Abstract
Glial cells are emerging as important regulators of synapse formation, maturation, and plasticity through the release of secreted signaling molecules. Here we use chromatin immunoprecipitation along with Drosophila genomic tiling arrays to define potential targets of the glial transcription factor Reversed polarity (Repo). Unexpectedly, we identified wingless (wg), a secreted morphogen that regulates synaptic growth at the Drosophila larval neuromuscular junction (NMJ), as a potential Repo target gene. We demonstrate that Repo regulates wg expression in vivo and that local glial cells secrete Wg at the NMJ to regulate glutamate receptor clustering and synaptic function. This work identifies Wg as a novel in vivo glial-secreted factor that specifically modulates assembly of the postsynaptic signaling machinery at the Drosophila NMJ.
Keywords: Drosophila, glia, NMJ, synapse, wnt/Wg
Introduction
Glial cells are intimately associated with neurons and exert significant control over neuronal development and function. Glia secrete a number of factors that potently modulate the formation and maturation of synapses (Ullian et al., 2004; Barres, 2008). Excitatory glutamatergic synapse formation is promoted by glia-derived thrombospondins (TSPs; Christopherson et al., 2005) and glypicans 4 and 6 in vivo (Allen et al., 2012). Glia can also regulate the formation of inhibitory synapses in vitro by acting on the assembly of postsynaptic GABAA receptors (Elmariah et al., 2005; Hughes et al., 2010). In vitro studies suggest that glial control of synapse formation is complex, requiring multiple factors that can differentially affect the assembly of presynaptic and postsynaptic structures (Christopherson et al., 2005; Elmariah et al., 2005; Hughes et al., 2010), and at least a subset of these proteinacious factors remain to be identified (Hughes et al., 2010).
The Drosophila nervous system is an attractive model to explore early aspects of glial cell fate induction, maturation, and control of neural circuit connectivity (Freeman and Doherty, 2006; Stork et al., 2012). The vast majority of newly born Drosophila glia express glial cells missing (gcm) gene, which encodes a transcription factor necessary and sufficient to induce the glial developmental program in the embryo (Hosoya et al., 1995; Jones et al., 1995; Vincent et al., 1996). Gcm transcriptionally activates a number of important glial genes including reversed polarity (repo; Akiyama et al., 1996), which encodes a homeodomain-containing transcription factor critical for both maturation of glial cell fate and active repression of neuronal fate (Campbell et al., 1994; Xiong et al., 1994). Null alleles of repo cause embryonic lethality, although most glia are initially specified and positioned correctly (Campbell et al., 1994; Halter et al., 1995). Interestingly, null repo mutant animals fail to activate a number of terminal differentiation genes in glia, including the EAAT1 and EAAT2 transporters that mediate reuptake of the neurotransmitter glutamate from synapses (Soustelle et al., 2002), and the regulator of G protein signaling locomotion defective (loco; Yuasa et al., 2003), a key mediator of blood–brain barrier formation (Granderath et al., 1999; Schwabe et al., 2005). Although the precise molecular basis for defects remains unclear, repo mutants exhibit profound alterations in neuronal physiology: in the repo1 mutant, retinal photoreceptor field potentials are completely reversed (Xiong et al., 1994).
Together, these observations argue that Repo plays a central role in activating programs essential for late steps in glial cell fate and neuron–glia signaling. We reasoned that the identification of direct targets of Repo would shed significant light on the signaling pathways activated during glial differentiation and early neuron–glia signaling events. In this study we used chromatin immunoprecipitation (ChIP) with Repo to identify an extensive collection of potential Repo target genes. This collection included many previously known glial genes and, unexpectedly, multiple components of the Wingless (Wg)/Wnt signaling pathway. Using the Drosophila neuromuscular junction (NMJ), we demonstrate that Repo can indeed modulate Wg levels in vivo and that glial-released Wg is a critical regulator of glutamate receptor (GluR) clustering and synaptic function.
Materials and Methods
Fly strains and constructs.
Flies of either sex were raised on standard Drosophila media at 25°C. For RNAi experiments, RNAi lines and controls were raised at 29°C. The following fly strains were used in this study: wild type (Canton-S); rl82-Gal4 (Sepp and Auld, 1999), repo-Gal4 (Lee and Jones, 2005), C380-Gal4 (Budnik et al., 1996), OK6-Gal4 (Marqués et al., 2002), moody-Gal4 (Bainton et al., 2005), PG-Gal4 (NP6293-Gal4) (Awasaki et al., 2008), nervana2-Gal4 [Nrv2-Gal4; Bloomington Stock Center (BSC), stock #6800; Sun et al., 1999], UAS-Wg-RNAi [stock #13352; Vienna Drosophila RNAi Center (VDRC)], UAS-Porc-RNAi (stock #47864; VDRC), UAS-Repo-RNAi (stock #10424; VDRC), UAS-mCD8-GFP (stock #5137; BSC), wg1cn1 (stock # 2987; BSC), repo1 (stock #4162; BSC), w1118; ry506 P[ry+t7.2=PZ] repo03702/TM3, ryRK Sb1 Ser1 (referred to here as repoPZ; stock #11604, BSC).
Immunolabeling, antibody source, and concentration.
Third instar Drosophila larvae were dissected in calcium-free saline (Jan and Jan, 1976) and fixed for 10 min with nonalcoholic Bouin's solution (5% acetic acid, 9% formaldehyde, 0.9% picric acid). Primary antibodies were used at the following dilutions: rabbit anti-Wg, 1:400 (Reichsman et al., 1996); mouse anti-GFP, 1:200 (Invitrogen); mouse anti-glutamate receptor IIA (anti-GluRIIA), 1:3 (Developmental Studies Hybridoma Bank); FITC or Texas Red-conjugated anti-HRP, 1:200 (Jackson ImmunoResearch). Secondary antibodies conjugated to FITC, Texas Red, or Cy5 (Jackson ImmunoResearch) were used at a concentration of 1:200.
Image quantification.
Samples were imaged using an Intelligent Imaging Innovations Everest spinning disc confocal system using a Zeiss Plan-Apochromat 63× 1.4 numerical aperture oil objective. Different genotypes were processed simultaneously and imaged using identical confocal acquisition parameters for comparison. Bouton number was quantified in larval abdominal segment A3, muscles 6 and 7, of wandering third instar larvae.
Fluorescence signal intensity was quantified by volumetric measurements of confocal stacks using Volocity 6.0 software (Improvision). For measurements of synaptic intensity, single boutons were selected from muscles 6 and 7, A3, and analyzed as three-dimensional volumes in Volocity. By thresholding the anti-HRP, anti-GluRIIA, and anti-Wg signals, we defined the presynaptic volume (HRP) and the signal in (presynaptic) and around (postsynaptic) the boutons for both GluRIIA and Wg. The mean signal intensity was then recorded for each bouton and pooled with simultaneously processed samples from the same genotype. For volume calculations, the volume of signal was obtained from the above analysis (total number of voxels), and this volume was normalized to that of its associated bouton. All samples and controls were normalized to wild type. Statistical analysis was performed using a Student's t test for pairwise comparisons or ANOVA with Tukey's post hoc for comparison of multiple genotypes.
Quantitative real-time PCR.
Total RNA was isolated from dissected third instar peripheral nerves and analyzed as in the study by Fuentes-Medel et al. (2012). The quantitative PCR was performed using the following Taqman (Applied Biosystems) primers: repo (assay ID Dm02134815_g1), wingless (assay ID Dm01803387_m1), and gapdh (assay ID Dm01841185_m1) as a housekeeping control.
Electrophysiology.
