Version Changes
Updated. Changes from Version 1
We have incorporated some of the suggestions of the reviewers in our revised manuscript. We have changed the title of the paper and have revised the name of our gun4 mutant. It is now named as gun4-II. We have cited the earlier identified Chlamydomonas gun4 mutant in our manuscript text as gun4-I. We have revised the first few sentences in the abstract. We are unable to obtain the gun4-I mutant and hence the comparative physiological experiment suggested by the reviewer, Dr. Jin, cannot be performed and is beyond the scope of our research. We have added two future biochemical experiments that can be performed on the gun4-II mutant. We have accommodated the suggestion of Dr. Yokthongwattana of categorizing some figures as supplementary figures. We have renamed Figure 6–Figure 10 as Figure S1–Figure S5, respectively. The numbering of Figure 11-Figure 17 has changed. We would like to keep the info on HYP1 and HYP2 as it is, in the manuscript. As we currently don’t know the exact insertion point of the pUC ori end of the pBC1 vector in the gun4-II genome, we have used HYP1 and HYP2 as marker genes to clarify the extent of deletion/genetic rearrangement surrounding the GUN4 mutation locus. The gene and protein sequences of HYP1 and HYP2 are available on the Phytozome database. The functions of these genes are unknown.
Abstract
The green micro-alga Chlamydomonas reinhardtii is an elegant model organism to study oxygenic photosynthesis. Chlorophyll (Chl) and heme are major tetrapyrroles that play an essential role in photosynthesis and respiration. These tetrapyrroles are synthesized via a common branched pathway that involves mainly enzymes, encoded by nuclear genes. One of the enzymes in the pathway is Mg chelatase (MgChel). MgChel catalyzes insertion of Mg 2+ into protoporphyrin IX (PPIX, proto) to form Magnesium-protoporphyrin IX (MgPPIX, Mgproto), the first biosynthetic intermediate in the Chl branch. The GUN4 (genomes uncoupled 4) protein is not essential for the MgChel activity but has been shown to significantly stimulate its activity. We have isolated a light sensitive mutant, 6F14, by random DNA insertional mutagenesis. 6F14 cannot tolerate light intensities higher than 90-100 μmol photons m -2 s -1. It shows a light intensity dependent progressive photo-bleaching. 6F14 is incapable of photo-autotrophic growth under light intensity higher than 100 μmol photons m -2 s -1. PCR based analyses show that in 6F14 the insertion of the plasmid outside the GUN4 locus has resulted in a genetic rearrangement of the GUN4 gene and possible deletions in the genomic region flanking the GUN4 gene. Our gun4 mutant has a Chl content very similar to that in the wild type in the dark and is very sensitive to fluctuations in the light intensity in the environment unlike the earlier identified Chlamydomonas gun4 mutant. Complementation with a functional copy of the GUN4 gene restored light tolerance, Chl biosynthesis and photo-autotrophic growth under high light intensities in 6F14. 6F14 is the second gun4 mutant to be identified in C. reinhardtii. Additionally, we show that our two gun4 complements over-express the GUN4 protein and show a higher Chl content per cell compared to that in the wild type strain.
Introduction
Chlamydomonas reinhardtii is a green micro-alga that can grow either heterotrophically using exogenous acetate as a carbon source or photo-autotrophically, using atmospheric CO 2. It possesses a photosynthetic apparatus very similar to higher plants, has a short and simple haplontic life cycle, can synthesize Chl both light dependently and light independently (unlike most angiosperms) and its genome has been sequenced 1. In addition, well developed molecular tools exist for genetic manipulations of its genome. All these traits make this alga an elegant model system for dissecting oxygenic photosynthesis 2, 3.
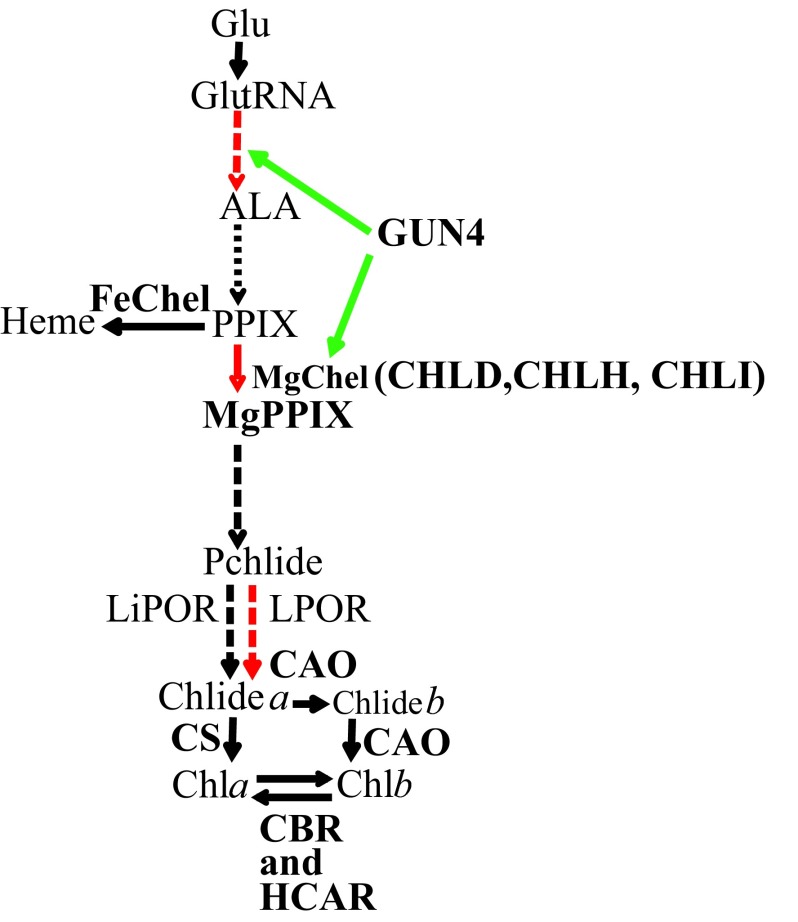
Chl, heme, siroheme, cobalamin, heme d1 and factor F430 are major tetrapyrroles that are involved in wide variety of essential life processes in all living organisms. Chl and heme are synthesized via a common branched pathway 4, 5 (outlined in Figure 1). Photosynthetic eukaryotes synthesize 5-aminolevulinic acid (ALA) from glutamine (Glu) bound to tRNA Glu through the C5 pathway consisting of two steps catalyzed by glutamyl-tRNA reductase and glutamate-1-semialdehyde aminotransferase 4, 5. ALA is subsequently converted in six steps to PPIX, the last common precursor for both Chl and heme biosynthesis 4, 5. Insertion of Fe 2+ into PPIX by ferrochelatase (FeChel) leads to heme. Insertion of Mg 2+ in PPIX by the heterotrimeric MgChel (comprised of three subunits: CHLD, CHLH and CHLI 6) leads to MgPPIX, the first biosynthetic intermediate in the Chl branch 6. MgPPIX is converted to Pchlide via three enzymatic steps. The reduction of Pchlide to form chlorophyllide (Chlide) can occur by two different mechanisms. One mechanism is catalyzed by the strictly light dependent enzyme NADPH:Pchlide oxidoreductase (LPOR) and occurs in all photosynthetic organisms; it is the only mechanism of Chl formation in angiosperms 7– 10. The second mechanism is catalyzed by the light independent NADPH:Pchlide oxidoreductase (LiPOR) and is present in anoxygenic bacteria, alga, ferns and gymnosperms 11– 20. The Chlide a undergoes a phytylation reaction, catalyzed by Chl synthase (CS), resulting in the formation of Chl a. In vascular plants and green algae a portion of the Chlide a is converted to Chlide b by Chlide a oxygenase (CAO) prior to phytylation 21– 24. Chl a is converted to Chl b by CAO via formation of 7-hydroxymethyl chlorophyll a (HCA) and Chl b can be converted back to Chl a via HCA by chlorophyll b reductase (CBR) and 7-hydroxymethyl chlorophyll a reductase (HCAR) 25. This inter-conversion of Chl a and Chl b, referred to as the “chlorophyll cycle”, plays an important role in greening, acclimation to light and senescence 25.
Figure 1. A simplified tetrapyrrole biosynthetic pathway.
Light regulated steps are in red. Dashed arrows denote multiple enzymatic steps and green arrows point to steps that are positively regulated by the GUN4 protein, respectively. Tetrapyrrole intermediates and enzymes are shown in black and bold black type, respectively. Readers are advised to look in the text for full names of tetrapyrrole intermediates and enzymes, which are abbreviated in this figure.
Stringent control of tetrapyrrole biosynthesis is especially essential for oxygenic photosynthetic organisms that are often prone to oxidative stress. Free Chl, heme and their immediate precursors are highly photo-toxic molecules and generate reactive oxygen species (ROS) under aerobic conditions 26. Hence most of the cellular Chls are usually bound to the light harvesting complex (LHC) and other photosystem (PS) proteins. Chl is made in the plastid. Most of these Chl binding proteins and enzymes of the tetrapyrrole biosynthetic pathways are encoded by the nuclear genes 5. Hence a tight coordination of biosynthesis of Chl with its apoprotein is necessary 27. Chl and heme biosynthesis in plants is under transcriptional, translational and post-translational control at multi level and is accomplished by a complex regulatory network among the chloroplasts, mitochondria and nucleus, that is not well understood 28– 30.
One of the major research interests of our laboratory is to identify components that play a role in the regulation of Chl biosynthesis under different irradiance conditions. We have generated a random DNA insertional Chlamydomonas mutant library and have screened it to isolate twenty one mutants that are either defective in Chl biosynthesis and/or are incapable of photo-autotrophic growth under different irradiance conditions. One of the isolated mutants ( 6F14) is a light sensitive mutant which shows a light intensity dependent progressive photo-bleaching and is incapable of photosynthesis under low light intensities (90–100 µmol m -2 s -1). Molecular analyses revealed that 6F14 is defective in the GUN4 (genome uncoupled 4) gene which codes for a protein that stimulates MgChel activity. 6F14 is the second gun4 mutant ( gun4-II) to be identified in Chlamydomonas 31. Transformation of 6F14 with a functional copy of the GUN4 gene restored the wild type phenotype. Western analyses show that the two isolated gun4-II complements are over-expressing the GUN4 protein. Chl analyses show that these gun4-II complements have 50–60% more Chl than that of the wild type strain. In this study, we present our molecular data on the identification of the mutation locus in 6F14 and its complementation.
