). Free and total LCB were analyzed by HPLC after fluorescent derivatization (Bach et al., 2008).
Supplementary Material
HIGHLIGHTS.
The acd11 mutant of Arabidopsis provides a genetic model for cell suicide in plants
ACD11 forms a GLTP-fold and functions as a ceramide-1-phosphate transfer protein
Apo-ACD11 is a unique GLTP-fold due to a π-bulge located near the lipid binding site
In acd11 mutants, ceramide-1-phosphate increases but phytoceramide rises acutely
Acknowledgments
This research was supported by NIH/NIGMS GM45928 (REB), NIH/NCI CA121493 (DJP & REB), Danish Strategic Research Council 09-067148 (JWM); NSF/MCB 0843312 (JEM); NIH/NCRR C06 RR018823 (JB/AB); Spanish Ministerio de Ciencia e Innovacion BFU2010-17711 (LM), Russian Foundation for Basic Research #12-04-00168 (JGM); Hormel Foundation (REB), Abby Rockefeller Mauze Trust (DJP), and Maloris Foundation (DJP). We thank J. Peter Slotte (Åbo Akademi Univ) for the IPC lipid, Helen Pike (UMN-Hormel Institute) for purifying protein used for transfer activity analyses and the staff of X-29 beamline at the National Synchrotron Light Source and ID-24-C/E beamlines at the Advanced Photon Source for help. The Lipidomics Shared Resource, MUSC is partially supported by P30 CA138313, HCC and P20 RR017677, SC COBRE in Lipidomics and Pathobiology.
Footnotes
ACCESSION NUMBERS
The atomic coordinates and structure factors for the crystal structures of Arabidopsis wt-ACD11 and mutants in complex with various lipids are deposited in the Protein Data Bank. The accession codes are: apo-ACD11 (4NT1), ACD11/lysoSM (4NT2), D60N-ACD11/12:0-C1P (4NTI), D60A-ACD11/12:0-C1P (4NTG) and D60A-ACD11/2:0-C1P (4NTO).
Supplemental Information includes seven figures, three tables, discussion, experimental procedures, and references and can be found with this article online at:
Author Contributions
DKS: all structural analyses and provided definitive evidence for C1P binding by ACD11; generated all ACD11 point mutants; wrote text. XZ: transfer analyses of wt-ACD11 and ACD11 point mutants; wrote text. DM and DH: prepared Arabidopsis mutants for sphingolipidomic analyses; wrote text. JEM: time-based sphingolipidomic analyses on Arabidopsis mutants; wrote text. JB/AB: sphingolipodomic analyses on cold-treated Arabidopsis mutants. JGM: synthesized fluorescent lipids. LM: contributed to structural data interpretation. JWM: directed set-up of Arabidopsis mutant analyses; finalized the write-up. DJP: directed ACD11 structural analyses; finalized the write-up. REB: directed functional and structural analyses, finalized the write-up, coordinated and integrated all section write-ups.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Airenne TT, Kidron H, Nymalm Y, Nylund M, West G, Mattjus P, Salminen TA. Structural evidence for adaptive ligand binding of glycolipid transfer protein. J Mol Biol. 2006;355:224–236. doi: 10.1016/j.jmb.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, Marion J, Da Costa M, Boutin JP, Miquel M, Tellier F, et al. The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA. 2008;105:14727–14731. doi: 10.1073/pnas.0805089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkey R, Bendigeri D, Xiao S. Sphingolipids and plant defense/disease: the “death” connection and beyond. Frontiers Plant Sci. 2012;3:68. doi: 10.3389/fpls.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski J, Pierce JS, Snider J, Rembiesa B, Szulc ZM, Bielawska A. Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods Mol Biol. 2009;579:443–467. doi: 10.1007/978-1-60761-322-0_22. [DOI] [PubMed] [Google Scholar]
- Boldyrev IA, Brown RE, Molotkovsky JG. An expedient synthesis of fluorescent labeled ceramide-1-phosphate analogues. Russ J Bioorg Chem. 2013;39:539–542. doi: 10.1134/S106816201305004X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Malinovsky FG, Hematy K, Newman MA, Mundy J. The role of salicylic acid in the induction of cell death in Arabidopsis acd11. Plant Physiol. 2005;138:1037–1045. doi: 10.1104/pp.105.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jorgensen LB, Brown RE, Mundy J. Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 2002;16:490–502. doi: 10.1101/gad.218202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RE, Mattjus P. Glycolipid transfer proteins. Biochim Biophys Acta. 2007;1771:746–760. doi: 10.1016/j.bbalip.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrel C, Vazquez T, Puyaubert J, Reze N, Lesch M, Kaiser WM, Dutilleul C, Guillas I, Zachowski A, Baudouin E. Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytol. 2011;189:415–427. doi: 10.1111/j.1469-8137.2010.03500.x. [DOI] [PubMed] [Google Scholar]
