Abstract
Protein phosphatases are integral components of the cellular signaling machinery in eukaryotes, regulating diverse aspects of growth and development. The genome of the filamentous fungus and model organism Neurospora crassa encodes catalytic subunits for 30 protein phosphatase genes. In this study, we have characterized 24 viable N. crassa phosphatase catalytic subunit knockout mutants for phenotypes during growth, asexual development, and sexual development. We found that 91% of the mutants had defects in at least one of these traits, whereas 29% possessed phenotypes in all three. Chemical sensitivity screens were conducted to reveal additional phenotypes for the mutants. This resulted in the identification of at least one chemical sensitivity phenotype for 17 phosphatase knockout mutants, including novel chemical sensitivities for two phosphatase mutants lacking a growth or developmental phenotype. Hence, chemical sensitivity or growth/developmental phenotype was observed for all 24 viable mutants. We investigated p38 mitogen-activated protein kinase (MAPK) phosphorylation profiles in the phosphatase mutants and identified nine potential candidates for regulators of the p38 MAPK. We demonstrated that the PP2C class phosphatase pph-8 (NCU04600) is an important regulator of female sexual development in N. crassa. In addition, we showed that the Δcsp-6 (ΔNCU08380) mutant exhibits a phenotype similar to the previously identified conidial separation mutants, Δcsp-1 and Δcsp-2, that lack transcription factors important for regulation of conidiation and the circadian clock.
Keywords: filamentous fungi, protein phosphorylation, functional genomics, serine-threonine protein phosphatases, tyrosine protein phosphatases
Phosphorylation of three amino acids—serine, threonine, and tyrosine—regulates myriad biological reactions in eukaryotic cells (Bauman and Scott 2002). Such regulatory cascades involve a cycle of phosphorylation via kinases and subsequent removal of the phosphate groups by phosphatases. A proper balance between kinases and phosphatases is essential for the maintenance of cell homeostasis. Because of their importance to cellular processes, kinases and phosphatases are among the most extensively studied enzymes (Cohen 2001; Hunter 1995). Phosphatases have been classified according to sequence homology, structural characteristics, and substrate specificity (Pao et al 2007; Shi 2009). Based on such properties, there are two major families of phosphatases: serine/threonine (S/T) protein phosphatases and protein tyrosine phosphatases. In general, protein phosphatases perform dephosphorylation in a mechanism involving nucleophilic attack on the phosphate ester moiety of the substrate (Sanvoisin and Gani 2001; Williams 2004). S/T phosphatases initiate the nucleophilic attack by means of a metal-activated water molecule in the catalytic groove, whereas protein tyrosine phosphatases use a catalytic cysteine residue as the nucleophile (McConnell and Wadzinski 2009).
S/T phosphatases are further classified into three main subfamilies, phosphoprotein phosphatases (PPPs), metal-dependent protein phosphatases (PPMs), and aspartate-based protein phosphatases, comprising the transcription factor IIF–interacting C-terminal domain phosphatase (FCP/SCP) and haloacid dehalogenase (HAD) classes (Shi 2009; Zhang et al. 2010). Protein tyrosine phosphatases are subdivided into classical protein-tyrosine phosphatases (PTPs), dual-specificity phosphatases (DSPs), low-molecular-weight phosphatases (LMW-PTP), and CDC25 class phosphatases (Andersen et al. 2001; Moorhead et al. 2007; Pao et al. 2007).
The PPP subfamily of S/T phosphatases has been implicated in a broad range of cellular processes, such as metabolism, cytoskeletal rearrangements, base excision repair, mitotic entry, and regulation of membrane receptors and ion channels (Burgess et al. 2010; Herzig and Neumann 2000; Lorca et al. 2010; Lu et al. 2004). Differences in relative inhibition by okadaic acid (Bialojan and Takai 1988) have led to subdivision of the PPP subfamily into PP2A, PP2B (calcineurin-A), and PP5 classes. PP2A is a highly conserved and ubiquitous protein phosphatase class, accounting for as much as 1% of total cellular proteins (Cohen 1990). The PP2A core enzyme consists of the PP2A catalytic or C subunit and the scaffolding A subunit [huntingtin-elongation-A subunit of PP2A TOR (HEAT) motifs] existing as a heterodimer that associates with the regulatory B subunit to constitute a heterotrimeric holoenzyme complex (Cho and Xu 2007; Xing et al. 2006; Xu et al. 2006). In mammals, PP2A class phosphatases dephosphorylate the microtubule-associated protein Tau and modulate mitogen-activated protein kinase (MAPK) signaling pathways by dephosphorylation of the component kinases (Janssens and Goris 2001). The PP2A class protein phosphatases PPP-1 and PPH-1 (catalytic subunits encoded by NCU00043; ppp-1 and NCU06630; pph-1) are important regulators of the circadian clock in Neurospora (Yang et al. 2004). Previous studies were successful in generating viable partial Repeat-Induced Point (RIP) mutants for ppp-1, but not for pph-1 (Yang et al. 2004). Although PPP-1 has been shown to influence the circadian clock by regulating the stability of the clock protein FRQ via dephosphorylation, in vitro studies demonstrated that the PP2A class holoenzyme dephosphorylates FRQ via the PPH-1 phosphatase but does not affect FRQ stability (Yang et al. 2004). Furthermore, dephosphorylation of the WCC (transcriptional activator of FRQ) has been shown to be dependent on the PP2A regulatory subunit RGB-1, leading to activation of WC-1 (Schafmeier et al. 2005).
The PPP class phosphatase calcineurin-A is a calcium-calmodulin–dependent enzyme (Bandyopadhyay et al. 2002; Klee et al. 1998). In animals, calcineurin dephosphorylates and activates the transcription factor NFATc, leading to T-cell differentiation via interleukin-2 expression (Crabtree 1999; Rusnak and Mertz 2000). Calcineurin is an essential gene involved in hyphal growth and maintenance of Ca+2 gradients in Neurospora crassa (Prokisch et al. 1997). The calcineurin holoenzyme is a heterodimer and consists of the large catalytic A subunit and a small regulatory B subunit (Klee et al. 1988). In Saccharomyces cerevisiae, cna1 and cna2 are functionally redundant catalytic subunits for calcineurin and, although cna1 cna2 double-mutants are viable, they are more sensitive to high levels of sodium, lithium, and other ions in the growth medium (Farcasanu et al. 1995; Garrett-Engele et al. 1995). Deletion of the cna-1 homolog is not lethal but results in a weakly growing mutant in Aspergillus nidulans (Cyert et al. 1991; Feng et al. 1991; Son and Osmani 2009). The regulatory subunit of calcineurin is encoded by the cnb1 gene in S. cereviseae and is required for adaptation to pheromone in vivo (Cyert and Thorner 1992), whereas in Neurospora the cnb-1 gene is required for normal vegetative growth (Kothe and Free 1998a).
The PPM subfamily of S/T phosphatases consists of PP2C enzymes with well-documented roles in cell-cycle progression (Cheng et al. 1999; Leroy et al. 2003; Lu and Wang 2008) and tumorgenicity (Ofek et al. 2003) in animals and act as negative regulators of the abscisic acid (ABA) signaling pathway in the model plant Arabidopsis thaliana (Ma et al. 2009). PP2C phosphatases act on a number of MAPK pathways (Arino et al. 2011). For example, Ptc1p in S. cerevisiae inactivates the high osmolarity glycerol (HOG) pathway by dephosphorylating the Hog1 MAPK (Warmka et al. 2001).
The FCP/SCP and HAD phosphatases are an aspartate-based class of S/T phosphatases with a shared DxDxT/V sequence motif. FCP1 is an essential protein phosphatase that dephosphorylates the C-terminal domain (CTD) of the largest subunit of RNA polymerase II (Archambault et al. 1998; Kobor et al. 1999). The HAD class of protein phosphatases contains important regulators of actin-cytoskeleton dynamics in mammals (Gohla et al. 2005; Seifried et al. 2013)
The PTP family is distinguished by a signature HC(X5)R catalytic motif. These proteins play important roles during meiosis and sporulation in yeast and cell adhesion, metabolism, and immune cell signaling in mammals (Elchebly et al. 1999; Mustelin et al. 2004; Zhan et al. 2000). Classical PTPs can be classified as receptor and nonreceptor PTPs and these phosphatases have functions in cell–substrate and cell–cell adhesion as well as insulin signaling in animals (Stoker 2005). DSPs dephosphorylate phosphotyrosine, phosphoserine, and phosphothreonine residues on substrates (Alonso et al. 2004; Tonks 2006). DSPs can be further classified on the basis of the presence (typical) or absence (atypical) of a MAPK-interacting domain, (Huang and Tan 2012; Jeffrey et al. 2007). For example, in Ustilago maydis, the DSP Rok1 is known to regulate mating and virulence by controlling the phosphorylation of Erk MAPKs Kpp2 and Kpp6 (Di Stasio et al. 2009). Among the other classes of PTPs, CDC25-type phosphatases have essential roles in mitotic entry (Gautier et al. 1991), whereas the LMW-PTP is less well-understood. In addition to these major classes of PTPs, SSU72 is a unique RNA polymerase II CTD phosphatase that shares high sequence similarity with PTPs (Ganem et al. 2003; Zhang et al. 2012). Y-phosphatases are a lesser studied class of PTPs that seem to be unique to filamentous fungi. In A. nidulans, AN4426 (Son and Osmani 2009) is a Y-phosphatase homologous to SIW14, a tyrosine phosphatase involved in endocytosis in S. cerevisiae (Sakumoto et al. 2002).
The filamentous fungus N. crassa is a model system for investigations of cell growth, development, gene silencing, the circadian clock, and stress responses in eukaryotic cells (Borkovich et al. 2004; Davis and Perkins 2002). N. crassa possesses 16 S/T phosphatases and 14 PTPs. Among the previously characterized protein phosphatases in N. crassa, the PP2A phosphatase pp2A (NCU06563) is involved in hyphal growth and cell–cell fusion (Fu et al. 2011; Pandey et al. 2004; Yatzkan et al. 1998), whereas another PP2A phosphatase, pph-1 (NCU06630), has so far been implicated in hyphal growth (Yang et al. 2004; Yatzkan et al. 1998). Mutation of the tangerine/tng gene (NCU03436), an ortholog of the cell-shape-control protein phosphatase cpp-1 in Fusarium verticillioides, leads to swollen hyphae and hyperbranching at the colony edge (McCluskey et al. 2011).
Taking advantage of the publicly available N. crassa genome sequence (Galagan et al. 2003) and the large-scale gene knockout project for ∼10,000 predicted genes (Park et al. 2011a), we have previously investigated the effects of mutating 86 S/T kinase genes in N. crassa (Colot et al. 2006; Park et al. 2011b). To elucidate the functions of protein phosphatases in N. crassa, we initiated a systematic analysis of the 30 predicted genes. In this study, we analyzed 24 viable phosphatase mutants for defects in basal growth, asexual development, and sexual development. Chemical sensitivity testing has proven to be a powerful method for identification of phenotypes for gene deletion mutants and genes of unknown function, as evident from previous studies in S. cerevisiae (Hillenmeyer et al. 2008) and our analysis of protein kinases (Park et al. 2011b). Accordingly, we tested the phosphatase mutants for altered sensitivity to several chemical stresses and growth under different nutritional regimens. We also measured phosphorylation of the p38 MAPK (OS-2) in all viable mutants to identify potential phosphatases acting on this pathway in N. crassa. The results reveal at least one defect for every phosphatase mutant analyzed, demonstrating the importance of these proteins to N. crassa biology. We present evidence linking the PP2C phosphatase pph-8 (NCU04600) and the HAD family phosphatase csp-6 (NCU08380) to important aspects of sexual and asexual development in N. crassa. We identified several protein phosphatases that influence basal or induced phosphorylation of the OS-2 MAPK.
Materials and Methods
Neurospora crassa strains and growth conditions
Wild-type strains ORS-SL6a [Fungal Genetics Stock Center (FGSC) 4200; mat a] and 74-OR23-IVA (FGSC 2489; mat A) and phosphatase mutants produced during the knockout project (Table 1) were obtained from the FGSC (Kansas City, MO). Knockout mutants for three phosphatase genes were not available as either homokaryons or heterokaryons (Table 1). Vegetative growth and asexual development (conidiation) were analyzed using Vogel minimal medium (VM) (Vogel 1956), whereas sexual development was assessed using synthetic crossing medium (SCM) (Westergaard and Mitchell 1947). Conidia used for inoculating cultures were propagated in VM agar flask cultures grown for 3 d at 30° in the dark and for 4 d at 25° in the light. Sorbose-containing medium (FGS) was used for isolation of colonies on plates and for ascospore germination assays (Davis and DeSerres 1970). When indicated, VM was supplemented with hygromycin (Calbiochem, San Diego, CA) at a concentration of 200 μg/ml.