Materials and methods
Algal media and cultures
Chlamydomonas strains 4A+ (a gift from Dr. Krishna Niyogi (UC, Berkeley), gun4-II and gun4-II complements (both generated by our laboratory) were grown either in Tris-Acetate Phosphate (TAP) heterotrophic media or in Sueoka’s High Salt (HS) photo-autotrophic media. TAP and HS liquid media and agar plates were prepared in the lab using reagents from Fisher Scientific (Pittsburgh, PA) according to the protocol given in Gorman and Levine (1965) 32 and Sueoka (1960) 33, respectively. The 4A+ strain and gun4-II complements were maintained on TAP agar plates and TAP + zeocin (Sigma, St. Louis, MO) plates, respectively under dim light intensities (10–15 µmol photons m -2 s -1) at 25°C. The final zeocin concentration was 15 µg/ml. The gun4-II mutant ( 6F14) was maintained in the dim light or in the dark on TAP 1.5% agar plates containing 10 µg/ml of paromomycin (Sigma, St. Louis, MO). Liquid algal cultures used for RNA and genomic DNA extractions and protein analyses were grown in 100 ml flasks on the New Brunswick Scientific Excella E5 platform shaker (Enfield, CT) in TAP media at 150 rpm in the dim light.
Generation of the 6F14 mutant
The purified pBC1plasmid from the DH5α Escherichia coli-pBC1 clone (obtained from Dr. Krishna Niyogi’s laboratory at UC, Berkeley) was used for random DNA insertional mutagenesis. This plasmid contains two antibiotic resistance genes: APHVIII and Amp R ( Figure 2). APHVIII confers resistance against the antibiotic paromomycin and was used as a selection marker for screening of Chlamydomonas transformants. Amp R was used as a selection marker for screening of E. coli clones harboring the pBC1 plasmid. E. coli was grown in 1 l of Luria Bertani (LB) broth containing 1% tryptone, 0.5% of yeast extract, 1% NaCl and ampicillin (final concentration of ampicillin:100 µg/ml). LB media was prepared in the laboratory using reagents purchased from Fisher (Pittsburgh, PA). Ampicillin was purchased from Fisher (Pittsburgh, PA). The culture was incubated at 37°C overnight. Plasmid purification from E. coli cells was facilitated by a Qiagen plasmid mega kit according to the protocol given in the technical manual (Qiagen, Valencia, CA). Once purified from E. coli, the circular pBC1 vector was linearized with the restriction enzyme KpnI (NEB, Beverly, MA) according to the protocol given in the technical manual. The linearized DNA was purified using a QIAEX II gel extraction kit (Qiagen, Valencia, CA) according to the protocol given in the technical manual. All agarose DNA gel electrophoresis was visualized by BioRad Molecular Imager Gel Doc XR+ (BioRad, Hercules, CA). Transformation of parental strain 4A+ by the linearized pBC1 vector was performed utilizing the glass bead transformation technique described by Kindle et al. (1989) 34 and Dent et al. (2005) 2. Transformants were plated onto fresh TAP agar plates containing 10 µg/ml paromomycin (TAP+P) in the dark. Single colonies of mutants were picked and transferred onto fresh TAP+P plates using a numbered grid layout. Screening of photosynthetic and pigment deficient mutants was done by visual inspection and monitoring of growth under different light intensities in heterotrophic, mixotrophic and photo-autotrophic conditions 2.
Figure 2. Linearized pBC1 plasmid used for random insertional mutagenesis.
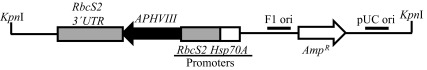
The cleavage site of Kpn1 restriction enzyme, used for linearization of the vector is shown. APHVIII is under the control of combo promoters which consist of the promoter of the gene encoding the small subunit of Rubisco (RbcS2) and the gene encoding the heat shock protein 70A (Hsp70A). pBC1 is a phagemid and its F1 origin (F1 ori) and pUC origin (pUC ori) are shown. The size of the plasmid is 4763 bp.
Genomic DNA and RNA extraction
4A+, gun4-II complements and gun4-II were grown in TAP liquid media in the dim light to a cell density of about 5 × 10 6 cells/ml of the culture. Genomic DNA was purified using a phenol-chloroform extraction method 35. RNA extraction was facilitated by TRIzol reagent from Invitrogen (Carlsbad, CA) following the protocol in the technical manual. DNA and RNA concentrations were measured using a Nanodrop 1000 spectrophotometer from Thermo Fisher Scientific (Wilmington, DE). DNase treatment was performed using Ambion’s TURBO DNA-free kit from Invitrogen (Carlsbad, CA) following the protocol in the technical manual to remove genomic DNA from the RNA preparation. Generation of cDNA was performed using Life Technologies Superscript III First-Strand Synthesis System from Invitrogen (Carlsbad, CA) following the protocol in the technical manual.
Thermal Asymmetric InterLaced PCR
TAIL (Thermal Asymmetric InterLaced) PCR was implemented, following the protocol of Dent et al. (2005) 2. HotStar Taq Plus DNA polymerase kit reagents (Qiagen, Valencia, CA) were used for PCR. The PCR reaction mixture consisted of 1 × PCR buffer, 200 µM of each dNTP, 1 × Q-solution, 2.5 units of HotStar Taq Plus DNA polymerase, 60 pmoles of the random degenerate primer RD1 and 5 pmol of the APHVIII specific primer. Primers were ordered from IDT (Skokie, IL; Table 1). Degenerate primer RD1 has an average T m of 51°C while the three APHVIII specific primers used had T m ranging from 58°C to 64°C. PCR cycling programs were created using the program given in Dent et al. (2005) 2. TAIL1 PCR product was diluted 10-fold and 2 µl of the diluted TAIL1 PCR product was used for TAIL2 PCR reactions. The TAIL2 PCR product was gel purified using a QIAEX II gel extraction kit (Qiagen, Valencia, CA) according to the protocol given in the technical manual. Purified TAIL2 PCR product was sequenced at the UC, Berkeley DNA Sequencing Facility (Berkeley, CA). All primer sequences are shown in Table 1.
Table 1. List of primers used for TAIL (Thermal Asymmetric InterLaced) PCR, verification of TAIL PCR product and DNA sequencing.
| Primer name | Sequence of primer | Location |
|---|---|---|
| RD1 | 5´-WNG GGS CNG CWT TT-3´ | Degenerate primer |
| 7F | 5´-ACG GAG GAT CGT TAC AAC CAA CAA-3´ | APHVIII 3´ UTR |
| 2R | 5´-CTC AAG TGC TGA AGC GGT AGC TTA-3´ | APHVIII 3´ UTR |
| 3R | 5´-TCT TCT GAG GGA CCT GAT GGT GTT-3´ | APHVIII 3´ UTR |
| 4R | 5´-GGG CGG TAT CGG AGG AAA AGC TG-3´ | APHVIII 3´ UTR |
Genomic and reverse transcription PCR
Primers were designed based on genomic DNA sequences available in the Chlamydomonas genome database in Phytozome. Amplifications of genomic DNA and cDNA were executed using the MJ Research PTC-200 Peltier Thermal Cycler (Watertown, MA). HotStar Taq Plus DNA polymerase kit (Qiagen, Valencia, CA) was used for PCR following the cycling conditions given in the Qiagen protocol booklet. Annealing temperature was between 55 and 60°C depending on the T m of the primers. Extension time was varied according to the size of the PCR product amplified. Final extension was set at 72°C for ten minutes. All genomic and reverse transcription PCR products were amplified for a total of thirty-five cycles. A 50–150 ng sample of genomic DNA or cDNA were used for PCR reactions. For semi-quantitative RT-PCR reactions, 3 µg of total RNA was converted into cDNA and then 150 ng of cDNA templates were used for RT-PCR. Sequences of primers used for genomic and RT-PCR are shown in Table 2– Table 4.
Table 2. List of GUN4 specific primers.
These primers were used for GUN4 (Cre05.g246800) genomic DNA PCR on 6F14 and 4A+ and also for DNA sequencing to generate the data in Figure S3 and Figure S4.
| Primer name | Sequence of primer | Location |
|---|---|---|
| 2R | 5´-AGTGTGTGTTTGGGCCAGCATTT-3´ | Exon1 |
| 3F | 5´-TGTGGAGAAGAAGAAGTCCGGCAA-3´ | Exon1 |
| 3R | 5´-TTGCCGGACTTCTTCTTCTCCACA-3´ | Exon1 |
| 14F | 5´-GATCCGCAGCCTCACGAG-3´ | Exon1 |
| 14R | 5´-CCTCGTGAGGCTGCGGATC-3´ | Exon1 |
| 7F | 5´-ACAACCCTTGACTTGCGACTCTGT-3´ | Exon2 |
| 7R | 5´-ACAGAGTCGCAAGTCAAGGGTTGT-3´ | Exon2 |
| 8F | 5´-ACCGCATCTTGCAAAGATTGCACC-3´ | Exon2 |
| 8R | 5´-GGTGCAATCTTTGCAAGATGCGGT-3´ | Exon2 |
| 10R | 5´-AGTCTTACACAGGCATACTGCAGCG-3´ | Exon2 |
| 11R | 5´-CTCTTTCAGTCTTACACAGGCATACTGC-3´ | Exon2 |
| 12F | 5´-AGCCGGACTGTTGCGTAATGTGAT-3´ | Exon2 |
| 12R | 5´-ATCACATTACGCAACAGTCCGGCT-3´ | Exon2 |
Table 3. List of primers used for checking the genomic region upstream of GUN4 (Cre05.g246800) and HYP2 [g5195] gene.