- Cartailler JP, Luecke H. Structural and functional characterization of pi bulges and other short intrahelical deformations. Structure. 2004;12:133–144. doi: 10.1016/j.str.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Chen MC, EB, Saucedo-García M, Plasencia J, Gavilanes-Ruíz M. Plant sphingolipids: structure, synthesis, function. In: Wada H, editor. Lipids in Photosynthesis. New York: Springer; 2009. pp. 77–115. [Google Scholar]
- Cooley RB, Arp DJ, Karplus PA. Evolutionary origin of a secondary structure: pi-helices as cryptic but widespread insertional variations of alpha-helices that enhance protein functionality. J Mol Biol. 2010;404:232–246. doi: 10.1016/j.jmb.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova ND, Badger JH, Robson GD, Wortman JR, Nierman WC. Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics. 2005;6 doi: 10.1186/1471-2164-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrst H, Saba JD. An update on sphingosine-1-phosphate and other sphingolipid mediators. Nat Chem Biol. 2010;6:489–497. doi: 10.1038/nchembio.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Heung LJ, Luberto C, Del Poeta M. Role of sphingolipids in microbial pathogenesis. Infect Immun. 2006;74:28–39. doi: 10.1128/IAI.74.1.28-39.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang O, Kim G, Jang YJ, Kim SW, Choi G, Choi HJ, Jeon SY, Lee DG, Lee JD. Synthetic phytoceramides induce apoptosis with higher potency than ceramides. Mol Pharmacol. 2001;59:1249–1255. doi: 10.1124/mol.59.5.1249. [DOI] [PubMed] [Google Scholar]
- Kamlekar RK, Simanshu DK, Gao YG, Kenoth R, Pike HM, Prendergast FG, Malinina L, Molotkovsky JG, Venyaminov SY, Patel DJ, et al. The glycolipid transfer protein (GLTP) domain of phosphoinositol 4-phosphate adaptor protein-2 (FAPP2): Structure drives preference for simple neutral glycosphingolipids. Biochim Biophys Acta. 2013;1831:417–427. doi: 10.1016/j.bbalip.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenoth R, Kamlekar RK, Simanshu DK, Gao Y, Malinina L, Prendergast FG, Molotkovsky JG, Patel DJ, Venyaminov SY, Brown RE. Conformational folding and stability of the HET-C2 glycolipid transfer protein fold: does a molten globule-like state regulate activity? Biochemistry. 2011;50:5163–5171. doi: 10.1021/bi200382c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenoth R, Simanshu DK, Kamlekar RK, Pike HM, Molotkovsky JG, Benson LM, Bergen HR, Prendergast FG, Malinina L, Venyaminov SY, et al. Structural Determination and Tryptophan Fluorescence of Heterokaryon Incompatibility C2 Protein (HET-C2), a Fungal Glycolipid Transfer Protein (GLTP), Provide Novel Insights into Glycolipid Specificity and Membrane Interaction by the GLTP Fold. J Biol Chem. 2010;285:13066–13078. doi: 10.1074/jbc.M109.093203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Kumagai K, Tomishige N, Yamaji T, Wakatsuki S, Nishijima M, Hanada K, Kato R. Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc Natl Acad Sci USA. 2008;105:488–493. doi: 10.1073/pnas.0709191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E. Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol. 2004;5:305–315. doi: 10.1038/nrm1358. [DOI] [PubMed] [Google Scholar]
- Liang H, Yao N, Song JT, Luo S, Lu H, Greenberg JT. Ceramides modulate programmed cell death in plants. Genes Dev. 2003;17:2636–2641. doi: 10.1101/gad.1140503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- Malinina L, Malakhova ML, Kanack AT, Lu M, Abagyan R, Brown RE, Patel DJ. The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure. PLoS Biol. 2006;4:e362. doi: 10.1371/journal.pbio.0040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ. Structural basis for glycosphingolipid transfer specificity. Nature. 2004;430:1048–1053. doi: 10.1038/nature02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Comm Mass Spectrom. 2007;21:1304–1314. doi: 10.1002/rcm.2962. [DOI] [PubMed] [Google Scholar]
- Markham JE, Li J, Cahoon EB, Jaworski JG. Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem. 2006;281:22684–22694. doi: 10.1074/jbc.M604050200. [DOI] [PubMed] [Google Scholar]