Table 1. Neurospora crassa phosphatase gene families and summary of phenotypes and p38 MAPK levels.
| Familya | Subfamilyb | Class/Domainc | NCUd | N. crassa Genee | S. cerevisiae Homologf | Phenotype Summary | Chemical Sensitivityj/Nutritionk | Phospho-p38 MAPKl | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Inviable | Linear Growthg | Asexual Developmenth | Sexual Developmenti | ||||||||
| S/T | PPP | PP2Ac | 00043 | ppp-1/ pph-3 | GLC7 | X | —m | — | — | — | — |
| S/T | PPP | PP2Ac | 03436 | tng | SIT4 | R | AH, C | PP, P, A | N/Sn | ||
| S/T | PPP | PP2Ac | 06563 | pp2A | PPG1 | C | PP, P, A | SC, S, B, M, FL, T, | B | ||
| S/T | PPP | PP2Ac | 07489 | pzl-1 | PPZ1 | R | AH, C | PP | SC, S, B, M, T, F, YE | ||
| S/T | PPP | PP2Ac | 08301 | pph-4 | PPH3 | R | AH | PP, A | C, M, T, YE, A | ||
| S/T | PPP | PP2Ac | 06630 | pph-1 | PPH21 | X | — | — | — | — | — |
| S/T | PPP | PP2Bc | 03804 | cna-1/ pph-2 | CMP2 | X | — | — | — | — | — |
| S/T | PPP | PP5c | 01433 | ppt-1 | PPT1 | I | T | ||||
| S/T | PPM | PP2Cc | 00958 | pph-7 | PTC7/AZR1 | FL | |||||
| S/T | PPM | PP2Cc | 01767 | pph-5 | PTC5 | R | AH* | T | |||
| S/T | PPM | PP2Cc | 03495 | pph-6 | PTC6 | I | T, F | ||||
| S/T | PPM | PP2Cc | 04600 | pph-8 | PTC2 | R | AH, C | PP, P, A | N/S | B | |
| S/T | PPM | PP2Cc | 00434 | pph-9 | PTC1 | nao | na | na | na | na | na |
| S/T | Asp-Based | HAD | 08948 | pph-11 | PSR1 | R | AH, C | PP, P, A | N/S | B | |
| S/T | Asp-Based | HAD | 08380 | csp-6 | PSR2 | R | AH, C | PP, P, A | SC, B, FL, T, F | ||
| S/T | Asp-Based | FCP/SCP | 09300 | fcp-1 | FCP1 | X | — | — | — | — | — |
| PTP | Classical | PTPc | 02257 | pty-2 | PTP1 | I | T, F | ||||
| PTP | Classical | PTPc | 05364 | pty-3 | PTP2/PTP3 | PP, P, A | S | B, I | |||
| PTP | Dual-specificity | DSPc | 03246 | cdc-14 | CDC14 | R | AH*, C | A | S, B, F | ||
| PTP | Dual-specificity | DSPc | 03426 | dsp-1 | PPS1 | AH* | T | ||||
| PTP | Dual-specificity | DSPc | 06252 | dsp-2 | MSG5 | AH, C | PP, P, A | S, T, A | |||
| PTP | Dual-specificity | DSPc | 06330 | dsp-3 | MSG5 | R | PP, P, A | ||||
| PTP | Dual-specificity | DSPc | 08158 | dsp-4 | YVH1 | PP, A | |||||
| PTP | Dual-specificity | DSPc | 05049 | dsp-5 | SDP1 | na | na | na | na | na | na |
| PTP | LM-PTP | LMWPc | 09841 | pty-4 | LTP1 | PP | M, F | B | |||
| PTP | CDC-25 type | CDC25 | 02496 | div-12 | MIH1 | PP, P, A | F | B, I | |||
| PTP | CDC-25 type | CDC25 | 06966 | pty-1 | YCH1 | C | B | ||||
| PTP | SSU72 | SSU72 | 03114 | pph-10 | SSU72 | AH* | PP, P, A | C | B | ||
| PTP | — | Y-phosphatase 3 | 01010 | pty-5 | — | C | |||||
| PTP | — | Y-phosphatase 2 | 03333 | pty-6 | SIW14 | F | |||||
Family abbreviations: S/T, serine/threonine; PTP, protein tyrosine phosphatase.
Subfamily abbreviations: PPP, phosphoprotein phosphatase; PPM, Mg2+ or Mn2+-dependent protein phosphatase; Asp-based, aspartate-based phosphatase; LMW-PTP, low-molecular-weight protein tyrosine phosphatase; CDC25 type, cell division cycle 25 type; SSU72, C-terminal domain RNA Pol II phosphatase.
Class/domain abbreviations: PP2Ac, protein phosphatase 2 A catalytic subunit; PP2Bc, protein phosphatase 2 B catalytic subunit; PP5 catalytic subunit, protein phosphatase 5 catalytic subunit; PP2Cc, protein phosphatase 2C catalytic subunit; HAD, haloacid dehalogenase; FCP/SCP, transcription factor IIF–interacting C-terminal domain phosphatase 1/ small C-terminal domain phosphatase; PTPc, protein tyrosine phosphatase catalytic subunit; DSPc, dual-specificity phosphatase catalytic subunit; LMWPc, low-molecular-weight phosphatase catalytic subunit; CDC25, cell division cycle; SSU72, C-terminal domain RNA polymerase II phosphatase; Y-phosphatase 3, tyrosine phosphatase 3; Y-phosphatase 2, tyrosine phosphatase 2.
Based on version 5 annotation of the Broad Institute’s Neurospora crassa database (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html).
Phosphatase gene names are consistent with the Neurospora e-Compendium Project at Leeds University (http://bmbpcu36.leeds.ac.uk/~gen6ar/newgenelist/genes/gene_list.html). All other gene names were given during this study in accordance with the e-Compendium system.
Yeast orthologs were obtained from literature or blastp search and are consistent the Saccharomyces Genome Database (http://www.yeastgenome.org/).
R, reduced growth; I, increased growth (significance testing performed with Student t test, paired, two-tailed; p<0.05, *p<0.1).
Asexual phenotypes are depicted by phenotypes in aerial hyphae (AH) or conidial development (C).
Sexual phenotypes are depicted by their occurrence during protoperithecial (PP), perithecial (P), or ascospore (A) development.
Chemical sensitivity phenotypes are represented based on the sensitivity or resistance to sodium chloride (SC), sorbitol (S), cytochalasin A (C), benomyl (B), tert-butyl hydroperoxide (T), menadione (M), FK506 (F), fludioxonil (FL), and yeast extract (YE).
Nutritional phenotypes are represented by increased growth on Avicel (A) as compared to wild-type.
p38 MAPK levels are represented relative to wild-type levels as elevated basal (B) or elevated induced (I).
Phenotypic analysis could not be performed because of inviability of the knockout mutants.
N/S, mutant strain was not analyzed for chemical screening because of poor growth compared to wild-type.
na, mutant not available.
In this study, the gene names for N. crassa phosphatases were taken from the literature (the e-compendium at Leeds University; http://bmbpcu36.leeds.ac.uk/~gen6ar/newgenelist/genes/gene_list.htm) or were assigned a name (Table 1).
Purification of homokaryotic phosphatase mutants from heterokaryons
Because N. crassa is multinucleate, primary transformants are often heterokaryons, with both mutant and wild-type nuclei (Dev and Maheshwari 2002; Paietta and Marzluf 1985; Park et al. 2011a). Therefore, transformants were crossed to wild-type to purify homokaryotic meiotic progeny for the knockout project (Colot et al. 2006; Park et al. 2011a). Using the aforementioned method, one phosphatase cassette did not yield transformants (Δpph-9; NCU000434) and viable ascospores could not be isolated for four phosphatase mutants: Δppp-1 (NCU00043); Δcna-1 (NCU03804); Δpph-1 (NCU06630); and Δdiv-12 (NCU02496). Homokaryotic mutants for Δdiv-12 (NCU02496) were purified after serial plating of conidia. Macroconidia were plated on FGS-hygromycin plates and incubated in the dark at 30°. The next day, one colony was picked and transferred to a VM-hygromycin agar slant and cultured for 5 d. Macroconidia were isolated from this slant and plated on a FGS-hygromycin plate; after incubation, a colony was transferred onto a fresh VM-hygromycin slant. These steps were repeated twice. Diagnostic PCR with gene-specific and hph primers (Supporting Information, Table S2) was used to test for the absence of the open-reading frame of the respective deleted gene (with wild-type as a positive control) and the simultaneous presence of the hph cassette in the purified strains (Colot et al. 2006). These experiments confirmed that the purified strains were homokaryons.
Analysis of growth and morphological and developmental phenotypes
The 24 viable N. crassa phosphatase knockout mutants (Table 1) were analyzed for phenotypes using methods reported previously (Turner 2011), with some modifications. Linear growth rates for the mutants (Table S1) were measured on VM at 25° in the dark using race tubes (Turner 2011). Mutants were grown on VM plates for 24 hr and hyphae at the colony edge were photographed using an Olympus SZX9 stereomicroscope with a C-4040 digital camera (Olympus, Lake Success, NY). VM slant tubes were inoculated with the mutant strains and grown for 3 d in the dark at 30°, for 4 d under constant light at 25°, and then scored for conidial production (Table S1). Aerial hyphal extension was measured in 2 ml VM (standing) liquid cultures. These cultures were inoculated at the liquid surface and incubated statically at 25° (in the dark) for 96 hr. The total height of aerial hyphae was measured in millimeters (Table S1). Data were subsequently tested for significance using Student t test (paired, two-tailed, independent means).
For analysis of female sexual fertility, strains were inoculated onto SCM slants and incubated under constant light for 7 d to 8 d at 25°. Cultures were scored for protoperithecia formation and then fertilized with conidia of the opposite mating type. Subsequent perithecia formation and ascospore development were scored 1 wk and 2 wk after fertilization, respectively. All scoring for female sexual fertility analysis was performed using the SZX9 stereomicroscope (Olympus). For visualizing unregulated protoperithecial formation in the Δpph-8 mutant (NCU04600), the strain was inoculated on VM and SCM agar plates and incubated under constant light or dark at 25° as indicated in Figure 3. A wild-type strain (FGSC 2489; mat A) was used as a control. Photographs were taken using the SZX9 stereomicroscope with a C-4040 digital camera (Olympus) at 5 d and 7 d after inoculation and 2 d after fertilization.
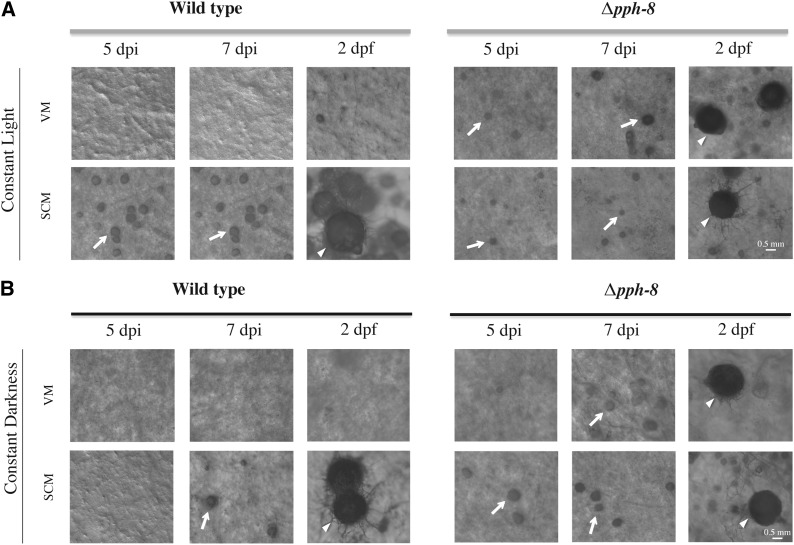
Figure 3.
The Δpph-8 mutant displays unregulated protoperithecial development on minimal medium. Wild-type and Δpph-8 strains were cultured on VM and SCM plates under constant light (A) or constant darkness (B) and photographed at 5 d postinoculation (dpi), 7 dpi, and 2 d postfertilization (dpf) with opposite mating-type conidia. The white arrows point to protoperithecia, whereas the white arrowheads indicate mature perithecia. Scale bar = 0.5 mm.
Conidial separation was investigated in wild-type (FGSC 4200; mat a), Δcsp-1 (NCU02713, FGSC 2555), Δcsp-2 (NCU06095, FGSC 2522), and Δcsp-6 (NCU08380) strains. Conidia were propagated by culturing strains in VM agar flasks for 3 d in the dark at 30° and for 4 d in the light at 25°. A small amount of conidia was withdrawn from the flask, suspended in 50 μl of sterile liquid VM, and 50 μl of calcofluor white (Eng Scientific, Clifton, NJ) was added to the suspension. A volume of 20 μl was placed on a glass slide and covered with a cover slip. Conidia were visualized using differential interference (DIC) microscopy with an IX71 inverted microscope (Olympus America, Center Valley, PA) using a 60× oil immersion objective. X-Cite 120PC Q (Lumen Dynamics, Ontario, Canada) was used as the fluorescence microscope light source with a DAPI filter cube on the microscope. Photographs were taken using a QIClick digital CCD camera (QImaging, Surrey, British Columbia, Canada).
Chemical sensitivity assays and nutritional phenotypes
Chemical sensitivity assays were restricted to viable phosphatase knockout mutants with growth rates at least 50% of the wild-type strain on VM as shown in Table S1 (Park et al. 2011b). The mutants were screened for responses to a variety of chemicals at concentrations that inhibited wild-type growth by ∼50–60% (Table S1). The chemicals included sodium chloride (0.35 M; EMD Chemicals, Gibbstown, NJ), sorbitol (0.8 M; Sigma, St. Louis, MO), cytochalasin A (40 ng/ml; Sigma), benomyl (92 ng/ml; Fluka, St. Louis, MO), tert-butyl hydroperoxide (t-BuOOH; 0.13 mM; Sigma), Menadione (100 μM; M5750; Sigma), FK-506 (50 ng/ml; LC Laboratories, Woburn, MA), and fludioxonil (2.75 ng/ml; a gift from Frank Wong and Allison Tally). Phosphatase mutants were also analyzed for nutritional phenotypes, including growth on VM supplemented with 2% yeast extract and utilization of crystalline cellulose (Avicel; PH-101; Sigma) as a carbon source (Haas et al. 1952; St. Lawrence et al. 1964; Znameroski et al. 2012). VM plates (60 mm × 15 mm) were supplemented with the respective chemicals and one edge of the plate was inoculated and radial colony growth was measured after 20–22 hr at 30°. A VM plate lacking chemical was used as a control for each of the tested strains. The percentage growth was calculated by dividing the radius with chemical by the radius in the absence of chemical for four biological replicates. Three independent experiments were performed. One-way ANOVA analysis (Bewick et al. 2004) was used for significance testing. Knockout mutants were considered sensitive/slower growing (S) or resistant/faster growing (R) (Table 3) if there was a difference in percent growth of wild-type in the presence of the chemical at p<0.05.
