Abstract
The etiology of chronic periodontitis clearly includes a heritable component. Our purpose was to perform a small exploratory genome-wide association study in adults ages 18–49 years to nominate genes associated with periodontal disease−related phenotypes for future consideration. Full-mouth periodontal pocket depth probing was performed on participants (N = 673), with affected status defined as two or more sextants with probing depths of 5.5 mm or greater. Two variations of this phenotype that differed in how missing teeth were treated were used in analysis. More than 1.2 million genetic markers across the genome were genotyped or imputed and tested for genetic association. We identified ten suggestive loci (p-value ≤ 1E-5), including genes/loci that have been previously implicated in chronic periodontitis: LAMA2, HAS2, CDH2, ESR1, and the genomic region on chromosome 14q21-22 between SOS2 and NIN. Moreover, we nominated novel loci not previously implicated in chronic periodontitis or related pathways, including the regions 3p22 near OSBPL10 (a lipid receptor implicated in hyperlipidemia), 4p15 near HSP90AB2P (a heat shock pseudogene), 11p15 near GVINP1 (a GTPase pseudogene), 14q31 near SEL1L (an intracellular transporter), and 18q12 in FHOD3 (an actin cytoskeleton regulator). Replication of these results in additional samples is needed. This is one of the first research efforts to identify genetic polymorphisms associated with chronic periodontitis-related phenotypes by the genome-wide association study approach. Though small, efforts such this are needed in order to nominate novel genes and generate new hypotheses for exploration and testing in future studies.
Keywords: GWAS, chronic periodontitis
More than one third of the dentate adult population in the United States suffers from chronic periodontitis (Albandar et al. 1999), a disease in which the host response to bacteria trapped between the tooth and gum destroys both the ligament holding the tooth to the bone and the bone itself. If left untreated, periodontal disease can result in impaired mastication and possible tooth loss. A number of factors increase risk of disease, including age, smoking, and diabetes. In addition, there is evidence of a genetic component to the disease.
Two studies have examined the heritability of chronic periodontitis. In a study of 117 same-sex adult twins from the Virginia Twin Registry, the heritability of examiner-determined chronic periodontitis ranged from 48% for a measure of attachment loss to 59% for the percentage of teeth with one or more sites having attachment loss ≥2 mm (Michalowicz et al. 2000). In a population study of 10,578 Swedish twin pairs, the heritability of self-reported diagnosis with chronic periodontitis or presence of loose teeth was estimated to be 39% and 33% for women and men, respectively (Mucci et al. 2005). Taken together, the two studies provide evidence that part of the risk of disease is genetic.
On the basis of this evidence, a number of studies have attempted to identify genes that might be implicated in the disease process. Recent reviews have summarized the set of genes considered to alter risk (Zhang et al. 2011b, Laine et al. 2012). Most previous studies have adopted a candidate gene approach based on knowledge of disease pathogenesis and phenotypes. Pathogenic players for which at least some support has been obtained include pro- and anti-inflammatory mediators, the vitamin D receptor, pattern recognition receptor genes, matrix metalloproteinases, and others. The candidate gene approach is a prudent strategy for investigating hypotheses about potential disease genes for which there exists a priori knowledge, but unbiased, genome-wide methods are needed to nominate novel loci.
The genome-wide association study (GWAS) approach previously has been used to study periodontitis-related phenotypes. In brief, this unbiased approach scans a multitude (typically millions) of genetic variants across the entire genome one-at-a-time for statistical evidence of association. Because of the complex correlational structure of the genome (i.e., linkage disequilibrium, LD), associated variants usually are not considered causal; rather, they are assumed to be proxies for unobserved causal variants. Therefore, GWAS results typically are viewed through a lens of biological plausibility, with implicated loci evaluated in terms their LD structure, the known biology of genes within an associated LD block, and of the location of these genes relative to specific associated variants. Novel candidate genes may be nominated on the basis of a combination of their physical proximity to associated variants and their compelling biological stories, although these candidate genes are not proven as disease genes via GWAS evidence alone. In this way, GWAS is extremely useful as a hypothesis-generating approach for diseases such as periodontitis, in which the genetic basis of disease is not fully understood.
Previously, the GWAS approach has been successfully used to study periodontitis-related phenotypes. A recent GWAS of the aggressive form of periodontitis has implicated GLT6D1 (Schaefer et al. 2010). In a GWAS of oral bacterial profile, 13 loci demonstrated suggestive evidence of association (Divaris et al. 2012). In a GWAS of chronic periodontitis in 4032 German subjects (ages 20−81 years), 10 loci showed suggestive evidence of association for four related phenotypes (Teumer et al. 2013). Finally, in a study of 4504 European Americans ages 53−74 years, GWAS identified six suggestive loci, three for moderate chronic periodontitis, and three for severe chronic periodontitis (Divaris et al. 2013). However, across these studies, there has been no overlap in genes identified. Therefore, more work is needed to identify and replicate the genetic variants associated with periodontal disease-related phenotypes.
One common strategy for elucidating the role of genetics in complex outcomes is to study disease or related phenotypes in younger individuals. This approach postulates that genetic factors may play a larger role in individuals who manifest disease at a younger age and that genetic liability may be minimally obscured by cumulative effects of environmental risk factors in younger individuals. We have followed this strategy in the current work, in which we report the results of a genome-wide association scan for genetic variants affecting chronic periodontitis-related phenotypes in a comparatively young population-based cohort.
