Abstract
Splicing of the c-src N1 exon is repressed by the polypyrimidine tract binding protein (PTB or PTBP1). During exon repression, the U1 snRNP binds properly to the N1 exon 5′ splice site but is made inactive by the presence of PTB. Examining the patterns of nuclease protection at this 5′ splice site, we find that the interaction of U1 is altered by the adjacent PTB. Interestingly, UV-crosslinking identifies a direct contact between the pre-mRNA-bound PTB and the U1 snRNA. EMSA, ITC and NMR studies show that PTB RRMs 1 and 2 bind the pyrimidine-rich internal loop of U1 snRNA stem loop 4. The PTB/U1 interaction prevents further assembly of the U1 snRNP with spliceosomal components downstream. This precise interaction between a splicing regulator and an snRNA component of the spliceosome points to a range of different mechanisms for splicing regulation.
INTRODUCTION
Most mammalian genes produce multiple mRNA and protein products through alternative patterns of pre-mRNA splicing (Nilsen and Graveley, 2010). Numerous pre-mRNA binding proteins have been identified that can alter splicing choices (Black, 2003; Chen and Manley, 2009; Matlin et al., 2005). However, the mechanisms by which these proteins act are not well understood. Precise contacts between regulatory proteins and components of the core splicing machinery have only rarely been identified.
The spliceosome assembles onto each intron through the sequential binding of the five small ribonucleoprotein particles (snRNPs) U1, U2, U4, U5, and U6, and multiple auxiliary proteins to form discreet RNP complexes termed E, A, B, and C (Wahl et al., 2009). Splicing catalysis takes place in the spliceosomal C complex, but a key question in the regulation of alternative splicing regards how the choice is made of which splice sites to pair within that later complex. Cross intron contacts between the U1 snRNP complex at the 5′ splice site and the U2 snRNP complex at the branchpoint and 3′ splice site are crucial in determining splice site pairing. On a simple single-intron pre-mRNA studied in vitro, such contacts are seen in the ATP-independent E complex. Commitment to splice site pairing is thought to occur during the formation of the subsequent A complex with the ATP-dependent base-pairing of the U2 snRNA to the branchpoint (Donmez et al., 2007; Kotlajich et al., 2009; Lim and Hertel, 2004; Michaud and Reed, 1993). However, a typical metazoan pre-mRNA containing multiple exons and introns will undergo a process of exon definition, allowing U1 and U2 to bind on each exon, prior to their interaction across introns. The molecular nature of the cross intron contacts between defined exons is not known, but they have been identified as targets for regulating splice site pairing choices (Bonnal et al., 2008; House and Lynch, 2006; Sharma et al., 2008).
The polypyrimidine tract binding protein (PTB) is a widely expressed splicing regulator controlling many alternative exons (Auweter and Allain, 2008; Spellman et al., 2005; Spellman and Smith, 2006). Earlier studies recognized PTB mainly as a splicing repressor, but recent genomewide identification of PTB target exons using micro-array and clip-seq methods show that PTB can also enhance inclusion of some exons (Boutz et al., 2007; Llorian et al., 2010; Xue et al., 2009). Exons repressed by PTB often contain multiple CU-rich binding elements: a high affinity site in the upstream intron and a second sometimes weaker site either within the exon or the downstream intron. In one well-studied example, PTB represses splicing of the N1 exon of the c-src pre-mRNA (Chan and Black, 1997). This regulation has been reconstructed in vitro using extracts from Hela cell nuclei containing high levels of PTB that inhibit N1 exon splicing (Chou et al., 2000; Markovtsov et al., 2000). In contrast, extracts from WERI retinoblastoma cells have reduced PTB, and contain the neuronal PTB homolog (nPTB, brPTB, PTBP2). In these extracts, the N1 exon is efficiently spliced. We previously showed that PTB binding to these CU-elements does not interfere with the recognition of the N1 exon 5′ splice site by the U1 snRNP (Sharma et al., 2005). Instead, PTB inhibits splice site pairing by blocking interaction of the U1 snRNP bound at the N1 exon 5′ splice site with the U2 snRNP complex at the downstream 3′ splice site (Sharma et al., 2008). This block to intron definition interactions prevents binding of the U4/U6-U5 tri-snRNP and the formation of an intronic spliceosomal B complex. The multiple PTB molecules assembled in the N1 exon complex interact, and are thought to create a RNA loop (Amir-Ahmady et al., 2005; Lamichhane et al., 2010; Spellman and Smith, 2006). In other systems, PTB also interacts with the corepressor protein Raver 1 to inhibit splicing (Rideau et al., 2006). However, in spite of the presence of snRNPs assembled on the repressed exons, direct interactions between PTB and the spliceosome have not been described.
We now examine how PTB prevents spliceosome assembly by the adjacent U1 snRNP. We find that the pre-mRNA-bound PTB directly contacts the U1 snRNA. This contact changes the interaction of the bound U1 with the 5′ splice site preventing its productive interaction with downstream spliceosomal components.
RESULTS
U1 snRNP/5′ splice site interactions are altered during splicing repression
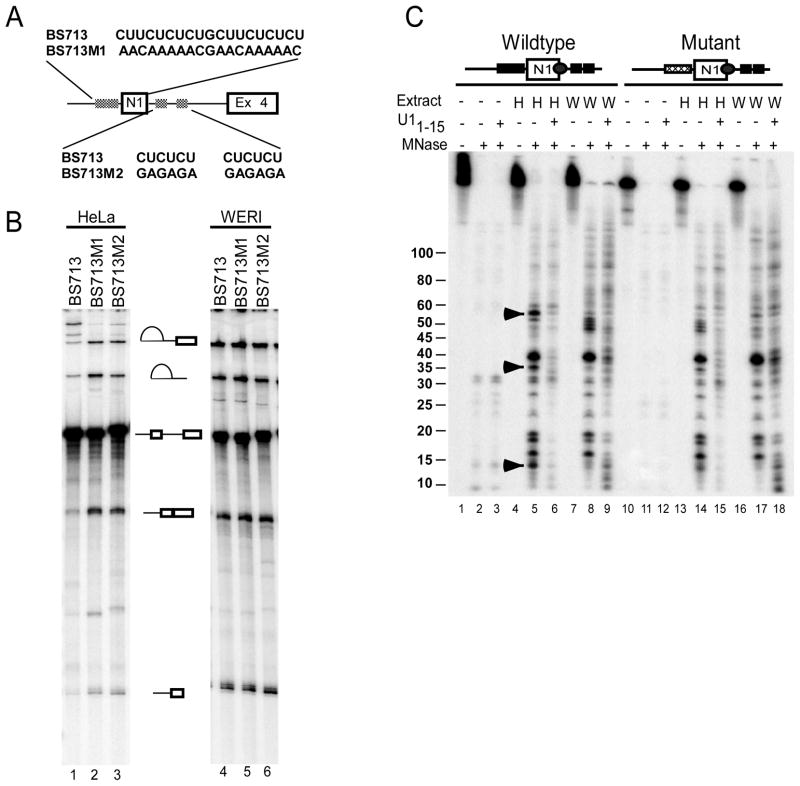