| Primer name | Sequence of primer | Location |
|---|---|---|
| ACF6 | 5´-ACATAGCAGCGAGACACACCACAT-3´ | Upstream of GUN4 region |
| ACF7 | 5´-AACAAATCCGCGAACGCCACTATG-3´ | Upstream of GUN4 region |
| ACR7 | 5´-CATAGTGGCGTTCGCGGATTTGTT-3´ | Upstream of GUN4 region |
| ACF11 | 5´-GCAACCGGTGTTTGGGCGTATTAT-3´ | Upstream of GUN4 region |
| ACR11 | 5´-ATAATACGCCCAAACACCGGTTGC-3´ | Upstream of GUN4 region |
| H3F | 5´-TCCCATGGTATCCCGAGCTTGAAA-3´ | 3´ end of HYP2 |
| H4F | 5´-TGAGGAAACTGGACTTGGCTGAGT-3´ | 3´ end of HYP2 |
| H5F | 5´-TACCAGCAGCATCTAAGCACCACA-3´ | 3´ end of HYP2 |
| H6R | 5´-TATTCTAATGCAGCACGGCAAGGC-3´ | 3´ end of HYP2 |
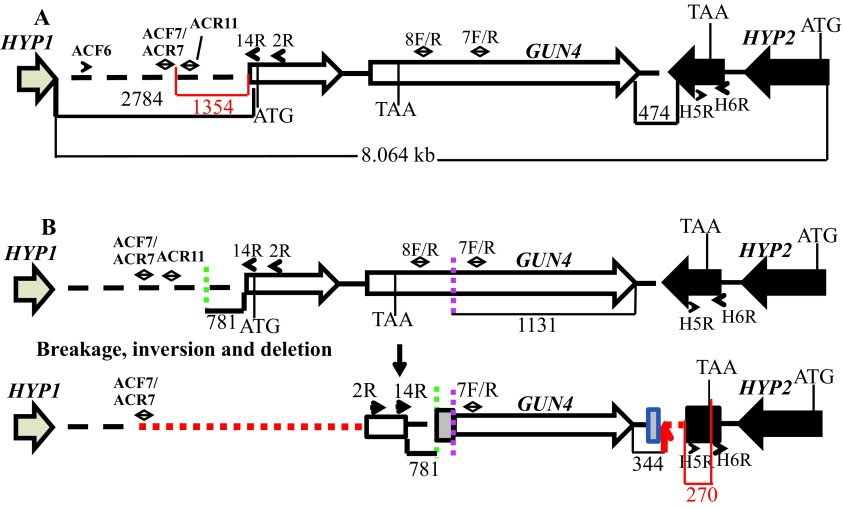
Figure 6. A schematic of the genetic rearrangement in 6F14.
( A) A schematic genomic map showing an 8.064 kb genomic DNA region spanning the GUN4 locus on chromosome 5. The numbers at the bottom of the map denote distances between respective points on the genomic DNA. The red highlighted region and number show the distance between primer ACF7 and the start of the GUN4 gene and the distance between the primer H5F and the end of the HYP2 gene, respectively. The two GUN4 exons are represented by white block arrows. The tan arrow and the black block arrow, denotes a part of HYP1 3´ UTR and HYP2 gene, respectively. ( B) An updated schematic diagram showing the rearrangement of the GUN4 locus based on PCR analyses and DNA sequencing. Two break points in the genome are denoted by green and pink dashed lines. The big and the small grey boxes, denote addition of 45 and 29 bp, respectively. The small black arrows denote primers that were used for genomic PCRs in Figure S4 and Figure S5. Red dashed lines denote possible deletions. The small red arrow indicates the point of insertion of the pBC1 plasmid. The black numbers at the bottom of the map denote distances between respective points on the genomic DNA. The red highlighted region and the corresponding number show the distance between the end of the primer H5F and the stop codon of the HYP2 gene.
Table 4. List of primers used for transcript analysis of GUN4 and GUN4 neighboring genes in 6F14.
These primers were used to generate the data in Figure 7. The gene loci numbers in Phytozome for the three neighboring genes of GUN4 on chromosome 5 and the control actin gene on chromosome 13 are: HYP1 [Cre05.g246750], HYP2 [g5195] and SOXE [Cre05.g246900] and Actin (Cre13.g603700), respectively.
| Primer name | Sequence of primer | Purpose |
|---|---|---|
| F2 | 5´-ACGACACCACCTTCAACTCCATCA-3´ | Actin |
| R2 | 5´-TTAGAAGCACTTCCGGTGCACGAT-3´ | Actin |
| NupF4 | 5´-TGTATGAACTCTGAGCAGGCGACA-3´ | HYP1 |
| Nup98R2 | 5´-CCTGCCGTATGTCGTGCACAAAC-3´ | HYP1 |
| 3F | 5´-TGTGGAGAAGAAGAAGTCCGGCAA-3´ | GUN4 |
| 8R | 5´-GGTGCAATCTTTGCAAGATGCGGT-3´ | GUN4 |
| HypF2 | 5´-TTCCTGGCTACTGCCGTATTCGCA-3´ | HYP2 |
| H6F | 5´-GCCTTGCCGTGCTGCATTAGAATA-3´ | HYP2 |
| PB120 | 5´-GCACGGATGGCAAGTACATG-3´ | SOXE |
| PB121 | 5´-CTACTTCACTGCCCTGGAGTTT-3´ | SOXE |
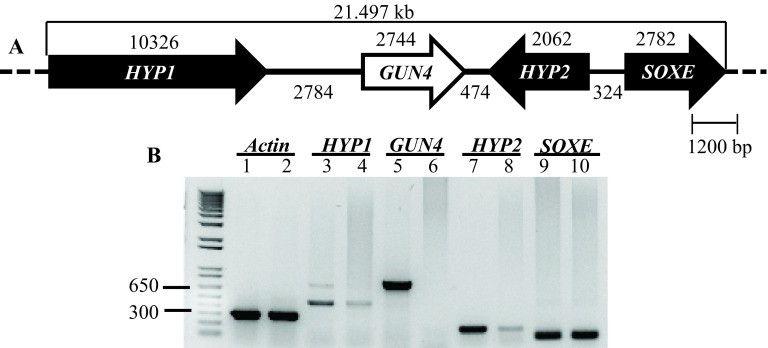
Figure 7. Transcript analyses of GUN4 and its neighboring genes.
( A) A schematic map of a 21.497 kb genomic region spanning the GUN4 locus on chromosome 5. HYP1 and HYP2 are genes located upstream and downstream of the GUN4 gene, respectively coding for hypothetical proteins. Sulfocyanin ( SOXE) codes for a blue copper protein. The top black number denotes size of a gene (bp) while the bottom black number denotes distance between genes (bp). ( B) Semi-quantitative RT-PCR results. Lanes: 1, 3, 5, 7, 9 denote 4A+ cDNA products. Lanes 2, 4, 6, 8, 10 denote gun4 cDNA products. Primer sequences are shown in Table 4. All primers span an intron. Actin was used as a control. Actin genomic product size: 527 bp; Actin cDNA product size: 305 bp. HYP1 genomic product size 726 bp; HYP1 cDNA product size: 459 bp. GUN4 genomic product size: 942 bp; GUN4 cDNA product size 775 bp. HYP2 genomic product size: 797 bp; HYP2 cDNA product size: 184bp. SOXE genomic product size: 517 bp; SOXE cDNA product size: 119 bp.
Cloning of the GUN4 gene in the pDBle vector
The pDBle vector (obtained from Dr. Saul Purton, University College London, UK) was double-digested with restriction enzymes EcoRI and NdeI (NEB, Beverly, MA) according to the protocol given in the technical manual. The GUN4 gene was amplified using primers given in Table 5. Ligation of the double digested ( NdeI and EcoRI digested) GUN4 gene and the NdeI/ EcoRI double-digested pDBle vector was done using the T4 ligase and 1 mM ATP (NEB, Beverly, MA). Chemically competent (CaCl 2 treated) E. coli cells were used for transformation. After transformation, E. coli cells were plated on LB+ampicillin (final concentration of ampicillin:100 µg/ml) plates and incubated at 37°C overnight. Single colonies were picked the next day and plasmids were isolated from these clones. Isolated plasmids were double-digested with EcoRI and NdeI to verify the cloning of the GUN4 gene. The GUN4-pDBle construct from the selected clone was sequenced by the UC, Berkeley DNA Sequencing Facility (Berkeley, CA). Chromas Lite ( http://technelysium.com.au/) and BLAST were used to analyze DNA sequences.
Table 5. List of primers used for cloning and complement testing.
These primers were used in the experiments that generated the data in Figure 8 and Figure 11 and were also used for GUN4 gene amplification for cloning.
| Primer name | Sequence of primer | Purpose |
|---|---|---|
| GUN4F1 | 5´-GGAATTCCATATGCTGGCCCAAACACACACT-3´ | Amplification of GUN4 for cloning |
| GUN4R1 | 5´-CCGGAATTCTTAGAACAGCGACTGTGTCCGCC-3´ | Amplification of
GUN4 for cloning
and for complement testing |
| PsaDF1 | 5´-CCACTGCTACTCACAACAAGCCCA-3´ | Complement testing |
Figure 8. A schematic figure of the pDBle vector used for complementation of gun4-II.
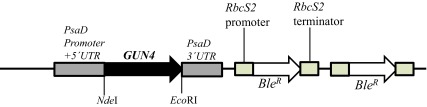
NdeI/ EcoRI double digested GUN4 gene (956 bp) was cloned into the NdeI/ EcoRI double digested pDBle plasmid. Primers used for amplification of the GUN4 gene are shown in Table 5. GUN4 expression is driven by the constitutive PsaD promoter. NdeI and EcoRI restriction sites are labeled. pDBle contains two copies of Ble R genes driven by the Rubisco ( RbcS2) promoter. The size of the GUN4- pDBle construct is 7653 bp. The black arrow and the white arrow, denotes GUN4 and Ble R genes, respectively. Grey and tan boxes denote UnTranslated Regions (UTRs).
Figure 11. Molecular analyses of gun4-II complements.