Materials and Methods
Participant recruitment
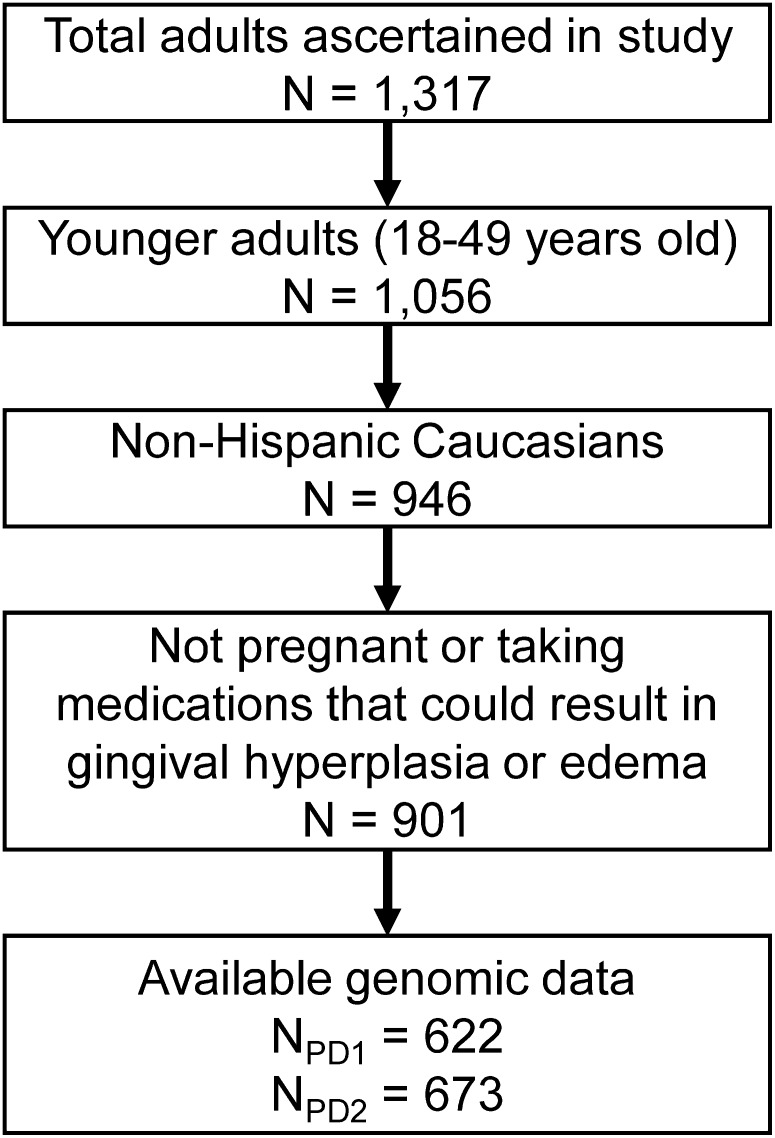
As previously described (Polk et al. 2008), the study population was ascertained through the Center for Oral Health Research in Appalachia, which recruited families from western Pennsylvania and northern West Virginia. To participate in the study, a family had to have at least one adult and at least one biologically related child between the ages of 1 and 18 who lived together. This resulted in the recruitment of 650 families. Everyone living in eligible households regardless of biological or legal relationship was invited to participate; recruitment was not based on periodontal disease or other oral health status. In the sample for this study, only adult participants (i.e., in most cases the parents of a recruited family) were included, resulting in 1317 adults, of whom 1056 were between the ages of 18 and 49 (Figure 1). Written informed consent was obtained from all adult participants. All study procedures and consent forms were approved by the Institutional Review Boards of the University of Pittsburgh and West Virginia University.
Figure 1.
Flowchart showing numbers of participants at each stage of the study.
Phenotype assessment and coding
Participants received a comprehensive orodental examination by a licensed dentist or dental hygienist in a well-equipped, modern dental operatory. To assess periodontal status, every tooth was evaluated except the third molars. The research dental hygienist walked the probe around the gingival crevice. At least six areas around each tooth were examined: mesiofacial, midfacial, distofacial, and the corresponding lingual/palatal areas. To develop periodontal disease-related phenotypes, the mouth was divided into sextants, and the probing depth for the deepest pocket in each sextant was recorded as shallow (i.e., <3.5 mm), moderate (i.e., 3.5−5.5 mm), or deep (i.e., ≥5.5 mm). If all teeth in a given sextant were missing (i.e., the sextant was completely edentulous), no observation was recorded for that sextant. Two periodontal disease-related phenotypes were created that differed in how we treated missing data: (1) PD1, where we assumed the missing teeth in edentulous sextants had not been affected by chronic periodontitis; (2) PD2, where we assumed the missing teeth in edentulous sextants had been affected by chronic periodontitis. In data analysis, we coded as affected participants with at least two sextants with a pocket probing depth of at least 5.5 mm or self-reported “gum surgery” (n = 14) and unaffected otherwise.
Genotyping, imputation, and quality control
Genotyping was performed at the Center of Inherited Disease Research of Johns Hopkins University as part of the GENEVA consortium using the Illumina Human610-Quadv1_B BeadChip (Illumina, San Diego, CA). Genotype imputation (inferring unobserved genotype data based on observed data) was performed using subjects from a HapMap Phase III reference panel (genetically-determined European ancestry) and resulted in approximately 1.4 million successfully imputed single-nucleotide polymorphisms (SNPs). The GENEVA consortium coordinating center at the University of Washington performed comprehensive data cleaning and quality assurance procedures (Laurie et al. 2010).