In previous work, we found that when splicing of the c-src N1 exon is repressed by PTB, the U1 snRNP is still bound to the N1 5′ splice site (Sharma et al., 2005). However, this U1 fails to stably interact with the exon 4 complex assembled downstream and progress into a higher order intronic spliceosome (Sharma et al., 2008). We used the PTB-dependent in vitro splicing system to examine how PTB was preventing this cross-intron interaction. We analyzed an RNA substrate containing the N1 exon and its flanking regulatory elements as well as the downstream exon 4 (BS713; Figure 1A). As seen for similar RNAs tested previously, this RNA was spliced in WERI extract (lanes 4–6), but repressed in Hela extract (lanes1–3). Mutating either PTB binding site allowed splicing in Hela extract (Figure 1B). Using this substrate, we examined whether the U1 interaction at the N1 5′ splice site differed between the active pre-spliceosome assembled in WERI extract and the PTB repressed complex assembled in Hela extract. In other studies, the presence of silencer elements was found to affect the extent of RNA surrounding the splice site that is protected by U1 snRNP from nuclease digestion (Yu et al., 2008). These differences in U1 interactions with the 5′ splice site were shown to correlate with the ability of the U1 snRNP to progress in spliceosome assembly. An RNA containing the N1 exon and its flanking CU elements was specifically labeled with 32P at the 5′ splice site and assembled into RNP complexes in Hela and WERI extracts. The assembled complexes were treated with micrococcal nuclease (MNase) as described (Maroney et al., 2000). The RNA fragments remaining after MNase treatment were extracted and visualized by urea-PAGE. Both a wildtype RNA and a mutant RNA with mutations in the upstream PTB binding site were nearly completely degraded by MNase in buffer (Figure 1C, lanes 2, 3, 11, and 12). However, after incubation in HeLa or WERI extract, specific RNA fragments encompassing the 5′ splice site were protected from MNase digestion (lanes 5 and 8). This protection was dependent on the U1 snRNP, as pre-incubation with an oligonucleotide complementary to the 5′ end of U1 snRNA led to loss of protected fragments (lanes 6, 9, 15 and 18). Interestingly, the pattern of protected fragments in HeLa extract was different from that in WERI extract, indicating that the U1/5′ splice site interaction was different under the two conditions. Fragments of approximately 55, 35, and 13 nucleotides were more strongly protected in HeLa extract than in WERI extract. Fragments common to both protection patterns included major bands at 38, 19, 18, and 15 nucleotides and some less prominent bands. In WERI extract, the protected fragments included a series of closely spaced bands between 45 and 50 nucleotides that were not seen in HeLa extract on the wildtype substrate. However, these bands varied in intensity between experiments (see lane 17) and are likely nuclease digestion intermediates.
Figure 1. U1 snRNP/5′ splice site interaction is altered during splicing repression.
A) Schematic maps of the wildtype and mutant N1 exon containing pre-mRNAs. B) CU-elements flanking the N1 exon are essential for splicing repression. In vitro splicing of the wildtype and mutant BS713 transcripts was carried out in HeLa (lanes 1–3) and WERI-1 (lanes 4–6) extract. The RNA splicing products and intermediates are shown to the right. B) C-src transcripts containing the N1 exon were site specifically 32P labeled at the N1 exon 5′ splice site. The labeled wildtype and PTB binding site mutant RNAs were incubated in Buffer DG (lanes 1–3 and 10–12), HeLa extract (lanes 5–7 and 13–15), and WERI extract (lanes 7–9 and 16–18). After incubation, reactions were treated with micrococcal nuclease (lanes 2, 3, 5, 6, 8, 9, 11, 12, 14, 15, 17 and 18). Extract were either mock treated or pre-incubated with oligonucleotide U11–15 prior to addition of the labeled N1 RNA. The protected fragments were extracted, using PCA, ethanol precipitated, separated using urea-PAGE, and visualized by phosphorimaging. The presence of a downstream 3′ splice site did not alter the MNase protection pattern in HeLa extract (see Figure S1).
Significantly, an RNA carrying a mutation in the upstream PTB binding site altered the nuclease protection pattern in HeLa extract to that seen in WERI extract (lane 14). Protection of the HeLa-specific 55, 35, and 13 nucleotide fragments was lost with the mutant RNA, but the 38, 19, 18, and 15 nucleotide fragments common to both extracts were still generated. With this mutant RNA, the sporadic group of fragments between 45 and 50 nucleotides was now seen in HeLa extract (lane 14), but not in WERI (lane 17). Thus, in the absence of PTB binding to the upstream site, the pattern of protected bands was very similar between the two extracts. A longer spliceable transcript containing the downstream exon 4 yielded a protection pattern in Hela extract that was the same as the shorter N1 exon RNA, indicating that complex at the N1 5′ splice site was not affected by the downstream 3′ splice site (Figure S1). We conclude that during splicing repression, PTB binding alters the interaction of the U1 snRNP with the 5′ splice site.
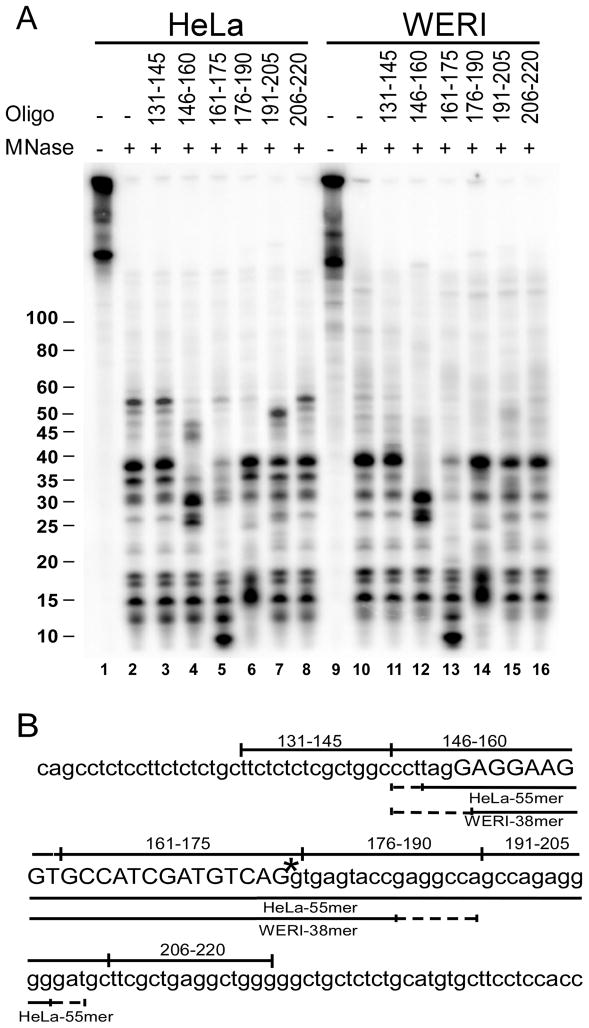
To map the RNA fragments protected by the U1 snRNP, we examined their patterns of cleavage by RNase H. Prior to electrophoresis, the RNA fragments remaining after MNase treatment were further incubated with RNase H and DNA oligos complementary to the pre-mRNA. RNase H cleavage sites within a DNA/RNA hybrid are variable, but cleavage induced by an oligo indicates that it is complementary to at least a portion of the RNA (Lapham et al., 1997). Since the RNA is labeled at a specific phosphate in the 5′ splice site, the size of the cleaved fragments can be used to determine the extent of protection surrounding this site. The 55, 38, and 35 nt fragments were all cleaved by the oligos complementary to RNA nucleotides 146 to 160 (Figure 2A, lane 4), generating products either 8 or 12 nucleotides shorter. This indicates that these three fragments each contain a 5′ end within the same region, which is at least 8 or 12 nucleotides from the 5′ end of the DNA oligo or between nucleotides 149 and 153 (Figure 2B). All the fragments were cleaved by the DNA oligo that encompasses the labeled phosphate at the 5′ splice site (nts 161–175; Figure 2A lane 5). Given its length and start site the 38 nt fragment should end between nucleotides 183 and 190. The lack of cleavage by the 176–190 oligo could result from the unpaired 5′ end of the DNA oligo, which can be a favored position for RNAse H cleavage (lane 6). The 55 nucleotide fragment is cleaved by two downstream oligos (complementary to nucleotides 176–190 and 191–205; Figure 2A lanes 6 and 7). Measuring 55 nucleotides from its possible start sites, this long HeLa specific fragment must end between nucleotides 200 and 203. In agreement with this, cleavage by the most downstream oligo generates a fragment approximately 50 nts long. RNase H mapping with other oligonucleotides, whose complementary regions are offset from this group, gave equivalent results (Figure S2).