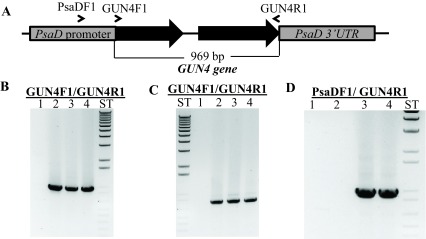
( A) A schematic diagram of the GUN4-pDBle construct. Primers used for PCR are shown on the map. ( B) Genomic DNA PCR analyses with GUN4 cloning primers (product size: 969 bp). Lane 1: gun4-II; Lane 2: 4A+; Lane 3: gun4-19; Lane 4: gun4-27. ( C) RT-PCR analyses with GUN4 cloning primers (product size: 802 bp). Lane 1: gun4; Lane 2: 4A+; Lane 3: gun4-19; Lane 4: gun4-27. ( D) Genomic PCR analyses using a PsaD 5′ UTR specific forward primer with a GUN4 cloning reverse primer (product size: 976 bp). Lane 1: gun4-II; Lane 2: 4A+; Lane 3: gun4-19; Lane 4: gun4-27. Primer sequences are shown in Table 5.
Generation and screening of gun4 complements
Complementation of the gun4-II was performed utilizing the glass bead transformation technique described by Kindle et al. 1989 34. 2 µg of the linearized GUN4-pDBle was used to complement 6F14. Transformed cells were plated onto fresh TAP plates containing 15 µg/ml zeocin (Z) and placed in the dark at 25°C. Single colonies were picked and transferred onto fresh TAP+Z plates using a numbered grid template for screening of potential gun4 complements. Screening of gun4-II complements was done by monitoring the Chl content and growth of complement strains either on TAP or HS plates under medium light (300 µmol photons m -2 s -1) in the presence or absence of antibiotics zeocin and paromomycin.
Cellular protein analysis
Chlamydomonas cells from different strains grown in TAP in the dim light were harvested, washed twice with fresh medium and resuspended in TEN buffer (10 mM Tris-HCl, 10 mM EDTA and 150 mM NaCl; pH 8). Gel lanes were loaded with an equal amount of Chl (4 µg Chl). Resuspended cell suspension was mixed in a 1:1 ratio with the sample solubilization buffer SDS-urea buffer (150 mM Tris-HCl, pH 6.8; 7% w/v SDS; 10% w/v glycerol; 2 M urea; bromophenol blue and 10% β-mercaptoethanol) and were incubated at room temperature for about thirty minutes, with intermittent vortexing. The sample solubilization buffer was prepared according to the protocol of Smith et al. (1990) 36 using reagents from Fisher (Pittsburgh, PA). After incubation, the solubilized protein samples were vortexed and spun at a maximum speed of 20,000 g in a 1.5 ml eppendorf tube (USA Scientific, Ocala, FL) for five minutes at 4°C. The soluble fraction was loaded on a “any kD™ Mini-PROTEAN ® TGX™ Precast Gel” (BioRad, Hercules, CA) and SDS-PAGE analysis was performed according to Laemmli (1970) 37 using a Page Ruler prestained molecular weight protein ladder (Fermentas, Glen Burnie, Maryland) at a constant current of 80 V for 2 hours. Gels were stained with colloidal Coomassie Gel Code blue stain reagent (Thermo Fisher Scientific, Rockford, IL) for protein visualization.
Western analysis
Electrophoretic transfer of the SDS-PAGE resolved proteins onto an Immobilon P–PVDF membrane (Millipore, Billerica, MA) was carried out for 2 hours at a constant current of 400 mA in the transfer buffer (25 mM Tris, 192 mM glycine and 20% methanol). The GUN4 polyclonal antibody was raised in rabbit against the full length Chlamydomonas GUN4 mature protein that lacks the first 45 amino acids corresponding to the predicted chloroplast transit peptide 31. This antibody was generated by Dr. Roberto Bassi’s laboratory (University of Verona, Italy) and was provided to us by Dr. Krishna Niyogi (UC, Berkeley). GUN4 primary antibodies were diluted to a ratio of 1:1000 before being used as a primary probe. The secondary antibodies used for Western blotting were conjugated to horseradish peroxidase (Pierce protein research product, Thermo Fisher Scientific, Rockford, IL) and diluted to a ratio of 1:20,000 with the antibody buffer. Western blots were developed by using the Supersignal West Pico chemiluminescent substrate kit (Pierce protein research product, Thermo Fisher Scientific, Rockford, IL).
Cell counts and chlorophyll extraction
Cell density (number of cells per ml of the culture) was calculated by counting the cells using a Neubauer ultraplane hemacytometer (Hausser Scientific, Horsham, PA). Pigments from intact cells were extracted in 80% acetone and cell debris was removed by centrifugation at 10,000 g for 5 minutes. The absorbance of the supernatant was measured with a Beckman Coulter DU 730 Life Science UV/Vis spectrophotometer (Brea, CA). Chl a and b concentrations were determined by Arnon (1949) 38 equations, with corrections as described by Melis et al. (1987) 39.
Results
Generation and identification of the mutant 6F14
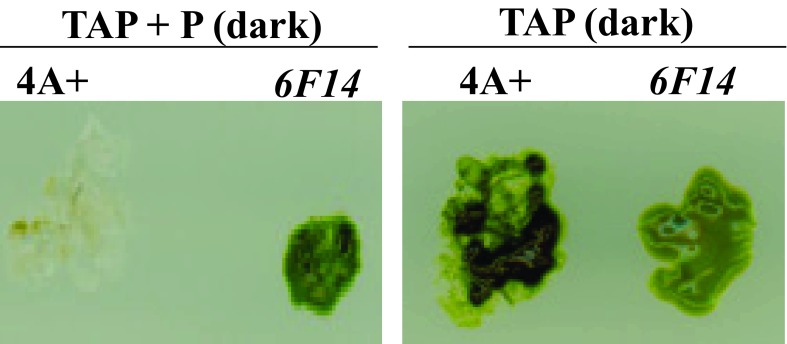
Mutant 6F14 was generated by random insertional mutagenesis of the C. reinhardtii wild type strain 4A+ (137c genetic background). 6F14 was identified as a slightly Chl deficient paromomycin resistant mutant on TAP+P plate in the dark ( Figure 3).
Figure 3. Identification of 6F14.
This figure shows the phenotypic difference of 6F14 compared to the parental strain, 4A+ on heterotrophic agar media (TAP) plates under two different growth conditions: dark + paromomycin (P) and dark.
Growth analyses of 6F14
Growth analyses in heterotrophic and photo-autotrophic liquid media revealed that 6F14 is light sensitive and shows progressive photo-bleaching with increase in light intensities ( Figure 4 and Figure 5). In mixotrophic conditions under 10–15 µmol photons m -2 s -1, 6F14 possesses 58% less Chl/cell than 4A+. At 40–50 µmol photons m -2 s -1, 6F14 has 72% less Chl/cell than the wild type. At 75–80 µmol photons m -2 s -1, 6F14 possesses 99% less Chl/cell than the wild type. At 75–80 µmol photons m -2 s -1, 6F14 starts to photo-bleach and turns yellow; it dies at light intensities 100–120 µmol photons m -2 s -1 in TAP ( Figure 4).
Figure 4. Heterotrophic and mixotrophic growth of 6F14 and the wild-type in TAP media.
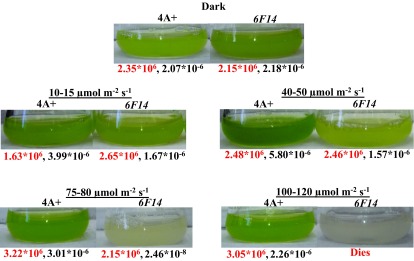
Dark adapted cells of 6F14 and 4A+ were shifted to different light intensities in this experiment. Light conditions and strains are labeled above the culture flasks. The cell density (cells/ml) and nmol chlorophyll (Chl) per cell are shown below the culture flasks in red and black numbers, respectively. For each light condition, experiments were performed on three biological replicates of each strain. Statistical error (±SD) was ≤ 10%.
Figure 5. Photo-autotrophic growth of 6F14 and wild-type in HS media.

Dark adapted cells of 6F14 and 4A+ were shifted to different light intensities in this experiment. The mean cell density (cells/ml) and the Chlorophyll (Chl) content (nmol Chl per cell) are shown below the culture flasks in red and black numbers, respectively. For each light condition, experiments were performed on three biological replicates of each strain. Statistical error (±SD) was ≤ 10%.
Figure 5 shows photo-autotrophic cultures of 6F14 and 4A+. 6F14 has the ability to grow photo-autotrophically in HS media in dim light (10–15 µmol of photons m -2 s -1). However, the mutant grows extremely slowly in comparison to the wild type. When grown at 10–15 µmol photons m -2 s -1 in HS media, 6F14 possesses 60% less Chl/cell than the wild type. At 40–50 µmol photons m -2 s -1 in HS media, 6F14 has 79% less Chl/cell than 4A+, and at 75–80 µmol photons m -2 s -1 in HS media, 6F14 possesses 83% less Chl/cell than the wild type. At 100–120 µmol photons m -2 s -1 in HS media, 6F14 fails to survive ( Figure 5).
Figure S1 demonstrates that when dim light adapted 6F14 was shifted to 40–50 µmol photons m -2 s -1 there was no significant change in Chl/cell content ( Figure 4). Dark adapted 6F14 showed a 50% reduction in Chl/cell when moved to 40–50 µmol photons m -2 s -1. When dim light adapted 6F14 was shifted to 75–80 µmol photons m -2 s -1, it showed a 98% reduction in Chl/cell while the dark adapted 6F14 failed to survive under 75–80 µmol photons m -2 s -1. Taken together, the results shown in Figure 4 and Figure S1 show that dark adapted 6F14 is more sensitive to the magnitude of light intensity changes in the environment than the dim light adapted 6F14 ( Figure S1).