Data analysis
To minimize the risk of an inflated type I error due to population stratification and to avoid a reduction in power due to genetic heterogeneity, we included in the GWAS only self-reported non-Hispanic Caucasians (n = 946; Figure 1). The self-reported race (ethnicity) variables showed excellent agreement with genetically determined ancestry (estimated by principal components analysis of genome-wide SNP data). To avoid other confounding factors, we excluded from our analyses those who were pregnant (n = 13) or who reported taking medications that could result in gingival hyperplasia or edema, including birth control pills (n = 24), estrogen-replacement therapy (n = 3), calcium channel blockers (n = 1), or phenytoin (n = 4). Because of the rural community-based recruitment strategy, our sample contained a minority of known and cryptic biological relatives, which could theoretically lead to genomic inflation (i.e., p-values biased away from the null hypothesis) in standard association tests. Because most samples in this study (>91%) were unrelated, we did not explicitly model the relatedness among individuals; instead, we guarded against any potential bias due to population structure (i.e., relatedness among participants) by closely monitoring genomic inflation factor. No bias was detected.
To test for genetic association between disease status and SNPs markers, we performed GWAS in PLINK (http://pngu.mgh.harvard.edu/~purcell/plink; Purcell et al. 2007) under the logistic regression model (–logistic option) while adjusting for age. The analyses were performed with both genotyped and imputed SNP data. Before analysis, HWE (p-value ≤ 1E-4) and minor allele frequency (MAF ≤ 0.02) filters were applied to exclude outlier or rare SNPs. We used variance components methods that condition on the known biological relationships (Almasy and Blangero 1998) in the sample to verify statistical significance of top hits. We explored all signals with “suggestive significance” (P-value ≤ 1E-5) using several statistical and bioinformatics tools including WGAViewer (http://compute1.lsrc.duke.edu/softwares/WGAViewer/; Ge et al. 2008) to search for genes near SNPs with suggestive significance, R statistical package (R Foundation for Statistical Computing, Vienna, Austria) to calculate genomic inflation factors (lambda) and generate Manhattan and quantile-quantile (Q-Q) plots, and LocusZoom (http://csg.sph.umich.edu/locuszoom/) to visualize regions around SNPs with suggestive significance. Likewise, we scrutinized SNPs in 31 previously implicated candidate genes—IL1B, IL1RN, IL6, IL10, VDR, CD14, TLR4, MMP1 (Laine et al. 2012), GLT6D1 (Divaris et al. 2012), NPY, WNT5A, NCR2, EMR1 (Divaris et al. 2013), EPHA3, RAB6C, C9orf150, IQSEC1, ERC2, CAMK4, MFSD1, LBP, ETS2, FAM180A (Teumer et al. 2013), KCNK1, FBXO38, UHRF2, IL33, RUNX2, TRPS1, CAMTA1, VAMP3 (Divaris et al. 2012)—for any evidence of association. To exclude the possibility of spurious signals caused by poor genotype calling, we also generated and visually inspected allele intensity plots for top associated SNPs.
Results
After filtering out the non-Caucasian individuals and those whose age was beyond our focus, we found that the sample size was 901 (62.2% female), with a mean age of 32.9 years (SD = 7.7, min = 18 years, max = 48.87 years). After taking into account genotyping availability, the sample size for PD1 was 622, with 93 participants being classified as affected (15.0%), whereas the sample size for PD2 was 673, with 176 participants being classified as affected (26.2%; Figure 1). The two approaches for classifying disease resulted in different classifications for 83 participants (12.3% of the total sample), 78 of whom were completely edentulous. Both PD1 (odds ratio [OR] 1.36, 95% confidence interval [95% CI] 1.08–1.72) and PD2 (OR 1.64, 95% CI 1.35–1.99) were significantly associated with greater age.
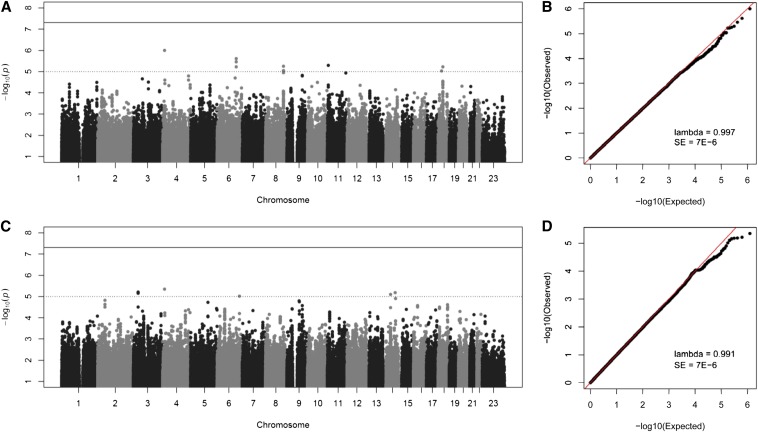
As shown in the Manhattan plots from the GWAS (Figure 2), we did not observe any associations meeting the genome-wide level of significance (i.e., P-value < 5E-8), which is not surprising because this threshold is incredibly conservative and our sample size is modest (in the context of GWAS). The genomic inflation factor, lambda, was 0.997 and 0.991 for PD1 and PD2, respectively, indicating that there was no inflation of P-values as the result of population structure or relatedness. Although no SNP passed the threshold for genome-wide significance, 10 suggestive loci, represented by 17 SNPs (10 genotyped and 7 imputed) with P-values between 1E-5 and 1E-7, were observed (Table 1 and Figure 3).