Figure 2. U1 snRNP protects a much longer region of the pre-mRNA under PTB-dependent repression.
A) The extent of protection of the labeled c-src RNAs was determined using oligonucleotide mediated RNase H cleavage. Site specifically labeled wildtype RNAs were incubated in HeLa extract (lanes 1–8) and WERI extract (lanes 9–16). After incubation, reactions were treated with micrococcal nuclease (lanes 2–8 and 10–16). The protected fragments were extracted using PCA and ethanol precipitated. The protected RNAs were then subject to RNase H cleavage in presence of 15 nucleotide long oligonucleotides as indicated. After RNase H treatment, the RNAs were separated using urea-PAGE and visualized by phosphorimaging. Oligos with complimentarity offset from those shown here gave equivalent results in both HeLa and WERI extracts (see Figure S2). B) Sequence of the N1 exon containing pre-mRNA. Positions of DNA oligonucleotides used for RNase H cleavage are indicated above the sequence. The lines below the sequence indicate the boundaries of nuclease protection in HeLa and WERI extracts.
The mixed cleavage patterns produced by RNase H prevents mapping the RNA fragment ends precisely to the nucleotide (Lapham et al., 1997). Nevertheless, from these data it is clear that the 5′ ends of the two major protected RNA fragments (55 and 38 nucleotides long) are nearly the same and are within nucleotides 149–153 of the N1 RNA (Figure 2B). The 3′ end is between nucleotides 183–190 for the 38 nt fragment and between nucleotides 200–203 for the 55 nt fragment. The 38 nucleotide fragments produced in both HeLa and WERI extract give identical patterns of cleavage. We conclude that the pattern of 5′ splice site protection by U1 is altered under PTB repression to produce a larger protected region within the downstream intron.
PTB interacts with the U1 snRNA
We showed previously that the U1 snRNA is correctly base-paired to the N1 exon 5′ splice site in both HeLa and WERI extract (Sharma et al., 2005). Thus, the extended protection of RNA seen in HeLa extract is not due to a shift in the U1 snRNA base pairing to a different position. The extended protection downstream of the 5′ splice site is also not likely due to PTB itself directly contacting that segment of RNA, as the nearest PTB binding site is approximately 20 nucleotides downstream from the end of the longest protected fragment. Instead, PTB could recruit additional factors to the protected region, or possibly a direct interaction of PTB with the U1 snRNP could alter its interaction with the 5′ splice site, as seen with a synthetic non-PTB dependent repressor element (Yu et al., 2008). To examine whether additional factors were being recruited to this region in HeLa extract, we performed site-specific labeling and UV-crosslinking experiments using 32P-labeled phosphates placed at nucleotides 198 and 208. No proteins were found to crosslink to position 198 (data not shown). The proteins crosslinking to position 208 included PTB and hnRNP H in HeLa extract and PTB, nPTB, and hnRNP H in WERI extract (Figure S3). These proteins are known to assemble at positions 218–231 (Chou et al., 2000; Markovtsov et al., 2000). Importantly, there was no difference in the crosslinking patterns between HeLa and WERI extracts and none of the U1 snRNP specific proteins crosslinked in either extract indicating that the extended protection seen in HeLa extract is not due to recruitment of new proteins to that region.
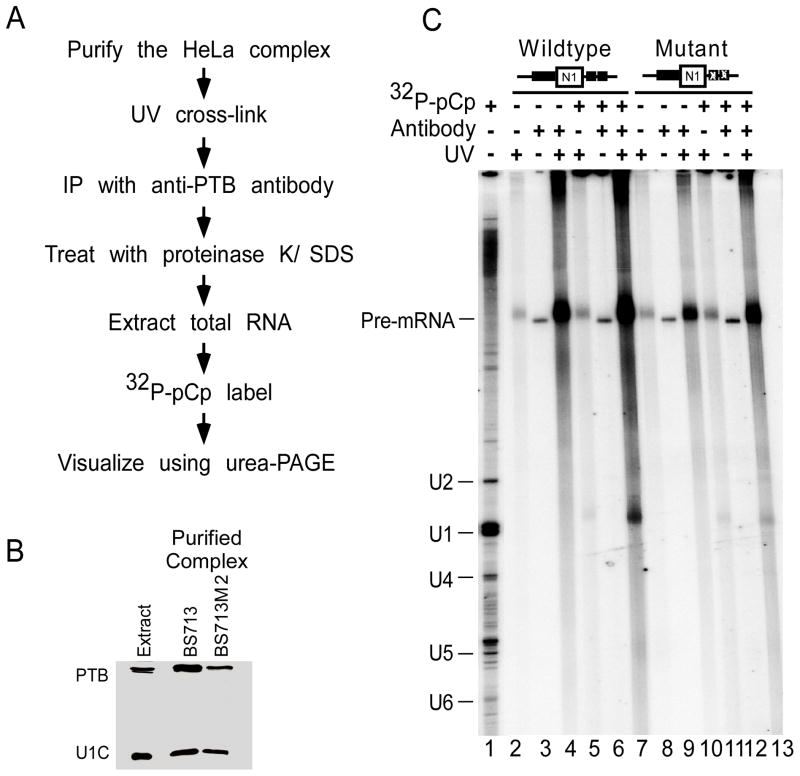
To examine whether PTB was interacting with the U1 snRNA and possibly changing its interaction with the 5′ splice site region, we UV-crosslinked N1 exon complexes purified from HeLa extract using a MS2 affinity tag (Figure 3A). The complex formed on the wildtype RNA was compared to that assembled on a mutant RNA lacking the PTB binding site downstream of the 5′ splice site. This mutation eliminates PTB dependent repression in Hela extract, but not PTB binding to the high affinity site upstream of the exon. Immunoblot analysis of complexes assembled on the wildtype RNA and mutant RNAs showed the presence of PTB and the U1 snRNP specific protein U1C in both complexes (Figure 3B). The purified complexes were exposed to 254nm UV light to cross-link proteins to RNA and the PTB within the complexes was isolated by immunoprecipitation (Figure 3A). After proteinase K digestion, total RNA was extracted, labeled with 32P-pCp, and visualized using urea-PAGE (Figure 3C). The uniformly labeled wildtype and mutant RNAs were both efficiently immunoprecipitated after crosslinking, relative to no UV and no antibody controls, indicating that PTB was binding and crosslinking to the upstream binding site in the mutant RNA (compare lanes 4 and 10 to lanes 2 and 3 or lanes 8 and 9). Most interestingly, labeling the RNA with 32P-pCp showed that in complexes containing the wildtype N1 RNA, the U1 snRNA was also crosslinked to PTB (lane 7). Moreover, mutation of the downstream PTB binding site nearly eliminated the crosslinking of PTB to U1 (lane 13). Thus, PTB directly contacts with the U1 snRNA in the repressed HeLa complex and mutation of the PTB binding site downstream can disrupt this interaction.