All experiments were performed as in the study by Ashley et al. (2005). Third instar larval body wall muscles were dissected in HL3 saline [70 mm NaCl, 5 mm KCl, 20 mm MgCl2, CaCl2 as indicated, 10 mm NaHCO3, 115 mm sucrose, 5 mm trehalose, 5 mm HEPES] containing 0.3 mm CaCl2. The brains were removed after carefully cutting the segmental nerves proximal to the ventral ganglion. During electrophysiological recordings, preparations were continuously superfused with HL3 saline containing 0.5 mm CaCl2. All recordings were performed at muscle 6, A3. Intracellular voltage recordings were obtained by impaling the muscle with sharp electrodes filled with 3 m KCl. Miniature excitatory junctional potentials (mEJPs) were continuously recorded for 4 min, and the average mEJP frequency in hertz was calculated by dividing the total number of mEJP events by 240 s. EJPs were evoked using a suction electrode to capture the segmental nerve; a 1 ms suprathreshold stimulus was applied at a rate of 0.3 pulses per second, and the resulting EJPs were averaged from a total of 4 min of recording for each larva. Quantal content was calculated by dividing the mean EJP amplitude by the mean mEJP amplitude for each sample.
Repo constructs.
To generate the pUAST-myc::Repo construct, the full-length repo cDNA (including the stop codon; 1839 nucleotides) was PCR amplified from GH05443 and cloned into N-terminal pUAST-myc using the BglII and XhoI sites. To generate the pUAST-Repo::myc construct, full-length repo (lacking the stop codon; 1836 nucleotides) was PCR amplified from GH05443 and cloned into C-terminal pUAST-myc using the BglII and SpeI sites.
S2 cell transfections.
Drosophila Schneider (S2) cells were seeded into T-75 cm2 flasks (1 × 106 cells/ml for 10 ml) and transfected with Effectene from Qiagen according to the manufacturer's protocol. pAc5.1C-Gal4 (Potter et al., 2010) was cotransfected with either pUAST-myc::repo or pUAST-repo::myc, and the cells were harvested 3 d after transfection.
Chromatin immunoprecipitation.
To cross-link chromatin with formaldehyde, S2 cells were harvested 3 d after transfection and fixed with formaldehyde solution (final concentration, 5 mm HEPES-KOH, pH 7.5, 10 mm NaCl, 0.1 mm EDTA, 0.05 mm EGTA, and 1.1% formaldehyde) and then quenched with glycine (final concentration, 125 mm). Cells were pelleted at 1100 g at 4°C, resuspended in lysis buffer 1 (50 mm HEPES-KOH, pH 7.5, 140 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1× protease inhibitor cocktail), incubated for 10 min at 4°C, and then pelleted via centrifugation. Cells were then resuspended in lysis buffer 2 (10 mm Tris-HCl, pH 8.0, 200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 1− protease inhibitor cocktail), incubated for 5 min at 4°C, and then pelleted again. Finally, cells were resuspended in lysis buffer 3 (10 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 0.1% sodium deoxycholate, 0.5% N-lauroylsarcosine, 1× protease inhibitor cocktail) and sonicated using a Bioruptor (Diagenode) with two 15 min rounds of pulsed sonication (30 s on, 30 s off). Lysate was mixed with 1/10 volume of 10% Triton X-100 and centrifuged at 20,000 × g for 10 min at 4°C to pellet debris, and the supernatant was precleared on Dynabeads (Life Technologies). The supernatant was then incubated overnight with Dynabeads plus anti-myc antibodies. Beads were washed with RIPA Buffer (50 mm HEPES-KOH, pH 7.5, 500 mm LiCl, 1 mm EDTA, 1.0% NP-40, 0.7% sodium deoxycholate) several times, and then once with TE (10 mm Tris-Cl, pH 7.5, 1 mm EDTA) with 50 mm NaCl. Samples were eluted with elution buffer (TE with 1% SDS), incubated at 65°C for 30 min, and then centrifuged at 16,000 × g for 1 min. The supernatant was then incubated at 65°C for 6 h to reverse the cross-linking. After heating, samples were treated with RNaseA (0.2 mg/ml final concentration) for 2 h at 37°C, and then treated with proteinase K (0.2 μg/ml final concentration) and incubated for 2 h at 55°C. Samples were then phenol/chloroform extracted using the Heavy Phase Lock gel tube (5 Prime) to separate the organic and aqueous phases. DNA was then ethanol precipitated and resuspended in TE buffer. Finally, the sample DNA was analyzed as per Menet et al. (2010) using a GeneChip Drosophila Tiling Array 2.0 (Affymetrix).
Results
Repo regulates multiple genes involved in neuron–glia signaling and specification of glial fate
We sought to identify novel potential transcriptional targets for Repo and explore their in vivo roles in glial cell development and function. We generated two constructs designed for expression of either an N- or C-terminal Myc-tagged version of Repo (Myc::Repo and Repo::Myc, respectively) in Drosophila S2 cells. We transfected S2 cells with either Myc::Repo or Repo::Myc and performed ChIPs using anti-Myc antibodies, and Repo-bound genomic regions were then identified by hybridization of isolates to Drosophila v2.0 tiling arrays (Affymetrix; Fig. 1A). Genomic regions exhibiting significant binding in ChIP assays were identified using the model-based tiling array (MAT) algorithm (Johnson et al., 2006) for both Myc::Repo and Repo::Myc separately. Data sets were cross-compared, and only those loci found to be significantly enriched in both experiments were selected. This approach led to the identification of 2041 loci exhibiting significant binding by Repo, which were defined as significant by having a p value of <10−4 in each ChIP experiment.
Figure 1.
Identification of potential Repo target genes. A, Tiling array data were obtained from two independent Repo ChIP experiments (myc::Repo and Repo::myc) after expression in Drosophila S2 cells. B, Data were analyzed using a MAT algorithm (Johnson et al., 2006) and converted to a linear scale to be viewed using the Affymetrix Integrated Genome Browser. For each gene, the genomic location and isoforms are shown. The relative amplitude of the peaks represents the probability of DNA binding (MAT score). C, Known glial or Wg/Wnt pathway signaling genes identified in Repo-ChIP experiments. D, RNA in situ hybridizations using a gs2 or Cp1 probe to control and repo-null mutant embryos.
Intriguingly, analysis of Repo-bound genomic fragments led to the identification of 16 genes already known to be glial genes, including loco, pointed, EAAT1, Glutamine synthetase 2, akap200, distalless, gliotactin, and dead ringer/retained (Fig. 1C; Freeman et al., 2003). Each of these genes exhibited binding in either 5′, intronic, and/or 3′ regions (Fig. 1B), and thus represented potential direct targets for Repo-dependent transcriptional activation or repression in vivo. At least two of these genes, EAAT1 and loco, have been shown previously to be positively regulated in vivo by Repo (Soustelle et al., 2002; Yuasa et al., 2003). To determine whether any of the new potential Repo targets were in fact regulated by Repo in vivo, we performed RNA in situ hybridizations for a number of genes to wild-type control and repo mutant embryos, including glutamine synthetase 2 (gs2) and Cysteine proteinase 1 (Cp1). Whereas control animals expressed gs2 in longitudinal glia from embryonic stage 14 onward, we could not detect expression of gs2 in glia at any stage in repo mutants (Fig. 1D). Similarly, whereas stage 13 embryos expressed Cp1 in the majority of glial cells, repo mutants exhibited a dramatically reduced pattern of Cp1 expression. Thus, gs2 and Cp1 represent new in vivo regulatory targets of Repo.