Table 3. Mutants with chemical sensitivity phenotypes.
| NCU | FGSC | Deleted Gene | Sodium Chloride | Sorbitol | Cytochalasin A | Benomyl | Tert-Butyl Hydroperoxide | Menadione | FK506 | Fludioxonil | Yeast Extract |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 06563 | 11546 | pp2a | S | S | R | S | S | R | |||
| 07489 | 11548 | pzl-1 | R | R | R | S | S | R | S | ||
| 08301 | 12454 | pph-4 | R | S | S | S | |||||
| 01433 | 15790 | ppt-1 | R | ||||||||
| 00958 | 19378 | pph-7 | S | ||||||||
| 01767 | 12451 | pph-5 | R | ||||||||
| 03495 | 16430 | pph-6 | S | S | |||||||
| 08380 | 20306 | csp-6 | S | R | S | R | S | ||||
| 02257 | 16060 | pty-2 | R | R | |||||||
| 05364 | 12444 | pty-3 | S | ||||||||
| 03246 | 13311 | cdc-14 | R | R | R | ||||||
| 03426 | 16425 | dsp-1 | R | ||||||||
| 06252 | 14464 | dsp-2 | S | S | |||||||
| 06330 | 15781 | dsp-3 | |||||||||
| 08158 | 19644 | dsp-4 | |||||||||
| 09841 | 18801 | pty-4 | R | R | |||||||
| 02496 | 16654 | div-12 | R | ||||||||
| 06966 | 14056 | pty-1 | |||||||||
| 03114 | 16337 | pph-10 | R | ||||||||
| 01010 | 16679 | pty-5 | |||||||||
| 0333 | 17653 | pty-6 | R |
One-way ANOVA analysis was performed to determine significance. These results reflect strains displaying chemical sensitivity phenotypes at p<0.05. Radial colony growth was measured and percentage growth was calculated as growth with chemical vs. growth without chemical. See Materials and Methods for details.
Mutants were classified as sensitive (S) or resistant (R) relative to the growth of wild-type.
p38 MAPK assays
For analysis of MAPK profiles of the phosphatase mutants, conidia were used to inoculate VM liquid cultures at an initial concentration of 1 × 106 conidia/ml as previously described (Jones and Borkovich 2010). Cultures were grown for 16 hr at 30° with shaking at 200 rpm and then treated with 0.8 M NaCl (for activation of p38 OS-2 MAPK) for 10 min. An untreated sample was used as an uninduced control (time zero). After treatment, the tissue was flash-frozen in liquid nitrogen and ground using 2-mm to 5-mm stainless steel beads (Qiagen) with the Qiagen Retsch TissueLyser system (Qiagen Retsch GmbH, Hannover, Germany). Depending on the amount of tissue, 300–700 μl extraction buffer (50 mM HEPES, pH 7.5; 2 mM EGTA; 2 mM EDTA; 1% SDS; 10% glycerol; 100 mM NaCl; 1 mM sodium orthovanadate; and 1 mM sodium fluoride) was added to the powdered fungal tissue, and the mixture was heated at 85° for 5 min. Afterwards, 10 μl of 100 mM PMSF and 1 μl of fungal protease inhibitor cocktail (Product #T8215; Sigma-Aldrich, St. Louis, MO) was added and the solution was centrifuged at 4000g for 15 min at 4°. The supernatant was collected and the protein concentration was determined using the BCA protein assay (Pierce Chemical, Rockford, IL). A volume of extract containing 30 μg protein was subjected to SDS-PAGE, followed by immunoblotting (Krystofova and Borkovich 2005). Commercial antibodies directed against mammalian or S. cerevisiae MAPKs were used to detect phospho-OS-2 (1:600 dilution; anti-phospho-p38 #9211; Cell Signaling Technology, Beverly, MA). Incubation with peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Sigma Chemical, St. Louis, MO) and subsequent chemiluminescence detection was performed as previously described (Krystofova and Borkovich 2005).
Results
Protein phosphatase catalytic subunit genes in the N. crassa genome
We utilized the amino acid sequences of known protein phosphatase catalytic subunit genes from the Saccharomyces Genome Database (http://www.yeastgenome.org/) as queries during a reciprocal BLAST approach for identifying corresponding homologs in the Broad Institute N. crassa database (http://www.broadinstitute.org/annotation/genome/neurospora) (Table 1). We then performed additional BLAST searches and CDD domain analysis at NCBI to eliminate spurious small-molecule phosphatases. We identified 30 catalytic subunit genes that were classified as S/T (PPP, PPM, and Asp-based subfamilies) or PTPs (classical, dual-specificity, LMW-PTP, Cdc25-type, and SSU72 subfamilies). Two genes in the tyrosine phosphatase family (NCU01010 and NCU03333) appeared to be unique to filamentous fungi.
We compared protein phosphatases across different eukaryotic species (Table 2), including humans (Moorhead et al. 2007, 2009), a model plant, A. thaliana (Kerk et al. 2008; Moorhead et al. 2007), baker’s yeast, S. cerevisiae (Breitkreutz et al. 2010), and the filamentous fungi A. nidulans (Son and Osmani 2009), and N. crassa. There are many more S/T phosphatases in A. thaliana and humans than in the three fungi (Table 2). Comparing the fungi, S. cerevisiae has the greatest number (21 genes), followed by A. nidulans with 17 genes and N. crassa with 16 genes (Table 2). The observation of lower numbers of S/T phosphatase genes in N. crassa compared to A. thaliana, humans, and baker’s yeast is consistent with the fact that N. crassa has fewer S/T kinase genes (Park et al. 2011b).
Table 2. Serine/threonine and protein tyrosine phosphatase genes in Homo sapiens, Arabidopsis thaliana, Saccharomyces cerevisiae, Neurospora crassa, and Aspergillus nidulans.
| Serine/Threonine Protein Phosphatase Genes | |||||
|---|---|---|---|---|---|
| Family | Homo sapiens (Moorhead et al. 2007) | Arabidopsis thaliana (Moorhead et al. 2007; Kerk et al. 2008) | Saccharomyces cerevisiae (Moorhead et al. 2007; Breitkreutz et al. 2010) | Neurospora crassa | Aspergillus nidulans (Son and Osmani 2009) |
| PPP | 13 | 26 | 11 | 8 | 8 |
| PPM | 18 | 76 | 7 | 5 | 6 |
| Asp-based | 13 | 23 | 3 | 3 | 3 |
| Total | 44 | 125 | 21 | 16 | 17 |
| Protein Tyrosine Phosphatase Genes | |||||
| Classical | 38 | 1 | 4 | 2 | 2 |
| Dual-specificity | 61 | 22 | 6 | 6 | 5 |
| LMW-PTP | 1 | 1 | 1 | 1 | 1 |
| CDC25 | 3 | — | 2 | 2 | 1 |
| SSU72 | 1 | 1 | 1 | 1 | 1 |
| Y-phosphatase | — | — | — | 2 | 1 |
| Total | 104 | 25 | 14 | 14 | 11 |
With regard to PTPs, the number of genes is similar in the three fungi but fewer than in humans and A. thaliana (Table 2). Despite the presence of tyrosine phosphatases, fungi do not possess recognizable tyrosine kinases (Borkovich et al. 2004; Kosti et al. 2010). The same is true for A. thaliana and several apicomplexan species whose genomes lack any true tyrosine kinases or receptor tyrosine kinases (Andreeva and Kutuzov 2008; Kerk et al. 2008; Moorhead et al. 2009). It is now believed that PTPs evolved before tyrosine kinases because of leaky phosphorylation of tyrosine residues by S/T kinases, thus providing a target for the tyrosine phosphatases (Moorhead et al. 2009).
As part of the Neurospora Genome Project, we attempted gene replacement of the 30 phosphatase genes (Colot et al. 2006). Transformants could not be recovered for ΔNCU00434 (ptc-1) and mutants for NCU05049 (dsp-5) were not available (Table 1). We were unable to purify four of the knockout mutants (ΔNCU03804; cna-1, ΔNCU00043; ppp-1, ΔNCU09300; fcp-1 and ΔNCU06630; pph-1) to homokaryons. As mentioned, cna-1 has been reported as an essential gene in N. crassa (Prokisch et al. 1997). The homolog for pph-1 has been shown to be essential in A. nidulans (Son and Osmani 2009) and the gene was also shown to be essential for cell survival in Neurospora (Yang et al. 2004). Previous work showed that homologs of ppp-1 and fcp-1 are essential in both A. nidulans (Son and Osmani 2009) and S. cerevisiae (Archambault et al. 1997; Feng et al. 1991), and our observation the two N. crassa mutants could not be purified to homokaryons supports ppp-1 and fcp-1 as essential genes in N. crassa. Thus, for phenotypic analyses and characterization, we were able to analyze a total of 24 viable protein phosphatase mutants.
Deletion of protein phosphatase genes leads to growth and developmental phenotypes in N. crassa
N. crassa is a heterothallic (self-sterile) fungus that spends most of its life cycle in the haploid state and grows vegetatively by apical extension of basal hyphae (Davis and Perkins 2002). The asexual phase of growth begins with germination of an asexual spore (conidium) that undergoes polarized growth to form hyphae. Hyphal fusion and branching give rise to the networked multicellular body of the organism, the mycelium. Different environmental stimuli, such as desiccation, heat, and/or nutrient deprivation, can stimulate the asexual sporulation pathway known as macroconidiation. This leads to the differentiation of aerial hyphae, which then bud from their tips, thus forming conidiophores and eventually giving rise to the free asexual spores, macroconidia or conidia (Springer 1993). Under nitrogen starvation, N. crassa enters into the sexual phase of development, inducing the formation of female reproductive structures known as protoperithecia (Raju and Leslie 1992). Chemotropic growth of a female hypha (trichogyne) toward a male cell (conidium) of opposite mating-type results in cell and nuclear fusion, followed by meiosis and enlargement of the protoperithecium into the fruiting body (perithecium). Perithecia contain the meiotic progeny known as ascospores that germinate to produce hyphae under appropriate environmental conditions (Raju and Leslie 1992).
To characterize N. crassa phosphatase genes, we began with phenotypic analyses of the 24 viable mutants. In terms of extension of basal hyphae, nine mutants showed reduced growth and three displayed increased growth as compared to wild-type (Figure 1 and Table 1; detailed phenotypic data in Table S1). A total of 14 mutants exhibited defects in asexual development. Among these strains, only one mutant (Δpph-5) possessed abnormalities in growth of basal hyphae and asexual development, but not in sexual development (Figure 1 and Table 1). The pph-5 homolog in S. cerevisiae (PTC5) is also required for normal vegetative growth (Yoshikawa et al. 2011). Mutants lacking the genes ppt-1, pph-6, and pty-2 displayed increased basal growth (compared to wild-type) as their only morphological phenotype. Interestingly, the S. cerevisiae pty-2 homolog, PTP1, is a negative regulator of filamentation (Fasolo et al. 2011). The faster hyphal growth observed in the N. crassa Δpty-2 mutant suggests that pty-2 and PTP1 may have similar functions in N. crassa and S. cerevisiae.
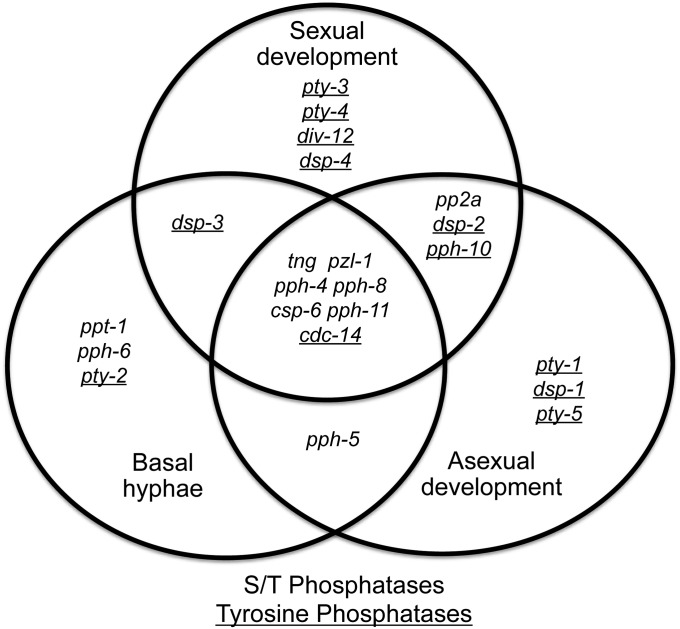
Figure 1.
Venn diagram displaying distribution of protein phosphatase mutants with growth and developmental phenotypes. The 22 viable protein phosphatase mutants exhibiting defects in at least one major growth/developmental pathway are indicated by the names for deleted genes. The underlined gene names correspond to tyrosine phosphatases, whereas the remaining are serine/threonine phosphatases.
Mutants lacking the DSP dsp-1, the CDC25 phosphatase pty-1, and the Y-phosphatase member pty-5 exhibited phenotypes in asexual development as their only morphological defect. Deletion of the dsp-1 homolog PPS1 in S. cerevisiae does not produce any adverse effects on growth, but overexpression of PPS1 results in growth arrest and aberrant DNA synthesis (Ernsting and Dixon 1997).
In our previous study of N. crassa kinases (Park et al. 2011b), we found that 32 out of 77 (42%) mutant strains exhibited defects in sexual development, with 30 out of 77 (39%) unable to produce ascospores (female-sterile; 94% of sexual phenotypes). In contrast, among the 24 phosphatase mutants, we found that 15 had a phenotype during sexual development (60%), with four strains (Δtng, Δpp2A, Δcsp-6, and Δdsp-2; 16% of mutants) being female-sterile, accounting for 26% of the sexual phenotypes in the phosphatase mutants (Table 1, Figure 1, and Table S1). These results demonstrate that although a greater proportion of phosphatases than kinases influence the sexual cycle, kinases are more critical for production of ascospores and absolute female fertility in N. crassa.