Figure 3. PTB interacts with the U1 snRNA during repression.
A) Schematic outline of the method used for studying U1 snRNA/PTB interaction. The MS2 hairpin tagged N1 exon RNAs were incubated in HeLa extract. The assembled complexes were purified using the MS2 affinity tag method. The purified complexes were exposed to UV-254 nm and immunoprecipitated using anti-PTB antibody, PTB-NT that was pre-bound to gamma-bind Sepharose. The immunoprecipitated complexes were then treated with SDS/proteinase K, while still bound to the gamma bind beads and total RNA was extracted, ethanol precipitated, labeled with 32P-pCp, and visualized using urea-PAGE. B) Western blot analysis of the proteins from the purified complexes. Proteins from complexes purified on wildtype RNA and mutant RNAs containing mutations in the downstrem PTB binding sites were separated using SDS-PAGE and probed using anti-PTB and anti-U1C antibodies. C) Analysis of RNA from purified complexes. RNA from complexes assembled and purified on wildtype (lanes 2–7) and mutant RNAs (lanes 8–13) were exposed to UV-254 nm (lanes 2, 4, 5, 7, 8, 10, 11, and 13) and subject to immunoprecipitation using beads alone (lanes 2, 5, 8, and 11) or with beads containing the anti-PTB antibody, PTB-NT (lanes 3, 4, 6, 7, 9, 10, 12, and 13). In Hela extract, no additional proteins were recruited to the region between the N1 exon 5′ splice site and the PTB binding site (see Figure S3).
Purified PTB interacts with U1 snRNA stem loop 4
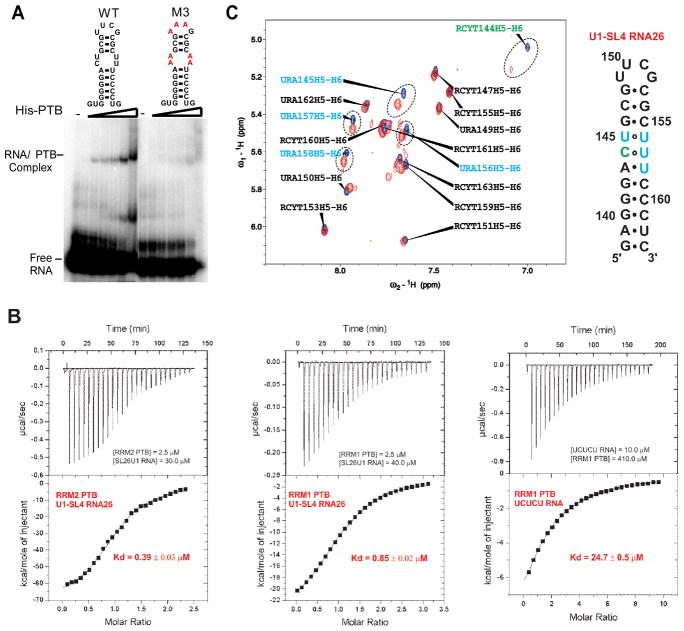
The U1 snRNP consists of the U1 snRNA, seven Sm core proteins, and three U1 specific proteins, U1 70k, U1C, and U1A. In the structures of the U1 snRNP determined by both cryoelectron microscopy and by X-ray crystallography, stem loop 4 (SL4) of the U1 snRNA is not bound by any U1 specific protein (Pomeranz Krummel et al., 2009; Stark et al., 2001). SL4 is a very stable GC-rich helix with a pyrimidine rich internal loop capped by a stable UUCG tetraloop. Although PTB recognizes single-stranded RNA sequences, several reports indicate that PTB can also bind bulged pyrimidine-nucleotides embedded in RNA duplexes or stem-loops, as seen in viral or cellular internal ribosome entry sites (IRES) (Bushell et al., 2006; Kafasla et al., 2009; Mitchell et al., 2005). We thus hypothesized that SL4 could be the binding site of PTB in U1 snRNA. To test this, we performed electrophoretic mobility shift assays (EMSA) of in vitro transcribed 32P-labeled SL4 RNA incubated with increasing concentrations of purified 6x-His tagged PTB (Figure 4A). The wildtype SL4 RNA assembled into a PTB complex that increased with increasing protein concentration. Mutation of the pyrimidines in the internal loop and the UUCG tetraloop regions (M3) led to a significant loss of PTB binding, indicating that PTB can bind specifically to SL4 of U1, and that binding requires interactions with pyrimidines. The binding of PTB to SL4 RNA did not go to completion, and thus did not appear to be of high affinity. Even at low affinity, this interaction is likely to occur more efficiently when the U1 snRNP and PTB are bound to adjacent sites in the pre-mRNA.
Figure 4. Recombinant PTB binds to U1-SL4 RNA.
A) Electrophoretic mobility shift assay to analyze binding of PTB to U1-SL4 RNA. Wildtype and mutant U1-SL4 RNAs at 50 nM were incubated with increasing concentrations of His-PTB (0, 0.1, 0.5, 0.75, 1.0, 2.5, and 5.0 μM). The complexes were separated on 8% Native-PAGE. B) Dissociation constants (Kd) of the individual domains PTB RRM1 and PTB RRM2 in complex with U1-SL4 RNA (26nts) and CU 5 mer RNA as determined by isothermal titration calorimetry at 30°C. The substrate at the lowest concentration is in the calorimeter cell. The upper panel displays the raw electrical power trace of the binding titration (baseline set at 0 μcal/s). The lower panel plots the integrated and normalized heat signal for each injection in the binding titration against the stoichiometry. C) On the left panel, overlay of 1H-1H TOCSY spectra of U1-SL4 RNA26 (26nts) free (blue peaks) and bound (red peaks) to PTB RRM2 at 40°C at a ratio 1 to 1 recorded on the Bruker Avance 900 MHz spectrometer and on the right panel, secondary structure of U1-SL4 RNA26 that highlights in green the cytidine and in blue the uridines that are the most shifted upon binding of PTB RRM2.