Wg expression in peripheral glia is regulated by Repo, and both neurons and glia are in vivo sources for NMJ Wg
Interestingly, in the above analysis we also found genes not previously associated with glia, such as members of Wnt signaling pathways (Fig. 1C). Wnt-1/Wingless is known to be released by motor neuron terminals at the larval NMJ and to regulate the development of both presynaptic and postsynaptic compartments through DFrizzled2 (DFz2) receptors localized at both sites (Packard et al., 2002; Mathew et al., 2005; Ataman et al., 2006; Miech et al., 2008). In the absence of Wnt signaling, the number of synaptic boutons is reduced, GluR subunits become distributed in abnormally broad clusters (Packard et al., 2002), and a subset of boutons (termed ghost boutons) lack postsynaptic proteins, postsynaptic structures, and presynaptic active zones (Packard et al., 2002; Ataman et al., 2006).
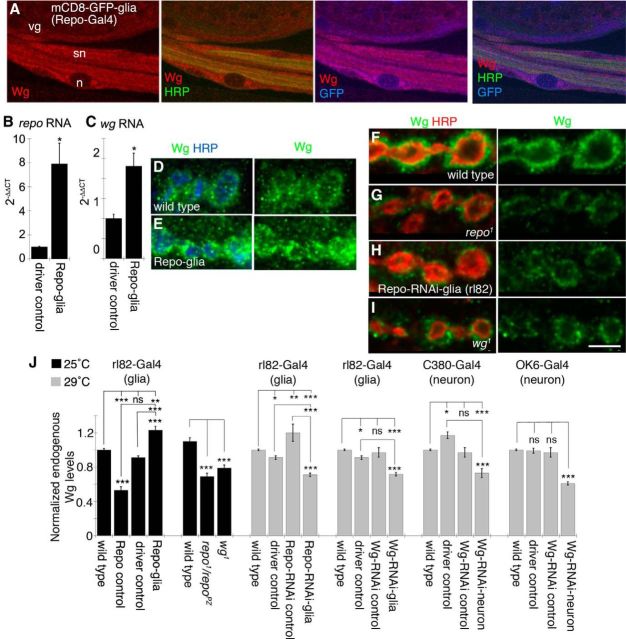
Given our previous studies demonstrating that glial engulfment function is required for normal NMJ development (Fuentes-Medel et al., 2009), we explored potential roles for glia in regulating NMJ development through release of Wg. We first used anti-Wg antibodies to label larval body wall muscle preparations and examined the segmental nerves, where peripheral glial cell bodies are located. Glial membranes were labeled by driving membrane-tethered GFP (mCD8-GFP) in all glia using the repo-Gal4 driver. Endogenous Wg signal was found throughout glial cell bodies, but was excluded from glial cell nuclei (Fig. 2A). To determine whether Repo altered wg transcript levels, we overexpressed Repo in peripheral glia using rl82-Gal4 and extracted RNA from dissected third instar larval segmental nerves, as peripheral glial cell bodies are the only cell bodies found along these nerves. Quantitative PCR revealed that overexpressing Repo in peripheral glial cells resulted in a significant increase in repo and wg transcript levels (Fig. 2B,C).
Figure 2.
Repo regulation of Wg in peripheral glia. A, Third instar larval segmental nerves expressing mCD8-GFP in glia and labeled with anti-HRP, anti-GFP, and anti-Wg. n, glial nucleus; sn, segmental nerve; vg, ventral ganglion. B, C, Real-time PCR from larval segmental nerve RNA showing that repo (B) and wg transcript (C) levels are increased when Repo is overexpressed in peripheral glia using repo-Gal4. Transcript fold changes were determined using the Δ-ΔCt method. D–I, Confocal images of third instar larval NMJ branches in preparations double labeled with anti-HRP and anti-Wg in wild-type controls (D, F), upon overexpressing Repo in peripheral glia (E), repo1 mutants (G), upon expressing Repo-RNAi in peripheral glia (H), and in wg1 mutants (I). J, Quantification of total Wg signal intensity divided by bouton volume in each of the indicated genotypes normalized to wild type. Gray and black bars indicate experiments performed at 29°C, to maximize RNAi expression, and 25°C, respectively. Error bars represent SEM. *p ≤ 0.05; **p ≤ 0.01; ***p < 0.001. Scale bar: (in I) A, 8 μm; D–I, 5 μm. The numbers of arbors quantified for normalized endogenous Wg levels are as follows: 25°C, wild type, 10; UAS-Repo/+ (Repo control), 10; rl82-Gal4/+ (driver control), 10; rl82-Gal4>Repo (Repo-glia), 10; repo1/repoPZ, 10; wg1, 10; 29°C, wild type, 47; rl82-Gal4/+ (driver control), 10; UAS-Repo-RNAi/+ (Repo-RNAi control), 10; rl82-Gal4>Repo-RNAi (Repo-RNAi-glia), 10; UAS-Wg-RNAi/+ (Wg-RNAi control), 10; rl82-Gal4>Wg-RNAi (Wg-RNAi-glia), 24; C380-Gal4/+ (driver control), 10; C380-Gal4>Wg-RNAi (Wg-RNAi-neuron), 13; OK6-Gal4/+ (driver control), 10; and OK6-Gal4>Wg-RNAi (Wg-RNAi-neuron), 21.
Overexpressing Repo in peripheral glia resulted in an increase in Wg protein levels at the NMJ (Fig. 2D,E,J). The opposite effect, a reduction in Wg protein level at the NMJ, was also observed in a hypomorphic repo allele, repo1, over another mutation in the repo locus (repo1/repoPZ), as well as by downregulating Repo in glia by RNAi (Fig. 2F–H,J). Combined with the ChIP and quantitative PCR results, these data strongly suggest that Repo regulates wg expression and raise the intriguing possibility that a pool of NMJ Wg is derived from glial cells. Expressing Wg-RNAi in neurons also led to a significant decrease in the intensity of the endogenous Wg signal at the NMJ, which was comparable to that observed in the hypomorphic wg1 allele (Fig. 2I,J). This suggests that NMJ Wg is derived from two sources, peripheral glia and presynaptic motor neurons.
Subperineurial glia can deliver Wg to the NMJ
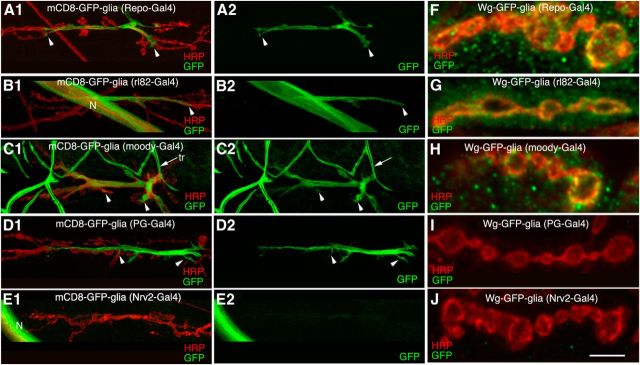
If glial Wg contributes directly to the NMJ Wg protein pool, then glial-expressed Wg should be delivered to the NMJ. To test this hypothesis, we expressed Wg-GFP in glia, using the pan-glial driver repo-Gal4, and examined the localization of Wg-GFP at the NMJ. Driving an mCD8-GFP reporter using repo-Gal4 resulted in GFP-labeled glial membrane extensions that associated with proximal regions of the NMJ, but, consistent with our previous observations (Fuentes-Medel et al., 2009), glial processes did not deeply infiltrate the NMJ (Fig. 3A1,A2, arrowheads). In contrast, expressing Wg-GFP with the repo-Gal4 driver led to Wg-GFP signal localizing to all synaptic boutons of the NMJ, both presynaptically and postsynaptically (Fig. 3F). Thus, glial-expressed Wg-GFP can be found throughout the NMJ despite the fact that glial membranes show only modest association with the NMJ. This observation indicates that peripheral glia are indeed able to deliver Wg to all synaptic boutons of the NMJ.
