Abstract
Cyclic dinucleotides (CDNs) have been previously recognized as important secondary signaling molecules in bacteria and, more recently, in mammalian cells. In the former case, they represent secondary messengers affecting numerous responses of the prokaryotic cell, whereas in the latter, they act as agonists of the innate immune response. Remarkable new discoveries have linked these two patterns of utilization of CDNs as secondary messengers and have revealed unexpected influences they likely had on shaping human genetic variation. This Review summarizes these recent insights and provides a perspective on future unanswered questions in this exciting field.
Cyclic di-GMP as a Second Messenger in Gram-Negative Bacteria
Cyclic dinucleotides (CDNs) were originally described more than 25 years ago as activators of bacterial cellulose synthase (Ross et al., 1987). These investigators defined cyclic di-GMP (cdG) as two GMP molecules linked in a heterocyclic configuration via two 3′-5′ phosphodiester bonds. However, only in 1998 did Tal and colleagues suggest that cdG may be important for other processes in bacteria (Tal et al., 1998). Since then, cdG has been implicated in central bacterial processes, including, but not limited to, virulence, stress survival, motility, antibiotic production, metabolism, biofilm formation, and differentiation (reviewed in detail in Römling et al. [2013]). In Gram-negative bacteria, cdG is now accepted as a universal bacterial secondary messenger in that genes encoding enzymes involved in synthesis and degradation of this CDN are recognizable in the genomes of all corresponding bacterial species.
The cytosolic level of cdG in bacterial cells is tightly controlled by the dual activity of diguanylate cyclases (DGCs) with GGDEF domains and phosphodiesterases (PDEs) with EAL or HD-GYP domains. The large number of enzymes that are involved in cdG-related pathways (for example, Vibrio cholerae encodes 72 DGC and PDE proteins) suggests tremendous complexity of cdG-mediated signaling in bacteria. Intriguingly, analyses show that genomes carry many genes encoding multidomain proteins that encode both DGC and PDE domains. However, in the majority of these cases, only one domain is active while the other one is involved in protein-protein or protein-RNA interactions. Even more diverse is the number of receptors that have been implicated in cdG sensing in ways that alter cellular activity by modifying transcription, translation, or protein activity (Römling et al., 2013). Thus far, we know that bacteria can sense their environment and respond by modulation of cdG levels to stimuli that include O2 and NO levels, light flux, redox state, and the stage of the cell cycle (Mills et al., 2011). However, our knowledge of the regulatory networks in which cdG (or other CDNs, see below) are involved is far from complete, and further research on environmental stresses, cdG-binding effectors, and the pathways that CDNs control is still needed. Newly developed methods such as fluorescence resonance energy transfer (FRET)-based biosensors that allow the monitoring of cdG concentrations within single bacterial cells (Christen et al., 2010), differential radial capillary action of ligand assay (DRaCALA) that allows rapid and high-throughput measuring of protein-CDN interactions (Roelofs et al., 2011), and development of similar tools for other CDNs will eventually lead to significant advances in the field.
Cyclic di-AMP in Gram-Positive Bacteria
In addition to the role of cdG as a central signaling molecule in Gram-negative bacteria, recent discoveries suggest that cyclic di-AMP (cdA) plays an important role, particularly in many Gram-positive bacteria. The diadenylate cyclase (DAC, DUF147) domain is found in almost 2,000 hypothetical proteins encoded by numerous bacterial species, and many of these DACs are fused to domains of unknown functions (Corrigan and Gründling, 2013), suggesting that signaling pathways for cdA are probably as complex and widespread as for cdG. The best-characterized DAC enzymes are from Bacillus subtilis: DisA and CdaS (YojJ) are required for sporulation, whereas another DAC CdaA (YbbP) is involved in cell wall biosynthesis (Mehne et al., 2013; Oppenheimer-Shaanan et al., 2011; Witte et al., 2008). Supporting evidence for the role of cdA in cell wall homeostasis comes from a study by Luo and Helmann (2012) that showed that overproduction of cdA phosphodiesterase gdpP (yybT) sensitizes B. subtilis to ampicillin, similar to a strain in which the gene encoding a cdA synthetase is deleted. A comparable antibiotic susceptibility phenotype was observed in Staphylococcus aureus and Listeria monocytogenes (Corrigan et al., 2011; Witte et al., 2012). In Mycobacterium smegmatis, overexpression of a DisA homolog was linked to changes in cell morphology and motility (Zhang and He, 2013).
Although the functions of cdA are far from being understood, observations that deletion of cdA synthetase are lethal in L. monocytogenes, Streptococcus pneumonia, S. aureus, and B. subtilis suggest that cdA controls central cellular pathways in many Gram-positive organisms. Corrigan and colleagues suggested that essentiality of the cdA oligonucleotide might be due to a cumulative effect on cdA receptors (Corrigan et al., 2013). Their recent work identified a potassium transporter, KtrA, a cation/proton antiporter, CpaA, a PII-like signal transduction protein A, PstA, and a sensor histidine kinase, KdpD, as specific receptors for cdA in S. aureus (Corrigan et al., 2013). It is possible that simultaneous inactivation of these and possibly other unidentified proteins under cdA control can, in sum, have a lethal effect on a cell that is experiencing depletion of cdA.
Cyclic AMP-GMP in V. cholerae
A new type of CDN called cyclic AMP-GMP (cAMP-GMP) was recently identified by Mekalanos and colleagues as a signaling molecule in Vibrio cholerae that is involved in virulence (Davies et al., 2012). This hybrid molecule is synthesized by the enzyme dinucleotide cyclase or DncV, a protein that shows similarity to eukaryotic 2′-5′ oligo-adenylate synthetase (OAS1). However, evidence suggests that DncV produces a CDN composed of AMP and GMP linked by two 3′-5′ phosphodiester bonds (Ablasser et al., 2013; Davies et al., 2012; Diner et al., 2013). Predicted orthologs of DncV are also recognizable in many other Gram-negative and -positive bacterial species (Davies et al., 2012). Interestingly, in addition to its function as synthase for cAMP-GMP, DncV can also synthesize cdA and cdG, thus raising a possibility that one enzyme can control the pool size of multiple CDNs in the bacterial cell. Thus, some CDN-binding effectors (and CDN degradation enzymes) might have specific or overlapping specificities toward cdA, cdG, and cAMP-GMP. Overexpression of DncV inhibits the chemotactic response of V. cholerae, a phenotype that has been linked to hyperinfectivity by Camilli and colleagues (Butler et al., 2006). Because chemotaxis is not altered by overexpression of a cdG synthetase in V. cholerae, these data suggest that different CDNs could alter different regulatory networks within the same cell. These networks are likely to be more complex in the light of the possibility that there might be spatially and temporally distinct pools of CDNs controlling any given phenotype (Lindenberg et al., 2013).