To examine the interaction of PTB with stem loop 4 more quantitatively we used isothermal titration calorimetry (ITC) and NMR spectroscopy with isolated RRM1 and RRM2 domains of PTB. These two domains were reported to prefer structured RNA over single-stranded RNA (Clerte and Hall, 2009). Both RRM1 and RRM2 show clear ITC binding curves to U1 stem loop 4 (Figure 4B). The Kd’s of SL4 for RRM 1 and 2 were 0.85 and 0.39 μM, respectively. Remarkably, the affinity of these domains for SL4 is much higher than the affinity of RRM1 for a short UCUCU single-stranded RNA (Kd of 24.7 μM). Thus, the two N-terminal RRMs of PTB bind to U1 SL4 with higher affinity than to a typical pyrimidine element found within splicing repressor elements of a pre-mRNA.
To map the interactions of RRM2 onto the SL4 RNA structure, we examined how the pyrimidines of SL4 RNA are affected upon binding of RRM2 using 1H-1H TOCSY NMR spectrometry (Figure 4C). The H5-H6 cross-peaks corresponding to C144, U145, U156, U157 and U158 within the pyrimidine internal loop of SL4 all show clear shifts upon the binding of RRM2 at a 1 to 1 stochiometric ratio. In contrast, the cross-peaks corresponding to pyrimidines in the tetraloop or in the base-paired portions of the stem were unaffected by RRM2 binding. Thus, PTB RRM2 binds specifically to the pyrimidine-rich internal loop of SL4 (with the integrity of the stem being maintained) rather than to the terminal loop or to the nine pyrimidine-tract (155–163) at the 3′ end of the SL4. This mode of RNA binding by the protein is likely to be different than RRM2 binding to single-stranded RNA (Oberstrass et al., 2005).
Excess U1-SL4 prevents the PTB induced change in the pre-mRNA/U1 interaction
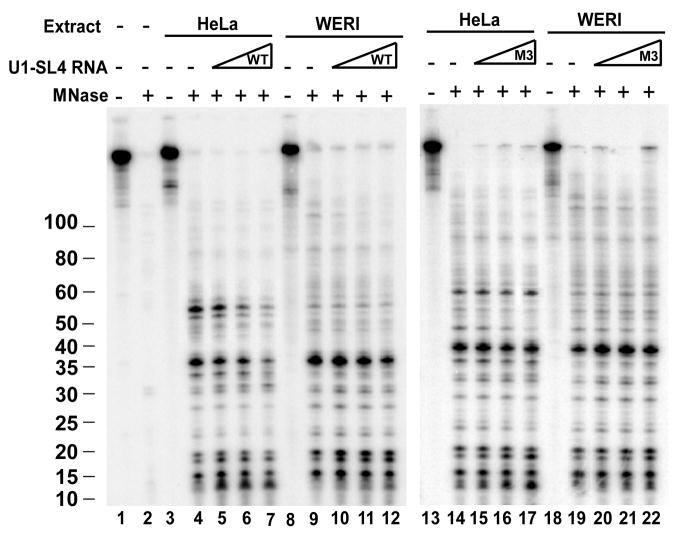
We next wanted to examine whether the alteration of U1 binding seen during splicing repression resulted from the PTB interaction with U1 SL4. If so, then an excess of free SL4 RNA might interfere with the interaction of PTB with U1 in the repressed HeLa complex. To test this, we titrated SL4 RNA into U1 assembly reactions (Figure 5). HeLa and WERI extracts were pre-incubated with the SL4 RNA prior to addition of N1 exon RNA labeled at the 5′ splice site. These reactions were treated with MNase, and the protected fragments analyzed as described above. Increasing concentrations of the SL4 RNA progressively reduced the formation of the 55 nucleotide protected RNA fragment that is specific to the repressed complex from Hela extract (lanes 5–7). At higher concentrations of SL4 RNA, the 38 and 35 nucleotide fragments were also reduced. In WERI extract, the pattern of 5′ splice site protection was largely unchanged by SL4 RNA (lanes 9–12). Similarly, the mutant SL4 RNA (M3) that does not bind PTB, did not significantly alter the pattern of protected 5′ splice site fragments in HeLa (lanes 14–17) or WERI (18–22) extract. Thus, in the presence of excess SL4 RNA, PTB does not alter the interaction of U1 snRNP with the 5′ splice site.
Figure 5. Competition with free U1-SL4 RNA shifts the interaction between the U1 snRNP and the pre-mRNA to that seen in the absence of PTB.
The c-src N1 exon containing transcripts were site specifically labeled at the N1 exon 5′ splice site. Prior to addition of the labeled transcript to HeLa and WERI extracts, the extracts were preincubated with increasing concentrations of free wildtype (lanes 1–12) and mutant (lanes 13–22) U1-SL4 RNAs at 0, 2.5, 5.0, and 10 μM. After pre-incubation the site specifically labeled transcript was added and incubation continued. The reactions were then treated with micrococcal nuclease. The protected fragments were extracted and visualized as described in figure 1.
DISCUSSION
We find that PTB bound to a pre-mRNA directly contacts the U1 snRNA within an exon complex that is repressed for splicing. PTB specifically interacts with the pyrimidine-rich internal loop of stem-loop 4 (SL4) of the U1 snRNA without affecting its secondary structure. We also find that when splicing is repressed by PTB, a pattern of nuclease protection is observed at the 5′ splice site that is different from that seen when PTB is absent and splicing is active. Competition experiments with excess SL4 RNA indicate that the PTB interaction with U1 is apparently required for the extended protection of the 5′ splice site region seen in a PTB repressed exon complex. Our previous investigations showed that binding of PTB to the pre-mRNA does not prevent 5′ splice site binding by the U1 snRNP, but alters the ability of this U1 to progress further in spliceosome assembly (Sharma et al., 2008). The contact of PTB with SL4 could force the U1 into a conformation that is incompatible with further assembly. This is similar to what has been proposed for the action of splicing silencer elements identified by in vitro selection (Yu et al., 2008). Alternatively, a specific contact between SL4 and another spliceosomal component may be required for later assembly steps and be blocked by PTB binding. For example, SL4 may mediate interactions between the U1 and U2 snRNPs during splice site pairing. A role for U1 SL4 during spliceosome assembly has not been described. Crystal and cryo-EM structures of purified or in vitro assembled U1 snRNP do not show interaction of the SL4 with any snRNP protein (Pomeranz Krummel et al., 2009; Stark et al., 2001). However, a previous study found that interactions of the phosphate backbone of the SL4 stabilize the association of the Sm ring with the U1 snRNA (McConnell et al., 2003). Blocking these interactions could alter the conformation of RNA-bound U1 or alter its interactions with other spliceosomal factors. A model for PTB mediated splicing repression is shown in Figure 6.
Figure 6. Model for N1 exon repression by PTB.