Figure 3.
Subperineurial glial membranes invade the NMJ and secrete Wg. A1–E2, Confocal images of third instar NMJs in preparations double labeled with anti-HRP and anti-GFP upon expressing mCD8-GFP in glial cell subtypes using repo-Gal4 (all glia; A1, A2), rl82-Gal4 (subperineurial glia; B1, B2), moody-Gal4 (subperineurial glia; C1, C2), PG-Gal4 (perineurial glia; D1, D2), and Nrv2-Gal4 (wrapping glia; E1, E2). F–J, Confocal images of NMJ branches labeled with anti-HRP and anti-GFP in larvae expressing Wg-GFP in subsets of glia using repo-Gal4 (F), rl82-Gal4 (G), moody-Gal4 (H), PG-Gal4 (I), and Nrv2-Gal4 (J). Scale bar: (in J) A1–J, 5 μm. Arrowheads represent extent of glial membrane infiltration into the NMJ, while arrow denotes tracheal cells (tr). N, nerve.
Peripheral glia can be subdivided into three categories: the perineurial glia (PGs) reside on the surface of the nerve; beneath these are the subperineurial glia (SPGs), which establish the blood–brain barrier; and finally, wrapping glia (WPGs) are found in the deepest region of the peripheral nerve, directly associating with axons of motor and sensory neurons (Stork et al., 2008). Several strains that express Gal4 in each peripheral glial subtype have been isolated (Stork et al., 2012), and we used these to determine precisely which subsets of peripheral glia interact with the NMJ and which were capable of delivering Wg-GFP to synaptic boutons. SPGs labeled by driving mCD8-GFP with rl82-Gal4 or moody-Gal4 (Schwabe et al., 2005) were found to elaborate frequent membrane extensions that associated with primarily proximal regions of the NMJ (Fig. 3B1–C2, arrowheads; note that the moody-Gal4 driver is also expressed in tracheal cells along the body wall, Fig. 3C1,C2, arrow). As was the case with repo-Gal4, expression of Wg-GFP using rl82- or moody-Gal4 resulted in Wg-GFP localization surrounding all synaptic boutons of the NMJ (Fig. 3G,H). These data are consistent with the notion that SPGs can release Wg to the NMJ.
Examination of PG-Gal4 (Awasaki et al., 2008), which expresses Gal4 in PGs, revealed that membrane extension from perineurial glia also reached the proximal region of the NMJ and became associated with a few boutons (Fig. 3D1,D2, arrowheads). However, unlike with SPGs, driving Wg-GFP in PGs did not result in Wg-GFP signal at the NMJ (Fig. 3I). Finally, we examined the distribution of WPG membranes by driving mCD8-GFP with Nrv2-Gal4 (Sun et al., 1999). Although bright mCD8-GFP signal was observed in the segmental nerves (Fig. 3E1), we never observed GFP positive membrane extensions associated with the NMJ (Fig. 3E2). In addition, Wg-GFP was not present at the NMJ when expressed with Nrv2-Gal4 (Fig. 3J). Together, these results provide compelling evidence that both SPGs and PGs, but not WPGs, extend membranes that interact with the NMJ. Furthermore, they suggest that SPG cells are uniquely specialized among the peripheral nerve glial subtypes to serve as a source of glia-derived Wg at the NMJ.
Subperineurial glia function is required for normal glutamate receptor cluster formation
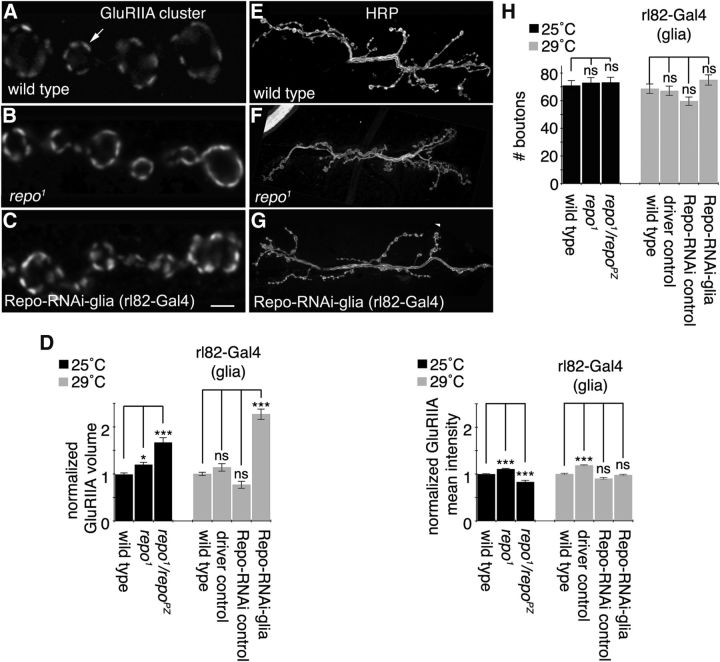
The finding that SPG cells can deliver Wg to the NMJ raised the possibility that glia are responsible for some of the structural and functional abnormalities observed upon interfering with Wnt signaling at the NMJ (Packard et al., 2002; Ataman et al., 2006). Consistent with this view, repo1, repo1/repoPZ, or expression of Repo-RNAi in glia with the SPG driver rl82-Gal4 resulted in an increase in the size of GluRIIA clusters (Fig. 4A–D). Although there were also significant changes in mean GluRIIA intensity across genotypes (Fig. 4D), these changes were relatively small. Thus, most likely, the increase in GluRIIA cluster size is correlated with an increase in GluRIIA number and is not simply the result of diluting the same number of GluRIIA receptors across a larger volume.
Figure 4.
Subperineurial glia are required for normal GluRIIA distribution. A–C, Confocal images of third instar NMJ branches in preparations double labeled with anti-GluRIIA in wild type (A, arrow denotes GluRIIA cluster), repo1 mutants (B), and upon expressing Repo-RNAi RNA in SPGs (C). D, Quantifications of GluRIIA volume divided by bouton volume and mean GluRIIA signal intensity in each of the indicated genotypes normalized to wild type. E–G, Confocal images of third instar larval NMJ arbors labeled with anti-HRP in wild type (E), repo1 mutants (F), and upon expressing Repo-RNAi RNA in SPGs (G). H, Quantification of total bouton number for each of the indicated genotypes. Gray and black bars indicate experiments performed at 29 and 25°C, respectively. Error bars represent SEM. *p ≤ 0.05; ***p < 0.001. Scale bar: (in C) A–C, 2 μm; E–G, 18 μm. The numbers of arbors quantified for GluRIIA parameters are as follows: 25°C, wild type, 10; repo1, 10; repo1/repoPZ, 10; 29°C, wild type, 32; rl82-Gal4/+ (driver control), 10; UAS-Repo-RNAi/+ (Repo-RNAi control), 10; and rl82-Gal4>Repo-RNAi (Repo-RNAi-glia), 10. The numbers of samples for total bouton number are as follows: 25°C, wild type, 42; repo1, 23; repo1/repoPZ, 14; 29°C, wild type, 166; driver control, 15; Repo-RNAi control, 19; and Repo-RNAi-glia, 18.
In contrast to Wnt signaling mutants, repo mutants or glial expression of Repo-RNAi did not change NMJ size, as determined by labeling body wall muscle preparations with anti-HRP antibodies and counting the number of synaptic boutons at the third instar larval stage (Fig. 4E–H).
