Abstract
Mitotic chromosome condensation is a prerequisite for the accurate segregation of chromosomes during cell division, and the conserved condensin complex a central player of this process. However, how condensin binds chromatin and shapes mitotic chromosomes remain poorly understood. Recent genome-wide binding studies showing that in most species condensin is enriched near highly expressed genes suggest a conserved link between condensin occupancy and high transcription rates. To gain insight into the mechanisms of condensin binding and mitotic chromosome condensation, we searched for factors that collaborate with condensin through a synthetic lethal genetic screen in the fission yeast Schizosaccharomyces pombe. We isolated novel mutations affecting condensin, as well as mutations in four genes not previously implicated in mitotic chromosome condensation in fission yeast. These mutations cause chromosome segregation defects similar to those provoked by defects in condensation. We also identified a suppressor of the cut3-477 condensin mutation, which largely rescued chromosome segregation during anaphase. Remarkably, of the five genes identified in this study, four encode transcription co-factors. Our results therefore provide strong additional evidence for a functional connection between chromosome condensation and transcription.
Keywords: condensin, fission yeast, synthetic lethality, mitotic chromosome condensation
Mitotic entry is characterized by the reorganization of long and entangled chromatin fibers into individualized, compact and rod-shaped mitotic chromosomes. This large-scale reorganization, called mitotic chromosome condensation, is essential for accurate chromosome segregation and is thought to serve at least two essential purposes: the removal of interchromosomal and interchromatids DNA catenations and the shortening of chromosome arms so that sister DNA molecules efficiently separate during anaphase. Although first described more than a century ago, the molecular mechanisms underlying mitotic chromosome condensation remain elusive.
From yeasts to human, mitotic chromosome condensation relies on two chromosomal components named topoisomerase II (topo II) and condensin (Baxter and Aragon 2012; Hirano 2012; Piazza et al. 2013). Condensin and/or topo II deficiency alters the structural integrity of chromosomes, leading to a failure to separate chromosomes arms and to the formation of chromatin bridges in anaphase. Although clear evidence now exists that condensin collaborates with topo II to remove catenations between chromatids (Baxter et al. 2011), how condensin associates with chromatin and, in this context, shapes mitotic chromosomes in a cell cycle−regulated manner remains poorly understood.
Condensin is a proteinaceous ring composed of two Smc ATPases, Cut3/Smc4/CAP-C and Cut14/Smc2/CAP-E, associated with three non-SMC auxiliary subunits (Hirano 2012; Piazza et al. 2013). Smc2 and Smc4 interact through their hinge domains, forming a V-shaped heterodimer with the two ATPase domains facing each other at the apices of two 50-nm-long coiled coil arms. The kleisin subunit Cnd2/Barren/CAP-H bridges the Smc2/Smc4 ATPase heads and recruits two additional subunits.
Condensin belongs to the family of structural maintenance of chromosomes (SMC) complexes, which includes the cohesin ring that mediates sister-chromatid cohesion. Like cohesin, condensin binds to chromosomes by encircling DNA, and this topological association appears important for chromosome condensation (Cuylen et al. 2011). On the basis of these observations, a structural model has been proposed whereby the role of condensin in chromosome condensation is to create and/or stabilize chromatin loops by encircling one or two chromatin fibers (Losada and Hirano 2005; Cuylen and Haering 2011). However, in human cells, ~80% of condensin I (the human counterpart of yeasts condensin) exchanges dynamically from mitotic chromosomes in early mitosis whereas the remaining ~20% are more stably bound (Gerlich et al. 2006), which suggests that at least two modes of interaction with chromatin may coexist and that condensin may drive chromosome condensation through a more dynamic and perhaps more complex mechanism. In line with this, condensin possesses an ATP-dependent DNA supercoiling activity stimulated by phosphorylation by the mitotic kinases cyclin-dependent kinase (CDK)1/Cdc2 and Polo (Kimura et al. 1998; St-Pierre et al. 2009; Piazza et al. 2013) and capable of driving decatenation by topo II during mitosis in vivo (Baxter et al. 2011). An enzymatic model proposes that condensin-mediated positive supercoiling of DNA is the driving force of mitotic chromosome condensation (Baxter and Aragon 2012). The different dynamics exhibited by chromosomal condensin complexes may reflect the coexistence of both an enzymatic and a more structural role.