Recognition of Pathogens by Host Innate Immune Receptors
Microbial organisms express molecules that can be chemically differentiated from those produced by host cells and, thus, can be targeted for recognition by receptors of the innate immune system. The term pathogen-associated molecular patterns (PAMPs) has been coined to define such microbial signaling agonists, and the receptors that recognize these agonists are referred to as pattern recognition receptors (PRRs) (Iwasaki and Medzhitov, 2010). Innate immune agonists include lipid A derived from lipopolysaccharides, peptides encoded within bacterial flagellin, peptidoglycan fragments, lipopeptides, microbial polysaccharides, and certain types of nucleic acids, among others molecules (Takeuchi and Akira, 2010). In some cases, host molecules can be recognized as “PAMP like” if they are present in an abnormal subcellular context (e.g., the presence of DNA in the cytosol rather than in the nucleus) (Barber, 2011a; Vance et al., 2009). This strategy for innate immune recognition can be rationalized by the host's need to detect the genetic products of pathogen replication within the cytosol, as well as the cellular disruptions caused by intracellular pathogens. Accordingly, host cells express a spectrum of PRRs in plasma membrane, vacuolar membranes, and cytosol in order to detect threats that have different anatomical and subcellular locations during pathogenesis (Palm and Medzhitov, 2009). The recognition of PAMPs by PRRs activates host cell signal transduction cascades that drive production of interferons (IFNs) and other proinflammatory cytokines, as well as antipathogen effector molecules, thereby triggering protective cell biological changes (e.g., inflammasome activation, apoptosis, and autophagy) needed to limit pathogen growth within infected cells (Baxt et al., 2013). The innate immune response also stimulates many elements of the adaptive immune system in order to marshal a specific counterattack on the detected microbial threat. Thus, PAMPs have been recognized for some time as potent adjuvants in the context of vaccines and immunotherapeutics (Duthie et al., 2011). The presence of CDNs in virtually all known bacteria and the fact that eukaryotes were not previously known to rely on these secondary messenger molecules suggested that CDNs might be PAMPs. Early evidence for this concept was provided by the observation that cdA and cdG had adjuvant and immunomodulatory activity in mice (Ebensen et al., 2011; araolis et al., 2007) and that STING was the innate immune receptor of bacterial CDNs (see below) (Burdette et al., 2011).
PRR Activation by Innate Immune Agonists
Several different types of PRRs are now recognized. In brief, Toll-like receptors (TLRs) play major roles in the recognition of PAMPs within the extracellular milieu and endosomal lumen (Kawai and Akira, 2011). TLR activation occurs after ligand (agonist) binding and leads to the recruitment of the adaptor molecules MyD88 and TRIF and subsequent activation of a kinase-driven signaling pathway, culminating in activation of NF-κB and IRF3/7 and thereby host gene expression (Dev et al., 2011). Nucleotide-binding domain leucine-rich repeat-containing proteins (NLRs) are PRRs that reside in the cytosol and most often are activated by bacterial products such as peptidoglycan fragments and peptides derived from flagellin, the type III secretion system rod components, toxins, bacterial, and viral double-stranded DNA (dsDNA) (Franchi et al., 2012). Besides NLRs, additional cytosolic innate receptors exist, including RNA helicases of the retinoic acid-inducible gene-1 (RIG-1)-like receptors (RLRs), which typically recognize cytosolic viral dsRNA and then recruit the adaptor IFN-β promoter stimulator 1 (IPS-1; also called MAVS, VISA, and Cardif), leading to phosphorylation of the transcription factors IRF-3 and IRF7 and expression of Type I IFN genes (Loo and Gale, 2011). Another non-NLR is absent in melanoma 2 (AIM2), an interferon-inducible protein that can bind dsDNA in the cytosol and induce autocleavage of caspase-1 and thereby inflammasome activation (Barber, 2011b). C-type lectin receptors (CLRs) such as Dectin-1 bind fungal cell wall components and trigger activation of NF-κB (Takeuchi and Akira, 2010). Given that PRRs can detect PAMPs associated with all pathogenic microorganisms, it seems likely that even prions might be detected by some element of the innate immune system because of the unusual β-amyloid structures they can form and the cellular disruptions that these polymers cause (Bradford and Mabbott, 2012; Sheedy et al., 2013).
Despite the presence of multiple PRRs in various cell compartments, both bacteria and viruses have developed mechanisms that allow them to avoid intracellular killing and subvert induction of the innate immunity. These include downregulation or modification of innate immune agonists such as flagellin (Gründling et al., 2004) or lipid A (Trent et al., 2006). In the case of bacteria, effectors delivered by type III and IV systems have also been shown to modify many different cellular pathways, including those involved in innate immune responses (Baxt et al., 2013; Galán, 2009). In some cases, bacterial effectors are similar in function to eukaryotic proteins that are involved in central cellular processes such as phosphorylation, ubiquitination, and Rho family GTPase signaling. In other cases, bacterial effectors are capable of targeting host proteins by modifying their function by dephosphorylation, acetylation, AMPylation, and N-myristoylation (Hicks and Galán, 2013). Among the myriad effects on host cells, bacterial effectors trigger blocks in signal transduction, avoidance of autophagy, interference with proinflammatory responses, and modifications of vesicular trafficking (Baxt et al., 2013) (Table 1). Similarly, viruses can suppress host IFN induction by various mechanisms, including, for example, inhibition of RIG-I signaling by blocking its ubiquitination by the TRIM25 ubiquitin E3 ligase (Rajsbaum et al., 2012) (Table 1). Thus, innate immune signaling is clearly a host response that pathogenic microorganisms have learned to manipulate through effector protein action as well as alterations in the production of innate immune agonists, including CDNs (see below).