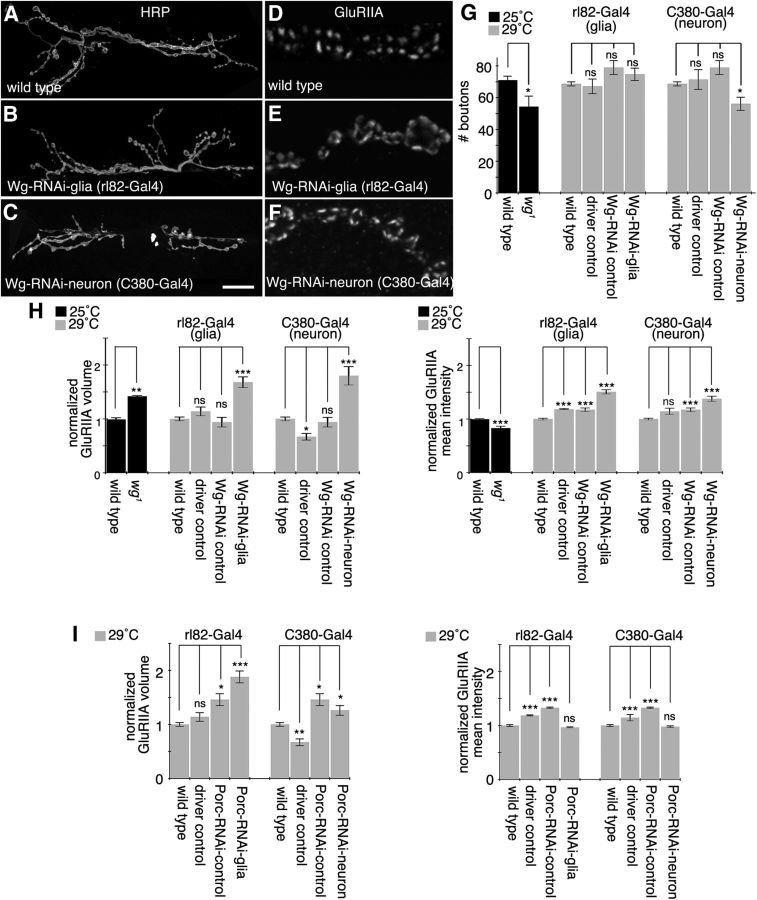
To determine whether the above phenotype in GluRIIA clustering was due to Wg function in SPGs, we examined NMJ size and organization of GluRIIA clusters upon selective downregulation of Wg in SPGs. Unlike the wg1 hypomorph, or expression of Wg-RNAi in neurons with C380-Gal4, in which a small but significant decrease in bouton number was observed (Fig. 5A,C,G), no change in bouton number was apparent when we expressed Wg-RNAi in SPGs (Fig. 5B,G). However, downregulating Wg in these glia led to a substantial increase in GluRIIA cluster size, similar to the repo mutant phenotype, and a significant increase in GluRIIA mean intensity (Fig. 5D,E,H). To determine whether the increase in GluRIIA cluster size/intensity was exclusively derived from Wg function in glia, we also downregulated Wg in neurons with C380-Gal4. Notably, expressing Wg-RNAi in neurons also resulted in a significant increase in the size and mean intensity of GluRIIA clusters (Fig. 5F,H). Although wg1 mutants showed a similar increase in GluRIIA cluster size, there was a small decrease in mean intensity (Fig. 5H). Thus, Wg operates in both glia and neurons to promote normal organization of GluRIIA clusters, and the expression of Wg in either of these cell types alone is not sufficient for normal assembly of GluRIIA clusters.
Figure 5.
SPG- and motor neuron-derived Wg regulate glutamate receptors. A–C, Confocal images of third instar larval NMJ arbors labeled with anti-HRP in wild type (A), upon expressing Wg-RNAi in SPGs (B), and upon expressing Wg-RNAi in neurons (C). D–F, Confocal images of third instar NMJ branches in preparations double labeled with anti-GluRIIA in wild type (D), upon expressing Wg-RNAi in SPGs (E), and upon expressing Wg-RNAi in motor neurons (F). G, Quantification of total bouton number for each of the indicated genotypes. H, I, Quantifications of GluRIIA volume divided by bouton volume and mean GluRIIA signal intensity in each of the indicated genotypes normalized to wild type. Gray and black bars indicate experiments performed at 29 and 25°C respectively. Error bars represent SEM. *p ≤ 0.05; **p ≤ 0.01; ***p < 0.001. Scale bar: (in C) A–C, 20 μm; D–F, 4 μm. The numbers of animals quantified for total bouton number are as follows: 25°C, wild type, 42; wg1, 13; 29°C, wild type, 166; rl82-Gal4/+ (driver control), 15; UAS-Wg-RNAi/+ (Wg-RNAi control), 9; rl82-Gal4>Wg-RNAi (Wg-RNAi-glia), 27; C380-Gal4/+ (driver control), 13; and C380-Gal4>Wg-RNAi (Wg-RNAi-neuron), 29. The numbers of arbors quantified for GluRIIA parameters are as follows: 25°C, wild type, 10; wg1, 10; 29°C, wild type, 32; rl82 driver control, 10; Wg-RNAi control, 10; Wg-RNAi-glia, 18; C380 driver control, 10; Wg-RNAi-neuron, 10; UAS-Porc-RNAi/+ (Porc-RNAi control), 10; rl82-Gal4>Porc-RNAi (Porc-RNAi-glia), 10; and C380-Gal4>Porc-RNAi (Porc-RNAi-neuron), 10.
To obtain further evidence for a role of SPG-derived Wg in the formation of normal GluR clusters, we downregulated Porcupine (Porc), an endoplasmic reticulum (ER) resident protein required for post-translational Wg modifications that are essential for Wg exit from the ER (van den Heuvel et al., 1993; Kadowaki et al., 1996; Tanaka et al., 2002). Downregulating Porc with rl82-Gal4 resulted in a significant increase in the size, but not the mean intensity, of GluRIIA clusters (Fig. 5I), reinforcing the idea that Wg secretion by glia is required for normal GluRIIA clustering. Similarly, downregulating Porc with C380-Gal4 resulted in an increase in GluRIIA size, but not intensity. These results provide strong evidence that both glial and motor neuron Wg are critical for correct NMJ development. In addition, our data argue that the functions of Wg in neurons versus glia are distinct: neuronal Wg is required to establish both normal NMJ size and GluRIIA clustering, whereas glial Wg regulates GluRIIA clustering, but does not appear to influence NMJ size.