Figure 2.
Manhattan and Q-Q plots for periodontal disease related phenotypes. (A) Manhattan plot for PD1. (B) Q-Q plot for PD1. (C) Manhattan plot for PD2. (D) Q-Q plot for PD2. All P-values are negative log10 transformed. Both genotyped and imputed SNPs are shown.
Table 1. Suggestive associations observed for periodontal disease-related phenotypes.
| Phenotype | Chr. | SNP | BP | MAF | OR | P-Value | Gene(s) within 400 kb | Corroborating Evidence |
|---|---|---|---|---|---|---|---|---|
| PD1 | 4p15 | rs733048 | 12851895 | 0.22 | 2.40 | 1.0E-6 | HSP90AB2P, RAB28, BOD1L, and NKX3-2 | None |
| 6q22-23 | rs10457525 | 129914659 | 0.80 | 2.33 | 3.5E-6 | LAMA2 and ARHGAP18 | LAMA2 is expressed in periodontal ligament and gingival fibroblasts (Han and Amar 2002) and is differentially expressed between chronically inflamed and healthy periodontal ligaments (Gersdorff et al. 2008) | |
| rs7749983 | 129916048 | 0.19 | 2.39 | 2.4E-6 | ||||
| rs10457526 | 129938194 | 0.78 | 2.26 | 6.0E-6 | ||||
| 8q24 | rs7816221 | 122765324 | 0.32 | 2.12 | 9.2E-6 | HAS2 and HAS2AS | HAS2 and HAS2AS regulate hyaluronan, which is produced during tissue repair and has been shown to promote adhesion and proliferation of periodontal ligament cells (Takeda et al. 2011), and inhibits periodontal pathogens (Rodrigues et al. 2010) | |
| rs3870371 | 122766313 | 0.32 | 2.15 | 5.6E-6 | ||||
| rs920455 | 122769378 | 0.68 | 2.11 | 9.2E-6 | ||||
| 11p15 | rs12799172 | 6706633 | 0.64 | 2.12 | 5.1E-6 | Many genes | none | |
| 18q11 | rs11659841 | 24040878 | 0.13 | 2.48 | 9.4E-6 | CDH2 | CDH2, a mediator of cell adhesion, is involved in periodontal ligament cell differentiation (Lin et al. 1999) | |
| 18q12 | rs8094794 | 32506747 | 0.79 | 2.17 | 5.9E-6 | FHOD3, TPGS2, and KIAA1328 | None | |
| PD2 | 3p22 | rs11713199 | 31950325 | 0.70 | 1.87 | 6.9E-6 | OSBPL10, ZNF860, GPD1L, CMTM8, and STT3B | None |
| rs12630254 | 31956231 | 0.30 | 1.90 | 6.7E-6 | ||||
| rs12630931 | 31956771 | 0.30 | 1.89 | 6.2E-6 | ||||
| 4p15 | rs733048 | 12851895 | 0.22 | 1.95 | 4.4E-6 | HSP90AB2P, RAB28, BOD1L, and NKX3-2 | None | |
| 6q25 | rs2297778 | 151686771 | 0.78 | 2.32 | 9.7E-6 | MTHFD1L, AKAP12, ZBTB2, RMND1, and ESR1 | ESR1 is involved in regulating alveolar bone formation and remodeling (Zhang et al. 2011a) and differentiation of periodontal ligament stem cells (Pan et al. 2011) | |
| 14q21 | rs3783412 | 49926391 | 0.53 | 1.85 | 7.9E-6 | SOS2, L2HGDH, ATP5S, CDKL1, MAP4K5, ATL1, SAV1, and NIN | This locus was the most significant association observed in an independent GWAS of severe chronic periodontitis in older adults (Divaris et al. 2013) | |
| 14q31 | rs12589327 | 81368926 | 0.39 | 2.13 | 6.6E-6 | SEL1L | None |
Chr, chromosome; SNP, single-nucleotide polymorphism; BP, base pair; MAF, minimum allele frequency; OR, odds ratio.