With the notable exception of budding and fission yeasts, most eukaryotic species possess a second condensin complex, called condensin II, that differs from the canonical condensin (also known as condensin I) by a distinct set of three auxiliary, non-SMC proteins (Hirano 2012). Condensin I and II do not colocalize along the longitudinal axes of sister-chromatids, differ in the spatiotemporal regulation of their localizations, in their dynamics of chromatin association (condensin II being more stably bound), and in their respective contribution to mitotic chromosome formation (Hirano 2012). Thus, non-SMC subunits must play a key role in determining where, when, and how condensins associate with chromatin. However, the underlying mechanisms remain poorly understood.
Chromatin environment can also influence the association and/or activity of condensin complexes (Xing et al. 2008; Tanaka et al. 2012). Human condensin II binds monomethylated H4-K20 and tends to colocalize with this mark on chromosomes (Liu et al. 2010). Both human condensin I and fission yeast condensin bind histone H2A or its variant H2A.Z, and these interactions are stimulated by the phosphorylation of the kleisin Cnd2/Barren/CAP-H by Aurora-B kinase (Tada et al. 2011). Although condensin associates with nucleosomes, its distribution along chromosomes is not homogenous. Genome-wide binding studies in yeasts and chicken DT40 cells have shown that yeast condensin and vertebrate condensin I are enriched at centromeres and near highly expressed genes along chromosome arms (D’Ambrosio et al. 2008; Kim et al. 2013). Condensin binding near transfer RNA genes and at the 35S ribosomal DNA (rDNA) relies on the transcription factors TFIIIC and Acr1, respectively (D’Ambrosio et al. 2008; Nakazawa et al. 2008; Tada et al. 2011). Furthermore, a physical interaction has been described between condensin and TFIIIC in budding and fission yeasts (Haeusler et al. 2008; Iwasaki et al. 2010). Yet, transcription has been shown to preclude condensin binding (Clemente-Blanco et al. 2009, 2011), and condensin accumulates on chromatin during mitosis, when transcription is generally down-regulated. The biological significance of this apparent paradox is not yet known.
Potentially adding further complexity to the picture are reports that depletion of Smc4 or Smc2 delays, but does not prevent, the formation of compacted metaphase chromosomes in Caenorhabditis elegans, chicken, and human cells (Hudson et al. 2009). However, those condensin-depleted chromosomes lack structural integrity and form extensive chromatin bridges during anaphase. These observations led to the proposal that condensin’s primary function is to preserve the structural integrity of the mitotic chromosome during anaphase while an as-yet-identified activity called regulator of chromosome architecture (RCA) ensures the initial compaction of chromatin when CDK activity is high (Vagnarelli et al. 2006). However, the finding that 5% of the wild-type level of Smc2 persist in Smc2-depleted chicken DT40 cells (Ohta et al. 2010) raises the alternative possibility that traces of condensin suffice to drive condensation from prophase to metaphase but not for providing the stiffness required for chromosome segregation upon anaphase onset.
Regardless of the exact mechanisms, condensin most likely functions with multiple cofactors to associate with chromatin and to ensure its reorganization in a cell cycle−regulated manner. To gain insights into the mechanisms of condensin binding and mitotic chromosome condensation, we searched for factors that collaborate with condensin through an unbiased genetic screen in fission yeast. Using a tester strain bearing a thermosensitive allele of the Cut3/Smc4 ATPase (Saka et al. 1994), we isolated mutations resulting in synthetic lethality at the permissive temperature. As expected, our screen resulted in the identification of mutations in other condensin subunits. In addition, we identified a class of mutations affecting chromatin modifiers linked to transcription, consistent with a close functional interplay between condensin and the transcription machinery.