The Δpp2A mutant failed to produce protoperithecia and also had reduced aerial hyphae extension (Table 1, Figure 1, and Table S1). These phenotypes are similar to those of mutants lacking components of the two MAPK pathways in N. crassa: MIK-1/MEK-1/MAK-1 (cell fusion and cell wall integrity) and NRC-1/MEK-2/MAK-2 (cell fusion) (Kothe and Free 1998b; Li et al. 2005; Park et al. 2011b). However, the HAD class phosphatase knockout Δcsp-6 was unique in that it had very few (and small) protoperithecia that were unable to mature into perithecia on fertilization with the opposite mating type.
Among the remaining strains with defects in sexual development, four mutants, Δpty-3, Δdiv-12, Δdsp-2, and Δpph-10, exhibited decreased numbers of protoperithecia and perithecia, as well as few or delayed shooting of ascospores (Table 1, Figure 1, and Table S1). The Δdsp-3 and Δdsp-4 each displayed abnormal and increased protoperithecia or perithecia formation and increased ascospore production (Table 1, Figure 1, and Table S1). It is interesting to note that all of these aforementioned genes are tyrosine phosphatases, suggesting that this phosphatase class is important for regulation of sexual development in N. crassa. Two more mutant strains (Δtng and Δpph-11) produced few protoperithecia and perithecia, and whereas one ejected no ascospores (Δtng), the other produced very few (Δpph-11). Another three mutants (Δpph-4, Δcdc-14, and Δpph-8) possessed defects in the timing or in the number of ascospores produced. The pph-4 mutant developed abnormal/small protoperithecia but normal-appearing perithecia, whereas Δpph-8 was precocious in protoperithecia formation, leading to perithecia that were embedded in the agar surface. A null mutation in the well-characterized phosphatase CDC14 (involved in mitotic exit and meiosis I spindle disassembly) is lethal in yeast (Taylor et al. 1997), whereas the N. crassa Δcdc-14 mutant is viable (but with defects in all three growth/developmental pathways). Deletion of cdc-14 in A. nidulans did not result in any obvious growth defects (Son and Osmani 2009).
Most of the 15 mutants that had phenotypes in sexual development also exhibited defects in basal hyphae extension and asexual differentiation. However, Δpty-1, Δdsp-1, and Δpty-5 demonstrated phenotypes only during asexual development and thus seem to be specific for aspects of conidiation in N. crassa (Figure 1, Table 1, and Table S1). Overall, our results show that 22 out of 24 mutants (91%) displayed a defect in at least one of three growth/developmental pathways analyzed in this study (Figure 1 and Table 1), with seven of the 24 mutants (29%) possessing phenotypes in all three stages. As a comparison, among the previously studied S/T protein kinase knockouts in N. crassa (Park et al. 2011b), 57% of the mutants possessed a defect in at least one of the growth/developmental stages, whereas 45% had overlapping defects in all three. This suggests that similar to kinases, phosphatases are also important regulators of growth and development in N. crassa.
Chemical sensitivity assays reveal additional phenotypes for protein phosphatase mutants
Various chemical and environmental stresses have been known to influence growth and developmental outcomes in eukaryotic cells. To gain a better understanding of the functions of the different protein phosphatases in N. crassa, we subjected the phosphatase mutants to a panel of chemical treatments (see Materials and Methods) and compared their relative sensitivity to each chemical to that of wild-type (Table 3; detailed results in Table S1). Strains with linear growth rates less than 50% of wild-type on minimal medium were excluded from this assay to avoid any bias attributable to their slow growth.
We analyzed the relative sensitivities of the phosphatase mutants to the reactive oxygen species (ROS) generating chemical menadione (Loor et al. 2010), whereas peroxide stress was introduced by exposure to t-BuOOH (Kim et al. 2012). Similar to our previous study of kinases (Park et al. 2011b), treatment with t-BuOOH yielded the greatest number of phenotypes, with a total of 10 strains displaying sensitivity or resistance to peroxide treatment. Three mutants (Δpp2A, Δpzl-1, and Δpph-4) showed increased sensitivity to both t-BuOOH as well as menadione (Table 3), whereas Δpph-6, Δcsp-6, and Δdsp-2 were exclusively sensitive to peroxide. The tyrosine phosphatase mutant Δpty-4 was resistant to menadione treatment, whereas Δppt-1, Δpph-5, Δpty-2, and Δdsp-1 were resistant to t-BuOOH. It is thus of particular interest to understand how these phosphatases might be regulating cellular responses to oxidative stress. Conidiation is known to be influenced by ROS in N. crassa (Hansberg et al. 1993; Toledo et al. 1994). In the case of the Δcsp-6, we have shown that this mutant is defective in conidial separation (Figure 4). The finding that it is also sensitive to peroxide stress reinforces the notion that csp-6 is an important component of the conidiation pathway in N. crassa.
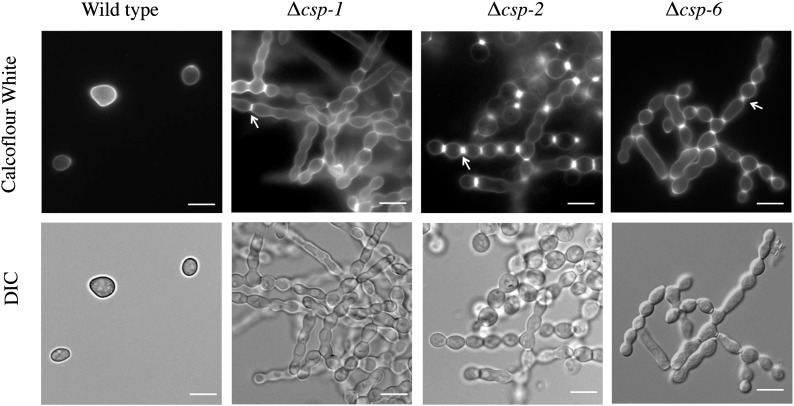
Figure 4.
The Δcsp-6 displays a conidiation separation defect most similar to Δcsp-1 strains. Wild-type, Δcsp-1, Δcsp-2, and Δcsp-6 strains were cultured on VM medium for 7 d under constant light and conidia were stained with calcofluor white to visualize developing crosswalls in the conidial chains. The arrow points to the conjoined conidia, indicating the separation defect. Scale bar size = 10 μ.
We used sodium chloride and sorbitol to induce salt/osmotic stress in the N. crassa phosphatase mutants. A total of six mutants exhibited phenotypes in these assays. The Δpp2A mutant was sensitive to both sorbitol and sodium chloride, suggesting it has important roles in osmotic stress resistance. The S. cerevisiae homolog of pzl-1, PPZ1, has been characterized as an important (negative) regulator of salt stress, halo tolerance, and pH homeostasis (Posas et al. 1995; Yenush et al. 2002). In our assays, we found that the Δpzl-1 mutant was resistant to both sorbitol and sodium chloride, providing evidence for similar functions for pzl-1 in N. crassa as observed in baker’s yeast. S. cerevisiae Psr1p and Psr2p are most similar to N. crassa PPH-11 and CSP-6, respectively (Siniossoglou et al. 2000) (Table 1). The slow growth rate of the N. crassa Δpph-11 mutant disqualified this strain for chemical sensitivity screening, but we observed that the Δcsp-6 mutant was sensitive to sodium chloride (Table 3). The observation that Δpsr1 and Δpsr2 single mutants are normal but that loss of both genes leads to sensitivity to salt stress in S. cerevisiae (Siniossoglou et al. 2000) illustrates the difference in genetic wiring between S. cerevisiae and N. crassa.
To decipher possible functions for protein phosphatases in cytoskeletal maintenance, we treated the mutants with cytochalasin A, which prevents polymerization and elongation of actin filaments (Cooper 1987) and benomyl, which binds to microtubules, thus inhibiting mitosis, meiosis, and cellular transport (Willhite 1983). Interestingly, the only phenotype observed using these chemicals was increased resistance. Two mutants, Δpph-4 and Δpph-10, were resistant to cytochalasin A, whereas four (Δpp2A, Δpzl-1, Δcdc-14, and Δcsp-6) showed enhanced growth as compared to wild-type with benomyl treatment (Table 3). It is possible that these missing phosphatases play important antagonistic roles in mitotic exit or in cell proliferation, perhaps through dephosphorylation of a mitotic/cell proliferation kinase. For example, cdc-14 is known to be an important regulator of the cell cycle and mitosis in fungi and deletion of the gene in N. crassa imparts resistance to benomyl. Deletion of cdc-14 leads to increased activity of cdk-1, which is known to promote cell proliferation and survival (in mammalian cells) via phosphorylation of the transcription factor FOXO1 (Liu et al. 2008; Stegmeier and Amon 2004). Resistance to benomyl in Δcdc-14 might be imparted via a similar mechanism, which in effect counteracts the inhibitory effects of the chemical.
FK506 is a macrolide lactone (Dumont et al. 1990) that binds the immunophilin FKBP12 (FK506 binding protein), inhibiting the S/T phosphatase calcineurin in the calcium-signaling pathway (Prokisch et al. 1997). Assays with this immunosuppressant drug revealed that six mutants (Δpzl-1, Δcsp-6, Δpty-2, Δcdc-14, Δpty-4, and Δpty-6) were resistant, whereas one (Δpph-6) was sensitive to FK506 (Table 3). Resistance to FK506 was the only phenotype for the Δpty-6 mutant.
Calcineurin A subunit mutants have been shown to have phosphatase activity with increased resistance to FKBP12-FK506 in mammalian cells (Kawamura and Su 1995). A similar effect is also observed in case of TOR pathway mutants in yeast (TOR1 and TOR2) that display resistance to a different macrolide, sirolimus, also known as rapamycin (Dumont et al. 1990). The TOR signaling pathway has also been implicated in regulation of microtubule structure/function and acts antagonistically to the calcineurin-signaling network (Choi et al. 2000; Mulet et al. 2006). Of the strains that were resistant to FK506, three mutants (Δpzl-1, Δcsp-6, and Δcdc-14) also displayed resistance toward benomyl. It is thus tempting to speculate that these three phosphatase knockout mutants with a common resistance to benomyl and FK506 might have overlapping roles in the TOR pathway and calcineurin function in N. crassa.
Fludioxonil is a phenylpyrrole class fungicide (Ochiai et al. 2001) that stimulates the OS-2 MAPK pathway, leading to increased glycerol production and cell death in N. crassa (Zhang et al. 2002). The OS MAPK module mutants (os-4/os-5/os-2) are resistant to fludioxonil but sensitive to sodium chloride and sorbitol (Park et al. 2011b; Zhang et al. 2002). We found that two mutants, Δcsp-6 and Δpph-7, are sensitive to fludioxonil, whereas the PP2A class phosphatase mutant Δpp2A and CDC25 phosphatase mutant Δdiv-12 were resistant. The resistance phenotype of the latter group suggests that these gene products might have important roles in the OS-2 MAPK signaling pathway. Incidentally, fludioxonil sensitivity was the only phenotype observed for the Δpph-7 mutant in this study.
We also analyzed the relative growth of the phosphatase mutants on medium supplemented with 2% yeast extract, which is rich in amino acids, peptides, and vitamins. Only two phosphatase mutants, Δpzl-1 and Δpph-4, exhibited a significant difference in growth relative to wild-type on 2% yeast extract. Both of these strains grow less well than wild-type, suggesting that nutrient sensing and/or utilization abilities are compromised in the mutants.
The 24 viable phosphatase mutants were also cultured on VM with Avicel (2%) substituted for sucrose as an alternate carbon source. We found that two phosphatase mutants (Δpph-4 and Δdsp-2) were better able to utilize Avicel than wild-type, consistent with the corresponding genes acting as negative regulators of cellulose utilization (Table 4). It is interesting to note that both of these mutants display a common sensitivity to t-BuOOH, and that Δpph-4 is also sensitive to menadione. It will be of interest in future studies to determine whether sensitivity to oxidative stress could prove beneficial in upregulating carbon metabolism genes in fungi and how these phosphatases could assist in the process.
Table 4. Mutants with altered growth on 2% Avicel.
| Strains | Sucrose (mm/day)a | Avicel (mm/d)b | % Growthc | SDd |
|---|---|---|---|---|
| Wild-type (mat a) | 33.6 | 18.2 | 54 | 0.106 |
| Δpph-4 | 18 | 14 | 78 | 0.114 |
| Δdsp-2 | 29.5 | 22.83 | 77 | 0.100 |
Radial growth of strains on minimal medium containing sucrose.
Radial growth of strains on minimal medium containing 2% Avicel.
% Growth = (radial growth on Avicel)/(radial growth on sucrose) × 100.
SD for three replicates.
Through our morphological testing, we determined that two of the 24 viable mutants had no obvious growth defects (Δpty-6 and Δpph-7). However, phenotypes were revealed for these two mutants through the chemical sensitivity assays, resulting in at least one phenotype for every phosphatase mutant analyzed. This supports the advantage of chemical testing for identifying defects for mutants that do not display growth or developmental phenotypes. Knockout strains for pzl-1 and pp2a possessed the greatest number of chemical sensitivity phenotypes and 17 of 21 tested strains exhibited at least one chemical sensitivity phenotype. Also, taking into account mutants that possessed either significant sensitivity or resistance to more than one chemical, we observed a total of 42 chemical sensitivity phenotypes for a set of 21 phosphatase mutants.