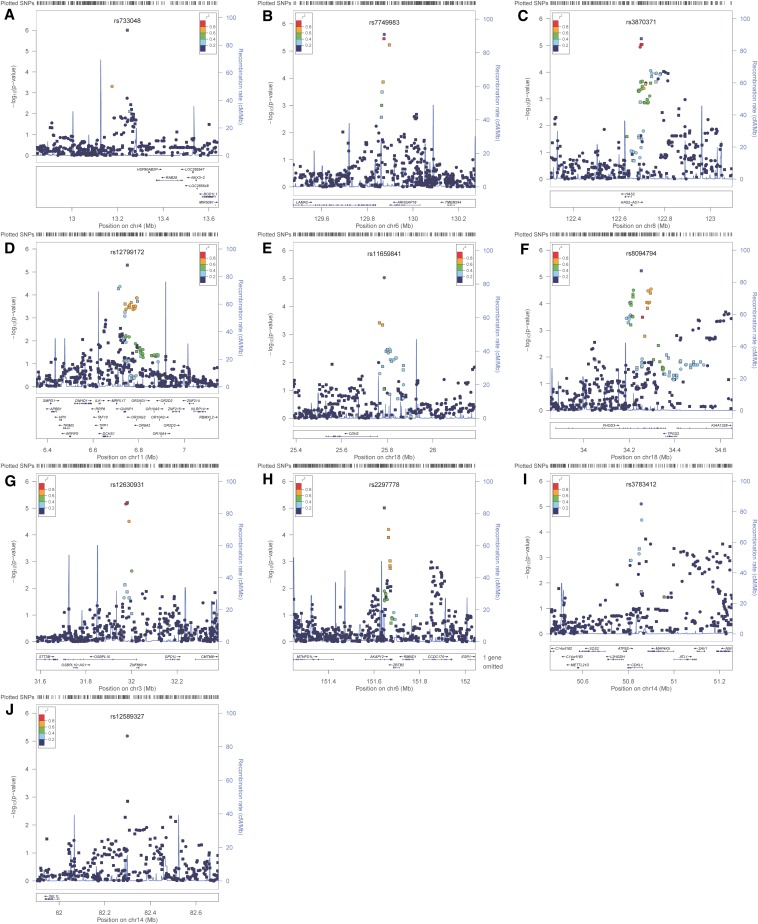
Figure 3.
Negative log10 transformed P-values and physical positions for SNPs in associated regions. Suggestive associations for PD1: (A) SNPs on chromosome 4 occurring near the HSP90AB2P, RAB28, BOD1L, and NKX3-2 genes. (B) SNPs on chromosome 6 occurring near the LAMA2 and ARHGAP18 genes. (C) SNPs on chromosome 8 occurring near the HAS2 and HAS2AS genes. (D) SNPs on chromosome 11p15. (E) SNPs on chromosome 18 occurring near the CDH2 gene. (F) SNPs on chromosome 18 near the FHOD3, TPGS2, and KIAA1328 genes. Suggestive associations for PD2: (G) SNPs on chromosome 3 near the OSBPL10, ZNF860, GPD1L, CMTM8, and STT3B genes. (H) SNPs on chromosome 6 near the MTHFD1L, AKAP12, ZBTB2, RMND1, and ESR1 genes. (I) SNPs on chromosome 14 near the SOS2, L2HGDH, ATP5S, CDKL1, MAP4K5, ATL1, SAV1, and NIN genes. (J) SNPs on chromosome 14 near the SEL1L gene. Colors indicate LD between the index SNP (purple) and other SNPs based on HapMap CEU data. The rug plot indicates regional SNP density. The recombination rate overlay (blue line, right x-axis) is based on HapMap CEU data. Gene positions and directions of transcription are annotated based on hg19/1000 Genomes Nov 2010 release.
Ten SNPs across six loci exhibited suggestive evidence of association with PD1. The top-ranking SNP was rs733048 in an intergenic region on chromosome 4p15 near HSP90AB2P, RAB28, BOD1L, and NKX3-2 genes. Other suggestive loci were observed on chromosome 6q22-23 near LAMA2 and ARHGAP18 genes, chromosome 8q24 near the HAS2 and HAS2AS genes, and chromosome 11p15 in a LD block containing many genes, including several olfactory receptor genes. Two separate loci were observed on chromosome 18q11 and 18q12, near the CDH2 gene, and near the FHOD3, TPGS2, and KIAA1328 genes, respectively.
Seven SNPs (four genotyped, three imputed) across five loci exhibited suggestive evidence of association with PD2. As with PD1, the top ranking SNP was rs733048 on chromosome 4p15. Other suggestive loci included chromosome 3p22 near the OSBPL10, ZNF860, GPD1L, CMTM8, and STT3B genes, and chromosome 6q25 near the MTHFD1L, AKAP12, ZBTB2, RMND1, and ESR1 genes. Two separate loci were observed on chromosome 14q21 near SOS2, L2HGDH, ATP5S, CDKL1, MAP4K5, ATL1, SAV1, and NIN genes, and on chromosome 14q31, near SEL1L.
Although not among our suggestive hits, we additionally inspected the strength of association for a priori candidate genes appearing in a recent review (IL1B, IL1RN, IL6, IL10, VDR, CD14, TLR4, MMP1; Laine et al. 2012) as well as hits from previous GWAS of aggressive periodontitis (GLT6D1; Schaefer et al. 2010), chronic periodontitis (NPY, WNT5A, NCR2, EMR1; Divaris et al. 2013), EPHA3, RAB6C, C9orf150, IQSEC1, ERC2, CAMK4, MFSD1, LBP, ETS2, FAM180A; Teumer et al. 2013), and periodontal bacterial colonization (KCNK1, FBXO38, UHRF2, IL33, RUNX2, TRPS1, CAMTA1, VAMP3; Divaris et al. 2012) in older individuals. Several SNPs among these genes demonstrated association at nominal levels of significance (i.e., P < 0.05), which is expected due to chance alone, given the issue of multiple comparisons. However, we observed stronger associations between PD2 and SNPs in CAMTA1 near the same exon in which association was previously reported (Divaris et al. 2012); two SNPs in CAMTA1 (rs1750817 and rs1193169) showed association with P -values = 2E-4, and several other SNPs in this gene showed association at nominal significance (i.e., P -values between 0.05 and 0.001). Likewise, 20 of 75 SNPs in RUNX2 showed nominal association (i.e., P-values between 0.05 and 0.001) with PD1, and 11 of 26 SNPs in ETS2 showed nominal (i.e., P-values between 0.05 and 0.001) with both PD1 and PD2. Although not meeting the burden of evidence necessary in a genome-wide context, given that CAMTA1, RUNX2, and ETS2 were already nominated in a previous GWAS, our results may be interpreted as evidence of replication.