Table 1. Examples of Bacterial and Viral Proinflammatory Response Modifications.
| Organism | Effector protein | Mechanism | Target | Reference |
|---|---|---|---|---|
| Bacillus anthracis | lethal factor | metalloprotease | MAPK kinase | (Duesbery et al., 1998) |
| Chlamydia trachomatis | ChlaDub1, 2 | DUB, deneddylase | IκBα | (Le Negrate et al., 2008b) |
| CT441 | protease | RelA | (Lad et al., 2007) | |
| Pathogenic E. coli; B. pseudomallei | Cif; CHBP | deaminase | ubiquitin and ubiquitin-like protein NEDD8 | (Cui et al., 2010; Jubelin et al., 2010) |
| Pathogenic E. coli | NleC | protease | RelA | (Yen et al., 2010) |
| Yersinia spp. | YopJ | acetyltransferase | RICK, TAK1 | (Meinzer et al., 2012) |
| YopM | inhibitor | caspase-1 | (LaRock and Cookson, 2012) | |
| Legionella pneumophila | LegK1 | Ser/Thr-kinase | IκBα, p100 | (Ge et al., 2009) |
| Salmonella Typhimurium | AvrA | acetyltransferase | IKKα/β | (Jones et al., 2008) |
| SseL | DUB | IκBα | (Le Negrate et al., 2008a) | |
| SspH1 | E3 ligase | PKN1 | (Ashida et al., 2010; Haraga and Miller, 2006) | |
| Shigella spp. | IpaH9.8 | E3 ligase | NEMO | (Ashida et al., 2010) |
| OspF | phosphothreonine lyase | MAPK kinases | (Li et al., 2007) | |
| OspG | Ser/Thr-kinase | UbcH5 (E2) | (Kim et al., 2005) | |
| HSV-1 | ICP27 | IκBα | (Kim et al., 2008) | |
| ICP0 | E3 ubiquitin ligase | IFI16 | (Orzalli et al., 2012) | |
| Influenza A | NS1 | TRIM25 and Riplet | (Rajsbaum et al., 2012) | |
| Vaccinia virus | B14R | IKKβ | (Chen et al., 2008) | |
| CP77 | SCF complex, p65 | (Chang et al., 2009) | ||
| N1L | TBK1 | (DiPerna et al., 2004) | ||
| K1L | IKK | (Shisler and Jin, 2004) |
Activation of the Type I Interferon Response by STING
Among the downstream effectors of the signaling cascade driven by activation of PRRs are specific cytokines and chemokines, including the type I interferons (IFN-α/β). These secreted cytokines can be produced in response to either viral or bacterial intracellular infections through signaling pathways that have only recently been revealed in molecular detail. For example, cytosolic DNA induces a type I IFN response that is dependent on a protein called STING (also known as ERIS, MITA, MPYS, and TMEM173), an endoplasmic reticulum (ER)-localized transmembrane protein (Ishikawa and Barber, 2008). The STING-dependent, type I IFN response appears to be critical to limit the replication of DNA viruses such as HSV- (Ishikawa and Barber, 2008). Although there is evidence that STING may directly recognize dsDNA with low affinity (Kd > 200 μM) (Abe et al., 2013), this is unlikely to be a critical property of STING in light of new information (see below). However, the pathway to type I IFN production after STING activation seems quite clear (Burdette et al., 2011). After a DNA recognition signal is received within the cytosol, STING is “activated,” and this corresponds with its relocation to discrete foci in the cell cytoplasm. These cell biological changes correlate with recruitment of the kinases TBK1 and IKK, which in turn activate IRF3, STAT6, and NF-κB, resulting in type I IFN induction (Cavlar et al., 2012) (Figure 1). The biochemical connection between cytosolic DNA recognition and activation of STING has been elegantly revealed by the recent work of Chen and colleagues (Sun et al., 2013; Wu et al., 2013) (see below).
Figure 1. Overview of STING-Dependent Interferon Induction.
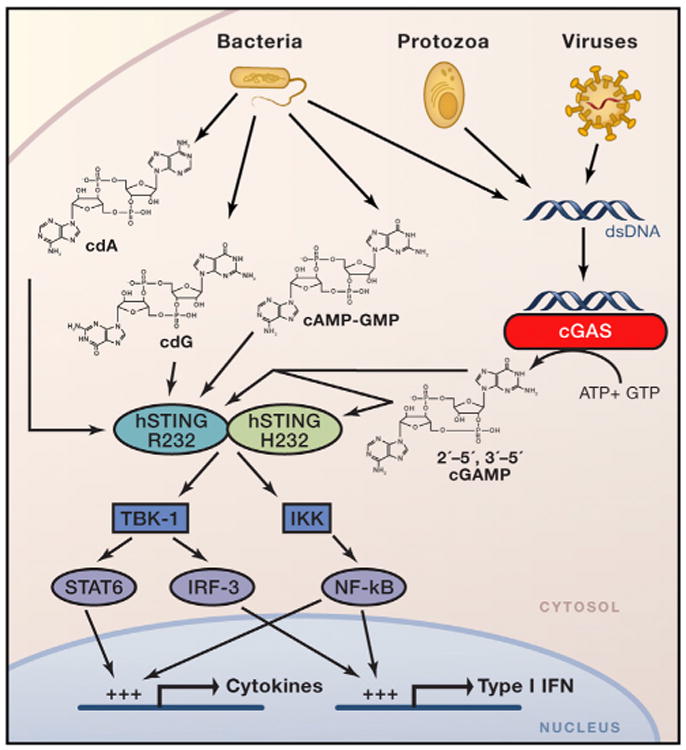
Infection of eukaryotic cells with viruses, protozoa, and bacteria leads to accumulation of extracellular DNA that signals the presence of pathogens to the cellular immune system. Cytosolic dsDNA is recognized by cGAS in a sequence-independent manner. cGAS induces production of 2′-5′, 3′-5′cGAMP to stimulate STING. At least some STING alleles (such as R232) can additionally be activated by CDNs produced by bacteria that include cdA, cdG, and 3′-5′, 3′-5′cGAMP (cAMP-GMP), whereas other STING alleles (such as H232) are not responsive to bacterial CDNs. Binding of STING to an activating ligand induces conformational change in STING, resulting in formation of a “closed form” in which ligand is tightly bound to the binding pocket. Following relocation of STING to discrete foci in the cell cytosol, activated STING recruits TBK1 and IKK kinases, which in turn activate IRF-3, STAT6, and NF-κB. Upon translocation of activated transcriptional factors to the nucleus, they bind to corresponding promoters, thus resulting in induction of type I IFN and cytokines.