The phospho-p38 MAPK level is elevated in a number of protein phosphatase mutants
MAPKs are a class of S/T kinases present in all eukaryotic cells. As a group, they are responsible for a wide variety of cellular responses toward stress and environmental stimuli and also regulate gene expression, metabolism, mitosis, apoptosis, cellular motility, and differentiation (Cargnello and Roux 2011; Gehart et al. 2010; Kukkonen-Macchi et al. 2011; Nelson and Fry 2001; Paliwal et al. 2007). MAPKs are highly conserved throughout evolution and also are one of the most widely studied groups of proteins for investigation of physiological responses (Widmann et al. 1999). The p38 MAPK homologs in S. cerevisiae (Hog1p) and N. crassa (OS-2) are involved in cellular responses to hyperosmolarity as well as oxidative stress (Banno et al. 2007; Lamb et al. 2012; Staleva et al. 2004). Previous studies have shown that loss of any of the three genes in the MAPK module (os-4, os-5, and os-2) in N. crassa does not appreciably affect basal hyphal growth but rather leads to fragile conidia, increased sensitivity to hyperosmotic conditions, resistance to the fungicide fludioxonil, and female sterility (Jones et al. 2007; Zhang et al. 2002).
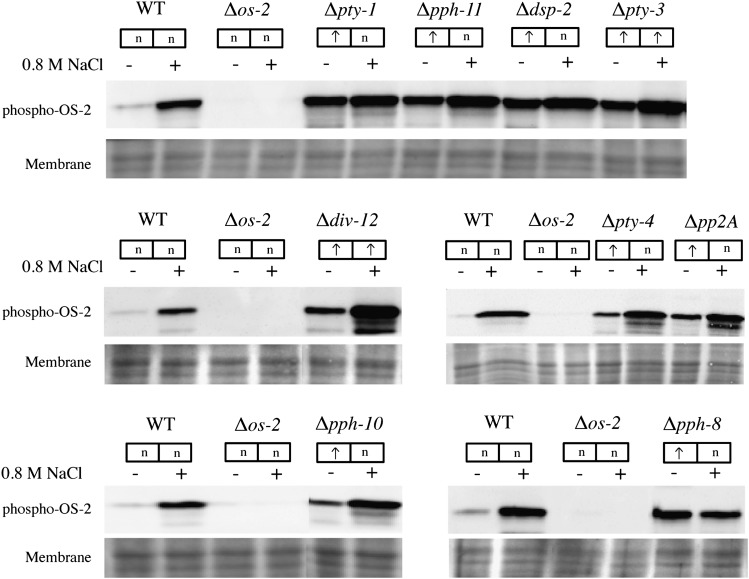
To identify protein phosphatases that may act on the OS MAPK cascade, we analyzed the phosphorylation status of OS-2 in the 24 viable phosphatase knockouts using 0.8 M NaCl for inducing osmotic stress in 16-hr liquid cultures (see Materials and Methods). For detecting the phosphorylated form of OS-2 in the protein samples from the cellular extracts, we used commercially available peptide antibodies raised against the mammalian MAPK phospho-p38. Similar to previous studies (Jones et al. 2007), this antibody was found to cross-react with a N. crassa phosphoprotein of ~41 kDa, which is near the predicted size of OS-2 (Figure 2). We found that nine mutants had elevated basal levels of phospho-OS-2: Δpty-1, Δpph-11, Δdsp-2, Δpty-3, Δdiv-12, Δpty-4, Δpp2A, Δpph-10, and Δpph-8 (Figure 2 and Table 1). However, only Δdiv-12 and Δpty-3 exhibited significantly higher levels of phospho-OS-2 than wild-type after induction using sodium chloride (Figure 2). As mentioned, Δdiv-12 also displays resistance to fludioxonil (Table 3), a phenotype similar to that of mutants lacking os-2, os-4, or os-5. Hence, DIV-12 (a CDC25-type PTP) may play a role in dephosphorylation of one or more of the component kinases of the OS pathway in N. crassa. In budding yeast, Mih1p (a DIV-12 homolog) is involved in dephosphorylation of CDC28 (Russell et al. 1989; Sia et al. 1996) and a role in Hog1p dephosphorylation has also been suggested (Clotet et al. 2006). The other CDC25-type phosphatase in N. crassa, PTY-1, also seems to have functions in the OS-2 pathway, because deletion of pty-1 leads to an increase in the basal levels of phospho-OS-2. We also found that Δpph-10 showed elevated basal phospho-OS-2. The yeast homolog SSU72 phosphatase is primarily involved in transcription termination via removal of phospho-Ser7 marks from the RNA Pol II CTD (Zhang et al. 2012). Our MAPK assays also implicate the SSU72 class of phosphatases in regulation of OS-2 phosphorylation, although such a function might also be imparted via regulation of the transcriptional machinery.
Figure 2.
Analysis of p38 MAPK phosphorylation. Conidia were used to inoculate shaken liquid cultures that were grown for 16 hr at 30°. Cultures were left untreated or brought to 0.8 M NaCl for 10 min to stimulate OS-2 phosphorylation. Phospho-OS-2 levels were analyzed by immunoblotting with a specific antiserum (top panels). A portion of the membrane was excised and stained using amido black to use as a loading control (bottom panels). The experiment was repeated at least three times and a representative blot is shown. The letter “n” signifies that the levels of phospho-OS-2 were similar to wild-type. The arrow signifies that the levels of phospho-OS-2 were elevated as compared to wild-type.
Deletion of the PTPs pty-3 and dsp-2 leads to increased basal levels of phospho-OS-2 in comparison to wild-type (Figure 2). The levels of phospho-OS-2 in the Δpty-3 strain are also elevated on induction using sodium chloride (as compared to wild-type). The yeast pty-3 homolog PTP3 is involved in dephosphorylation of both Hog1p and Slt2p in the cell wall integrity pathway (Hahn and Thiele 2002; Wurgler-Murphy et al. 1997). In contrast, Msg5p, the yeast homolog of DSP-2, is implicated in dephosphorylation of Slt2p and Fus3p in the pheromone-sensing pathway (Andersson et al. 2004; Flandez et al. 2004; Marin et al. 2009). Our results suggest that DSP-2 is also required for inactivation of the OS-2 pathway, and further study of the Erk class MAPKs MAK-1 and MAK-2 in N. crassa may uncover similar functions for this phosphatase as seen in yeast. It is also possible that there is a high degree of crosstalk between the different MAPK cascades, leading to an overlap of function.
Our study of the LMW-PTP pty-4 is especially unique because, so far, a cellular role has not been demonstrated in S. cerevisiae (Ostanin et al. 1995). The N. crassa Δpty-4 mutant exhibits decreased production of protoperithecia and resistance toward menadione and FK-506 (Table 1, Table 3, and Table S1). This suggests that PTY-4 negatively regulates pathways involved in activation of oxidative stress responses. Deletion of pty-4 leads to an increased basal level of phospho-OS-2 as compared to wild-type (Figure 2). These results suggest that PTY-4 may regulate sexual development and ROS sensitivity via OS-2 phosphorylation in N. crassa.
Among the PP2A class of S/T phosphatases, Δpp2A was the only mutant exhibiting elevated basal phospho-OS-2 levels (Figure 2). The finding that the pp2A mutant was sensitive to sodium chloride and sorbitol but resistant to fludioxonil treatment also supports a role as a major phosphatase in the OS-2 pathway (Table 3). The PP2C class phosphatase mutant Δpph-8 and the HAD class mutant Δpph-11 also exhibited elevated levels of basal phospho-OS-2 (Figure 2). The S. cerevisiae homolog of pp2A is PPG1 and that for pph-8 is PTC2; the PPG1 and PTC2 gene products are required for glycogen accumulation and dephosphorylation of Hog1p, respectively (Posas et al. 1993; Young et al. 2002). These results suggest that similar to their homologs in yeast, these phosphatases are bona fide regulators of OS-2 dephosphorylation and have important roles in the MAPK signaling cascade.
Taken together, our assays revealed that 9 out of 24 viable mutants exhibit altered p38 MAPK phosphorylation. Information for a number of these phosphatases in S. cerevisiae indicates that there is considerable crosstalk and/or overlap in function for some of these phosphatases with other MAPK cascades, such as the cell wall integrity pathway (Andersson et al. 2004; Gonzalez et al. 2006; Hahn and Thiele 2002; Isoda et al. 2009). Our results suggest that a similar commonality in function may also exist in N. crassa.
Deletion of the PP2C class protein phosphatase gene pph-8 leads to unregulated protoperithecial development in N. crassa
In N. crassa, the formation of female sexual structures (protoperithecia) is stimulated by growth on SCM containing low nitrogen (Sommer et al. 1987). While performing phenotypic analysis of the phosphatase mutants, we observed that a PP2C class phosphatase knockout mutant (Δpph-8) displayed inappropriate protoperithecial formation on VM medium containing high nitrogen (Figure 3). To further assess this unregulated protoperithecial formation, Δpph-8 was cultured on VM and SCM agar plates in constant light or constant darkness for 5 d to 7 d and then fertilized with an opposite mating type wild-type strain. At 5 d under constant light conditions, the Δpph-8 mutant displayed small protoperithecia that were embedded under the agar surface of both media. At 7 d, Δpph-8 protoperithecia differentiated on VM had enlarged to the same size seen in the wild-type strain at 7 d on SCM medium, whereas Δpph-8 protoperithecia produced on SCM were smaller than wild-type (Figure 3A). After fertilization, the Δpph-8 mutant was able to form mature perithecia (Figure 3A) and shoot ascospores 10 days after fertilization (data not shown). Previous studies have shown that blue light is necessary for photo-induction of protoperithecial development (Innocenti et al. 1983). When cultured in constant darkness, wild-type produced protoperithecia after 7 d on SCM (but not VM), a delay of 2 d relative to constant light conditions (Figure 3B). The Δpph-8 mutant exhibited no delay in protoperithecial development on either VM or SCM in constant darkness (Figure 3B). Interestingly, Δpph-8 protoperithecia formed in constant darkness were slightly larger than those formed in light (Figure 3). Perithecia were produced in wild-type on SCM and the Δpph-8 mutant on both SCM and VM medium in constant darkness (Figure 3). When formed, perithecia from wild-type and the Δpph-8 mutant produced ascospores by 10 d after fertilization (data not shown). However, in contrast to wild-type, Δpph-8 ascospore progeny from VM cultures (light or dark conditions) did not germinate (data not shown).
The results presented suggest that loss of pph-8 significantly affects nitrogen sensing and the sexual development pathway in N. crassa. This mutant also exhibited multiple defects in hyphal growth and asexual sporulation (Table 1 and Table 3). PPH-8 shares a high degree of homology to Ptc2p in S. cerevisiae (Young et al. 2002). Ptc2p dephosphorylates Hog1p as well as Cdc28p, and is also implicated in functioning with proteins such as RAD53 to regulate DNA damage checkpoint pathways (Cheng et al. 1999; Marsolier et al. 2000; Young et al. 2002). As seen from the MAPK assays, Δpph-8 has a high basal level of phospho-OS-2, and levels after treatment with sodium chloride are similar to those of treated wild-type. This suggests that PPH-8 is involved in dephosphorylation of the OS-2 MAPK in N. crassa (Figure 2). Other studies have shown that the os-4/os-5/os-2 mutants are unable to produce protoperithecia (Jones et al. 2007), a phenotype in opposition to that observed for Δpph-8. Hence, it is plausible that the PPH-8 phosphatase regulates protoperithecial development via modulation of the OS-2 MAPK pathway.
The protein phosphatase mutant Δcsp-6 displays a conidial separation defect most similar to Δcsp-1 mutants
Two conidial separation mutants, Δcsp-1 and Δcsp-2, form major constriction chains with double crosswalls in developing conidiophores but no free macroconidia (Selitrennikoff et al. 1974). Genetic and molecular studies that have implicated a number of genes in the macroconidiation pathway place csp-1 and csp-2 downstream of other genes, including acon-2 and fl (Bailey-Shrode and Ebbole 2004; Springer and Yanofsky 1989). The csp-1 gene encodes a light-inducible zinc finger transcription factor, and deletion of csp-1 leads to shortening of the period length for the circadian clock by approximately 1 hr (Lambreghts et al. 2009; Schneider et al. 2009). Recent evidence showed that CSP-1 is a transcription repressor, with its function and abundance coupled to the circadian activity of the white collar complex (WCC), thus constituting an important output for the clock (Sancar et al. 2011; Smith et al. 2010). CSP-1 is primarily involved in ergosterol biosynthesis, modulating the lipid composition of membranes (Sancar et al. 2011; Smith et al. 2010). In contrast to csp-1, deletion of the grainy head transcription factor gene csp-2 lengthens the clock period by 1.5 hr in N. crassa (Brody et al. 2010; Pare et al. 2012). Among its functions, CSP-2 influences expression of genes involved in construction and remodeling of the cell wall (Pare et al. 2012).
Microscopic observation of conidia revealed that Δcsp-6, lacking a HAD class S/T phosphatase, appeared to possess a conidial separation defect reminiscent of Δcsp-1 and Δcsp-2 mutants. Similar to the Δcsp-1 and Δcsp-2 strains, when slant cultures of Δcsp-6 are agitated, no free conidia are released. In addition to a conidial separation defect, the Δcsp-6 mutant exhibited reduced basal hyphal growth and produced few, small protoperithecia that did not develop into mature perithecia after fertilization during sexual development (Table 1 and Table S1). This contrasts with knockout mutants lacking csp-1 or csp-2, which have reduced hyphal growth but do not possess defects in female sexual development (Broad database). To more accurately compare and contrast the conidial separation defects of Δcsp-1, Δcsp-2, and Δcsp-6 strains, we used the fluorescent stain calcofluor white to visualize the cell wall (Fig. 4). On staining with calcofluor white, we found that the Δcsp-2 mutant is able to form numerous double-doublets at interconidial junctions (Fig. 4), consistent with results from a previous study (Springer and Yanofsky 1989). This suggests that the Δcsp-2 mutant is blocked at the double-doublet stage before connective formation takes place. Consistent with previous results, we also observed that the Δcsp-1 mutant displays fewer double-doublets and sometimes does not form septa between macroconidial compartments (Figure 4) (Springer and Yanofsky 1989). In the case of the Δcsp-6 mutant, we observed double-doublets (Fig. 4), but such structures were not as extensive as in Δcsp-2 strains. In addition, the csp-6 mutant sometimes lacked septa between macroconidial compartments in conidiophores (Figure 4). Hence, the conidial separation defect of Δcsp-6 is more similar to Δcsp-1 than to Δcsp-2. This conclusion supports CSP-1 and CSP-6 acting in the same pathway to regulate growth and conidiation, perhaps through dephosphorylation of phosphorylated CSP-1 transcription factor (or a regulated target) by the CSP-6 protein phosphatase.