Wnt signaling is required in both glia and neurons for proper NMJ function
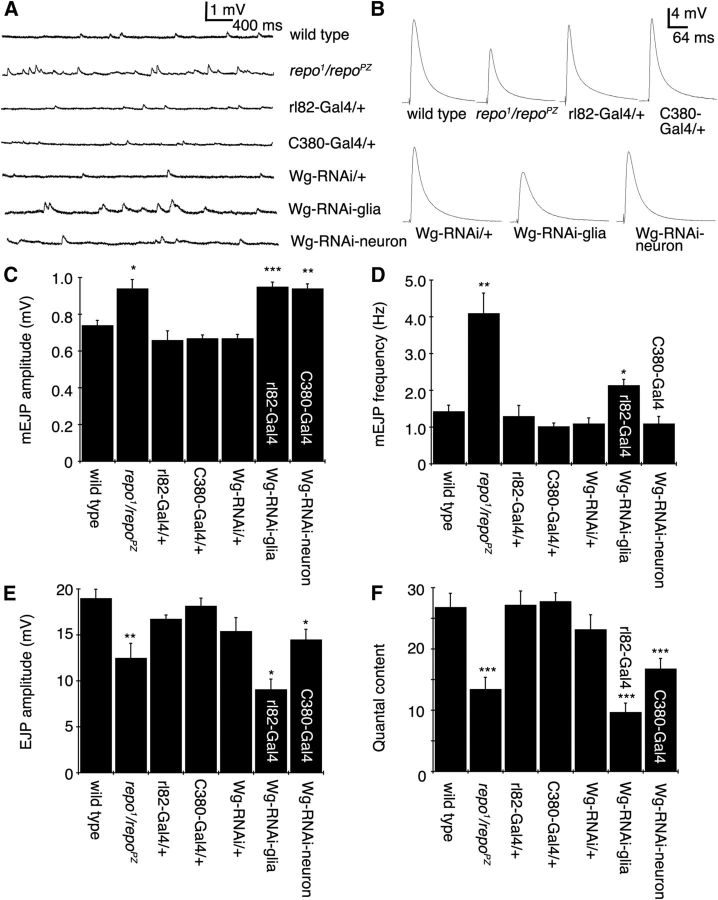
Previous studies suggest that an increase in size of GluRIIA clusters upon disrupting Wg signaling at the NMJ is reflected by a change in neurotransmission, particularly an increase in the amplitude of mEJPs (Speese et al., 2012). To determine whether interfering with Wg function in glia also mimicked this phenotype, we recorded evoked (EJP) and spontaneous (mEJP) synaptic potentials in larvae expressing Wg-RNAi in SPGs with rl82-Gal4. We found that the amplitude of mEJPs was significantly increased when Wg-RNAi was expressed in SPGs (Fig. 6A,C). A similar phenotype was observed in the repo1/repoPZ transallelic mutant (Fig. 6A,C). As expected from the similar effects of disrupting Wg in glia or neurons for normal GluRIIA cluster formation, mEJP amplitude was also increased upon downregulating Wg in neurons (Fig. 6A,C). Nevertheless, the amplitude of evoked EJPs was decreased in all of the above genotypes (Fig. 6B,E), which resulted in significantly reduced quantal content (Fig. 6F).
Figure 6.
Synaptic transmission is altered in repo mutants and upon Wg decrease in glia or neurons. A, B, Representative mEJP (A) and evoked EJP traces (B) in the indicated genotypes. C–F, Quantification of mEJP amplitude (C), mEJP frequency (D), evoked EJP amplitude (E), and quantal content (F). Error bars represent SEM. *p ≤ 0.05; **p ≤ 0.01; ***p < 0.001. The numbers of animals quantified are as follows: wild type, 23; repo1/repoPZ, 5; rl82-Gal4/+ (driver control), 8; C380-Gal4/+ (driver control), 7; UAS-Wg-RNAi/+ (Wg-RNAi control), 12; rl82-Gal4>Wg-RNAi (Wg-RNAi-glia), 30; and C380-Gal4>Wg-RNAi (Wg-RNAi-neuron), 11.
Interestingly, although neuronal Wg-RNAi had no effect on mEJP frequency, repo mutants and glial Wg-RNAi animals exhibited a marked increase in mEJP frequency (Fig. 6A,D). Thus both glia- and neuron-derived Wg regulate multiple aspects of NMJ neurotransmission in vivo, but a unique feature of glia-derived Wg is its modulation of mEJP frequency.
Discussion
In the past decade, glial cells have emerged as important regulators of neural circuit assembly and function. In particular, glia have been shown to exert significant control over synapse formation, growth, and plasticity, but glia-derived factors capable of regulating neural development and physiology in vivo are only beginning to be defined (Christopherson et al., 2005; Barres, 2008; Eroglu et al., 2009; Eroglu and Barres, 2010; Allen et al., 2012; Fuentes-Medel et al., 2012). By initiating the discovery of transcriptional targets of the glial transcription factor Repo, we identified the Wnt family protein Wg as a glia-derived factor promoting synapse growth. We showed that Repo can bind the wg locus in cultured cells, and that Repo can regulate synaptic levels of Wg in vivo. Furthermore, we demonstrated that glia-derived Wg is an important in vivo regulator of synapse formation and physiology: downregulation of glia-derived Wg at the NMJ leads to defects in the distribution of glutamate receptor clusters, an increase in mEJP amplitude and frequency, and a decrease in evoked EJPs. Wg is thus a novel prosynaptomorphic molecule released by glia that modulates the organization of postsynaptic structures and NMJ function.
Repo regulates a broad class of genes involved in neuron–glia signaling
The diversity of genes directly regulated by Repo—a critical transcriptional regulator of glial cell development in Drosophila—has not been thoroughly explored. Our ChIP studies from Drosophila S2 cells identified several potential Repo targets that have been shown to govern fundamental aspects of glial development or function. For example, known targets were identified that actively promote glial cell fate specification (e.g., pointed, distalless; Klaes et al., 1994; Freeman et al., 2003), blood–brain barrier formation (e.g., gliotactin, loco, coracle, Nrv1; Granderath et al., 1999; Banerjee and Bhat, 2007), engulfment activity (e.g., dCed-6; Awasaki et al., 2006), neurotransmitter metabolism (e.g., EAAT1, Gs2; Freeman et al., 2003; Rival et al., 2004), ionic homeostasis (e.g., fray; Leiserson et al., 2000, 2011), and neuron–glia signaling during nervous system morphogenesis (e.g., Pvr; Learte et al., 2008). For at least two potential targets, gs2 and Cp1, we demonstrated a key requirement for Repo in their transcriptional activation during development.
Given the broad roles of this collection of genes in glial cell biology, our work supports the hypothesis that Repo transcriptionally regulates a diverse class of genes that modulate many aspects of glial cell development. For instance, Pointed, which is now a predicted Repo target, is a key glial factor that activates glial fate at very early developmental stages (Klaes et al., 1994). Likewise, Repo appears to regulate Gliotactin, Coracle, and Nrv1, which are molecules essential for formation of the pleated septate junction-based blood–brain barrier at mid to late embryogenesis in Drosophila (Schwabe et al., 2005; Banerjee and Bhat, 2007). At the same time, EAAT1 and GS2 are activated late in the embryonic glial program, with expression being retained even in fully mature glia, and these transporters are critical for synaptic neurotransmitter recycling (Freeman and Doherty, 2006). Since EAAT1 and GS2 are both activated by Repo, and primarily expressed in CNS glia, our data argue that Repo is directly upstream of multiple key glial factors required for glutamate clearance from CNS synapses.
Glia to synapse signaling through Wingless/Wnt
Mammalian excitatory glutamatergic synapse formation is modulated by multiple soluble glia-derived factors including TSPs (Christopherson et al., 2005), Hevin/Sparc (Kucukdereli et al., 2011), and glypicans 4 and 6 (Allen et al., 2012). These factors, along with other secreted glial factors that remain to be identified, are essential for initial synapse formation and (with the exception of TSPs) can promote postsynaptic differentiation through membrane insertion and clustering of AMPA receptors (Kucukdereli et al., 2011). In this study, we identified Wg as a novel glia-derived factor essential for postsynaptic structure and function in vivo at the Drosophila glutamatergic NMJ. Combined with our previous findings that NMJ glia can also release a TGF-β family member to regulate presynaptic growth in a retrograde manner (Fuentes-Medel et al., 2012), these studies provide compelling evidence that Drosophila glia function as a major integrator of synaptic signals during development.