Discussion
The current study is one of the first to investigate chronic periodontitis-related phenotypes by the GWAS approach. Neither our study, nor the three previously reported GWAS studies of chronic periodontitis-related phenotypes identified any genome-wide significant associations (Divaris et al. 2012, 2013; Teumer et al. 2013), which is not surprising, given that this threshold is extremely conservative, and power to detect such effects may be low given the sample sizes of these studies. Such hits could be used to nominate disease genes on the basis of statistical evidence alone. However, in the absence of such overwhelming statistical evidence, we have interpreted our suggestive associations (which also show very strong statistical evidence), in the context of the known biology of implicated genes. We identified 10 loci that were associated with our phenotypes, defined as two or more sextants with periodontal probing depths deeper than 5.5 mm. Scrutiny of many of these loci yielded corroborating evidence linking them periodontal health.
For instance, one of the suggestive loci implicated in PD1 was an LD-block containing LAMA2 and ARHGAP18. LAMA2 codes laminin alpha 2, a member of the multifunctional laminin family of proteins known to be expressed in periodontal ligament and gingival fibroblasts (Han and Amar 2002). A gene expression study comparing in vivo cells from chronically inflamed and healthy human periodontal ligaments showed nearly 3-fold down-regulation of LAMA2 in inflamed cells (Gersdorff et al. 2008). Other laminins (i.e., LAMA4, -B1, -B2, -B3, and -C3) also showed differential expression (between 2- and 10-fold). The in vivo differential expression observed for LAMA2 (and other laminins) supports our finding that genetic variation in this gene may affect periodontal health.
Other suggestive loci identified in the analysis of PD1 have previously documented relationships with the periodontium, including HAS2, HAS2AS, and CDH2. HAS2 codes hyaluronan synthase 2, an enzyme that facilitates the transfer of hyaluronan across the cell membrane. HAS2AS (HAS2 antisense mRNA) regulates HAS2 mRNA levels and inhibits hyaluronan biosynthesis (Chao and Spicer 2005). Hyaluronan is produced during wound healing and tissue repair to provide a framework for the ingrowth of blood vessels and fibroblasts (Slevin et al. 2002). Consistent with this, hyaluronan speeds up the healing of intra-oral wounds (Hammad et al. 2011), promotes the adhesion and proliferation of periodontal ligament cells (Takeda et al. 2011), and significantly reduces the growth of two periodontal pathogens (Rodrigues et al. 2010). It also facilitates reductions in probing depth and bleeding on probing over 12 weeks following scaling and root planning (Johannsen et al. 2009) and inhibits plaque growth (Rodrigues et al. 2010). Thus, there is evidence that hyaluronan facilitates healing in the oral cavity and gingiva, supporting the biological plausibility of these genes.
CDH2 codes cadherin-2 (i.e., N-cadherin), which is involved in mediating calcium-ion-dependent cell adhesion (Takeichi 1987). In vitro, N-cadherin facilitates cell-to-cell interactions during periodontal ligament cell differentiation (Lin et al. 1999). In vitro, a proteinase produced by a periodontal pathogen induces cleavage of N-cadherin, and this cleavage is associated with loss of cell adhesion (Sheets et al. 2005). Thus, evidence supports the biological plausibility of CDH2.
For the PD2 phenotype, the most exciting suggestive association was on chromosome 14q21, which overlaps with a region previously implicated in a GWAS of chronic periodontitis in an older cohort. In the GWAS by Divaris et al., the SNP rs12883458 in NIN was the top hit for the severe chronic periodontitis phenotype (Divaris et al. 2013). Although we observed association for a different SNP approximately 350 kb upstream of NIN, our hit was in the same LD block and our association signal was unusually broad, encompassing NIN and other genes in this region. It is unclear how NIN, which is important for anchoring microtubules for centrosomal function, may affect periodontitis. A more plausible gene in this region, which is in fact closer to our associated SNP, is SOS2. Although SOS2 has not previously been implicated in periodontal health, disruption of its closely related homolog, SOS1, is responsible for hereditary gingival fibromatosis (Hart et al. 2002), suggesting a plausible role of SOS genes (Ras-specific exchange factors) in periodontal health. Additional studies are needed to determine the specific gene and causal variant driving association at this replicated periodontal risk locus.
Another suggestive locus identified in the analysis of PD2 was a genomic region, including the estrogen receptor (i.e., ESR-α coded by ESR1), which has a previously documented relationship with the periodontium. By binding with receptors in periodontal tissue, including periodontal ligament stem cells, estrogen regulates the remodeling of alveolar bone, promotes bone formation, and inhibits bone resorption (e.g., Zhang et al. 2011a). Estrogen induces periodontal ligament stem cells to differentiate into osteoblast-like cells via ESR-α (Pan et al. 2011). This differentiation can be down-regulated by a lentivirus-mediated siRNA targeting ESR-α (Zhang et al. 2011a). Finally, estrogen reverses the stimulatory effects of lipopolysaccharide on proinflammatory cytokine expression by human periodontal ligament cells (Shu et al. 2008). Thus ESR1 may play a role in chronic periodontitis.