Discussion
In this study, we examined the role of protein phosphatases in growth and development and regulation of p38 MAPK dephosphorylation in the filamentous fungus, N. crassa. We have identified 30 protein phosphatase genes in the N. crassa genome and found that these genes are highly conserved among humans, plants, and other fungi. In particular, N. crassa phosphatases are in number similar to A. nidulans. Two phosphatases (pty-5 and pty-6) showed little or no homology to genes in yeast, animals, or plants, whereas similar genes are present in A. nidulans, suggesting that these are specific for filamentous fungi. Our results demonstrated that Δpty-5 mutants possess defects in conidiation, whereas strains lacking pty-6 are resistant to fludioxonil. Conidiation is observed in many filamentous fungi, but not baker’s yeast. Likewise, in contrast to many filamentous fungal species, S. cerevisiae is naturally resistant to fludioxonil, apparently lacking the cellular target of this fungicide (Motoyama et al. 2005; Zhang et al. 2002). Future studies will shed light on the cellular pathways impacted by these two tyrosine phosphatases in filamentous fungi.
A majority of protein phosphatase knockouts (91%) exhibited defects in basal growth, asexual development, or sexual development. We found that three mutants (Δpty-2, Δpph-6, and Δppt-1) actually displayed increased basal growth rates compared to wild-type. This is in contrast to our previous study with kinase mutants (Park et al. 2011b), in which all mutants with a basal hyphae growth defect exhibited reduced growth. Because, in general, phosphatases impart their roles by dephosphorylation of their targets, it is likely that phosphatase mutants with increased growth may experience constitutive phosphorylation of targets, leading to unregulated cell proliferation. Interestingly, the Δpty-2 and Δppt-1 mutants were resistant to t-BuOOH, perhaps suggestive of a link between increased growth and oxidative stress resistance.
Morphological analyses of two development pathways in N. crassa showed that certain phosphatases are specific for sexual or asexual development (Figure 1). In particular, four tyrosine phosphatases (pty-3, pty-4, div-12, and dsp-4) are restricted to sexual development, whereas another three tyrosine phosphatases (pty-1, dsp-1, and pty-5) are only involved in asexual differentiation. In contrast, S/T phosphatases seem to have broader roles in fungal development (Figure 1). Deletion of various S/T kinases in N. crassa led to a high proportion (40%) of female-sterile strains (Park et al. 2011b). In contrast, only four phosphatase mutants (Δtng, Δpp2A, Δcsp-6, and Δdsp-2) were female-sterile, representing 16% of the viable phosphatase mutants. This may reflect the antagonistic roles of protein phosphatases and kinases, with constitutive phosphorylation of targets in phosphatase mutants less likely to result in female sterility. Opposing functions for kinases and phosphatases are also manifested by the chemical sensitivity phenotypes. Treatment of S/T kinase mutants with t-BuOOH only revealed strains with increased sensitivity (Park et al. 2011b), whereas 40% of the affected phosphatase mutants displayed a resistant phenotype.
Two of the analyzed protein phosphatase mutants (Δpph-7 and Δpty-6) did not have obvious growth or developmental phenotypes. As in our previous study of kinases, chemical sensitivity assays proved to be an effective tool in assigning a function for such mutants lacking a morphological phenotype. Deletion of pty-6 resulted in increased resistance to the calcineurin inhibitor FK506, whereas the absence of pph-7 rendered the strain sensitive to fludioxonil (which stimulates the OS-2 pathway) (Table 3). In S. cerevisiae, deletion of the pty-6 homolog SIW14 leads to cytoskeletal abnormalities and defective endocytosis (Care et al. 2004). Hence, it is possible that pty-6 and its related phosphatases might regulate cytoskeletal organization in concert with the calcineurin-mediated signaling pathways in N. crassa. In the case of pph-7, an intron in the mRNA of the S. cerevisiae homolog PTC7 is alternatively spliced, producing two protein isoforms (Juneau et al. 2009). The protein derived from the spliced mRNA is localized to the mitochondrion, whereas that produced from the unspliced mRNA is found on the nuclear envelope. The mitochondrial protein is modified in a carbon source–dependent fashion, whereas mutants lacking the version on the nuclear envelope are more sensitive to latrunculin (a chemical that disrupts actin filaments) than wild-type (Juneau et al. 2009). In contrast to its closest yeast homolog, N. crassa pty-6 lacks an intron in the ORF and biochemical studies have localized the protein to the mitochondrion (Keeping et al. 2011). Furthermore, we did not observe altered sensitivity of the Δpty-6 mutant to cytochalasin A, but instead to fludioxonil, which has been shown to activate the OS-2 MAPK pathway, leading to glycerol production. No proteins involved in fludioxonil sensitivity have been localized to the mitochondrion. Our findings support a scenario in which loss of pty-6 leads to elevated production of glycerol in N. crassa. This likely occurs at a point downstream of the OS-2 MAPK, because we observed that Δpty-6 mutants possessed normal basal and induced levels of phospho-OS-2. Concomitant loss of pty-6 and inappropriate activation of the OS-2 MAPK by fludioxonil would render the mutant more sensitive than wild-type.
A number of studies in N. crassa investigating utilization of cellulose as an alternate carbon source have shown that there is an upregulation of lignocellulolytic enzymes when N. crassa is switched from sucrose to cellulose (Sun and Glass 2011; Tian et al. 2009; Wu et al. 2013; Znameroski et al. 2012). Previous work has identified the zinc finger transcription factor CRE-1 as a carbon catabolite repressor, whereby deletion of cre-1 leads to increased expression of cellulolytic genes when N. crassa is grown on the microcrystalline cellulose source, Avicel (Sun and Glass 2011). We have identified two protein phosphatase mutants, Δpph-4 and Δdsp-2, that display increased growth on Avicel, consistent with roles as negative regulators of cellulose utilization. Therefore, it is possible that these phosphatases may operate in the same pathway or play parallel roles with CRE-1 in regulating the transcriptional machinery or downstream events to influence cellulolytic activity in N. crassa.
The phosphatases pp2A (NCU06563) and pzl-1 (NCU07489) belong to the PP2A class of S/T phosphatases, a highly conserved family of proteins with several important functions in cellular signaling, from mammals to fungi (Du et al. 2013; Erental et al. 2007; Seshacharyulu et al. 2013). It is therefore not surprising that deletion of these genes led to several defects in growth and development and also yielded the highest number of chemical sensitivity phenotypes (six for Δpp2A and seven for Δpzl-1). The Δpp2A mutant was sensitive to osmotic stresses, peroxide stress, and ROS, as well as displaying resistance to fludioxonil and benomyl (Table 3). All of these chemical sensitivity phenotypes were observed with high significance and very low p-values, and the p38 MAPK assays further reaffirm the authenticity of the responses for this mutant to the respective chemicals (Table S1). From the p38 MAPK assays, we found that Δpp2A displays elevated levels of basal phospho-OS-2 (Figure 2), and its function in the OS-2 pathway is also reflected by the resistance of the mutant to fludioxonil, similar to the os mutants (Park et al. 2011b; Zhang et al. 2002). It remains to be investigated whether this phosphatase plays a direct role in dephosphorylation of the terminal MAPK OS-2 or is acting on the upstream MAPKKK (OS-4) or MAPKK (OS-5). This is additionally interesting when considering that OS-2 is so tightly connected to circadian rhythm and the WCC. It is already known that WCC is able to exert transcriptional control on phospho-OS-2 expression (Lamb et al. 2011). It is likely that both the catalytic subunit and the regulatory (RGB-1) subunit of PP2A are able to control expression of the phospho-OS-2 MAPK via the WCC, thus providing additional layers of regulation of phospho-OS-2 MAPK expression.
Recent evidence from Sordaria macrospora suggests a role for the pp2A homolog SmPP2Ac in regulating cell–cell fusion and sexual development as an integral component of the STRIPAK complex (Bloemendal et al. 2012). However, the exact role of SmPP2Ac in control of these developmental outcomes remains to be deciphered. It will be interesting to further investigate whether the PP2A has any role in MAK-2 phosphorylation, a protein that is a major component of cell–cell fusion in Neurospora (Fu et al. 2011).
Another phosphatase mutant with interesting phenotypes as well as elevated phospho-OS-2 levels in this study is the PP2C class protein phosphatase mutant Δpph-8. When grown on minimal medium, the mutant displayed inappropriate formation of protoperithecia (Figure 3), similar to the S/T kinase mutant Δime-2 (Hutchison et al. 2012; Hutchison and Glass 2010). One possible scenario is that IME-2 and PPH-8 regulate two different target phosphoproteins with opposing functions on protoperithecial development. Whereas one target protein could inhibit protoperithecial development on dephosphorylation by PPH-8, the other one could repress it on being phosphorylated by IME-2. Genetic epistasis studies should provide further insight into understanding the underlying mechanism of how ime-2 and pph-8 regulate protoperithecial formation in N. crassa.
With the success of the N. crassa gene knockout project, we have focused on analysis of phenotypes for large groups of genes with crucial roles in cellular homeostasis, including transcription factors, S/T protein kinases, and, now, S/T and tyrosine protein phosphatases. Before our study, most of the protein phosphatases in N. crassa had not been characterized. In numerous cases, we now have important clues to their functions. Further studies of these protein phosphatases should provide a greater understanding of how these proteins are able to regulate important cellular roles in N. crassa, related fungi, and other eukaryotic organisms.
Supplementary Material
Acknowledgments
We thank Liubov Litvinkova and Lorena Altamirano for technical assistance during early stages of this work. We acknowledge Gloria Turner, Richard Weiss, and undergraduate students in the Neurospora Genetics and Genomics Summer Research Institute at UCLA for phenotypic analysis of many mutants and many helpful discussions. We thank Michael Plamann, Kevin McCluskey, and Aric Wiest at the Fungal Genetics Stock Center for maintenance of N. crassa knockout mutants. Fludioxonil was a gift from Frank Wong and Allison Tally. This work was supported by P01GM068087 (to K.A.B.).