The generation of STING mutant mice (Goldenticket or Gtmice) has greatly facilitated the identification of other STING-dependent pathways for induction of type I IFNs, implicating this molecule in the sensing of PAMPs produced by intracellular bacterial pathogens (Jin et al., 2011a; Sauer et al., 2011). STING was eventually shown to directly bind to cdG, and STING mutants defective in such binding lose their ability to induce the production of type I IFNs in response to bacterial CDNs (Burdette et al., 2011). Consistent with this discovery, there are now several lines of evidence suggesting that bacterial CDNs can gain access to the host cell cytosol during infection. For example, the cytosolic intracellular pathogen L. monocytogenes utilizes an efflux pump to secrete cdA directly into the host cytosol, where it triggers a STING-dependent production of type I IFNs in mouse cells (Woodward et al., 2010). Mutants of L. monocytogenes that are confined to the vacuolar lumen do not induce production of IFN-β, suggesting that this pathogen must escape the endosome to deliver cdA to the cytosol. More recently, it was shown that concentration of cdA in cytosol of Chlamydia-infected mouse macrophages is greatly increased compared to uninfected cells and that this correlated with STING-dependent activation of the type I IFN response (Barker et al., 2013). Thus, evidence is accumulating that cytosolic bacterial pathogens can modulate innate immune responses through the CDNs they produce. However, given that some human STING alleles do not respond to bacterial CDNs (see below), more detailed studies are needed to fully understand the bacterial CDN, STING-dependent IFN response.
Synthesis of cGAMP by the Mammalian DNA Sensor cGAS Provides the Missing Link between DNA Sensing and STING Activation
The role of STING as the common downstream regulator of the type I IFN response induced by both cytoplasmic DNA and CDNs has been recently explained through the discovery by Chen and colleagues of the enzyme cyclic GMP-AMP synthase or cGAS (also known as C6orf150 and MB21D1) and its ability to synthesize a CDN ligand that activated STING (Sun et al., 2013; Wu et al., 2013). In brief, Chen and colleagues showed that, in the presence of dsDNA, cGAS selectively catalyzed the synthesis of cAMP-GMP (which they termed cGAMP) and that, after STING binds this endogenous CDN, it activates the type I IFN response (Sun et al., 2013; Wu et al., 2013). Like most major advances, the discovery of cGAS gave rise to a new series of questions. How is cGAS activated by recognition of only dsDNA in a sequence-independent fashion? Given that cGAS belongs to the same nucleotidyltransferase (NTase) family as DncV, is it making exactly the same hybrid CDN as the bacterial enzyme? If so, how could this conclusion be reconciled with the existence of a mutant STING allele (H231) that was not responsive to cdG and cdA but still able to sense dsDNA? Do all naturally occurring STING alleles recognize all CDNs with equal affinity? Recently, a series of seven papers have been published that answer many of these questions (Ablasser et al., 2013; Civril et al., 2013; Diner et al., 2013; Gao et al., 2013a, 2013b; Kranzusch et al., 2013; Zhang et al., 2013).
Crystal structures of the nucleotidyl transferase (NT) domain of porcine cGAS (residues 135-497) (Civril et al., 2013), mouse cGAS (147–507) (Gao et al., 2013a), and human cGAS (residues 157–522) (Kranzusch et al., 2013) have been solved with or without dsDNA ligand and nucleotide substrates. cGAS is a 60 kDa protein that consists of protease-sensitive, unstructured, and poorly conserved ∼150 amino-acid-long N-terminal domain and a protease-resistant, conserved NTase C-terminal domain. cGAS dinucleotide synthase activity is dependent on the presence of dsDNA, Mg2+ or Mn2+, ATP, and GTP. The crystal structures revealed that the catalytic NTase domain fold shows high similarity to an RNA sensor 2′-5′ oligo-adenylate synthetase 1 (OAS1) (Donovan et al., 2013). cGAS interacts with the sugar-phosphate backbone along the minor groove of DNA, thus determining its specificity toward dsDNA in a sequence-independent manner. In comparison to other NTase-containing enzymes, vertebrate cGAS contains a zinc-ribbon domain with an atypical H(X5)CC(X6)C motif that is essential for a metal coordination and interaction with the major groove of DNA, suggesting that it functions as a molecular “ruler” that determines specificity of cGAS toward dsDNA (Civril et al., 2013; Kranzusch et al., 2013). Binding of DNA to cGAS induces structural changes that result in rearrangement of catalytic residues in the NTase active site and enzyme activation. Given the similarity in the NTase fold of OAS1 and cGAS and the fact that both enzymes make secondary messenger molecules in response to detected nucleic acids, it seems likely that cGAS and OAS constitute a novel evolutionary conserved group of PRRs, the OAS-like second messenger receptors (OLRs), that evolved to detect the nucleic acids of cytosolic pathogens (Kranzusch et al., 2013).
Discovery of the Second “Missing Link” Corresponding to a Unique Bond in Mammalian cGAMP
Further characterization of cGAS showed that the enzyme produces a CDN that has a very rare in nature 2′-5′ phosphodiester linkage between GMP and AMP followed by a 3′-5′ return linkage from AMP to GMP (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013a; Kranzusch et al., 2013; Zhang et al., 2013). This CDN isomer (designated here 2′-5′, 3-5′cGAMP for simplicity) is different from all characterized bacterial CDNs, including cAMP-GMP synthesized by DcnV, which is apparently a 3′-5′, 3′-5′ isomer (Ablasser et al., 2013; Davies et al., 2012; Diner et al., 2013). Thus, the phosphodiester linkages of the cGAS product are different in structure than the initially proposed 3′-5′, 3′-5′cGAMP isomer (Wu et al., 2013). The cGAS product has been confirmed to be the 2′-5′, 3′-5′cGAMP isomer by reverse-phase high-performance liquid chromatography (HPLC) analysis (Ablasser et al., 2013; Gao et al., 2013a; Zhang et al., 2013), high-resolution tandem mass spectrometry (MS/MS) spectra of cGAS product (Ablasser et al., 2013; Zhang et al., 2013), and nuclease digestions (Ablasser et al., 2013; Diner et al., 2013) using a chemically synthesized cGAMP isomers as a gold standard. NMR analysis (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013a; Zhang et al., 2013) and CD spectrum (Zhang et al., 2013) also indicated that the cGAS product had 2′-5′ as well as 3′-5′ phosphodiester bonds. Furthermore, crystallization of cGAS in the presence of ATP and GMP clearly showed that a linear oligonucleotide corresponding to pppG(2′-5′)pA was bound to its catalytic pocket, suggesting that ring closure occurs by formation of a phosphodiester bond between the 5′ phosphate of G and the 3′ OH of A (Gao et al., 2013a). Thus, combined evidence from several groups has confirmed that cGAS produces the 2′-5′, 3′-5′cGAMP isomer.