- Markham JE, Lynch DV, Napier JA, Dunn TM, Cahoon EB. Plant sphingolipids: function follows form. Curr Opin Plant Biol. 2013;16:350–357. doi: 10.1016/j.pbi.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Mattjus P, Molotkovsky JG, Smaby JM, Brown RE. A fluorescence resonance energy transfer approach for monitoring protein-mediated glycolipid transfer between vesicle membranes. Anal Biochem. 1999;268:297–304. doi: 10.1006/abio.1998.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson LVN, JA . Sphingolipid signaling in plants. In: Munnik T, editor. Lipid signaling in plants, Plant cell monographs. Heidelberg; New York: Springer; 2010. pp. 307–321. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Palma K, Thorgrimsen S, Malinovsky FG, Fiil BK, Nielsen HB, Brodersen P, Hofius D, Petersen M, Mundy J. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 2010;6:e1001137. doi: 10.1371/journal.ppat.1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M, Clave C. The fungus-specific HET domain mediates programmed cell death in Podospora anserina. Eukaryot Cell. 2007;6:2001–2008. doi: 10.1128/EC.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pata MO, Hannun YA, Ng CK. Plant sphingolipids: decoding the enigma of the Sphinx. New Phytol. 2010;185:611–630. doi: 10.1111/j.1469-8137.2009.03123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer M, Stegmann M, Mueller MJ, Waller F. Pseudomonas syringae infection triggers de novo synthesis of phytosphingosine from sphinganine in Arabidopsis thaliana. FEBS Lett. 2010;584:4053–4056. doi: 10.1016/j.febslet.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Petersen NH, McKinney LV, Pike H, Hofius D, Zakaria A, Brodersen P, Petersen M, Brown RE, Mundy J. Human GLTP and mutant forms of ACD11 suppress cell death in the Arabidopsis acd11 mutant. FEBS J. 2008;275:4378–4388. doi: 10.1111/j.1742-4658.2008.06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reape TJ, McCabe PF. Apoptotic-like programmed cell death in plants. New Phytol. 2008;180:13–26. doi: 10.1111/j.1469-8137.2008.02549.x. [DOI] [PubMed] [Google Scholar]
- Samygina VR, Ochoa-Lizarralde B, Popov AN, Cabo-Bilbao A, Goni-de-Cerio F, Molotkovsky JG, Patel DJ, Brown RE, Malinina L. Structural insights into lipid-dependent reversible dimerization of human GLTP. Acta Crystallogr D Biol Crystallogr. 2013;69:603–616. doi: 10.1107/S0907444913000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samygina VR, Popov AN, Cabo-Bilbao A, Ochoa-Lizarralde B, Goni-de-Cerio F, Zhai X, Molotkovsky JG, Patel DJ, Brown RE, Malinina L. Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein. Structure. 2011;19:1644–1654. doi: 10.1016/j.str.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu DK, Kamlekar RK, Wijesinghe DS, Zou X, Zhai X, Mishra SK, Molotkovsky JG, Malinina L, Hinchcliffe EH, Chalfant CE, et al. Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids. Nature. 2013;500:463–467. doi: 10.1038/nature12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritz R, Hickey MJ, Lin AH, Hadwiger P, Sah DW, Neuwelt EA, Mueller BM, Kruse CA. FAPP2 gene downregulation increases tumor cell sensitivity to Fas-induced apoptosis. Biochem Biophys Res Commun. 2009;383:167–171. doi: 10.1016/j.bbrc.2009.03.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, Dixit VD. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nature Med. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Wang W, Yang X, Tangchaiburana S, Ndeh R, Markham JE, Tsegaye Y, Dunn TM, Wang GL, Bellizzi M, Parsons JF, et al. An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell. 2008;20:3163–3179. doi: 10.1105/tpc.108.060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G, Viitanen L, Alm C, Mattjus P, Salminen TA, Edqvist J. Identification of a glycosphingolipid transfer protein GLTP1 in Arabidopsis thaliana. FEBS J. 2008;275:3421–3437. doi: 10.1111/j.1742-4658.2008.06498.x. [DOI] [PubMed] [Google Scholar]
- Zhai X, Malakhova ML, Pike HM, Benson LM, Bergen HR, 3rd, Sugar IP, Malinina L, Patel DJ, Brown RE. Glycolipid acquisition by human glycolipid transfer protein dramatically alters intrinsic tryptophan fluorescence: insights into glycolipid binding affinity. J Biol Chem. 2009;284:13620–13628. doi: 10.1074/jbc.M809089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.