Footnotes
Communicating editor: J. C. Dunlap
Literature Cited
- Alonso A., Sasin J., Bottini N., Friedberg I., Osterman A., et al. , 2004. Protein tyrosine phosphatases in the human genome. Cell 117: 699–711 [DOI] [PubMed] [Google Scholar]
- Andersen J. N., Mortensen O. H., Peters G. H., Drake P. G., Iversen L. F., et al. , 2001. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 21: 7117–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J., Simpson D. M., Qi M., Wang Y., Elion E. A., 2004. Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J. 23: 2564–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A. V., Kutuzov M. A., 2008. Protozoan protein tyrosine phosphatases. Int. J. Parasitol. 38: 1279–1295 [DOI] [PubMed] [Google Scholar]
- Archambault J., Chambers R. S., Kobor M. S., Ho Y., Cartier M., et al. , 1997. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94: 14300–14305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J., Pan G., Dahmus G. K., Cartier M., Marshall N., et al. , 1998. FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J. Biol. Chem. 273: 27593–27601 [DOI] [PubMed] [Google Scholar]
- Arino J., Casamayor A., Gonzalez A., 2011. Type 2C protein phosphatases in fungi. Eukaryot. Cell 10: 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Shrode L., Ebbole D. J., 2004. The fluffy gene of Neurospora crassa is necessary and sufficient to induce conidiophore development. Genetics 166: 1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay J., Lee J., Lee J. I., Yu J. R., Jee C., et al. , 2002. Calcineurin, a calcium/calmodulin-dependent protein phosphatase, is involved in movement, fertility, egg laying, and growth in Caenorhabditis elegans. Mol. Biol. Cell 13: 3281–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno S., Noguchi R., Yamashita K., Fukumori F., Kimura M., et al. , 2007. Roles of putative His-to-Asp signaling modules HPT-1 and RRG-2, on viability and sensitivity to osmotic and oxidative stresses in Neurospora crassa. Curr. Genet. 51: 197–208 [DOI] [PubMed] [Google Scholar]
- Bauman A. L., Scott J. D., 2002. Kinase- and phosphatase-anchoring proteins: harnessing the dynamic duo. Nat. Cell Biol. 4: E203–E206 [DOI] [PubMed] [Google Scholar]
- Bewick V., Cheek L., Ball J., 2004. Statistics review 9: one-way analysis of variance. Crit. Care 8: 130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialojan C., Takai A., 1988. Inhibitory effect of a marine-sponge toxin, okadaic acid, on protein phosphatases. Specificity and kinetics. Biochem. J. 256: 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemendal S., Bernhards Y., Bartho K., Dettmann A., Voigt O., et al. , 2012. A homologue of the human STRIPAK complex controls sexual development in fungi. Mol. Microbiol. 84: 310–323 [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., et al. , 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68: 1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., et al. , 2010. A global protein kinase and phosphatase interaction network in yeast. Science 328: 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody S., Oelhafen K., Schneider K., Perrino S., Goetz A., et al. , 2010. Circadian rhythms in Neurospora crassa: Downstream effectors. Fungal Genet. Biol. 47: 159–168 [DOI] [PubMed] [Google Scholar]
- Burgess A., Vigneron S., Brioudes E., Labbe J. C., Lorca T., et al. , 2010. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. USA 107: 12564–12569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A., Vousden K. A., Binley K. M., Radcliffe P., Trevethick J., et al. , 2004. A synthetic lethal screen identifies a role for the cortical actin patch/endocytosis complex in the response to nutrient deprivation in Saccharomyces cerevisiae. Genetics 166: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello M., Roux P. P., 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75: 50–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Ross K. E., Kaldis P., Solomon M. J., 1999. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 13: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho U. S., Xu W., 2007. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445: 53–57 [DOI] [PubMed] [Google Scholar]
- Choi J. H., Adames N. R., Chan T. F., Zeng C., Cooper J. A., et al. , 2000. TOR signaling regulates microtubule structure and function. Curr. Biol. 10: 861–864 [DOI] [PubMed] [Google Scholar]
- Clotet J., Escote X., Adrover M. A., Yaakov G., Gari E., et al. , 2006. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J. 25: 2338–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P., 1990. The structure and regulation of protein phosphatases. Adv. Second Messenger Phosphoprotein Res. 24: 230–235 [PubMed] [Google Scholar]
- Cohen P., 2001. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur. J. Biochem. 268: 5001–5010 [DOI] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. A., 1987. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105: 1473–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree G. R., 1999. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell 96: 611–614 [DOI] [PubMed] [Google Scholar]
- Cyert M. S., Thorner J., 1992. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12: 3460–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert M. S., Kunisawa R., Kaim D., Thorner J., 1991. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. USA 88: 7376–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., DeSerres F. J., 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17: 79–143 [Google Scholar]
- Davis R. H., Perkins D. D., 2002. Timeline: Neurospora: a model of model microbes. Nat. Rev. Genet. 3: 397–403 [DOI] [PubMed] [Google Scholar]
- Dev K., Maheshwari R., 2002. Transformation in heterokaryons of Neurospora crassa is nuclear rather than cellular phenomenon. Curr. Microbiol. 44: 309–313 [DOI] [PubMed] [Google Scholar]
- Di Stasio M., Brefort T., Mendoza-Mendoza A., Munch K., Kahmann R., 2009. The dual specificity phosphatase Rok1 negatively regulates mating and pathogenicity in Ustilago maydis. Mol. Microbiol. 73: 73–88 [DOI] [PubMed] [Google Scholar]
- Du Y., Shi Y., Yang J., Chen X., Xue M., et al. , 2013. A serine/threonine-protein phosphatase PP2A catalytic subunit is essential for asexual development and plant infection in Magnaporthe oryzae. Curr. Genet. 59: 33–41 [DOI] [PubMed] [Google Scholar]
- Dumont F. J., Staruch M. J., Koprak S. L., Melino M. R., Sigal N. H., 1990. Distinct mechanisms of suppression of murine T cell activation by the related macrolides FK-506 and rapamycin. J. Immunol. 144: 251–258 [PubMed] [Google Scholar]
- Elchebly M., Payette P., Michaliszyn E., Cromlish W., Collins S., et al. , 1999. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 283: 1544–1548 [DOI] [PubMed] [Google Scholar]
- Erental A., Harel A., Yarden O., 2007. Type 2A phosphoprotein phosphatase is required for asexual development and pathogenesis of Sclerotinia sclerotiorum. Mol. Plant Microbe Interact. 20: 944–954 [DOI] [PubMed] [Google Scholar]
- Ernsting B. R., Dixon J. E., 1997. The PPS1 gene of Saccharomyces cerevisiae codes for a dual specificity protein phosphatase with a role in the DNA synthesis phase of the cell cycle. J. Biol. Chem. 272: 9332–9343 [DOI] [PubMed] [Google Scholar]
- Farcasanu I. C., Hirata D., Tsuchiya E., Nishiyama F., Miyakawa T., 1995. Protein phosphatase 2B of Saccharomyces cerevisiae is required for tolerance to manganese, in blocking the entry of ions into the cells. Eur. J. Biochem. 232: 712–717 [PubMed] [Google Scholar]
- Fasolo J., Sboner A., Sun M. G., Yu H., Chen R., et al. , 2011. Diverse protein kinase interactions identified by protein microarrays reveal novel connections between cellular processes. Genes Dev. 25: 767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z. H., Wilson S. E., Peng Z. Y., Schlender K. K., Reimann E. M., et al. , 1991. The yeast GLC7 gene required for glycogen accumulation encodes a type 1 protein phosphatase. J. Biol. Chem. 266: 23796–23801 [PubMed] [Google Scholar]
- Flandez M., Cosano I. C., Nombela C., Martin H., Molina M., 2004. Reciprocal regulation between Slt2 MAPK and isoforms of Msg5 dual-specificity protein phosphatase modulates the yeast cell integrity pathway. J. Biol. Chem. 279: 11027–11034 [DOI] [PubMed] [Google Scholar]
- Fu C., Iyer P., Herkal A., Abdullah J., Stout A., et al. , 2011. Identification and characterization of genes required for cell-to-cell fusion in Neurospora crassa. Eukaryot. Cell 10: 1100–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D., et al. , 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868 [DOI] [PubMed] [Google Scholar]
- Ganem C., Devaux F., Torchet C., Jacq C., Quevillon-Cheruel S., et al. , 2003. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 22: 1588–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Engele P., Moilanen B., Cyert M. S., 1995. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H(+)-ATPase. Mol. Cell. Biol. 15: 4103–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J., Solomon M. J., Booher R. N., Bazan J. F., Kirschner M. W., 1991. Cdc25 is a specific tyrosine phosphatase that directly activates P34cdc2. Cell 67: 197–211 [DOI] [PubMed] [Google Scholar]
- Gehart H., Kumpf S., Ittner A., Ricci R., 2010. MAPK signalling in cellular metabolism: stress or wellness? EMBO Rep. 11: 834–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohla A., Birkenfeld J., Bokoch G. M., 2005. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat. Cell Biol. 7: 21–29 [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Ruiz A., Serrano R., Arino J., Casamayor A., 2006. Transcriptional profiling of the protein phosphatase 2C family in yeast provides insights into the unique functional roles of Ptc1. J. Biol. Chem. 281: 35057–35069 [DOI] [PubMed] [Google Scholar]
- Haas F., Mitchell M. B., Ames B. N., Mitchell H. K., 1952. A series of histidineless mutants of Neurospora crassa. Genetics 37: 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J. S., Thiele D. J., 2002. Regulation of the Saccharomyces cerevisiae Slt2 kinase pathway by the stress-inducible Sdp1 dual specificity phosphatase. J. Biol. Chem. 277: 21278–21284 [DOI] [PubMed] [Google Scholar]
- Hansberg W., de Groot H., Sies H., 1993. Reactive oxygen species associated with cell differentiation in Neurospora crassa. Free Radic. Biol. Med. 14: 287–293 [DOI] [PubMed] [Google Scholar]
- Herzig S., Neumann J., 2000. Effects of serine/threonine protein phosphatases on ion channels in excitable membranes. Physiol. Rev. 80: 173–210 [DOI] [PubMed] [Google Scholar]
- Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., et al. , 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. Y., Tan T. H., 2012. DUSPs, to MAP kinases and beyond. Cell Biosci. 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T., 1995. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80: 225–236 [DOI] [PubMed] [Google Scholar]
- Hutchison E. A., Bueche J. A., Glass N. L., 2012. Diversification of a protein kinase cascade: IME-2 is involved in nonself recognition and programmed cell death in Neurospora crassa. Genetics 192: 467–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison E. A., Glass N. L., 2010. Meiotic regulators Ndt80 and ime2 have different roles in Saccharomyces and Neurospora. Genetics 185: 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti F. D., Pohl U., Russo V. E., 1983. Photoinduction of protoperithecia in Neurospora crassa by blue light. Photochem. Photobiol. 37: 49–51 [DOI] [PubMed] [Google Scholar]
- Isoda M., Kanemori Y., Nakajo N., Uchida S., Yamashita K., et al. , 2009. The extracellular signal-regulated kinase-mitogen-activated protein kinase pathway phosphorylates and targets Cdc25A for SCF beta-TrCP-dependent degradation for cell cycle arrest. Mol. Biol. Cell 20: 2186–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Goris J., 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353: 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey K. L., Camps M., Rommel C., Mackay C. R., 2007. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat. Rev. Drug Discov. 6: 391–403 [DOI] [PubMed] [Google Scholar]
- Jones C. A., Borkovich K. A., 2010. Analysis of mitogen-activated protein kinase phosphorylation in response to stimulation of histidine kinase signaling pathways in Neurospora. Methods Enzymol. 471: 319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. A., Greer-Phillips S. E., Borkovich K. A., 2007. The response regulator RRG-1 functions upstream of a mitogen-activated protein kinase pathway impacting asexual development, female fertility, osmotic stress, and fungicide resistance in Neurospora crassa. Mol. Biol. Cell 18: 2123–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau K., Nislow C., Davis R. W., 2009. Alternative splicing of PTC7 in Saccharomyces cerevisiae determines protein localization. Genetics 183: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura A., Su M. S., 1995. Interaction of FKBP12-FK506 with calcineurin A at the B subunit-binding domain. J. Biol. Chem. 270: 15463–15466 [DOI] [PubMed] [Google Scholar]
- Keeping A., Deabreu D., Dibernardo M., Collins R. A., 2011. Gel-based mass spectrometric and computational approaches to the mitochondrial proteome of Neurospora. Fungal Genet. Biol. 48: 526–536 [DOI] [PubMed] [Google Scholar]
- Kerk D., Templeton G., Moorhead G. B., 2008. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 146: 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Chan K. L., Faria N. C., Martins Mde L., Campbell B. C., 2012. Targeting the oxidative stress response system of fungi with redox-potent chemosensitizing agents. Front. Microbiol. 3: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee C. B., Draetta G. F., Hubbard M. J., 1988. Calcineurin. Adv. Enzymol. Relat. Areas Mol. Biol. 61: 149–200 [DOI] [PubMed] [Google Scholar]
- Klee C. B., Ren H., Wang X., 1998. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 273: 13367–13370 [DOI] [PubMed] [Google Scholar]
- Kobor M. S., Archambault J., Lester W., Holstege F. C., Gileadi O., et al. , 1999. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 4: 55–62 [DOI] [PubMed] [Google Scholar]
- Kosti I., Mandel-Gutfreund Y., Glaser F., Horwitz B. A., 2010. Comparative analysis of fungal protein kinases and associated domains. BMC Genomics 11: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe G. O., Free S. J., 1998a Calcineurin subunit B is required for normal vegetative growth in Neurospora crassa. Fungal Genet. Biol. 23: 248–258 [DOI] [PubMed] [Google Scholar]
- Kothe G. O., Free S. J., 1998b The isolation and characterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa. Genetics 149: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystofova S., Borkovich K. A., 2005. The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gbetagamma dimer required for normal female fertility, asexual development, and galpha protein levels in Neurospora crassa. Eukaryot. Cell 4: 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukkonen-Macchi A., Sicora O., Kaczynska K., Oetken-Lindholm C., Pouwels J., et al. , 2011. Loss of p38gamma MAPK induces pleiotropic mitotic defects and massive cell death. J. Cell Sci. 124: 216–227 [DOI] [PubMed] [Google Scholar]