Molecular characterization of the mutation in 6F14
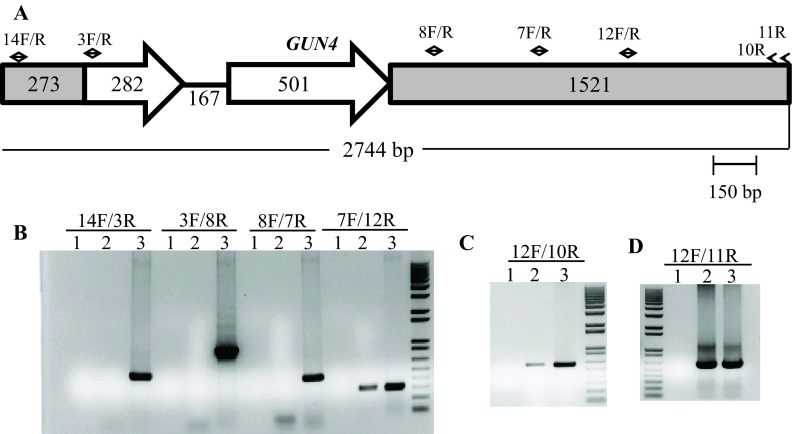
The linearized pBC1 plasmid was used to generate 6F14 ( Figure 2). To find the insertion of the APHVIII end of the plasmid in 6F14, TAIL PCR method was employed. Figure S2A shows the position of the vector specific TAIL PCR primers and also shows the arbitrary position of the random degenerate primer. A 2.9 kb DNA product from TAIL2 PCR was purified from the agarose gel ( Figure S2B, Table 1). This purified DNA product was used for further PCR using internal primers specific to the 3´ UnTranslated Region (UTR) of the APHVIII gene. The PCR results confirmed that the 2.9 kb DNA product contains the 3´ UTR of the APHVIII gene ( Figure S2C). Sequencing of the 2.9 kb TAIL2 PCR product revealed that the APHVIII end of the plasmid has been inserted 344 bp away from the GUN4 gene (Cre05.g246800) on chromosome 5. The GUN4 locus was cleaved at least at two places ( Figure S3). The first cleavage was about 781 bp away from the 5′ end of the GUN4 gene and the second cleavage was 1131 bp away from the 3´ end of the GUN4 gene. These cleavages were followed by the inversion of the cleaved genomic DNA which then ligated to the 3´ UTR of the GUN4 gene ( Figure S3). Plasmid insertion also led to an addition of 29 bp at the APHVIII end of the plasmid. An addition of 45 bp was found at the breakage point in the 3´ UTR of the GUN4 gene ( Figure S3).
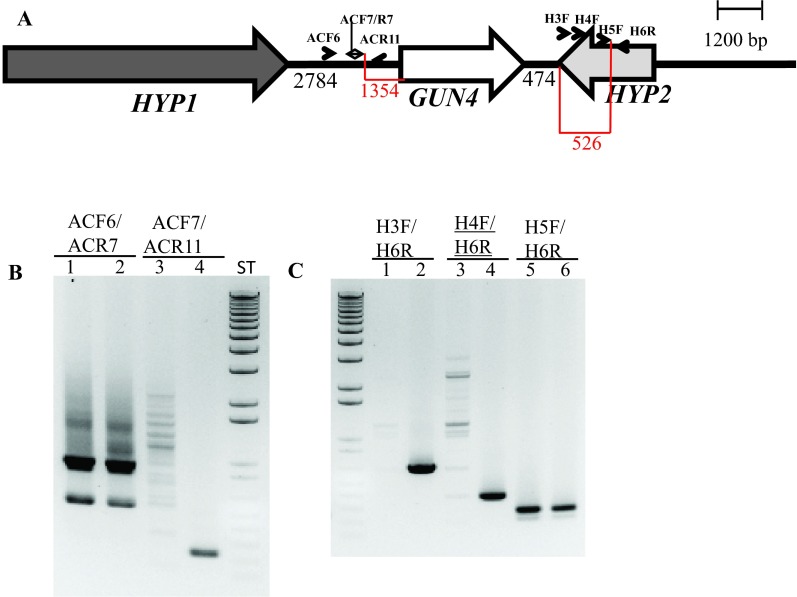
Further genomic DNA PCR analyses with GUN4 specific primers confirmed that the 3´ part of the GUN4 first exon and the 5′ part of the GUN4 second exon were deleted or displaced ( Figure S4). We also used primers specific to the genomic region upstream of the GUN4 gene and primers specific to the 3´ UTR of a hypothetical gene, HYP2, (g5195) located downstream of GUN4 to see the extent of deletion on either side of the GUN4 gene. Our PCR analyses show that a 1.354 kb genomic DNA region, located upstream of GUN4 was deleted/displaced. Additionally, there is a deletion of approximately 526 bp in the 3´ UTR of the downstream HYP2 gene ( Figure S5 and Figure 6). Taken together the data show that plasmid insertion in the 6F14 genome has rearranged the GUN4 locus and has affected a part of the 3´ UTR of the HYP2 gene. We do not yet know the exact location of the pUC ori end of the plasmid in the 6F14 genome ( Figure 2).
Checking for the absence/presence of the transcript of the GUN4 and three neighboring genes of GUN4
Transcript levels of GUN4 and the neighboring genes ( HYP1 [Cre05.g246750]; HYP2 [g5195] and SOXE [Cre05.g246900]) were checked using semi-quantitative RT-PCR using GUN4, HYP1, HYP2 and SOXE specific primers, respectively ( Figure 7). Reduced levels of HYP1 and HYP2 transcripts were observed in 6F14 compared to that in the wild type ( Figure 7). GUN4 transcript is missing in 6F14 as expected ( Figure 7). The transcript level of SOXE, the second gene downstream of GUN4, was not affected. Cre05.g246750 and g5195 are genes in the Chlamydomonas database coding for hypothetical proteins. We have named these genes as HYP1 and HYP2 arbitrarily for our study. The SOXE gene codes for sulfocyanin, a blue copper protein. Readers are requested to identify GUN4 and its neighboring genes by the gene locus number (Cre or the g number) in the Phytozome database.
Complementation of gun4-II
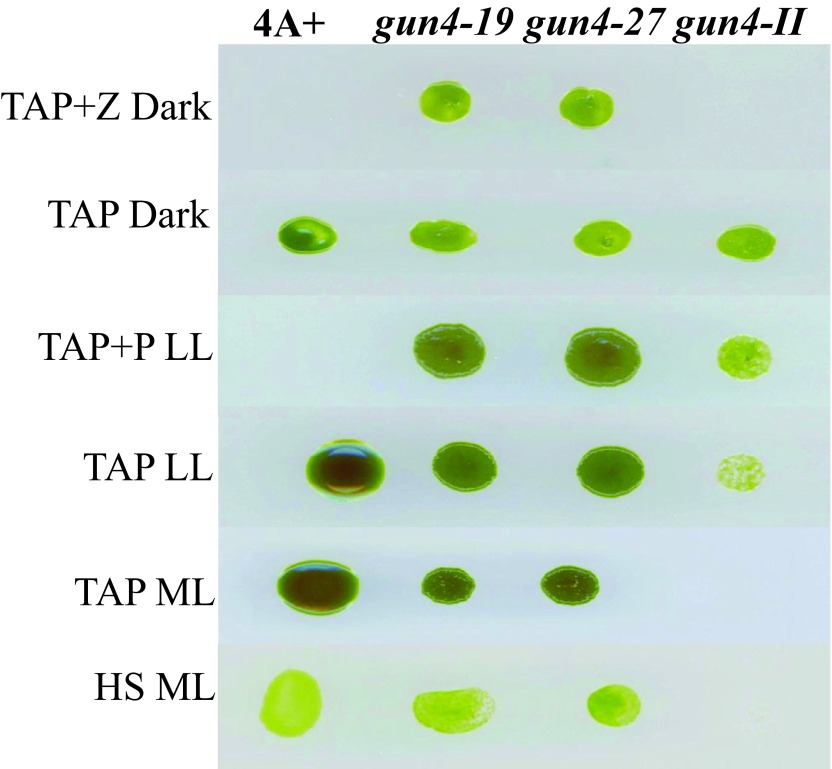
We will be referring to 6F14 as gun4-II from here onward. As gun4-II specifically lacks a functional GUN4 gene, we cloned the GUN4 gene in the pDBle vector to transform 6F14 ( Figure 8, Table 5). The trans GUN4 expression is driven by the constitutive PsaD promoter in the GUN4-pDBle construct. pDBle has two Ble genes that confer resistance to the antibiotic zeocin. Figure 9 shows growth phenotypes of two gun4-II complements ( gun4-19 and gun4-27), 6F14 and 4A+. gun4-II complements are not light sensitive and are able to grow and photosynthesize under medium light intensities (300 µmol photons m -2 s -1) without photo-bleaching ( Figure 9). As gun4-II complements harbor the Ble gene (from the pDBle vector) and APHVIII gene (derived from the parental strain gun4-II), they can grow both on zeocin and paromomycin media plates unlike gun4-II and 4A+ ( Figure 9).
Figure 9. Growth phenotype analysis of gun4-II complements.
gun4-19 and gun4-27 complements, gun4-II and 4A+ were grown for a week under six different growth conditions: TAP + Z (zeocin) in the dark, TAP in the dark, TAP + P (paromomycin) low light (LL; 50 µmol photons m -2 s -1), TAP LL, TAP medium light (ML; 300 µmol photons m -2 s -1) and HS ML.
Chl analyses show that under heterotrophic conditions both gun4-II complements have 65–68% more Chl than that of the wild type cells ( Figure 10). Under photo-autotrophic conditions gun4-II complement cells possess 50–60% more Chl than that of the wild type cells ( Figure 10). Figure 11A shows a schematic figure of the trans GUN4 gene used for complementation. PCR analyses using the genomic DNA show that the gun4-II complements possess the functional trans GUN4 gene ( Figure 11B, 11C and 11D). Figure 12A shows a stained protein gel that was loaded on equal Chl basis. Western analyses of the two gun4-II complements with a Chlamydomonas GUN4 specific antibody show that the GUN4 protein is absent in the gun4-II mutant but present in the gun4 complements ( Figure 12B). Western analyses also show that the two gun4-II complements have higher levels of the GUN4 protein compared to that of the wild type ( Figure 12B).
Figure 10. Mixotrophic and photo-autotrophic growth of gun4-II complements.