It remains a mystery why cGAS has evolved to make the 2′-5′, 3′-5′cGAMP. It is possible that the 2′-5′ phosphodiester linkage promotes a greater stability to cGAMP product, thus allowing stronger and more prolonged signal amplification (Gao et al., 2013a). Furthermore, production of 2′-5′, 3′-5′cGAMP instead of 3′-5′, 3′-5′cGAMP might be a defense mechanism of eukaryotic cells that allows them to avoid subversion of innate immune response by bacteria because bacterial cells might not be able to degrade 2′-5′ phosphodiester linkages. While cGAMP phosphodiesterases are currently unknown both in bacteria and eukaryotes, determining how 2′-5′, 3′-5′cGAMP turnover occurs in eukaryotic cells is a particularly exciting challenge in the field. It would be interesting if such enzymes (both in prokaryotes and eukaryotes) have substrate specificity that could differentiate 3′-5′, 3′-5′ from 2′-5′, 3′-5′ linkages and if they are strictly specific toward cGAMP molecules or also recognize other CDNs such as cdG and cdA. Such mammalian phosphodiesterases would be potentially interesting targets for altering CDN homeostasis in the context of immunity and inflammation. However, as discussed below, variation in the responsiveness of naturally occurring alleles of STING to bacterial CDNs, which have exclusively 3′-5′ phosphodiester bonds, may provide clues to the evolutionary selection that drove cGAS to make a distinctly different CDN.
The Product Specificity of the cGAS Enzyme and the Response Selectivity of STING Variants and Mutants
Several groups have now confirmed that 2′-5′, 3′-5′cGAMP is a strong inducer of the type I IFN response in a STING-dependent manner (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013b; Zhang et al., 2013). Isothermal titration calorimetry (ITC) experiments showed that 2′ -5′, 3′ -5′cGAMP binds to human STING (hSTING), and this reaction is endothermic compared to the 3′-5′, 3′-5′cdG binding reaction that is exothermic; these results suggest that STING undergoes structural rearrangements exclusively upon binding to 2′-5′, 3′-5′cGAMP (Zhang et al., 2013). However, the controversy over STING preference toward 2′-5′, 3′-5′cGAMP or bacterial 3′-5′, 3′-5′cGAMP still remains. For example, (Zhang et al., 2013) concluded that hSTING (carrying the more common R232 allele) has much higher affinity for 2′-5′, 3′-5′cGAMP than 3′-5′, 3′-5′CDNs based on titrating different CDNs into the hSTING-3′-5′, 3′-5′cdG complex. In their hands, the affinity of 2′-5′, 3′-5′cGAMP for STING was almost 300-fold higher than the affinity seen with 3′-5′, 3′-5′cdG and 3′-5′, 3′-5′cGAMP. In contrast, Patel and his group did ITC experiments and directly measured binding of mSTING R231 and A231 alleles and hSTING H232 and R232 alleles (position 231 in mSTING is identical to 232 in hSTING) to cGAMP (Gao et al., 2013a). They were able to show that all alleles of STING bind to 2′-5′, 3′-5′ and 3′-5′, 3′-5′cGAMP with similar affinity and observed only a 2-fold difference between 2′-5′, 3′-5′cGAMP and 3′-5′, 3′-5′cGAMP in STING activation. Although 2′-5′, 3′ -5′cGAMP was 10- to 20-fold more potent at inducing an interferon response in cells than 3′-5′, 3′-5′cdG, this variation could reflect different efficiencies of cellular uptake of these two different CDNs because digitonin was used to permeabilize cells. These binding experiments correlate well with the in vitro data of Zhang and colleagues showing that both 2′-5′, 3′-5′cGAMP and 3′-5′, 3′-5′cGAMP can induce a strong IFN signaling response in the mouse cell line L929 carrying the R231 allele (Zhang et al., 2013), similar to what is seen with the hSTING R232 allele (Diner et al., 2013). Curiously, neither Diner et al. (2013) nor Ablasser et al. (2013) observed activation ofthe hSTING H232 allele by the DncV product (presumably 3′-5′, 3′-5′cGAMP), although this hSTING allele does respond to the 2′-5′, 3′-5′cGAMP cGAS product. The different experimental approaches and STING alleles used by various investigators make it difficult to draw consensus conclusions regarding a correlation between binding affinity and signaling strength for different CDNs and their isomers. In contrast, new STING structural studies have advanced our understanding in this area quite dramatically.
Crystal structures of both mouse STING (mSTING) and hSTING R232 (residues 139–379) revealed that 2′-5′, 3′-5′ and 3′-5′, 3′-5′cGAMP induce a “closed” conformation, whereas 3′-5′, 3′-5′cdG is bound in a more “open” STING conformation (Figure 2) (Gao et al., 2013b; Zhang et al., 2013). In the “closed” conformation, the binding pocket for cGAMP is slightly deeper and allows coordination of 2′-5′, 3′-5′ and 3′-5′, 3′-5′cGAMP isomers via extensive hydrophobic and polar interactions with STING residues. Furthermore, crystal structures of mSTING R231 and hSTING H232 bound to 2′-5′, 3′ -5′cGAMP were found to be identical, thus confirming that both of these STING alleles bind this CDN in a “closed” conformation. In contrast, cdG only induced the “open” conformation in hSTING H232, perhaps explaining why this CDN fails to significantly activate this human STING variant (Ablasser et al., 2013; Burdette et al., 2011; Diner et al., 2013; Gao et al., 2013b). Thus, both mSTING and R232 and H232 alleles of hSTING are responsive to 2′-5′, 3′-5′cGAMP, but only mSTING and the R232 allele of hSTING respond to 3′-5′, 3′-5′cdG and 3′-5′, 3′-5′cGAMP (Burdette et al., 2011; Zhang et al., 2013). Although the closed conformation was observed earlier for hSTING solved with cdG (Huang et al., 2012), this may be a rare conformation for the 3′-5′, 3′-5′ CDN complex that depends more on crystallization conditions. The hSTING H232 allele binds 2′-5′, 3′-5′cGAMP with slightly lower affinity than the hSTING R232 allele, but the signaling response selectivity for the 2′ -5′ bond by hSTING H232 defines a clear difference between it and hSTING R232 (Diner et al., 2013; Gao et al., 2013b). Furthermore, because mSTING (R231) binds 3′-5′, 3′-5′CDNs in a nearly identical closed conformation (Gao et al., 2013b), the responsiveness of these two STING alleles to the bacterial CDNs connects the closed conformation (rather than binding affinity per se) with downstream signaling that induces the type I IFN response. Crystal structures of hSTING H232 allele with 3′-5′, 3′-5′ CDNs (which do not fully activate this protein) should provide a definitive answer to this hypothesis. Together, these data suggest a view of how various alleles of STING might function in innate immune recognition, depending on the source of the CDN signal (Figure 1). It is also worth noting that different alleles of STING might also be activated in some cells by binding to the DExD/H-box helicase DDX41, which has been implicated in binding of DNA and triggering a type I IFN response (Parvatiyar et al., 2012; Zhang et al., 2011), but the role of cGAS and 2′-5′, 3′-5′cGAMP in this alternative pathway for STING activation has not yet been carefully addressed. Furthermore, a recent extensive biochemical analysis has detected numerous proteins that may contribute to the innate immune response against cytosolic or foreign nuclear DNA (Lee et al., 2013), but these host proteins have not been evaluated for the role of cGAS, CDNs, or STING in their observed biochemical interactions. Thus, there is still much to learn about how cGAS and STING integrate their activity with multiple host responses to foreign or mislocalized DNA.