One SNP, rs733048, was implicated in GWAS of both PD1 and PD2. However, all other SNPs exhibiting suggestive association (i.e., P-value < 1E-5) in only one scan were also nominally significant (with P-values on the order of 1E-3 to 1E-5), and had comparable effect sizes (odds ratios), in the other scan.
Aside from the SOS2 to NIN region on chromosome 14, none of the other nine implicated loci overlapped with results from published GWAS of periodontitis (Schaefer et al. 2010; Divaris et al. 2012, 2013; Teumer et al. 2013). This may be due to the possible heterogeneity between different cohorts, especially differences in ages, and more importantly, the usage of different phenotypes among studies. In particular, aggressive periodontitis and periodontal pathogen colonization may have separate etiologies than the periodontal disease−related phenotypes of the current study. Even among the three GWAS studies of chronic periodontitis, which altogether considered eight different periodontitis-related phenotypes, there was no overlap in trait definitions. Nevertheless, modest evidence of replication was observed for previously nominated genes CAMTA1, RUNX2, and ETS2.
This study possesses several strengths, including high-quality genotyping and imputation data generated by Center of Inherited Disease Research and the GENEVA coordinating center and complementary phenotype definitions using different considerations for missing data. However, several limitations warrant further discussion. First, the pocket probing depth measure of affected status is a nonclinical measure of periodontal health. Because it uses pocket probing depth to estimate attachment loss rather than measuring attachment loss directly, it may underestimate disease in populations, especially older people with severe gingival recession (Landry and Jean 2002). Given that our population is younger, however, severe gingival recession is unlikely to be a problem. Thus, estimates based on the pocket probing depth measure may be a good surrogate for true disease status. In contrast, the clinical significance of genetic associations with our phenotype, given the young age of our sample, may not necessarily be generalizable to older, high-risk populations. Indeed, it is currently unknown whether the same genetic liabilities affect periodontal health throughout adulthood, although we speculate that at least some of the genetic factors that are important for chronic periodontitis in older individuals also affect periodontal health in younger individuals. Finally, our results lack genome-wide significance, that is, they do not meet the conservative threshold of significance needed to unequivocally prove association based on statistical evidence alone. This result is not surprising, given the limited power in our comparatively small sample size (for a GWAS) to detect the small effects of individual genetic variants. However, this limitation was lessened by interpreting the observed suggestive loci through a lens of biological plausibility, as statistical evidence alone was insufficient to fully implicate these loci. Targeted replication studies and functional analysis of nominated genes are highly desirable, as some of the loci identified via GWAS are likely to be false positive signals. Nevertheless we feel that the strong statistical evidence combined with plausible biology gives credence to our nomination of these genes as possible periodontitis genes. Unfortunately, there is currently a paucity of comparable or adequately large chronic periodontitis studies available for such replication studies.
In summary, using the GWAS approach, we identified 10 genetic loci associated with periodontal disease-related phenotypes at the suggestive significance level. Although approximately half of the implicated loci harbor genes with known functions potentially related to chronic periodontitis, the other loci/genes are novel. Hypothesis-generating studies, such as this, that seek to nominate novel genes as possible periodontitis-related loci, are much needed. These findings are important because they provide the foundation for the exploration of novel pathways through which chronic periodontitis may occur.
Acknowledgments
We thank the staff and participants of the Center for Oral Health Research in Appalachia and GENEVA study for their important contributions. This work was funded through the following grants awarded by the National Institutes of Health (NIH), National Institute of Dental Craniofacial Research (NIDCR), and National Human Genomics Research Institute (NHGRI): R01-DE014899, U01-DE018903, U01-HG004446, and K23-DE114736.