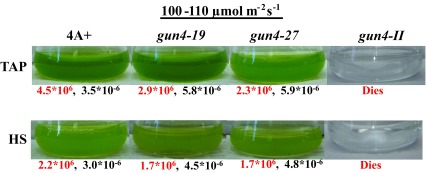
Light intensity is labeled above the culture flask. Growth media is labeled to the left of the culture flask. The mean cell density (cells/ml) and the Chlorophyll (Chl) content (nmol Chl per cell) are shown below the culture flasks in red and black numbers, respectively. For each light condition and growth condition, experiments were performed on three biological replicates of each strain. Statistical error (±SD) was ≤ 10%.
Figure 12. SDS-PAGE and Western analyses.
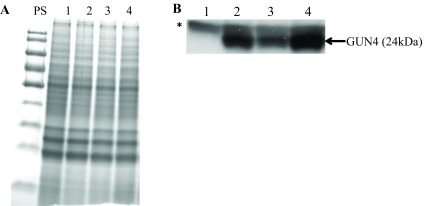
( A) A stained protein gel. Lanes 1, 2, 3 and 4 represent gun4-II, gun4-19, 4A+ and gun4-27, respectively. PS denotes prestained molecular weight protein ladder. Total cell extract of different strains were loaded on equal Chlorophyll (Chl) basis (4 µg of Chl). ( B) Western analyses using a GUN4 antibody generated against the Chlamydomonas mature full length GUN4 protein. Lanes 1, 2, 3 and 4 represent gun4-II, gun4-19, 4A+ and gun4-27, respectively. GUN4 (24 kDa) protein detected by the antibody is labeled. * denotes a 25 kDa protein detected non-specifically by the GUN4 antibody.
The cultures were tested under a range of light intensites. The light intensity shown under the light column denotes the lower limit of the light intensity range in µmol photons m-2s-1. 10 denotes a range of 10-15 µmol photons m-2s-1; 40 denotes a range of 40-50 µmol photons m-2s-1; 75 denotes a range of 75-80 µmol photons m-2s-1 and 100 denotes a range of 100-110 µmol photons m-2s-1. 6F14 does not survive under 100-110 µmol photons m-2s-1. The cultures were tested shifted to a range of light intensity. The light intensity shown under the light column denotes the lower limit of the light intensity range in µmol photons m-2s-1. 40 denotes a range of 40-50 µmol photons m-2s-1 and 75 denotes a range of 75-80 µmol photons m-2s-1. Dark adapted 6F14 shifted to 75-80 µmol photons m-2s-1 did not survive. The cultures were tested under a range of light intensity. The light intensity shown under the light column denotes the lower limit of the light intensity range in µmol photons m-2s-1.100 means 100-110 µmol photons m-2s-1. 6F14 fails to survive under 100-110 µmol photons m-2s-1 in TAP and HS media.
Discussion
Plastid development and gene expressions are largely under nuclear “anterograde” control 40. Additionally, chloroplast functional and developmental states can regulate expression of nuclear genes encoding chloroplast localized proteins via retrograde signaling 40. The first evidence for the involvement of Chl biosynthetic precursors in retrograde signaling came from the work in Chlamydomonas 41. In Arabidopsis MgPPIX was hypothesized to be a retrograde signal from the chloroplast to the nucleus on the basis of data obtained with mutants that are defective in the norflurazon (NF) induced down-regulation of transcription of light harvesting complex protein B (LHCB) [gun (genomes uncoupled) phenotype] 40, 42. Six gun mutants are known; five of which directly influence tetrapyrrole biosynthesis (gun2-gun6) 43, 44. The gun4 mutation is localized to a porphyrin binding protein GUN4. GUN4 enhances the sensitivity of MgChel to Mg 2+ at physiologically low Mg 2+ concentration 31, 45. Cyanobacterial and higher plant GUN4 directly interacts with the CHLH subunit of MgChel and binds PPIX and MgPPIX, the substrate and the reaction product of the MgChel 30, 46– 50. Although GUN4 is not an essential component of the MgChel complex, the presence of GUN4 markedly improves the enzyme activity in vitro by increasing the apparent substrate-binding capacity of CHLH for PPIX, particularly under low Mg 2+ concentrations 45, 51, 52. It is proposed that GUN4 upon porphyrin binding, stabilizes interactions between the catalytic subunit of MgChel and the chloroplast membranes, the site of Chl biosynthesis 46, 47. This enables MgChel to interact with enzyme complexes involved in the further downstream steps in the pathway 46, 47. Apart from its role in substrate channeling into the Chl synthesizing branch of tetrapyrrole biosynthesis, GUN4 has also been implicated in providing photo-protection under increasing light intensities 30, 46, 47. The porphyrin binding property of GUN4 has been implicated in ROS attenuation but conclusive experimental support is lacking 47. In higher plants, GUN4 has been implicated as an essential component in a post-translational feedback regulation mechanism that modulates ALA biosynthesis in response to enzymatic activities of the Mg branch of tetrapyrrole biosynthesis as well as to the accumulating Mg porphyrin levels 30 ( Figure 1).
6F14 is the second gun4 mutant ( gun4-II) to be identified in C. reinhardtii. The first C. reinhardtii gun4 mutant was identified and characterized in 2012 by Formighieri et al. 31. In this gun4 mutant, 184 bp of the second exon of the GUN4 gene is deleted. In gun4-II the plasmid insertion outside the GUN4 gene has caused a genetic rearrangement of the GUN4 gene that prevented gene expression ( Figure 6). Transcripts of GUN4 and the neighboring genes of GUN4 in gun4-II were checked by performing semi-quantitative reverse transcription PCR. In gun4-II, the transcript level of the first downstream hypothetical ( HYP2) gene was lower than that in the wild type ( Figure 7). The plasmid insertion in gun4-II has led to a deletion of part of the 3´ UTR region of the HYP2 gene (270 bp away from the stop codon of the coding region of HYP2; Figure 6). The 3´ UTR is usually responsible for the stability of the transcript. Hence the nature of the deletion in HYP2 explains the decrease in transcript levels of HYP2. GUN4 and the upstream gene, HYP1, are separated from each other by 2.784 kb ( Figure S5). There is a possible deletion/genetic rearrangement in the 5′ genomic region upstream of the GUN4 which does not extend into the HYP1 gene ( Figure S5 and Figure 6). Although the transcription of HYP1 was not hampered, the HYP1 transcript level is lower in our gun4 compared to that in the wild type ( Figure 7). Based on the RT-PCR analyses, it is speculated there might be some uncharacterized downstream regulatory sequences present in the 2.784 kb region that might regulate HYP1 transcription. In future, quantitative real time-PCR experiments can be used to accurately quantify transcript levels of HYP1 and HYP2 in gun4-II.
The photosensitive phenotype of our gun4-II mutant resembles that of the earlier identified Chlamydomonas gun4 mutant which we will refer from here onward, as gun4-I. Over-accumulation of photo-excitable PPIX leads to photo-oxidative damage to the cells in presence of light and oxygen 4, 26, 53. The light sensitivity of gun4-II is most probably due to an over-accumulation of the PPIX which occurs due to the inactivity of MgChel enzyme as has been shown by Formighieri et al. (2012) 31 in the gun4-I mutant. Future HPLC (High Performance Liquid Chromatography) analyses of steady state tetrapyrrole intermediates in gun4-II will confirm this hypothesis. Formighieri et al. (2012) 31 explored four light conditions (dark, 6-, 50-, and 500 µmol photons m -2 s -1) and showed that the gun4-I Chlamydomonas mutant dies under high light (500 µmol photons m -2 s -1). These researchers did not explore or clarify the maximum light irradiance condition that can be tolerated by the Chlamydomonas gun4-I mutant in heterotrophic and photosynthetic growth conditions. In this study, we found that gun4-II photo-bleached at 75–80 µmol photons m -2 s -1 and could not tolerate light intensity above 100 µmol photons m -2 s -1 ( Figure 4 and Figure 5). The earlier identified C. reinhardtii gun4-I mutant is able to grow in continuous light slightly better than in photoperiodic shifts 31. In Arabidopsis, the gun4 mutant is seen to exhibit significant improved growth in continuous light compared to periodic shifts in light 30. In this study, gun4-II and the wild type were adapted to dark or dim light and then shifted to two different light irradiances (40–50 µmol photons m -2 s -1 and 75–80 µmol photons m -2 s -1). Cultures exposed to light shifts showed a significant reduction in the Chl content than those grown under a constant light intensity ( Figure 4 and Figure S1). Additionally, dark adapted gun4-II showed a significant reduction in the Chl content compared to the dim light adapted gun4-II, when cells were shifted to similar light intensities ( Figure 6). These results show that gun4-II is very sensitive to the magnitude of light intensity fluctuations in the environment unlike the earlier reported Chlamydomonas gun4-I mutant 31. Our light shift experimental results support the findings in cyanobacterial and Arabidopsis gun4 mutants 30, 48– 50.
By spectrophotometric analysis we have shown that in the dark gun4-II possesses almost similar Chl content like that in the wild type ( Figure 4). This phenotype is very different from that of gun4-I, which possesses 50% of the wild type level of Chl/cell 31 in the dark. Variation in Chl/cell in the dark between the two C. reinhardtii gun4 mutants could possibly be due to a variation of the parental strain’s ability to synthesize Chl in the dark. The parental strain used by Formighieri et al. (2012) 31 was cw15mt-. However, the 50% decrease in Chl seen in the the gun4-I mutant was determined through HPLC analyses. Hence the discrepancy in Chl content in the two gun4 mutants could be due to the sensitivity of the HPLC method compared to that of the spectrophotometric method used for Chl assays.
Steady state tetrapyrrole analyses by HPLC can be performed to check the various tetrapyrrole intermediate accumulation in gun4-II under different light conditions. Measurements of ALA biosynthesis rate in gun4-II can show if GUN4 also regulates earlier steps in the tetrapyrrole biosynthetic pathway, as suggested by some researchers 48.