Materials and Methods
Media, molecular genetics, and strains
Media and molecular genetics methods were as described previously (Moreno et al. 1991). Complete medium was YES+A. The synthetic medium was PMG unless otherwise stated. Gene deletions were performed using a polymerase chain reaction (PCR)-based method, as described (Bahler et al. 1998). All deletions were confirmed by PCR. Strains used in this study are listed in Supporting Information, Table S1.
Construction of the tester strain nmt41-cut3 cut3-477
The cut3 open reading frame was PCR-amplified from genomic DNA using Pfu ultra DNA polymerase with primers GTTGTCGCGAACACACCTCTTTTCACGAC and TATACCGCGGCTTTGCGCAGATTTTACAG, digested with NruI and SacII (sites are underlined in primer sequences), and cloned into the replicative expression vector pJR2-41XL (Moreno et al. 2000). The recombinant plasmid, pREP41-cut3, was linearized by cutting within ars1 with MluI and inserted into the genome of a thermosensitive cut3-477 mutant strain, presumably at the ars1 locus. A stable transformant that exhibited thiamine-dependent thermosensitivity for growth at 36° was selected and backcrossed to give rise to the tester strain nmt41-cut3 cut3-477. The genes cut3-477 and nmt41-cut3 segregated independently of each other in crosses.
Mutagenesis and screening
The tester strain was mutagenized with ethylmethane sulfonate 2% (w/v) during one generation at 32° in EMM2 synthetic medium without thiamine, as described (Moreno et al. 1991). Mutagenized cells were plated on PMG medium without thiamine at 32°, and colonies were replicated onto PMG plus phloxine B without or with (20 μM) thiamine. Colonies that stained dark red onto PMG phloxine B plus thiamine at 32° were selected.
Reverse Transcriptase (RT)-quantitative (q)PCR
Total RNA was extracted from 108 cells by standard hot-phenol method. Reverse transcription was performed on 500 ng of total RNA using Superscript II (Invitrogen) and random hexamers. RT-qPCR analyses were performed with a Rotor-Gene PCR cycler.
Genetic mapping
Genetic mapping was performed as described (Anders et al. 2008). Mutations were assigned to a given chromosome through homozygous rec12Δ crosses. Subchromosomal mapping, when necessary, was performed using swi5Δ.
Immunofluorescence
Immunofluorescence was performed as described previously (Bernard et al. 2010) with the use of anti-α tubulin Tat1 antibody (Woods et al. 1989). Spindle length was measured with Image J software.
Chromatin immunoprecipitation (ChIP)
Cells arrested in prometaphase at 17° were fixed with 1% formaldehyde for 30 min and processed for chromatin immunoprecipitation as described (Bernard et al. 2010) using the anti-GFP antibody A11122. Immune complexes were collected with Dynabeads protein A. Real time-qPCR analyses were performed using Rotor-Gene PCR cycler. Primers sequences are available upon request.
Sequencing
Next-generation sequencing was performed by using Illumina HiSeq 2000. A total of 20 μg of genomic DNA was prepared from the mutant strains slc129, slc174, slc185, or sup122, sonicated using BioRuptor for 35 min at 320W (30 sec ON, 30 sec OFF) and processed using the TruSeq DNA Sample Prep Kit (Illumina). DNA samples were sequenced with a coverage varying between 180 and 230X and aligned to the S. pombe reference genome using the CASAVA 1.8 software. Single-nucleotide polymorphisms (SNPs) and single-nucleotide insertions or deletions were sought using CASAVA 1.8. Mutations of interest were confirmed by PCR amplification of candidate gene using Phusion DNA polymerase and Sanger sequencing of the PCR product.