Previous work has demonstrated that Wg/Wnt signaling potently modulates the coordinated assembly of both presynaptic and postsynaptic structures at the Drosophila NMJ (Speese and Budnik, 2007). Loss of Wg, or its receptor DFz2, leads to a dramatic decrease in synaptic boutons and disrupted clustering of postsynaptic glutamate receptors (Packard et al., 2002). Although previous studies supported evidence implicating motor neurons in Wg release (Packard et al., 2002), the presence of alternative cellular sources remained an open and important question. Our surprising discovery of Wg as a candidate Repo target gene by ChIP led us to explore the possibility that NMJ glia could act as an additional in vivo source of NMJ Wg. Consistent with this idea, we found that peripheral glia expressed Wg, SPGs were able to deliver Wg::GFP to the NMJ, and knockdown of SPG Wg significantly reduced NMJ Wg levels and led to a partial phenocopy of wg mutant phenotypes.
Interestingly, we found that loss of glia-derived Wg could account for some, but not all, wg loss-of-function phenotypes. For example, whereas depletion of glia-derived Wg disrupted clustering of postsynaptic glutamate receptors, it had no effect on the formation of synaptic boutons. In contrast, depletion of neuronal Wg led to defects in both glutamate receptor clustering as well as bouton formation. Although only neuronal Wg regulated bouton growth, these data argue that both glial and neuronal Wg are capable of modulating the assembly of glutamate receptor complexes. Thus, we have identified two in vivo sources of Wg at the NMJ: the presynaptic neuron and local glial cells.
Regarding the modulation of neurotransmission, we found that both glial and neuronal Wg had important roles, which, as in the case of the development of synaptic structure, were only partially overlapping. Loss of glial or neuronal Wg resulted in postsynaptic defects in neurotransmission, including increased mEJP amplitude (a postsynaptic property), decreased nerve-evoked EJPs, and decreased quantal content. Consistent with Repo regulating glial Wg expression, these phenotypes were mimicked by loss of repo function. The most notable difference in functional requirements for glial versus neuronal Wg is in mEJP frequency (a presynaptic function): depletion of glial Wg resulted in a dramatic increase in mEJP frequency, whereas manipulating neuronal Wg had no effect. Thus both glial and neuronal Wg are critical regulators of synaptic physiology in vivo, likely modulating NMJ neurotransmission in a combinatorial fashion, although glial Wg has the unique ability to modulate presynaptic function.
The increase in mEJP amplitude is consistent with our findings that GluR cluster size was increased upon loss of glia- or neuron-derived Wg, and that in general this was accompanied by minor changes in GluRIIA signal intensity. A potential explanation is that neuron- and glia-derived Wg regulate the levels of GluRIIA subunits. Previously, it was demonstrated that downregulation of the postsynaptic Frizzled Nuclear Import (FNI) pathway also increased GluRs at the NMJ (Speese et al., 2012). This suggests that glia- and neuron-derived Wg may act in concert via the FNI pathway to stabilize the synapse by regulating GluR expression.
An important property of the larval NMJ is the ability to maintain constant synaptic function throughout development via structural and functional modifications. The combined functions of glial and neuronal Wg likely contribute to this mechanism, as together they positively regulate synaptic growth and function as well as organize postsynaptic machinery. However, a previous study suggested that the transcription factor Gooseberry (Gsb), in its role as positive regulator of synaptic homeostasis in neurons, may be antagonized by Wg function (Marie et al., 2010). Mutations in gsb block the increase in neurotransmitter release observed when postsynaptic GluRs are downregulated. Furthermore, Marie et al. (2010) showed that the gsb mutant defect can be rescued by a heterozygous wg mutant allele. However, the specific role of Gsb in this process is unclear, as rapid synaptic homeostasis was normal in the mutant, and defects appeared restricted to a long-term decrease in GluR function (Marie et al., 2010). It will be important to define the specific role of Gsb in synaptic homeostasis and to manipulate Wg function in alternative ways before a clear relationship between Wg and Gsb can be established.
How could neuronal versus glial Wg differ in regulating NMJ development and physiology? One possibility is that the level or site of Wg delivery by each cell type is different. For example, since SPGs invade the NMJ only intermittently (Fuentes-Medel et al., 2009), it is possible that they release most of their Wg outside of the NMJ, whereas the presynaptic neuron, which is embedded in the muscle cell, delivers it more efficiently and directly to the postsynaptic muscle cell. Alternatively, the Wg morphogen released by glia versus that released by neurons could be qualitatively different through alternative post-translational modifications such as glycosylation. Either mechanism would allow for glia to modulate specific aspects of NMJ physiology independently from neuronal Wg, perhaps in an activity-dependent manner.
Although glia-derived Wg does not modulate NMJ growth, Drosophila glia can indeed regulate synaptic growth at the NMJ in vivo. We demonstrated previously that Drosophila glia release the TGF-β ligand Maverick to modulate TGF-β/BMP retrograde signaling at the NMJ and thereby the addition of new synaptic boutons (Fuentes-Medel et al., 2012). Our discovery that glia-derived Wg can exert significant control over the physiological properties of NMJ synapses expands the mechanisms by which Drosophila glia can control NMJ synapse development and function. In the future it will be important to understand how glial Wg and TGF-β signaling integrate to promote normal NMJ growth, physiology, and plasticity.
Footnotes
This work was supported by NIH Grants NS053538 (M.R.F.), NS060808 (V.B., M.R.F.), and RO1MH070000 (V.B.). M.R.F. is an Investigator of the Howard Hughes Medical Institute. We thank the Rosbash Lab for guidance and resources to perform the ChIP-chip. We thank Roland Bainton, Bradley Jones, Takashi Awasaki, and Vanessa Auld for fly stocks and reagents.