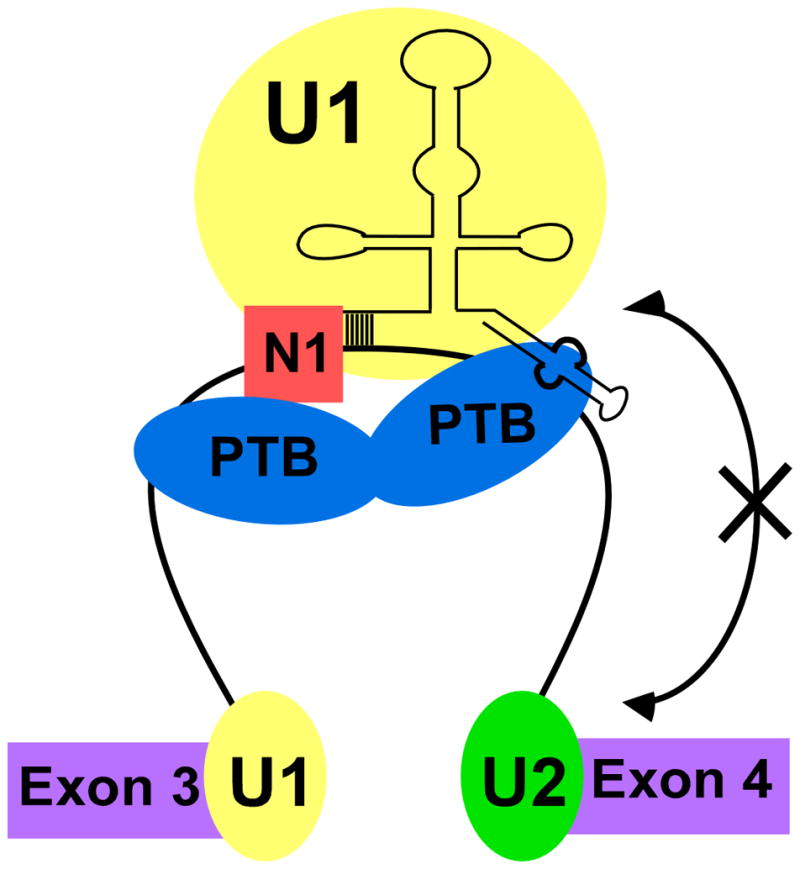
Binding of PTB to the CU rich elements flanking the N1 exon allow its interaction with SL4 of U1 snRNA. This prevents U1 from interacting with the complex at the downstream 3′ splice site.
Interestingly, the affinity of PTB RRMs 1 and 2 for U1 SL4 (Kd of 0.85 μM for RRM1) is higher than that measured for a short pyrimidine oligonucleotide (UCUCU) found in typical splicing repressor elements (Kd of 24.7 μM for RRM1) (Auweter et al., 2007; Oberstrass et al., 2005). SL4 must contain specific structural features that are recognized by PTB RRM1 and RRM2. This indicates that PTB can engage in modes of binding that differ from those reported for single-stranded RNA (Oberstrass et al., 2005). On the other hand, the affinity of the individual PTB RRMs for the U1 SL4 is lower than the affinity of full length PTB for longer splicing silencer elements. It is likely that free PTB in nuclear extract does not bind to the U1 snRNP, but binding of the two molecules to the same pre-mRNA allows the contact with SL4 to occur. Such contact may occur on other transcripts where simultaneous binding of PTB and U1 snRNP to the same regulatory region has been reported. In fact, exonic binding of PTB has been shown to stimulate U1 binding to the Fas exon 6 5′ splice site (Izquierdo et al., 2005). During repression of exon 6 of β-Tropomyosin, U1 snRNP also binds in the vicinity of the PTB binding site (Sauliere et al., 2006). Similarly, in CT/CGRP pre-mRNA, both PTB and U1 snRNP bind an intronic enhancer downstream of exon 4 to activate polyadenylation (Lou et al., 1999). The regulation of pre-mRNA processing in these systems may also involve direct contacts between PTB and the U1 snRNP.
Studies indicate that splicing repression by PTB requires multiple binding sites and that multiple PTB molecules are present in RNA/PTB complexes (Amir-Ahmady et al., 2005; Cherny et al., 2010; Chou et al., 2000; Clerte and Hall, 2009; Kafasla et al., 2009). After assembly with the pre-mRNA there are likely free RRM domains available to interact with U1-SL4. Analysis of PTB binding to the encephalomyocarditis virus (ECMV) IRES showed that the individual RRMs bind to separate stem-loop domains to stabilize the IRES structure (Kafasla et al., 2009). We find that the PTB binding site downstream of U1 is required for PTB/U1 crosslinking and that both RRM1 and RRM2 can bind U1-SL4 with similar affinity. These two RRMs are connected by flexible linkers with the rest of the protein and could adopt a variety of conformations to contact a nearby snRNA. It needs to be determined which of these two RRMs contacts U1 in the repressed N1 exon/PTB complex and whether this RRM must be placed downstream of the 5′ splice site. RRM3 and RRM4 presumably bind the pre-mRNA rather than U1-SL4 through their preferential affinity for single stranded RNA (Clerte and Hall, 2009). If both of these domains engage the pre-mRNA, they will generate an RNA loop through their interdomain interaction. One of these domains will likely anchor the protein in position downstream from U1, while the other may interact with an adjacent element or with a portion of the repressor element upstream of the exon (Lamichhane et al., 2010; Oberstrass et al., 2005).
It will also be interesting to investigate whether this interaction between PTB and U1-SL4 also plays a role in enhancement of exon inclusion by PTB. Tethering experiments with chimeric PTB-MS2 proteins indicate that recruiting RRM2 along with its adjacent linker to the pre-mRNA is sufficient for both repression and enhancement of regulated exons (Llorian et al., 2010; Rideau et al., 2006; Robinson and Smith, 2006). Similar studies may allow identification of the PTB domains important for interaction with U1-SL4.
Other splicing repressors are also found not to interfere with the recognition of splice sites but to prevent later steps in spliceosome assembly. Studies of CD45 exons 4 and 5 showed that hnRNP L binding to an exonic splicing silencer does not prevent recognition of the 5′ and 3′ splice sites by the U1 and U2 snRNPs (House and Lynch, 2006). However, the resulting exon definition complex is inactive and does not bind the U4/U6-U5 tri-snRNP. Repression of Fas exon 6 by RBM5 and PTB also results in the formation of a non-functional exon definition complex (Bonnal et al., 2008; Izquierdo et al., 2005). Direct interactions of hnRNP L and RBM5 with a U1 or U2 snRNP component have not been identified. However, as seen here, these exon complexes must either have a conformation that is not optimal for further spliceosome assembly, or be blocked for a specific interaction needed in the next assembly step.