Figure 2. Model of STING Binding to CDNs.
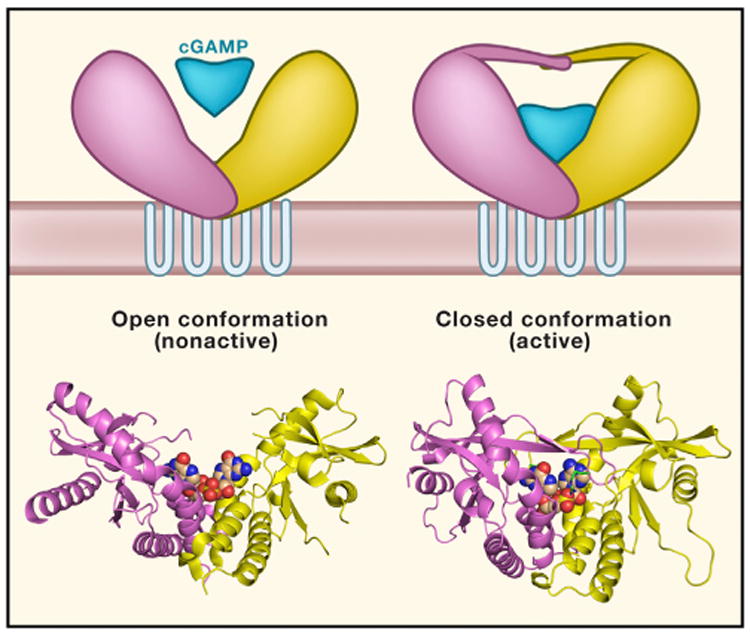
In its “open” form, STING does not fully encapsulate cdG and possibly other bacterial 3′-5′, 3′-5′CDNs. STING structure is more flexible; partially disoriented loops are covering the binding pocket. Binding of 2′-5′, 3′-5′cGAMP to STING happens at a deeper pocket compared to cdG and results in formation of the “closed” form. STING it its closed form is more compact, and the binding pocket is covered by a four-stranded β sheet cap. RCSB Protein Data Bank coordinates: cdG-hSTING H232 complex (4EF4) and 2′-5′, 3′-5′cGAMP-hSTING H232 complex (4LOH).
Impact of Allele Variations in STING on Human Fitness
Variations in the hSTING locus have been documented by Jin et al. (2011b). Approximately 18% of humans in two large cohorts, totaling over 1,000 individuals, were found to be heterozygous for the H232 allele, with the more prevalent allele being R232 (thus, it should be considered a wild-type allele). Given that the H232 allele is nonresponsive to 3′-5′, 3′-5′CDNs, it will be interesting to know whether this allele is dominant (that is, whether heterodimers with the R232 allele produce STING that is nonresponsive to cdG). Given that certain alleles of human STING (e.g., H232) have likely lost the specific ability to respond to bacterial CDNs while retaining their ability to respond to the human endogenous messenger 2′ -5′, 3′-5′cGAMP produced by cGAS, it is tempting to speculate that there was a strong selective pressure to lose responsiveness to bacterial CDNs during human evolution (Diner et al., 2013). The need to preserve a robust innate immune response to various viruses, including the smallpox virus and retroviruses, provides a reasonable explanation for retaining STING responsiveness to the endogenous 2′-5′, 3′-5′cGAMP produced by cGAS (Gao et al., 2013c). However, understanding the selective pressure to “blind” some hSTING alleles to bacterial CDNs is more challenging (Monroe et al., 2009). The frequency and penetrance for immune dysfunction of such hSTING alleles in the global human population and their geographic distribution may provide clues to the nature of the selective process. One can envision at least two types of selective pressure that may have been driving this human STING genetic variation: (1) bacterial CDN-driven susceptibility or enhanced pathology associated with bacterial diseases of high prevalence in certain human populations and (2) bacterial CDN-driven alterations of nutritional or metabolic states that significantly affected human mortality.
Selective pressure under hypothesis 1 would include exposure to bacterial pathogens whose replication (and thus virulence) was enhanced by a type I IFN response. This could occur by the known ability of type I IFNs to polarize the adaptive immune response as well as cause increased pathology by driving excessive levels of inflammation. Bacterial invasive diseases with high-inflammatory symptoms such as tuberculosis, bubonic plague, shigellosis, meningitis, enteric fever, and pneumonias might be considered as selective drivers under hypothesis 1 if their causative agents expose host cells to bacterial CDNs during infections. Mycobacterium tuberculosis specifically induces a type I IFN response by releasing bacterial DNA into the host cell cytosol after permeabilization of a phagosomal membrane via the activity of its virulence-associated ESX secretion system (Manzanillo et al., 2012). The bacterial CDNs produced by M. tuberculosis apparently do not contribute to this response (Manzanillo et al., 2012), and thus this pathogen may have evolved this new way to activate STING by stimulating production of the endogenous cGAS synthesized 2′-5′, 3′-5′cGAMP. Mice defective in type I IFN signaling are considerably more resistant to M. tuberculosis (Manzanillo et al., 2012), and induction of type I IFNs exacerbates pulmonary tuberculosis in an IFN-α/β receptor-dependent fashion (Antonelli et al., 2010). Thus, one could easily imagine that the human host would be under selective pressure to lower its steady-state level of STING activation in order to counteract this pathogen's immune modulation strategy. As noted earlier, other pathogens such as L. monocytogenes actively secrete CDNs into the cytosol of cultured cells and directly activate the type I IFN response by a STING-dependent pathway (Woodward et al., 2010). However, the level of STING activation in different host tissues is largely unknown both before and after bacterial infections. Understanding whether susceptibility to replication or host injury by bacterial pathogens can be altered in mice by replacement of mSTING with the hSTING H232 allele could reveal how some bacteria may have utilized the host response to bacterial CDNs to enhance their own replication or fitness within the human host.