Footnotes
Communicating editor: V. Vieland
Literature Cited
- Albandar J. M., Brunelle J. A., Kingman A., 1999. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988–1994. J. Periodontol. 70: 13–29 [DOI] [PubMed] [Google Scholar]
- Almasy L., Blangero J., 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62: 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao H., Spicer A. P., 2005. Natural antisense mRNAs to hyaluronan synthase 2 inhibit hyaluronan biosynthesis and cell proliferation. J. Biol. Chem. 280: 27513–27522 [DOI] [PubMed] [Google Scholar]
- Divaris K., Monda K., North K. E., Olshan A. F., Lange E. M., et al. , 2012. Genome-wide association study of periodontal pathogen colonization. J. Dent. Res. 91: 21S–28S [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divaris K., Monda K. L., North K. E., Olshan A. F., Reynolds L. M., et al. , 2013. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Hum. Mol. Genet. 22: 2312–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D., Zhang K., Need A. C., Martin O., Fellay J., et al. , 2008. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 18: 640–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersdorff N., Miro X., Roediger M., Geffers R., Toepfer T., et al. , 2008. Gene expression analysis of chronically inflamed and healthy human periodontal ligament cells in vivo. Dent. Res. J. 5: 1–11 [Google Scholar]
- Hammad H. M., Hammad M. M., Abdelhadi I. N., Khalifeh M. S., 2011. Effects of topically applied agents on intra-oral wound healing in a rat model: a clinical and histomorphometric study. Int. J. Dent. Hyg. 9: 9–16 [DOI] [PubMed] [Google Scholar]
- Han X., Amar S., 2002. Identification of genes differentially expressed in cultured human periodontal ligament fibroblasts vs. human gingival fibroblasts by DNA microarray analysis. J. Dent. Res. 81: 399–405 [DOI] [PubMed] [Google Scholar]
- Hart T. C., Zhang Y., Gorry M. C., Hart P. S., Cooper M., et al. , 2002. A mutation in the SOS1 gene causes hereditary gingival fibromatosis type 1. Am. J. Hum. Genet. 70: 943–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen A., Tellefsen M., Mikesjo U., Johannsen G., 2009. Local delivery of hyaluronan as an adjunct to scaling and root planing in the treatment of chronic periodontitis. J. Periodontol. 80: 1493–1497 [DOI] [PubMed] [Google Scholar]
- Laine M. L., Crielaard W., Loos B. G., 2012. Genetic susceptibility to periodontitis. Periodontol. 2000 58: 37–68 [DOI] [PubMed] [Google Scholar]
- Landry R. G., Jean M., 2002. Periodontal screening and recording (PSR) index: precursors, utility, and limitations in a clinical setting. Int. Dent. J. 52: 35–40 [DOI] [PubMed] [Google Scholar]
- Laurie, C. C., K. F. Doheny, D. B. Mirel, E. W. Pugh, L. J. Bierut, et al., and GENEVA Investigators, 2010 Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 34: 591–602. [DOI] [PMC free article] [PubMed]
- Lin W. L., Chien H. H., Cho M. I., 1999. N-cadherin expression during periodontal ligament cell differentiation in vitro. J. Periodontol. 70: 1039–1045 [DOI] [PubMed] [Google Scholar]
- Michalowicz B. S., Diehl S. R., Gunsolley J. C., Sparks B. S., Brooks C. N., et al. , 2000. Evidence of a substantial genetic basis for risk of adult periodontitis. J. Periodontol. 71: 1699–1707 [DOI] [PubMed] [Google Scholar]
- Mucci L. A., Bjorkman L., Douglass C. W., Pedersen N. L., 2005. Environmental and heritable factors in the etiology of oral diseases—a population-based study of Swedish twins. J. Dent. Res. 84: 800–805 [DOI] [PubMed] [Google Scholar]
- Pan F., Zhang R., Wang G., Ding Y., 2011. Oestrogen receptors are involved in the osteogenic differentiation of periodontal ligament stem cells. Biosci. Rep. 31: 117–124 [DOI] [PubMed] [Google Scholar]
- Polk D. E., Weyant R. J., Crout R. J., McNeil D. W., Tarter R. E., et al. , 2008. Study protocol of the Center for Oral Health Research in Appalachia (COHRA) etiology study. BMC Oral Health 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., et al. , 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues S. V., Acharya A. B., Bhadbhade S., Thakur S. L., 2010. Hyaluronan-containing mouthwash as an adjunctive plaque-control agent. Oral Health Prev. Dent. 8: 389–394 [PubMed] [Google Scholar]
- Schaefer A. S., Richter G. M., Nothnagel M., Manke T., Dommisch H., et al. , 2010. A genome-wide association study identifies GLT6D1 as a susceptibility locus for periodontitis. Hum. Mol. Genet. 19: 553–562 [DOI] [PubMed] [Google Scholar]
- Sheets S. M., Potempa J., Travis J., Fletcher H. M., Casiano C. A., 2005. Gingipains from Porphyromonas gingivalis w38 induce cell adhesion molecule cleavage and apoptosis in endothelial cells. Infect. Immun. 73: 1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L., Guan S. M., Fu S. M., Guo T., Cao M., et al. , 2008. Estrogen modulates cytokine expression in human periodontal ligament cells. J. Dent. Res. 87: 142–147 [DOI] [PubMed] [Google Scholar]
- Slevin M., Kumar S., Gaffney J., 2002. Angiogenic oligosaccharides of hyaluronan induce multiple signaling pathways affecting vascular endothelial cell mitogenic and wound healing responses. J. Biol. Chem. 277: 41046–41059 [DOI] [PubMed] [Google Scholar]
- Takeda K., Sakai N., Shiba H., Nagahara T., Fujita T., et al. , 2011. Characteristics of high-molecular-weight hyaluronic acid as a brain-derived neurotrophic factors scaffold in periodontal tissue regeneration. Tissue Eng. Part A 17: 955–967 [DOI] [PubMed] [Google Scholar]
- Takeichi M., 1987. Cadherins: a molecular family essential for selective cell-cell adhesion and animal morphogenesis. Trends Genet. 3: 213–217 [Google Scholar]
- Teumer A., Holtfreter B., Volker U., Petersmann A., Nauck M., et al. , 2013. Genome-wide association study of chronic periodontitis in a general German population. J. Clin. Periodontol. 40: 977–985 [DOI] [PubMed] [Google Scholar]
- Zhang B., Li Y., Zhou Q., Ding Y., 2011a Estrogen deficiency leads to impaired osteogenic differentiation of periodontal ligament stem cells in rats. Tohoku J. Exp. Med. 223: 177–186 [DOI] [PubMed] [Google Scholar]
- Zhang J., Sun X., Xiao L., Xie C., Zuan D., et al. , 2011b Gene polymorphisms and periodontitis. Periodontol. 2000 56: 102–124 [DOI] [PubMed] [Google Scholar]