Many proteins have been identified that bind to pre-mRNA and control alternative splicing patterns. However, few direct interactions are known between splicing regulators and the spliceosome and these regulatory interactions are with protein components of the spliceosome. In the classic example, SR proteins promote 5′ and 3′ splice site use through interactions with the SR domains of the U1 70k and U2AF proteins (Graveley, 2000; Kohtz et al., 1994; Wu and Maniatis, 1993). Similarly, the TIA1 protein can help recruit the U1 snRNP to a 5′ splice site via interactions with the U1C protein (Forch et al., 2002). Potential regulatory interactions with snRNA’s are also known. The hnRNP C protein was found to bind a uridine containing tetraloop in the 5′ stem loop of U2 snRNA (Temsamani and Pederson, 1996). Another study showed that PSF and p54nrb associate with the U4/U6-U5 tri-snRNP via an interaction with the U5 snRNA stem Ib (Peng et al., 2002). In these cases, a role for the protein/snRNA interaction in splicing regulation has not yet been demonstrated. However, from the results presented here, it is clear that a relatively small contact on the snRNA can have a significant effect on a choice of splicing pattern. Most known splicing regulators contain multiple RNA binding domains that recognize short RNA elements potentially found in one of the spliceosomal snRNAs. The structural flexibility of RNA would allow it to adopt conformations providing protein contacts that might not occur in normal assembly and function. Such protein interactions stabilizing RNA conformations that are non-productive for further spliceosome assembly can have dramatic effects on the competition between two splice site choices. Thus, it seems likely that snRNAs will prove to be targets of other splicing regulators.
Experimental Procedures
Site specific labeling
Site specifically 32P-labeled c-src RNAs were made as described previously (Reed and Chiara, 1999). Transcription templates for the 5′ and 3′ RNA fragments were generated by PCR. The RNA fragments were synthesized in standard T7 RNA polymerase transcription reactions. The 5′ fragment reaction contained the 5′ CAP nucleotide analog, G(5′)ppp(5′)G, where as the reaction for the 3′ fragment did not. Fifteen pmols of the 3′ RNA was first dephosphorylated using shrimp alkaline phosphatase (USB). The dephosphorylated RNA was extracted with phenol:chloroform:isoamyl alcohol (PCA) and ethanol precipitated. 32P-labeling was carried out in a 15 μl reaction containing the 1 μl of [γ-32P]ATP (6000 Ci/mmol), 1.5 μl of T4 polynucleotide kinase (10,000 units/ml), and 1x T4 PNK buffer (NEB). After labeling, the RNA was extracted with PCA and 15 pmols of the 5′ RNA and the 40-mer bridging oligonucleotide were added. This mix was then ethanol precipitated. The precipitate was then resuspended in 7 μl of water and denatured. Ligation was carried out at 25 °C for 3–12 hrs in a 15 μl reaction containing 1.5 μl of T4 DNA ligase (400,000 units/ml), 0.5 μl of RNAguard, and 1x T4 DNA ligase buffer (NEB). Ligated RNA was separated and extracted using urea-PAGE.
MNase protection assay
The micrococcal nuclease protection assay was carried out as described by Maroney et al (Maroney et al., 2000). The site specifically 32P-labeled RNA (10,000 cpm) was incubated at 30 °C for 30 minutes in a 25 μl splicing reaction that contained 15 μl of HeLa or WERI nuclear extract, 0.4 mM ATP, 20 mM creatine phosphate, 2.2 mM MgCL2, and 10 units of RNAguard. MNase (NEB) and CaCl2 were then added to a final concentration of 5 units/μl and 5 mM, respectively. MNase digestion was carried out at 25°C for 20 minutes. Digestion was stopped by adding EDTA to a final concentration of 20 mM. The reactions were then treated with SDS and proteinase K. Total RNA was extracted using PCA and ethanol precipitated. The protected RNA fragments were separated on a 10% urea-PAGE gel and visualized using Phophorimager. When treated with the U11–15 oligonucleotide, the reactions were preincubated with 2 pmols of the oligo and 10 units of RNase H (Ambion) for 30 minutes at 30°C prior to addition of the site specifically labeled RNA.
Electrophoretic Mobility Shift Assay
Recombinant 6x-His tagged PTB was purified using Ni-NTA agarose (Invitrogen) according to manufacturer’s instructions. The purified protein was dialyzed and stored in Buffer DG (20 mM HEPES-KOH pH 7.9, 80 mM K. glutamate, 20% glycerol, 2.2 mM MgCl2, 1 mM DTT, and 0.1 mM PMSF). The wildtype and mutant U1SL4 RNAs were transcribed in vitro using T7 RNA polymerase. For EMSA, the binding reactions were 10 μl and contained 50 nM RNA, 2.2 mM MgCL2, and increasing concentrations of His-PTB (0, 0.1, 0.5, 0.75, 1.0, 2.5, and 5.0 μM) and sufficient Buffer DG to bring its volume to 60% of the reaction volume. After incubation at room temperature for 20 minutes, the complexes were separated on an 8% native-PAGE gel using 25 mM Tris-glycine buffer at 4°C.
Purification and analysis of N1 RNA complexes
RNA complexes on in vitro transcribed and uniformly 32P-labeled MS2 tagged N1 RNAs were assembled in 500 μl reactions that contained 10 nM pre-mRNA, 100 nM MS2-MBP fusion protein, 0.4 M ATP, 20 mM CP, 2.2 mM MgCl2, 10U RNAguard and 300 μL of nuclear extract (Sharma et al., 2005). The MS2 tagged RNAs were preincubated with MS2-MBP prior to the addition of nuclear extract. The reactions were incubated at 30 °C for 30 minutes and the assembled complexes were purified using amylose resin as described previously (Sharma et al., 2005). The purified complexes were then UV-crosslinked using the Stratalinker to total energy of 1600 mJoules. After crosslinking the complexes were subject to immunoprecipitation using anti-PTB antibody (PTB-NT) coupled to gamma-bind Sepharose. The immunoprecipitated complexes were digested with SDS and proteinase K. Total RNA was then extracted using PCA, ethanol precipitated, and 3′ end labeled using 5′ 32P-pCp and T4 RNA ligase (NEB). The labeled RNA was separated on 8% urea-PAGE gels and visualized by Phosphorimager. Total RNA from HeLa extracts was also labeled with 5′ 32P-pCp.
Sample preparation for ITC measurements and NMR spectroscopy
The RRM1 (residues 41–163) and RRM2 (residues 172–316) domains of PTB (Acc. No. X62006) were subcloned in pTYB11 vector (N-terminal fusion vector), which is part of IMPACT-CN system from New England Biolab, Inc. The principle of this system consists of fusing the protein of interest with an Intein tag that self-cleaves under specific buffer conditions when loaded on the Chitin column. The subcloned vectors were transformed into E. coli strain BL21 (DE3) codonPlus-RIL®. The expression was carried out in M9 minimal medium containing 15 N isotopically labeled NH4Cl and 100 μg/ml of ampicillin at 37 °C until induction. At OD600 ~ 0.6, expression was induced with 1 mM IPTG and cells were grown at 20°C overnight. Purification has been performed according to the IMPACT-CN manual of New England Biolabs, Inc. To eliminate any RNAse and protease activity, we added SUPERase-In RNase inhibitor (Ambion) and Protease Inhibitor Cocktail (Roche) after sample concentration. In order to eliminate the peptide generated from self-cleavage reaction, the sample was then purified by size-exclusion chromatography using a Superdex™ 75 10/300 GL column. The RNA was synthesized by in vitro transcription using T7 RNA polymerase and synthetic DNA templates. The oligonucleotide products were then purified by anion exchange chromatography in denaturing conditions (6M urea at pH 8.0 and 85°C). The purified RNA was precipitated by butanol extraction to eliminate urea and salts. The precipitate was resuspended in 3 ml of deionised water and precipitated subsequently by butanol extraction. This operation was repeated three times before lyophilization overnight. The RNA samples were then dissolved in aqueous solution containing 20 mM NaCl and 10 mM NaH2PO4 adjusted at pH 6.5.