Selective pressure under hypothesis 2 might include any STING-driven process that interferes with nutrition and thus increases infant or child mortality through secondary effects that could include susceptibility to severe diarrheal disease. In this regard, recent data suggest that mutations in innate immune pathways are enriched in patients that appear more susceptible to cholera (Karlsson et al., 2013). Inflammatory states in the gut are known to decrease nutrient uptake; however, little is known about the role of STING in contributing to mucosal inflammatory states. The large quantity of bacterial mass typically contained within the human intestine might be a source of bacterial CDNs that somehow find their way into host cells and produce significant STING activation within the local intestinal epithelium. Because the gut microbiome is also thought to play a role in metabolic syndromes such as obesity and diabetes (Gross, 2013), it is possible that alterations in the response CDNs and other PAMPs derived from commensal bacterial cells could be the selected outcome of human evolution that was seeking nutritional fitness in the face of famine. The fact that mutations in receptors associated with the innate immune system have been linked to colitis and other inflammatory conditions (Cario, 2010) underlines how little we know about the selective forces that drove mutational changes in humans that were challenged with pathogens as well as the need for immune homeostasis when stimulated by a complex intestinal milieu.
In conclusion, the newly recognized cGAS-STING-type I IFN pathway for innate immune responses to cytosolic DNA will likely lead to exciting new investigations on the role of bacterial CDNs in altering host susceptibility to pathogens as well as inflammatory conditions linked to the human microbiota or disease states that lead to leakage of nuclear DNA into the host cell cytosol. Targeting both cGAS and STING with either activating or inhibitory drugs may be a promising new strategy for treatment of chronic infection, inflammatory states, and other diseases in which immunomodulation of the type I IFN pathway might show therapeutic benefit.
Acknowledgments
This work was supported in part by the National Institutes of Health grants AI-26289 and AI-018045 to J.J.M. The authors would like to thank Michaela Gack, Daniel Portnoy, and Russell Vance for their helpful comments. J.J.M. is on the scientific advisory board of Aduro BioTech, which is developing dicyclic nucleotide-based adjuvants for vaccines and immunotherapeutics.
References
- Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, Barber GN. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′ -5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli LR, Gigliotti Rothfuchs A, Gonçalves R, Roffê E, Cheever AW, Bafica A, Salazar AM, Feng CG, Sher A. Intranasal Poly-IC treatment exacerbates tuberculosis in mice through the pulmonary recruitment of a pathogen-permissive monocyte/macrophage population. J Clin Invest. 2010;120:1674–1682. doi: 10.1172/JCI40817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKK-gamma to dampen the host NF-kappaB-mediated inflammatory response. Nat Cell Biol. 2010;12:66–73. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. Cytoplasmic DNA innate immune pathways. Immunol Rev. 2011a;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- Barber GN. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr Opin Immunol. 2011b;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JR, Koestler BJ, Carpenter VK, Burdette DL, Waters CM, Vance RE, Valdivia RH. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. MBio. 2013;4:e00018–e13. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxt LA, Garza-Mayers AC, Goldberg MB. Bacterial subversion of host innate immune pathways. Science. 2013;340:697–701. doi: 10.1126/science.1235771. [DOI] [PubMed] [Google Scholar]
- Bradford BM, Mabbott NA. Prion disease and the innate immune system. Viruses. 2012;4:3389–3419. doi: 10.3390/v4123389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SM, Nelson EJ, Chowdhury N, Faruque SM, Calderwood SB, Camilli A. Cholera stool bacteria repress chemotaxis to increase infectivity. Mol Microbiol. 2006;60:417–426. doi: 10.1111/j.1365-2958.2006.05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario E. Toll-like receptors in inflammatory bowel diseases: a decade later. Inflamm Bowel Dis. 2010;16:1583–1597. doi: 10.1002/ibd.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavlar T, Ablasser A, Hornung V. Induction of type I IFNs by intracellular DNA-sensing pathways. Immunol Cell Biol. 2012;90:474–482. doi: 10.1038/icb.2012.11. [DOI] [PubMed] [Google Scholar]
- Chang SJ, Hsiao JC, Sonnberg S, Chiang CT, Yang MH, Tzou DL, Mercer AA, Chang W. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-kappaB activation by tumor necrosis factor alpha but is independent of its host range function. J Virol. 2009;83:4140–4152. doi: 10.1128/JVI.01835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RA, Ryzhakov G, Cooray S, Randow F, Smith GL. Inhibition of IkappaB kinase by vaccinia virus virulence factor B14. PLoS Pathog. 2008;4:e22. doi: 10.1371/journal.ppat.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science. 2010;328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Gründling A. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 2013;11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Gründling A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA. 2013;110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Yao Q, Li S, Ding X, Lu Q, Mao H, Liu L, Zheng N, Chen S, Shao F. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science. 2010;329:1215–1218. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev A, Iyer S, Razani B, Cheng G. NF-κB and innate immunity. Curr Top Microbiol Immunol. 2011;349:115–143. doi: 10.1007/82_2010_102. [DOI] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPerna G, Stack J, Bowie AG, Boyd A, Kotwal G, Zhang Z, Arvikar S, Latz E, Fitzgerald KA, Marshall WL. Poxvirus protein N1L targets the I-kappaB kinase complex, inhibits signaling to NF-kappaB by the tumor necrosis factor superfamily of receptors, and inhibits NF-kappaB and IRF3 signaling by toll-like receptors. J Biol Chem. 2004;279:36570–36578. doi: 10.1074/jbc.M400567200. [DOI] [PubMed] [Google Scholar]
- Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Nat Acad Sci USA. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- Duthie MS, Windish HP, Fox CB, Reed SG. Use of defined TLR ligands as adjuvants within human vaccines. Immunol Rev. 2011;239:178–196. doi: 10.1111/j.1600-065X.2010.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebensen T, Libanova R, Schulze K, Yevsa T, Morr M, Guzmán CA. Bis-(3′,5′)-cyclic dimeric adenosine monophosphate: strong Th1/Th2/ Th17 promoting mucosal adjuvant. Vaccine. 2011;29:5210–5220. doi: 10.1016/j.vaccine.2011.05.026. [DOI] [PubMed] [Google Scholar]