Interestingly, it has been shown by Formighieri et al. (2012) 31 that Chlamydomonas gun4-I complements expressing more GUN4 protein grow better under high light and that there is no correlation between the accumulation of PPIX and the ability to grow better under high light 31. However these gun4-I complements were not over-expressers of the GUN4 protein compared to the wild type strain cw15, used in their experiments 31. Our two gun4-II complements ( gun4-19 and gun4-27) are over-expressing the GUN4 protein compared to the wild type strain 4A+ in the dim light ( Figure 12B). These two gun4-II complements open up new avenues to test if GUN4 has a distinct photo-protective role that is independent from the PPIX-induced GUN4 photo-protective role proposed by several researchers 46, 47. Comparative growth studies, quantitative measurements of GUN4 transcripts by Real Time PCR, GUN4 protein levels by Western analyses and PPIX content by HPLC analyses of the high light-(500 µmol photons m -2 s -1) and dim light-(15–20 µmol photons m -2 s -1) adapted gun4-II complements and the wild type strain will help to confirm if GUN4 has a distinct photo-protective role that is independent of tetrapyrrole metabolism.
Taken together our work reconfirms the results of other researchers who have studied GUN4 in other photosynthetic organisms 30, 31, 46, 47, 49, 50. Although loss of GUN4 caused a perturbation in Chl biosynthesis in gun4-II mutant, the effect is not as dramatic as it is in Arabidopsis, where the loss of GUN4 results in a nearly albino mutant 51. The earlier identified Chlamydomonas gun4-I mutant phenotypically resembles our gun4-II mutant 31. Therefore it seems that in C. reinhardtii Chl biosynthesis is less dependent on the GUN4 function. One explanation for this difference in the mutant phenotype could be that Chlamydomonas is capable of synthesizing Chl in the dark unlike the angiosperms. GUN4 interacts with PPIX and acts at the branch point in the tetrapyrrole biosynthetic pathway where PPIX is diverted to heme and Chl biosynthesis. Hence, although GUN4 has a conserved physiological role in all oxygenic photosynthetic organisms, it might have a different role in different evolutionary groups depending on the channelization of PPIX into the heme and Chl branch in the pathway.
Acknowledgements
We would like to thank Dr. Krishna K. Niyogi (UC, Berkeley) for providing the 4A+ strain and the pBC1 plasmid that were used for mutagenesis and the GUN4 antibody for Western analyses. We are grateful to Dr. Saul Purton (University College London, UK) for providing the complementation vector pDBle. We are grateful to Dr. Bernhard Grimm and Dr. Pawel Brzezowski (Humboldt University, Berlin, Germany) for providing us the SOXE gene specific primers for RT-PCR analyses. We would also like to thank Dr. Leos Kral (University of West Georgia) for allowing us to use his nano spectrophotometer and Dr. Anastasios Melis (UC, Berkeley) for allowing us to perform the protein and Western analyses at his laboratory.
Funding Statement
This project was supported by several grants awarded to Dr. Mautusi Mitra. These are: the start-up grant of the University of West Georgia (UWG), the Faculty Research Grant by the UWG College of Science and Mathematics, the Internal Development Grant by the UWG office of Research and Sponsored Project, the Research Incentive grant by the UWG College of Science and Mathematics, the UWG Student Research Assistance Program (SRAP) grant and the UWise-BOR-STEM II grant from UWG.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
v2; ref status: indexed
Supplementary materials
Figure S1. Effect of light shift on the growth of 6F14 and wild type.
6F14 was adapted to dim light (10–15 µmol photons m -2 s -1) or dark for one week in TAP media. Dark and dim light adapted cultures were then shifted to 40–50 or 75–80 µmol photons m -2 s -1. The mean cell density (cells/ml) and the Chlorophyll (Chl) content (nmol Chl per cell) are shown below the culture flasks in red and black numbers, respectively. For each light condition, experiments were performed on three biological replicates of 6F14. Statistical error (±SD) was ≤ 10%. The average Chl content in the dim light and dark adapted 6F14 was 1.7 × 10 -6 and 2.18 × 10 -6 nmol/cell.
Figure S2. Locating the APHVIII flanking genomic sequence in 6F14.
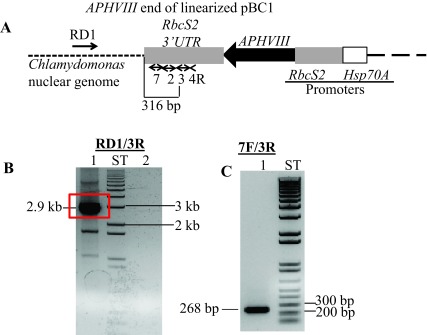
( A) A diagram showing a truncated pBC1 illustrating the APHVIII end of the linearized pBC1 vector. Primers used for PCR and DNA sequencing are shown by numbered black arrows. Thermal Asymmetric InterLaced1 (TAIL1) PCR was performed using primers 4R and RD1 (a random degenerate primer). ( B) TAIL2 PCR was performed using primers 3R and RD1. In lane 1, 10-fold diluted TAIL1 PCR product was used for TAIL2 PCR; Lane 2 is a zero DNA control lane. The 2.9 kb TAIL2 PCR product used for DNA sequencing is highlighted in the red box. Initial DNA sequencing was performed using vector specific primers 2R and 3R ( Table 1). ( C) Gel purified DNA product (2.9 kb) from TAIL2 PCR was used to verify if the product is specific to the APHVIII gene. PCR primer names are labeled on the top of the gel. PCR product size is labeled. F and R stand for forward and reverse primers, respectively. All primer sequences are shown in Table 1. ST stands for 1 kb plus ladder (Invitrogen, Carlsbad, CA). DNA samples were run on a 1% agarose gel.
Figure S3. A schematic of the genetic rearrangement in 6F14 based on TAIL PCR analyses.
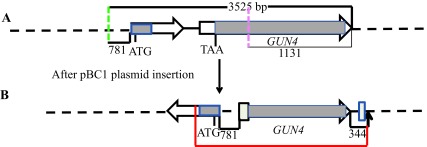
( A) A schematic genomic map showing a 3525 bp genomic DNA region spanning the GUN4 locus. The number at the bottom of the map denotes distance between respective points on the genomic DNA. The two GUN4 exons are represented by white block arrows. Grey boxes in GUN4 denote UnTranslated Regions (UTRs). The two break points are shown by the dashed pink and green lines. ( B) A schematic diagram showing the rearrangement of the GUN4 locus after the insertion of the plasmid. The big and the small tan boxes, denote addition of 45 and 29 bp, respectively. The genomic DNA sequence obtained by sequencing the 2.9 kb Thermal Asymmetric InterLaced2 (TAIL2) PCR product is highlighted in red. The bold black small arrow indicates insertion point of the pBC1 plasmid. DNA sequencing was performed using GUN4 specific primers 2R, 7F and 7R, 12R and 14R. F and R stand for forward and reverse primers, respectively. Primer sequences are shown in Table 2.
Figure S4. Genomic DNA analysis of 6F14 and 4A+.
( A) A schematic of the GUN4 gene. Small black arrows denote GUN4 specific primers. The two GUN4 exons are represented by white block arrows. Grey boxes in GUN4 denote UnTranslated Regions (UTRs). ( B), ( C) and ( D) DNA gels showing DNA products obtained from genomic DNA PCR using GUN4 specific primers. Lanes 1, 2 and 3 are the zero DNA controls, 6F14 and 4A+, respectively. PCR primer names are labeled on the top of the gel. 14F/3R gives a 517 bp product while 3F/8R gives a 942 bp product. 8F/7R gives a 449 bp product. 7F/12R gives a 313 bp product. 12F/10R gives a 606 bp product and 12F/11R gives a 623 bp product. F and R stand for forward and reverse primers, respectively. Primer sequences are shown in Table 2.
Figure S5. PCR using primers specific to the upstream region of GUN4 and the 3´ UTR of the HYP2 gene.
( A) A schematic diagram showing GUN4 (white arrow) and its two neighboring genes HYP1 (black arrow) and HYP2 (grey arrow). Black numbers at the bottom denote distances between respective genes. The two red highlighted regions and corresponding numbers show the distance between primer ACF7 and the start of the GUN4 gene and the distance between the primer H5F and the end of the HYP2 gene, respectively. Primers used for PCR are labeled. ( B) A DNA gel showing the genomic DNA amplified using GUN4 upstream region specific primers. Lanes 1, 3 and 5: 6F14; Lanes 2, 4 and 6: 4A+. ( C) A DNA gel showing the genomic DNA amplified using primers spanning the 3´ UTR region of HYP2 gene. Lanes 1, 3 and 5: 6F14; Lanes 2, 4 and 6: 4A+. Product size of ACF6/ACR7: 937 bp (this primer set also produces a nonspecific 550 bp product); product size of ACF7/ACR11: 244 bp; product size of H3F/H6R: 667 bp; product size of H4F/H6R: 391 bp; product size of H5F/H6R: 278 bp. F and R stand for forward and reverse primers, respectively. Primer sequences are shown in Table 3.