- Lamb T. M., Finch K. E., Bell-Pedersen D., 2012. The Neurospora crassa OS MAPK pathway-activated transcription factor ASL-1 contributes to circadian rhythms in pathway responsive clock-controlled genes. Fungal Genet. Biol. 49: 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb T. M., Goldsmith C. S., Bennett L., Finch K. E., Bell-Pedersen D., 2011. Direct transcriptional control of a p38 MAPK pathway by the circadian clock in Neurospora crassa. PLoS ONE 6: e27149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambreghts R., Shi M., Belden W. J., Decaprio D., Park D., et al. , 2009. A high-density single nucleotide polymorphism map for Neurospora crassa. Genetics 181: 767–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy C., Lee S. E., Vaze M. B., Ochsenbein F., Guerois R., et al. , 2003. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 11: 827–835 [DOI] [PubMed] [Google Scholar]
- Li D., Bobrowicz P., Wilkinson H. H., Ebbole D. J., 2005. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 170: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Kao T. P., Huang H., 2008. CDK1 promotes cell proliferation and survival via phosphorylation and inhibition of FOXO1 transcription factor. Oncogene 27: 4733–4744 [DOI] [PubMed] [Google Scholar]
- Loor G., Kondapalli J., Schriewer J. M., Chandel N. S., Vanden Hoek T. L., et al. , 2010. Menadione triggers cell death through ROS-dependent mechanisms involving PARP activation without requiring apoptosis. Free Radic. Biol. Med. 49: 1925–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorca T., Bernis C., Vigneron S., Burgess A., Brioudes E., et al. , 2010. Constant regulation of both the MPF amplification loop and the Greatwall-PP2A pathway is required for metaphase II arrest and correct entry into the first embryonic cell cycle. J. Cell Sci. 123: 2281–2291 [DOI] [PubMed] [Google Scholar]
- Lu G., Wang Y., 2008. Functional diversity of mammalian type 2C protein phosphatase isoforms: new tales from an old family. Clin. Exp. Pharmacol. Physiol. 35: 107–112 [DOI] [PubMed] [Google Scholar]
- Lu X., Bocangel D., Nannenga B., Yamaguchi H., Appella E., et al. , 2004. The p53-induced oncogenic phosphatase PPM1D interacts with uracil DNA glycosylase and suppresses base excision repair. Mol. Cell 15: 621–634 [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., et al. , 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Marin M. J., Flandez M., Bermejo C., Arroyo J., Martin H., et al. , 2009. Different modulation of the outputs of yeast MAPK-mediated pathways by distinct stimuli and isoforms of the dual-specificity phosphatase Msg5. Mol. Genet. Genomics 281: 345–359 [DOI] [PubMed] [Google Scholar]
- Marsolier M. C., Roussel P., Leroy C., Mann C., 2000. Involvement of the PP2C-like phosphatase Ptc2p in the DNA checkpoint pathways of Saccharomyces cerevisiae. Genetics 154: 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey K, Wiest AE, Grigoriev IV, Lipzen A, Martin J, et al. 2011. Rediscovery by whole genome sequencing: classical mutations and genome polymorphisms in Neurospora crassa G3 (Bethesda) 1: 303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J. L., Wadzinski B. E., 2009. Targeting protein serine/threonine phosphatases for drug development. Mol. Pharmacol. 75: 1249–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G. B., Trinkle-Mulcahy L., Ulke-Lemee A., 2007. Emerging roles of nuclear protein phosphatases. Nat. Rev. Mol. Cell Biol. 8: 234–244 [DOI] [PubMed] [Google Scholar]
- Moorhead G. B., De Wever V., Templeton G., Kerk D., 2009. Evolution of protein phosphatases in plants and animals. Biochem. J. 417: 401–409 [DOI] [PubMed] [Google Scholar]
- Motoyama T., Ohira T., Kadokura K., Ichiishi A., Fujimura M., et al. , 2005. An Os-1 family histidine kinase from a filamentous fungus confers fungicide-sensitivity to yeast. Curr. Genet. 47: 298–306 [DOI] [PubMed] [Google Scholar]
- Mulet J. M., Martin D. E., Loewith R., Hall M. N., 2006. Mutual antagonism of target of rapamycin and calcineurin signaling. J. Biol. Chem. 281: 33000–33007 [DOI] [PubMed] [Google Scholar]
- Mustelin T., Alonso A., Bottini N., Huynh H., Rahmouni S., et al. , 2004. Protein tyrosine phosphatases in T cell physiology. Mol. Immunol. 41: 687–700 [DOI] [PubMed] [Google Scholar]
- Nelson J. M., Fry D. W., 2001. Akt, MAPK (Erk1/2), and p38 act in concert to promote apoptosis in response to ErbB receptor family inhibition. J. Biol. Chem. 276: 14842–14847 [DOI] [PubMed] [Google Scholar]
- Ochiai N., Fujimura M., Motoyama T., Ichiishi A., Usami R., et al. , 2001. Characterization of mutations in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest Manag. Sci. 57: 437–442 [DOI] [PubMed] [Google Scholar]
- Ofek P., Ben-Meir D., Kariv-Inbal Z., Oren M., Lavi S., 2003. Cell cycle regulation and p53 activation by protein phosphatase 2C alpha. J. Biol. Chem. 278: 14299–14305 [DOI] [PubMed] [Google Scholar]
- Ostanin K., Pokalsky C., Wang S., Van Etten R. L., 1995. Cloning and characterization of a Saccharomyces cerevisiae gene encoding the low molecular weight protein-tyrosine phosphatase. J. Biol. Chem. 270: 18491–18499 [DOI] [PubMed] [Google Scholar]
- Paietta J. V., Marzluf G. A., 1985. Gene disruption by transformation in Neurospora crassa. Mol. Cell. Biol. 5: 1554–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal S., Iglesias P. A., Campbell K., Hilioti Z., Groisman A., et al. , 2007. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature 446: 46–51 [DOI] [PubMed] [Google Scholar]
- Pandey A., Roca M. G., Read N. D., Glass N. L., 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3: 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao L. I., Badour K., Siminovitch K. A., Neel B. G., 2007. Nonreceptor protein-tyrosine phosphatases in immune cell signaling. Annu. Rev. Immunol. 25: 473–523 [DOI] [PubMed] [Google Scholar]
- Pare A., Kim M., Juarez M. T., Brody S., McGinnis W., 2012. The functions of grainy head-like proteins in animals and fungi and the evolution of apical extracellular barriers. PLoS ONE 7: e36254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G., Colot H. V., Collopy P. D., Krystofova S., Crew C., et al. , 2011a High-throughput production of gene replacement mutants in Neurospora crassa. Methods Mol. Biol. 722: 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G., Servin J. A., Turner G. E., Altamirano L., Colot H. V., et al. , 2011b Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot. Cell 10: 1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F., Bollen M., Stalmans W., Arino J., 1995. Biochemical characterization of recombinant yeast PPZ1, a protein phosphatase involved in salt tolerance. FEBS Lett. 368: 39–44 [DOI] [PubMed] [Google Scholar]
- Posas F., Clotet J., Muns M. T., Corominas J., Casamayor A., et al. , 1993. The gene PPG encodes a novel yeast protein phosphatase involved in glycogen accumulation. J. Biol. Chem. 268: 1349–1354 [PubMed] [Google Scholar]
- Prokisch H., Yarden O., Dieminger M., Tropschug M., Barthelmess I. B., 1997. Impairment of calcineurin function in Neurospora crassa reveals its essential role in hyphal growth, morphology and maintenance of the apical Ca2+ gradient. Mol. Gen. Genet. 256: 104–114 [DOI] [PubMed] [Google Scholar]
- Raju N. B., Leslie J. F., 1992. Cytology of recessive sexual-phase mutants from wild strains of Neurospora crassa. Genome 35: 815–826 [DOI] [PubMed] [Google Scholar]
- Rusnak F., Mertz P., 2000. Calcineurin: form and function. Physiol. Rev. 80: 1483–1521 [DOI] [PubMed] [Google Scholar]
- Russell P., Moreno S., Reed S. I., 1989. Conservation of mitotic controls in fission and budding yeasts. Cell 57: 295–303 [DOI] [PubMed] [Google Scholar]
- Sakumoto N., Matsuoka I., Mukai Y., Ogawa N., Kaneko Y., et al. , 2002. A series of double disruptants for protein phosphatase genes in Saccharomyces cerevisiae and their phenotypic analysis. Yeast 19: 587–599 [DOI] [PubMed] [Google Scholar]
- Sancar G., Sancar C., Brugger B., Ha N., Sachsenheimer T., et al. , 2011. A global circadian repressor controls antiphasic expression of metabolic genes in Neurospora. Mol. Cell 44: 687–697 [DOI] [PubMed] [Google Scholar]
- Sanvoisin J., Gani D., 2001. Protein phosphatase 1 catalyses the direct hydrolytic cleavage of phosphate monoester in a ternary complex mechanism. Bioorg. Med. Chem. Lett. 11: 471–474 [DOI] [PubMed] [Google Scholar]
- Schafmeier T., Haase A., Kaldi K., Scholz J., Fuchs M., et al. , 2005. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122: 235–246 [DOI] [PubMed] [Google Scholar]
- Schneider K., Perrino S., Oelhafen K., Li S., Zatsepin A., et al. , 2009. Rhythmic conidiation in constant light in vivid mutants of Neurospora crassa. Genetics 181: 917–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifried A., Schultz J., Gohla A., 2013. Human HAD phosphatases: structure, mechanism, and roles in health and disease. FEBS J. 280: 549–571 [DOI] [PubMed] [Google Scholar]
- Selitrennikoff C. P., Nelson R. E., Siegel R. W., 1974. Phase-specific genes for macroconidiation in Neurospora crassa. Genetics 78: 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshacharyulu P., Pandey P., Datta K., Batra S. K., 2013. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 335: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., 2009. Serine/threonine phosphatases: mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Sia R. A., Herald H. A., Lew D. J., 1996. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell 7: 1657–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Hurt E. C., Pelham H. R., 2000. Psr1p/Psr2p, two plasma membrane phosphatases with an essential DXDX(T/V) motif required for sodium stress response in yeast. J. Biol. Chem. 275: 19352–19360 [DOI] [PubMed] [Google Scholar]
- Smith K. M., Sancar G., Dekhang R., Sullivan C. M., Li S., et al. , 2010. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for Neurospora white collar complex. Eukaryot. Cell 9: 1549–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T., Degliinnocenti F., Russo V. E. A., 1987. Role of nitrogen in the photoinduction of protoperithecia and carotenoids in Neurospora crassa. Planta 170: 205–208 [DOI] [PubMed] [Google Scholar]
- Son S., Osmani S. A., 2009. Analysis of all protein phosphatase genes in Aspergillus nidulans identifies a new mitotic regulator, fcp1. Eukaryot. Cell 8: 573–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. L., 1993. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. Bioessays 15: 365–374 [DOI] [PubMed] [Google Scholar]
- Springer M. L., Yanofsky C., 1989. A morphological and genetic analysis of conidiophore development in Neurospora crassa. Genes Dev. 3: 559–571 [DOI] [PubMed] [Google Scholar]
- Staleva L., Hall A., Orlow S. J., 2004. Oxidative stress activates FUS1 and RLM1 transcription in the yeast Saccharomyces cerevisiae in an oxidant-dependent manner. Mol. Biol. Cell 15: 5574–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A., 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38: 203–232 [DOI] [PubMed] [Google Scholar]
- St. Lawrence P., Maling B. D., Altwerger L., Rachmeler M., 1964. Mutational alteration of permeability in Neurospora: effects on growth and the uptake of certain amino acids and related compounds. Genetics 50: 1383–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker A. W., 2005. Protein tyrosine phosphatases and signalling. J. Endocrinol. 185: 19–33 [DOI] [PubMed] [Google Scholar]
- Sun J., Glass N. L., 2011. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS ONE 6: e25654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G. S., Liu Y., Baskerville C., Charbonneau H., 1997. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J. Biol. Chem. 272: 24054–24063 [DOI] [PubMed] [Google Scholar]
- Tian C., Beeson W. T., Iavarone A. T., Sun J., Marletta M. A., et al. , 2009. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. USA 106: 22157–22162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo I., Aguirre J., Hansberg W., 1994. Enzyme inactivation related to a hyperoxidant state during conidiation of Neurospora crassa. Microbiology 140(Pt 9): 2391–2397 [DOI] [PubMed] [Google Scholar]
- Tonks N. K., 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7: 833–846 [DOI] [PubMed] [Google Scholar]
- Turner G. E., 2011. Phenotypic analysis of Neurospora crassa gene deletion strains. Methods Mol. Biol. 722: 191–198 [DOI] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora. Microbial. Genet. Bull. 13: 42–46 [Google Scholar]
- Warmka J., Hanneman J., Lee J., Amin D., Ota I., 2001. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 21: 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. Neurospora-V - a Synthetic Medium Favoring Sexual Reproduction. Am. J. Bot. 34: 573–577 [Google Scholar]
- Widmann C., Gibson S., Jarpe M. B., Johnson G. L., 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79: 143–180 [DOI] [PubMed] [Google Scholar]
- Willhite C. C., 1983. Benomyl. J. Appl. Toxicol. 3: 261–264 [DOI] [PubMed] [Google Scholar]
- Williams N. H., 2004. Models for biological phosphoryl transfer. Biochim. Biophys. Acta 1697: 279–287 [DOI] [PubMed] [Google Scholar]
- Wu W., Hildebrand A., Kasuga T., Xiong X., Fan Z., 2013. Direct cellobiose production from cellulose using sextuple beta-glucosidase gene deletion Neurospora crassa mutants. Enzyme Microb. Technol. 52: 184–189 [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy S. M., Maeda T., Witten E. A., Saito H., 1997. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol. Cell. Biol. 17: 1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Xu Y., Chen Y., Jeffrey P. D., Chao Y., et al. , 2006. Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell 127: 341–353 [DOI] [PubMed] [Google Scholar]
- Xu Y., Xing Y., Chen Y., Chao Y., Lin Z., et al. , 2006. Structure of the protein phosphatase 2A holoenzyme. Cell 127: 1239–1251 [DOI] [PubMed] [Google Scholar]
- Yang Y., He Q., Cheng P., Wrage P., Yarden O., et al. , 2004. Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes Dev. 18: 255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatzkan E., Szoor B., Feher Z., Dombradi V., Yarden O., 1998. Protein phosphatase 2A is involved in hyphal growth of Neurospora crassa. Mol. Gen. Genet. 259: 523–531 [DOI] [PubMed] [Google Scholar]
- Yenush L., Mulet J. M., Arino J., Serrano R., 2002. The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity and cell cycle progression. EMBO J. 21: 920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K., Tanaka T., Ida Y., Furusawa C., Hirasawa T., et al. , 2011. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast 28: 349–361 [DOI] [PubMed] [Google Scholar]
- Young C., Mapes J., Hanneman J., Al-Zarban S., Ota I., 2002. Role of Ptc2 type 2C Ser/Thr phosphatase in yeast high-osmolarity glycerol pathway inactivation. Eukaryot. Cell 1: 1032–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X. L., Hong Y., Zhu T., Mitchell A. P., Deschenes R. J., et al. , 2000. Essential functions of protein tyrosine phosphatases PTP2 and PTP3 and RIM11 tyrosine phosphorylation in Saccharomyces cerevisiae meiosis and sporulation. Mol. Biol. Cell 11: 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. W., Mosley A. L., Ramisetty S. R., Rodriguez-Molina J. B., Washburn M. P., et al. , 2012. Ssu72 Phosphatase-dependent Erasure of Phospho-Ser7 Marks on the RNA Polymerase II C-terminal Domain Is Essential for Viability and Transcription Termination. J. Biol. Chem. 287: 8541–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Liu J., Kim Y., Dixon J. E., Pfaff S. L., et al. , 2010. Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1. Protein Sci. 19: 974–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lamm R., Pillonel C., Lam S., Xu J. R., 2002. Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl. Environ. Microbiol. 68: 532–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Znameroski E. A., Coradetti S. T., Roche C. M., Tsai J. C., Iavarone A. T., et al. , 2012. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc. Natl. Acad. Sci. USA 109: 6012–6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.