Results
The condensin mutant cut3-477 is defective for chromosome condensation at 32°
To identify factors that collaborate with condensin, we performed a genetic screen for mutants unable to survive when condensin is partly deficient. We chose the thermosensitive cut3-477 mutation to weaken condensin because this mutation in the Cut3/Smc4 ATPase has been shown to cause synthetic lethality when combined with top2-250 at the permissive temperature of 30° (Saka et al. 1994), suggesting that cut3-477 mutant cells survive with a partly defective condensin complex. In agreement, we observed chromatin bridges and chromatin masses trailing on the mitotic spindle in cut3-477 mutant cells cultured at 25°, 32° and 37° (Figure S1A). Growth of the cut3-477 mutant strain at 32° (see Figure 1B) despite the presence of chromosome segregation defects in ~60% of anaphases may be explained by the resolution of those chromosome segregation defects by telophase, an event that might escape our detection focused on anaphase cells. We also assessed condensin binding to chromatin by ChIP. Cells expressing the GFP-tagged Cnd2 subunit of condensin were arrested in prometaphase by using the cryosensitive nda3-KM311 mutation and processed for ChIP against GFP. Mitotic arrest was achieved with similar efficiency in cnd2-GFP and cnd2-GFP cut3-477 strains, as indicated by the septation indexes and the percentages of cells showing Cnd2-GFP enriched in the nucleus (Figure S1B) (Sutani et al. 1999). In good agreement with recent results (Tada et al. 2011), we found that the binding of Cnd2-GFP to centromere, rDNA, and several sites distributed along chromosome arms was reduced in a cut3-477 background (Figure S1B). These data indicate that condensin binding and mitotic chromosome condensation are impaired in a cut3-477 genetic background at 25° and 32°, although the cut3-477 mutant strain is alive at these temperatures. This most likely renders the cut3-477 strain hypersensitive to additional nonlethal mutations that alter the condensation process, such as top2-250. Therefore, screening for mutations synthetically lethal with cut3-477 may lead to the identification of factors required for mitotic chromosome condensation.
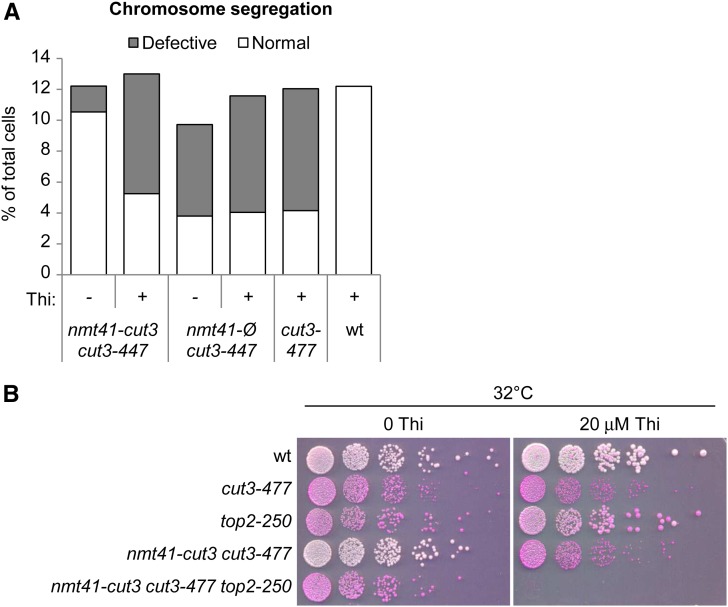
Figure 1.
The nmt41-cut3 cut3-477 tester strain. (A) Cell cultures in synthetic medium without thiamine (Thi) at 25° were split in two, and Thi (20 μM final) was added to one half. Cells were incubated at 25° for three doublings, shifted to 36° for three additional doublings, fixed, and stained with Hoechst 33342 to visualize DNA. Binucleated cells were examined for chromosome segregation defects (n > 100). (B) Cells grown in synthetic medium without Thi were serially diluted and spotted on synthetic medium plus phloxine B supplemented or not with Thi (20 μM final).