ITC measurement and analysis
ITC experiments were performed using a VP-ITC (Microcal, Inc) and normalized heat signals were calculated using the bundled Origin software. Prior to each experiment, both the PTB RRM1 and RRM2 domains and the RNA constructs were dialyzed overnight against 5 L of 20mM NaCl and 10mM NaH2PO4 adjusted at pH 6.5. For the ITC experiments, the titrant at a concentration from 30 to 410 μM was injected with an injection size of 10 μL into a cell containing 2.5 10 μM of the substrate. For all experiments, an injection interval of 3 min was set to allow for complete equilibration. The RNA and protein concentrations were measured respectively by UV spectroscopy and NMR spectroscopy using 1D PULCON experiment (Wider and Dreier, 2006). All ITC data were analyzed and plotted in Origin software using a 1 to 1 binding model.
NMR spectroscopy
Titration of U1-SL4 RNA with PTB RRM2 was carried out on the 900 MHz Bruker Avance spectrometer at 40°C. We acquired a series of 2D 1H-1H TOCSY (50 ms mixing time) to monitor proton chemical shift changes upon protein binding. The pyrimidine resonances of the U1-SL4 RNA were assigned in the free state based on a 2D 1H-1H NOESY, a natural abundance 1H-13C HSQC and 2D 1H-1H TOCSY.
Supplementary Material
Acknowledgments
We thank Tim Nilsen and members of the Black lab for helpful discussion and comments. This work was supported by grants from the Swiss National Science Foundation (Nr. 3100ab-133134) and the SNF-NCCR Structural Biology to F.H.T.A., and by NIH grant RO1:GM49662 to DLB. DLB is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amir-Ahmady B, Boutz PL, Markovtsov V, Phillips ML, Black DL. Exon repression by polypyrimidine tract binding protein. Rna. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Allain FH. Structure-function relationships of the polypyrimidine tract binding protein. Cell Mol Life Sci. 2008;65:516–527. doi: 10.1007/s00018-007-7378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, Allain FH. Solving the structure of PTB in complex with pyrimidine tracts: an NMR study of protein-RNA complexes of weak affinities. J Mol Biol. 2007;367:174–186. doi: 10.1016/j.jmb.2006.12.053. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Bonnal S, Martinez C, Forch P, Bachi A, Wilm M, Valcarcel J. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell. 2008;32:81–95. doi: 10.1016/j.molcel.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M, Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Chan RC, Black DL. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny D, Gooding C, Eperon GE, Coelho MB, Bagshaw CR, Smith CW, Eperon IC. Stoichiometry of a regulatory splicing complex revealed by single-molecule analyses. Embo J. 2010;29:2161–2172. doi: 10.1038/emboj.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MY, Underwood JG, Nikolic J, Luu MH, Black DL. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- Clerte C, Hall KB. The domains of polypyrimidine tract binding protein have distinct RNA structural preferences. Biochemistry. 2009;48:2063–2074. doi: 10.1021/bi8016872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Hartmuth K, Kastner B, Will CL, Luhrmann R. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol Cell. 2007;25:399–411. doi: 10.1016/j.molcel.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. Embo J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. Rna. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House AE, Lynch KW. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat Struct Mol Biol. 2006;13:937–944. doi: 10.1038/nsmb1149. [DOI] [PubMed] [Google Scholar]
- Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Kafasla P, Morgner N, Poyry TA, Curry S, Robinson CV, Jackson RJ. Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol Cell. 2009;34:556–568. doi: 10.1016/j.molcel.2009.04.015. [DOI] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Kotlajich MV, Crabb TL, Hertel KJ. Spliceosome assembly pathways for different types of alternative splicing converge during commitment to splice site pairing in the A complex. Mol Cell Biol. 2009;29:1072–1082. doi: 10.1128/MCB.01071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane R, Daubner GM, Thomas-Crusells J, Auweter SD, Manatschal C, Austin KS, Valniuk O, Allain FH, Rueda D. RNA looping by PTB: Evidence using FRET and NMR spectroscopy for a role in splicing repression. Proc Natl Acad Sci U S A. 2010;107:4105–4110. doi: 10.1073/pnas.0907072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapham J, Yu YT, Shu MD, Steitz JA, Crothers DM. The position of site-directed cleavage of RNA using RNase H and 2′-O-methyl oligonucleotides is dependent on the enzyme source. Rna. 1997;3:950–951. [PMC free article] [PubMed] [Google Scholar]
- Lim SR, Hertel KJ. Commitment to splice site pairing coincides with A complex formation. Mol Cell. 2004;15:477–483. doi: 10.1016/j.molcel.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, Smith CW. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Helfman DM, Gagel RF, Berget SM. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol Cell Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Romfo CM, Nilsen TW. Nuclease protection of RNAs containing site-specific labels: a rapid method for mapping RNA-protein interactions. Rna. 2000;6:1905–1909. doi: 10.1017/s1355838200001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- McConnell TS, Lokken RP, Steitz JA. Assembly of the U1 snRNP involves interactions with the backbone of the terminal stem of U1 snRNA. Rna. 2003;9:193–201. doi: 10.1261/rna.2136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S, Reed R. A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev. 1993;7:1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- Mitchell SA, Spriggs KA, Bushell M, Evans JR, Stoneley M, Le Quesne JP, Spriggs RV, Willis AE. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Peng R, Dye BT, Perez I, Barnard DC, Thompson AB, Patton JG. PSF and p54nrb bind a conserved stem in U5 snRNA. Rna. 2002;8:1334–1347. doi: 10.1017/s1355838202022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Chiara MD. Identification of RNA-protein contacts within functional ribonucleoprotein complexes by RNA site-specific labeling and UV crosslinking. Methods. 1999;18:3–12. doi: 10.1006/meth.1999.0751. [DOI] [PubMed] [Google Scholar]
- Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Huttelmaier S, Singer RH, Matthews S, Curry S, Smith CW. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat Struct Mol Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- Robinson F, Smith CW. A splicing repressor domain in polypyrimidine tract-binding protein. J Biol Chem. 2006;281:800–806. doi: 10.1074/jbc.M510578200. [DOI] [PubMed] [Google Scholar]
- Sauliere J, Sureau A, Expert-Bezancon A, Marie J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the beta-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol Cell Biol. 2006;26:8755–8769. doi: 10.1128/MCB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol. 2008;15:183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW. Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- Spellman R, Smith CW. Novel modes of splicing repression by PTB. Trends Biochem Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- Temsamani J, Pederson T. The C-group heterogeneous nuclear ribonucleoprotein proteins bind to the 5′ stem-loop of the U2 small nuclear ribonucleoprotein particle. J Biol Chem. 1996;271:24922–24926. doi: 10.1074/jbc.271.40.24922. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wider G, Dreier L. Measuring protein concentrations by NMR spectroscopy. J Am Chem Soc. 2006;128:2571–2576. doi: 10.1021/ja055336t. [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.