- Franchi L, Muñoz-Planillo R, Núñez G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol. 2012;13:325–332. doi: 10.1038/ni.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013a;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013b;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013c doi: 10.1126/science.1240933. Published online August 8, 2013. 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Xu H, Li T, Zhou Y, Zhang Z, Li S, Liu L, Shao F. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc Natl Acad Sci USA. 2009;106:13725–13730. doi: 10.1073/pnas.0907200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M. Does the gut microbiome hold clues to obesity and diabetes? Curr Biol. 2013;23:R359–R362. doi: 10.1016/j.cub.2013.04.047. [DOI] [PubMed] [Google Scholar]
- Gründling A, Burrack LS, Bouwer HG, Higgins DE. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc Natl Acad Sci USA. 2004;101:12318–12323. doi: 10.1073/pnas.0404924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A, Miller SI. A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell Microbiol. 2006;8:837–846. doi: 10.1111/j.1462-5822.2005.00670.x. [DOI] [PubMed] [Google Scholar]
- Hicks SW, Galán JE. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol. 2013;11:316–326. doi: 10.1038/nrmicro3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011a;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Xu LG, Yang IV, Davidson EJ, Schwartz DA, Wurfel MM, Cambier JC. Identification and characterization of a loss-of-function human MPYS variant. Genes Immun. 2011b;12:263–269. doi: 10.1038/gene.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Wu H, Wentworth C, Luo L, Collier-Hyams L, Neish AS. Salmonella AvrA Coordinates Suppression of Host Immune and Apoptotic Defenses via JNK Pathway Blockade. Cell Host Microbe. 2008;3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- Jubelin G, Taieb F, Duda DM, Hsu Y, Samba-Louaka A, Nobe R, Penary M, Watrin C, Nougayrède JP, Schulman BA, et al. Pathogenic bacteria target NEDD8-conjugated cullins to hijack host-cell signaling pathways. PLoS Pathog. 2010;6:e1001128. doi: 10.1371/journal.ppat.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, Philpott D, Schroeder JT, Hyodo M, Hayakawa Y, et al. Bacterial c-di-GMP is an immunostimulatory molecule. J Immunol. 2007;178:2171–2181. doi: 10.4049/jimmunol.178.4.2171. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Harris JB, Tabrizi S, Rahman A, Shlyakhter I, Patterson N, O'Dushlaine C, Schaffner SF, Gupta S, Chowdhury F, et al. Natural selection in a bangladeshi population from the cholera-endemic ganges river delta. Sci Transl Med. 2013;5:192ra186. doi: 10.1126/scitranslmed.3006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci USA. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Lee SY, Kim SY, Kim JK, Kim HJ, Lee HM, Choi MS, Min JS, Kim MJ, Choi HS, Ahn JK. HSV-1 ICP27 suppresses NF-kappaB activity by stabilizing IkappaBalpha. FEBS Lett. 2008;582:2371–2376. doi: 10.1016/j.febslet.2008.05.044. [DOI] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad SP, Li J, da Silva Correia J, Pan Q, Gadwal S, Ulevitch RJ, Li E. Cleavage of p65/RelA of the NF-kappaB pathway by Chlamydia. Proc Natl Acad Sci USA. 2007;104:2933–2938. doi: 10.1073/pnas.0608393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock CN, Cookson BT. The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe. 2012;12:799–805. doi: 10.1016/j.chom.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Negrate G, Faustin B, Welsh K, Loeffler M, Krajewska M, Hasegawa P, Mukherjee S, Orth K, Krajewski S, Godzik A, et al. Salmonella secreted factor L deubiquitinase of Salmonella typhimurium inhibits NF-kappaB, suppresses IkappaBalpha ubiquitination and modulates innate immune responses. J Immunol. 2008a;180:5045–5056. doi: 10.4049/jimmunol.180.7.5045. [DOI] [PubMed] [Google Scholar]
- Le Negrate G, Krieg A, Faustin B, Loeffler M, Godzik A, Krajewski S, Reed JC. ChlaDub1 of Chlamydia trachomatis suppresses NF-kappaB activation and inhibits IkappaBalpha ubiquitination and degradation. Cell Microbiol. 2008b;10:1879–1892. doi: 10.1111/j.1462-5822.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- Lee MN, Roy M, Ong SE, Mertins P, Villani AC, Li W, Dotiwala F, Sen J, Doench JG, Orzalli MH, et al. Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat Immunol. 2013;14:179–185. doi: 10.1038/ni.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 2013;32:2001–2014. doi: 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 2012;83:623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stülke J. Cyclic di-AMP homeostasis in bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem. 2013;288:2004–2017. doi: 10.1074/jbc.M112.395491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinzer U, Barreau F, Esmiol-Welterlin S, Jung C, Villard C, Léger T, Ben-Mkaddem S, Berrebi D, Dussaillant M, Alnabhani Z, et al. Yersinia pseudotuberculosis effector YopJ subverts the Nod2/RICK/TAK1 pathway and activates caspase-1 to induce intestinal barrier dysfunction. Cell Host Microbe. 2012;11:337–351. doi: 10.1016/j.chom.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Mills E, Pultz IS, Kulasekara HD, Miller SI. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell Microbiol. 2011;13:1122–1129. doi: 10.1111/j.1462-5822.2011.01619.x. [DOI] [PubMed] [Google Scholar]
- Monroe KM, McWhirter SM, Vance RE. Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PLoS Pathog. 2009;5:e1000665. doi: 10.1371/journal.ppat.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA. 2012;109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMPand cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villán E, García-Sastre A, Gack MU. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012;8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci USA. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy FJ, Grebe A, Rayner KJ, Kalantari P, Ramkhelawon B, Carpenter SB, Becker CE, Ediriweera HN, Mullick AE, Golenbock DT, et al. CD36 coordinates NLRP3 inflammasome activation by facilitating intracellular nucleation of soluble ligands into particulate ligands in sterile inflammation. Nat Immunol. 2013;14:812–820. doi: 10.1038/ni.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisler JL, Jin XL. The vaccinia virus K1L gene product inhibits host NF-kappaB activation by preventing IkappaBalpha degradation. J Virol. 2004;78:3553–3560. doi: 10.1128/JVI.78.7.3553-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, et al. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent MS, Stead CM, Tran AX, Hankins JV. Diversity of endotoxin and its impact on pathogenesis. J Endotoxin Res. 2006;12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Hartung S, Büttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Witte CE, Archer KA, Rae CS, Sauer JD, Woodward JJ, Portnoy DA. Innate immune pathways triggered by Listeria monocytogenes and their role in the induction of cell-mediated immunity. Adv Immunol. 2012;113:135–156. doi: 10.1016/B978-0-12-394590-7.00002-6. [DOI] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H, Ooka T, Iguchi A, Hayashi T, Sugimoto N, Tobe T. NleC,atype III secretion protease, compromises NF-κB activation by targeting p65/RelA. PLoS Pathog. 2010;6:e1001231. doi: 10.1371/journal.ppat.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, He ZG. Radiation-sensitive gene A (RadA) targets DisA, DNA integrity scanning protein A, to negatively affect its cyclic-di-AMP synthesis activity in Mycobacterium smegmatis. J Biol Chem. 2013;288:22426–22436. doi: 10.1074/jbc.M113.464883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