Construction of a tester strain, nmt41-cut3 cut3-477
To screen for mutations synthetically lethal with cut3-477, we created the tester strain nmt41-cut3 cut3-477, which possesses the cut3-477 mutation at the cut3 locus plus a wild-type cut3 gene whose expression is driven by the thiamine-repressible promoter nmt41 (no message in thiamine) (Basi et al. 1993), inserted into the genome at an ectopic location. Because the cut3-477 mutation is recessive (Saka et al. 1994), the tester strain should behave as a wild-type strain when grown in the absence of thiamine (nmt41-cut3 expressed), but like a cut3-477 mutant in the presence of thiamine (nmt41-cut3 repressed). An asynchronous population of exponentially growing fission yeast cells contains ~10% of mitotic cells. As shown in Figure 1A, nmt41-cut3 cut3-477 cells cultured in the absence of thiamine at 36° exhibited less than 2% of defective anaphases, but the frequency increased to 7% in the presence of thiamine, reaching a level similar to the cut3-477 mutant. In contrast, the addition of thiamine to the growing medium did not modify the frequency of defective anaphases exhibited by nmt41-Ø cut3-477 cells, bearing an empty nmt41 plasmid. These data indicate that the nmt41-cut3 gene complements, at least partly, the cut3-477 mutation at 36° in the absence of thiamine, but not in the presence of 20 μM thiamine. Furthermore, using the tester strain we observed a thiamine-dependent synthetic lethal genetic interaction between top2-250 and cut3-477 at 32° (Figure 1B). Thus, the strain nmt41-cut3 cut3-477 behaves as a wild-type strain when it is cultured in the absence of thiamine, but like a cut3-477 condensin mutant strain in the presence of thiamine.
Screening for mutations synthetically lethal with cut3-477
The tester strain was mutagenized with ethylmethane sulfonate and mutant colonies viable on synthetic medium without thiamine but dead on synthetic medium plus thiamine at 32° were selected. Mutants were subsequently backcrossed at least three times. The first backcross was performed with a wild-type strain (lacking both nmt41-cut3 and cut3-477). This allowed us to eliminate secondary mutation in the cut3-477 gene that were lethal at 32° because mutants bearing this type of mutation cannot produce cut3-477 progenies viable at 32° in the absence of nmt41-cut3. This first backcross also allowed the elimination of mutations that conferred thiamine hypersensitivity, i.e., mutants that produced progenies hypersensitive to thiamine regardless of nmt41-cut3. From ~180,000 colonies, 15 slc mutants (synthetically lethal with cut3) were selected (see Table 1). Representative examples are shown in Figure 2A. During the course of the backcrosses, we noticed that all slc mutants, except slc129, produced cut3-477 slc progenies devoid of the nmt41-cut3 gene at 25°. Those cut3-477 slc double mutants were viable at 25° but dead at 32° (Figure 2B). Thus, the synthetically lethal interactions between the vast majority of slc mutations and cut3-477 are thermo-dependent.
Table 1. Genes identified in the screen.
| Gene Name | cut14 | cnd1 | cnd3 | ark1 | arp9 | snf21 | cph2 | ulp2 | nut2 |
|---|---|---|---|---|---|---|---|---|---|
| Function | Condensin Smc2 | Condensin non-Smc | Condensin non-Smc | Aurora-B kinase | Swi/Snf and RSC chromatin remodeling | RSC chromatin remodeling | Clr6S Histone deacetylase | Sumo decon-jugating enzyme | Mediator |
| slc mutations | slc71 | slc175 | slc184 | slc190 | slc127 | slc129 | slc174 | slc185 | sup122 |
| slc85 | slc181 | ||||||||
| slc90 | slc182 | ||||||||
| slc173 | |||||||||
| slc179 | |||||||||
| slc180 | |||||||||
| Allele names | cut14-71 | cnd1-175 | cnd3-184 | ark1-190 | arp9-127 | snf21-129 | cph2-174 | ulp2-185 | nut2-122 |
| cut14-85 | cnd1-181 | ||||||||
| cut14-90 | cnd1-182 | ||||||||
| cut14-173 | |||||||||
| cut14-179 | |||||||||
| cut14-180 | |||||||||
| Protein | 71: G397D | 175: G667E | E195K | R241H | Q13a | E524K | C453Y | R81C | M1I |
| 85: D656N | 181: G912D | ||||||||
| 90: T652M | 182: R1117C | ||||||||
| 173: T652M | |||||||||
| 179: T652M | |||||||||
| 180: C631Y |
RSC, remodels the structure of chromatin.