References
- Akiyama Y, Hosoya T, Poole AM, Hotta Y. The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc Natl Acad Sci U S A. 1996;93:14912–14916. doi: 10.1073/pnas.93.25.14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, Barres BA. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley J, Packard M, Ataman B, Budnik V. Fasciclin II signals new synapse formation through amyloid precursor protein and the scaffolding protein dX11/Mint. J Neurosci. 2005;25:5943–5955. doi: 10.1523/JNEUROSCI.1144-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataman B, Ashley J, Gorczyca D, Gorczyca M, Mathew D, Wichmann C, Sigrist SJ, Budnik V. Nuclear trafficking of Drosophila Frizzled-2 during synapse development requires the PDZ protein dGRIP. Proc Natl Acad Sci U S A. 2006;103:7841–7846. doi: 10.1073/pnas.0600387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LT, Schwabe T, DeSalvo M, Gaul U, Heberlein U. moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell. 2005;123:145–156. doi: 10.1016/j.cell.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Bhat MA. Neuron-glial interactions in blood-brain barrier formation. Annu Rev Neurosci. 2007;30:235–258. doi: 10.1146/annurev.neuro.30.051606.094345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Budnik V, Koh YH, Guan B, Hartmann B, Hough C, Woods D, Gorczyca M. Regulation of synapse structure and function by the Drosophila tumor suppressor gene dlg. Neuron. 1996;17:627–640. doi: 10.1016/S0896-6273(00)80196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G, Göring H, Lin T, Spana E, Andersson S, Doe CQ, Tomlinson A. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development. 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Elmariah SB, Oh EJ, Hughes EG, Balice-Gordon RJ. Astrocytes regulate inhibitory synapse formation via Trk-mediated modulation of postsynaptic GABAA receptors. J Neurosci. 2005;25:3638–3650. doi: 10.1523/JNEUROSCI.3980-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu C, Barres BA. Regulation of synaptic connectivity by glia. Nature. 2010;468:223–231. doi: 10.1038/nature09612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu C, Allen NJ, Susman MW, O'Rourke NA, Park CY, Ozkan E, Chakraborty C, Mulinyawe SB, Annis DS, Huberman AD, Green EM, Lawler J, Dolmetsch R, Garcia KC, Smith SJ, Luo ZD, Rosenthal A, Mosher DF, Barres BA. Gabapentin receptor alpha2delta-1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell. 2009;139:380–392. doi: 10.1016/j.cell.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, Doherty J. Glial cell biology in Drosophila and vertebrates. Trends Neurosci. 2006;29:82–90. doi: 10.1016/j.tins.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/S0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- Fuentes-Medel Y, Logan MA, Ashley J, Ataman B, Budnik V, Freeman MR. Glia and muscle sculpt neuromuscular arbors by engulfing destabilized synaptic boutons and shed presynaptic debris. PLoS Biol. 2009;7:e1000184. doi: 10.1371/journal.pbio.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Medel Y, Ashley J, Barria R, Maloney R, Freeman M, Budnik V. Integration of a retrograde signal during synapse formation by glia-secreted TGF-beta ligand. Curr Biol. 2012;22:1831–1838. doi: 10.1016/j.cub.2012.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granderath S, Stollewerk A, Greig S, Goodman CS, O'Kane CJ, Klämbt C. loco encodes an RGS protein required for Drosophila glial differentiation. Development. 1999;126:1781–1791. doi: 10.1242/dev.126.8.1781. [DOI] [PubMed] [Google Scholar]
- Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, Technau GM. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development. 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Takizawa K, Nitta K, Hotta Y. glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82:1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- Hughes EG, Elmariah SB, Balice-Gordon RJ. Astrocyte secreted proteins selectively increase hippocampal GABAergic axon length, branching, and synaptogenesis. Mol Cell Neurosci. 2010;43:136–145. doi: 10.1016/j.mcn.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Properties of the larval neuromuscular junction in Drosophila melanogaster. J Physiol. 1976;262:189–214. doi: 10.1113/jphysiol.1976.sp011592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BW, Fetter RD, Tear G, Goodman CS. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Wilder E, Klingensmith J, Zachary K, Perrimon N. The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes Dev. 1996;10:3116–3128. doi: 10.1101/gad.10.24.3116. [DOI] [PubMed] [Google Scholar]
- Klaes A, Menne T, Stollewerk A, Scholz H, Klämbt C. The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell. 1994;78:149–160. doi: 10.1016/0092-8674(94)90581-9. [DOI] [PubMed] [Google Scholar]
- Kucukdereli H, Allen NJ, Lee AT, Feng A, Ozlu MI, Conatser LM, Chakraborty C, Workman G, Weaver M, Sage EH, Barres BA, Eroglu C. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc Natl Acad Sci U S A. 2011;108:E440–449. doi: 10.1073/pnas.1104977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learte AR, Forero MG, Hidalgo A. Gliatrophic and gliatropic roles of PVF/PVR signaling during axon guidance. Glia. 2008;56:164–176. doi: 10.1002/glia.20601. [DOI] [PubMed] [Google Scholar]
- Lee BP, Jones BW. Transcriptional regulation of the Drosophila glial gene repo. Mech Dev. 2005;122:849–862. doi: 10.1016/j.mod.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Leiserson WM, Harkins EW, Keshishian H. Fray, a Drosophila serine/threonine kinase homologous to mammalian PASK, is required for axonal ensheathment. Neuron. 2000;28:793–806. doi: 10.1016/S0896-6273(00)00154-9. [DOI] [PubMed] [Google Scholar]
- Leiserson WM, Forbush B, Keshishian H. Drosophila glia use a conserved cotransporter mechanism to regulate extracellular volume. Glia. 2011;59:320–332. doi: 10.1002/glia.21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Pym E, Bergquist S, Davis GW. Synaptic homeostasis is consolidated by the cell fate gene gooseberry, a Drosophila pax3/7 homolog. J Neurosci. 2010;30:8071–8082. doi: 10.1523/JNEUROSCI.5467-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqués G, Bao H, Haerry TE, Shimell MJ, Duchek P, Zhang B, O'Connor MB. The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function. Neuron. 2002;33:529–543. doi: 10.1016/S0896-6273(02)00595-0. [DOI] [PubMed] [Google Scholar]
- Mathew D, Ataman B, Chen J, Zhang Y, Cumberledge S, Budnik V. Wingless signaling at synapses is through cleavage and nuclear import of receptor DFrizzled2. Science. 2005;310:1344–1347. doi: 10.1126/science.1117051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miech C, Pauer HU, He X, Schwarz TL. Presynaptic local signaling by a canonical wingless pathway regulates development of the Drosophila neuromuscular junction. J Neurosci. 2008;28:10875–10884. doi: 10.1523/JNEUROSCI.0164-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard M, Koo ES, Gorczyca M, Sharpe J, Cumberledge S, Budnik V. The Drosophila wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation. Cell. 2002;111:319–330. doi: 10.1016/S0092-8674(02)01047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichsman F, Smith L, Cumberledge S. Glycosaminoglycans can modulate extracellular localization of the wingless protein and promote signal transduction. J Cell Biol. 1996;135:819–827. doi: 10.1083/jcb.135.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rival T, Soustelle L, Strambi C, Besson MT, Iché M, Birman S. Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr Biol. 2004;14:599–605. doi: 10.1016/j.cub.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in Drosophila. Cell. 2005;123:133–144. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Auld VJ. Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics. 1999;151:1093–1101. doi: 10.1093/genetics/151.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustelle L, Besson MT, Rival T, Birman S. Terminal glial differentiation involves regulated expression of the excitatory amino acid transporters in the Drosophila embryonic CNS. Dev Biol. 2002;248:294–306. doi: 10.1006/dbio.2002.0742. [DOI] [PubMed] [Google Scholar]
- Speese SD, Budnik V. Wnts: up-and-coming at the synapse. Trends Neurosci. 2007;30:268–275. doi: 10.1016/j.tins.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, Moore MJ, Budnik V. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Engelen D, Krudewig A, Silies M, Bainton RJ, Klämbt C. Organization and function of the blood-brain barrier in Drosophila. J Neurosci. 2008;28:587–597. doi: 10.1523/JNEUROSCI.4367-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork T, Bernardos R, Freeman MR. Analysis of glial cell development and function in Drosophila. Cold Spring Harb Protoc. 2012;2012:1–17. doi: 10.1101/pdb.top067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Xu P, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proc Natl Acad Sci U S A. 1999;96:10438–10443. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kitagawa Y, Kadowaki T. Drosophila segment polarity gene product porcupine stimulates the posttranslational N-glycosylation of wingless in the endoplasmic reticulum. J Biol Chem. 2002;277:12816–12823. doi: 10.1074/jbc.M200187200. [DOI] [PubMed] [Google Scholar]
- Ullian EM, Christopherson KS, Barres BA. Role for glia in synaptogenesis. Glia. 2004;47:209–216. doi: 10.1002/glia.20082. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Harryman-Samos C, Klingensmith J, Perrimon N, Nusse R. Mutations in the segment polarity genes wingless and porcupine impair secretion of the wingless protein. EMBO J. 1993;12:5293–5302. doi: 10.1002/j.1460-2075.1993.tb06225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S, Vonesch JL, Giangrande A. Glide directs glial fate commitment and cell fate switch between neurones and glia. Development. 1996;122:131–139. doi: 10.1242/dev.122.1.131. [DOI] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- Yuasa Y, Okabe M, Yoshikawa S, Tabuchi K, Xiong WC, Hiromi Y, Okano H. Drosophila homeodomain protein REPO controls glial differentiation by cooperating with ETS and BTB transcription factors. Development. 2003;130:2419–2428. doi: 10.1242/dev.00468. [DOI] [PubMed] [Google Scholar]