References
- 1.Merchant SS, Prochnik SE, Vallon O, et al. : The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250 10.1126/science.1143609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dent RM, Haglund CM, Chin BL, et al. : Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 2005;137(2):545–556 10.1104/pp.104.055244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutman BL, Niyogi KK: Chlamydomonas and Arabidopsis. A dynamic duo. Plant Physiol. 2004;135(2):607–610 10.1104/pp.104.041491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beale SI: Green genes gleaned. Trends Plant Sci. 2005;10(7):309–312 10.1016/j.tplants.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 5.Grossman AR, Lohr M, Im CS: Chlamydomonas reinhardtii in the landscape of pigments. Annu Rev Genet. 2004;38:119–173 10.1146/annurev.genet.38.072902.092328 [DOI] [PubMed] [Google Scholar]
- 6.Walker CJ, Willows RD: Mechanism and regulation of Mg-chelatase. Biochem J. 1997;327(Pt 2):321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong GA, Runge S, Frick G, et al. : Identification of NADPH:protochlorophyllide oxidoreductases A and B: a branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol. 1995;108(4):1505–1517 10.1104/pp.108.4.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oosawa N, Masuda T, Awai K, et al. : Identification and light-induced expression of a novel gene of NADPH-protochlorophyllide oxidoreductase isoform in Arabidopsis thaliana. FEBS Lett. 2000;474(2–3):133–136 10.1016/S0014-5793(00)01568-4 [DOI] [PubMed] [Google Scholar]
- 9.Reinbothe S, Reinbothe C: Regulation of chlorophyll biosynthesis in angiosperms. Plant Physiol. 1996;111(1):1–7 10.1104/pp.111.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su Q, Frick G, Armstrong G, et al. : POR C of Arabidopsis thaliana: a third light- and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol Biol. 2001;47(6):805–813 10.1023/A:1013699721301 [DOI] [PubMed] [Google Scholar]
- 11.Fujita Y, Takahashi Y, Chuganji M, et al. : The nifH-like ( frxC) gene is involved in the biosynthesis of chlorophyll in the filamentous cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1992;33(1):81–92 Reference Source [Google Scholar]
- 12.Fujita Y, Matsumoto H, Takahashi Y, et al. : Identification of a nifDK-like gene ( ORF467) involved in the biosynthesis of chlorophyll in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1993;34(2):305–314 [PubMed] [Google Scholar]
- 13.Fujita Y, Takagi H, Hase T: Identification of the chlB gene and the gene product essential for the light-independent chlorophyll biosynthesis in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1996;37(3):313–323 [DOI] [PubMed] [Google Scholar]
- 14.Fujita Y, Takagi H, Hase T: Cloning of the gene encoding a protochlorophyllide reductase: the physiological significance of the co-existence of light-dependent and -independent protochlorophyllide reduction systems in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1998;39(2):177–185 [DOI] [PubMed] [Google Scholar]
- 15.Fujita Y, Bauer CE: Reconstitution of light-independent protochlorophyllide reductase from purified bchL and BchN-BchB subunits. In vitro confirmation of nitrogenase-like features of a bacteriochlorophyll biosynthesis enzyme. J Biol Chem. 2000;275(31):23583–23588 10.1074/jbc.M002904200 [DOI] [PubMed] [Google Scholar]
- 16.Fujita Y, Bauer C: The light-independent protochlorophyllide reductase: a nitrogen-like enzyme catalyzing a key reaction for greening in dark. In: Kadish KM, Smith KM, Guilard R, eds, The Porphyrin Handbook, Amsterdam: Academic Press.2013; 13:109–156 [Google Scholar]
- 17.Nomata J, Swem LR, Bauer CE, et al. : Overexpression and characterization of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. Biochim Biophys Acta. 2005;1708(2):229–237 10.1016/j.bbabio.2005.02.002 [DOI] [PubMed] [Google Scholar]
- 18.Nomata J, Ogawa T, Kitashima M, et al. : NB-protein (BchN-BchB) of dark-operative protochlorophyllide reductase is the catalytic component containing oxygen-tolerant Fe-S clusters. FEBS Lett. 2008;582(9):1346–1350 10.1016/j.febslet.2008.03.018 [DOI] [PubMed] [Google Scholar]
- 19.Shui J, Saunders E, Needleman R, et al. : Light-dependent and light-independent protochlorophyllide oxidoreductases in the chromatically adapting cyanobacterium Fremyella diplosiphon UTEX 481. Plant Cell Physiol. 2009;50(8):1507–1521 10.1093/pcp/pcp095 [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto H, Kurumiya S, Ohashi R, et al. : Oxygen sensitivity of a nitrogenase-like protochlorophyllide reductase from the cyanobacterium Leptolyngbya boryana. Plant Cell Physiol. 2009;50(9):1663–1673 10.1093/pcp/pcp111 [DOI] [PubMed] [Google Scholar]
- 21.Sakuraba Y, Yamasato A, Tanaka R, et al. : Functional analysis of N-terminal domains of Arabidopsis chlorophyllide a oxygenase. Plant Physiol Biochem. 2007;45(10–11):740–749 10.1016/j.plaphy.2007.07.016 [DOI] [PubMed] [Google Scholar]
- 22.Tanaka A, Ito H, Tanaka R, et al. : Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc Natl Acad Sci U S A. 1998;95(21):12719–12723 10.1073/pnas.95.21.12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka R, Tanaka A: Effects of chlorophyllide a oxygenase overexpression on light acclimation in Arabidopsis thaliana. Photosynth Res. 2005;85(3):327–340 10.1007/s11120-005-6807-z [DOI] [PubMed] [Google Scholar]
- 24.Yamasato A, Nagata N, Tanaka R, et al. : The N-terminal domain of chlorophyllide a oxygenase confers protein instability in response to chlorophyll b accumulation in Arabidopsis. Plant Cell. 2005;17(5):1585–1597 10.1105/tpc.105.031518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meguro M, Ito H, Takabayashi A, et al. : Identification of the 7–hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. Plant Cell. 2011;23(9):3442–3453 10.1105/tpc.111.089714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moulin M, Smith AG: Regulation of tetrapyrrole biosynthesis in higher plants. Biochem Soc Trans. 2005;33(Pt 4):737–742 10.1042/BST0330737 [DOI] [PubMed] [Google Scholar]
- 27.Beck CF: Signaling pathways from the chloroplast to the nucleus. Planta. 2005;222(5):743–756 10.1007/s00425-005-0021-2 [DOI] [PubMed] [Google Scholar]
- 28.Papenbrock J, Grimm B: Regulatory network of tetrapyrrole biosynthesis--studies of intracellular signaling involved in metabolic and developmental control of plastids. Planta. 2001;213(5):667–681 10.1007/s004250100593 [DOI] [PubMed] [Google Scholar]
- 29.Papenbrock J, Mishra S, Mock HP, et al. : Impaired expression of the plastidic ferrochelatase by antisense RNA synthesis leads to a necrotic phenotype of transformed tobacco plants. Plant J. 2001;28(1):41–50 10.1046/j.1365-313X.2001.01126.x [DOI] [PubMed] [Google Scholar]
- 30.Peter E, Grimm B: GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol Plant. 2009;2(6):1198–1210 10.1093/mp/ssp072 [DOI] [PubMed] [Google Scholar]
- 31.Formighieri C, Ceol M, Bonente G, et al. : Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii. Mol Plant. 2012;5(6):1242–1262 10.1093/mp/sss051 [DOI] [PubMed] [Google Scholar]
- 32.Gorman DS, Levine RP: Cytochrome f and plastocyanin – their sequence in photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1965;54(6):1665–1669 10.1073/pnas.54.6.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sueoka N: Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1960;46(1):83–91 10.1073/pnas.46.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kindle KL, Schnell RA, Fernandez E, et al. : Stable nuclear transformation of chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989;109(6 Pt 1):2589–2601 10.1083/jcb.109.6.2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies JP, Weeks DP, Grossman AR: Expression of the arylsulfatase gene from the beta 2–tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 1992;20(12):2959–2965 10.1093/nar/20.12.2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith BM, Morrissey PJ, Guenther JE, et al. : Response of the Photosynthetic Apparatus in Dunaliella salina (Green Algae) to Irradiance Stress. Plant Physiol. 1990;93(4):1433–1440 10.1104/pp.93.4.1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laemmli UK: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 38.Arnon DI: Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24(1):1–15 10.1104/pp.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melis A, Spangfort M, Andersson B: Light-absorption and electron-transport balance between photosystem-II and photosystem-I in spinach-chloroplasts. Photochem Photobiol. 1987;45(1):129–136 10.1111/j.1751-1097.1987.tb08413.x [DOI] [Google Scholar]
- 40.Nott A, Jung HS, Koussevitzky S, et al. : Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol. 2006;57:739–59 10.1146/annurev.arplant.57.032905.105310 [DOI] [PubMed] [Google Scholar]
- 41.Johanningmeier U, Howell SH: Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardi. Possible involvement of chlorophyll synthesis precursors. J Biol Chem. 1984;259(21):13541–13549 [PubMed] [Google Scholar]
- 42.Susek RE, Ausubel FM, Chory J: Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74(5):787–799 10.1016/0092-8674(93)90459-4 [DOI] [PubMed] [Google Scholar]
- 43.Strand Å, Asami T, Alonso J, et al. : Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421(6918):79–83 10.1038/nature01204 [DOI] [PubMed] [Google Scholar]
- 44.Woodson JD, Perez-Ruiz JM, Chory J: Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr Biol. 2011;21(10):897–903 10.1016/j.cub.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davison PA, Schubert HL, Reid JD, et al. : Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry. 2005;44(21):7603–7612 10.1021/bi050240x [DOI] [PubMed] [Google Scholar]
- 46.Adhikari ND, Orler R, Chory J, et al. : Porphyrins promote the association of GENOMES UNCOUPLED 4 and a Mg-chelatase subunit with chloroplast membranes. J Biol Chem. 2009;284(37):24783–24796 10.1074/jbc.M109.025205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adhikari ND, Froehlich JE, Strand DD, et al. : GUN4–porphyrin complexes bind the ChlH/GUN5 subunit of Mg-chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell. 2011;23(4):1449–1467 10.1105/tpc.110.082503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peter E, Wallner T, Wilde A, et al. : Comparative functional analysis of two hypothetical chloroplast open reading frames (ycf) involved in chlorophyll biosynthesis from Synechocystis sp. PCC6803 and plants. J Plant Physiol. 2011;168(12):1380–1386 10.1016/j.jplph.2011.01.014 [DOI] [PubMed] [Google Scholar]
- 49.Sobotka R, Dühring U, Komenda J, et al. : Importance of the cyanobacterial Gun4 protein for chlorophyll metabolism and assembly of photosynthetic complexes. J Biol Chem. 2008;283(38):25794–25802 10.1074/jbc.M803787200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilde A, Mikolajczyk S, Alawady A, et al. : The gun4 gene is essential for cyanobacterial porphyrin metabolism. FEBS Lett. 2004;571(1–3):119–123 10.1016/j.febslet.2004.06.063 [DOI] [PubMed] [Google Scholar]
- 51.Larkin RM, Alonso JM, Ecker JR, et al. : GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003;299(5608):902–906 10.1126/science.1079978 [DOI] [PubMed] [Google Scholar]
- 52.Verdecia MA, Larkin RM, Ferrer JL, et al. : Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding. PLoS Biol. 2005;3(5):e151 10.1371/journal.pbio.0030151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka R, Tanaka A: Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 2007;58:321–346 10.1146/annurev.arplant.57.032905.105448 [DOI] [PubMed] [Google Scholar]