STOP codon.
Figure 2.
slc mutations are synthetically lethal with cut3-477. Strains of indicated genotypes were serially diluted and spotted onto (A) synthetic medium without thiamine (0 Thi) or supplemented with thiamine (20 μM final) to repress transcription of the nmt41-cut3 gene, or (B) onto complete medium.
Genetic analyses
All slc mutations segregate 2:2 in crosses. They are recessive and organized into eight complementation groups, defining eight loci (Table 1). To test for allelism with expected hits, each slc locus was assessed for genetic linkage with chromosome condensation genes: condensin subunits cut14, cnd1, cnd2, and cnd3, top2, aurora-B and polo kinases, survivin, incenp, pcs1, and mde4 (Tada et al. 2011). We observed a tight linkage between 3 slc loci (representing 10 mutations) and cut14, cnd1, or cnd3, and between a fourth slc locus and ark1, which encodes Aurora-B kinase. Sanger sequencing of those genes in candidate slc mutant backgrounds confirmed allelism. The 11 slc mutations were therefore renamed following the rule gene name-mutation number, e.g., the slc71 mutation in the cut14 gene was renamed cut14-71 (Table 1). For the remaining slc127, slc129, slc174, and slc185 loci, we observed no genetic linkage with any tested condensation gene, raising the possibility that these four loci could correspond to unknown chromosome condensation genes.
Phenotypic analyses
slc mutations might alter mitotic chromosome condensation. To test this, we examined chromosome segregation in slc single-mutant cells. Mitotic cells were identified by the presence of a mitotic spindle and examined for chromosome segregation defects. In wild-type cells, the migration of centromeres to the opposite poles of the mitotic spindle upon anaphase onset, and the movement of chromosome arms that ensues, lead to the formation of a mini chromatin bridge, which rapidly disappears as cells progress into late anaphase (Tada et al. 2011). We observed mini chromatin bridges on short mitotic spindles with length comprised between 3 and 4 microns in wild-type cells (Figure 3A). However, these chromatin bridges disappeared as mitotic spindles increased to 5 microns in length, and passed this stage, chromosomes were almost always fully separated and clustered at the spindle poles (Figure 3, A and B). In sharp contrast, the four mutants, slc127, slc129, slc174, and slc185, exhibited frequent chromatin bridges and/or chromatin masses trailing on long anaphase spindle (>5 microns in length; Figure 3, A and B). Thus, chromosome migration during anaphase is altered in slc127, slc129, slc174, and slc185 single-mutant cells.
Figure 3.
Defective chromosome migration during anaphase in slc mutant cells. (A) Chromosome segregation in cells fixed and processed for immunofluorescence against α-tubulin (Tubulin). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Bar: 5 μm. (B) Frequencies of chromosome segregation defects in late anaphase. Cells were fixed, processed for tubulin staining, and examined for chromosome segregation defects in late anaphase (spindle >5 μm, n > 100). Left panel: cells were cultured and fixed at 32°. Right panel: cells cultured at 32° were shifted at 36° for 2.5 hr and fixed at 36°.
Cut3 is an essential gene, and slc mutations may interfere with its expression. Similarly, a reduced expression of top2 would most likely kill a cut3-477 mutant strain. We therefore assessed by RT-qPCR the expression levels of each subunit of condensin and of top2 in slc127, slc129, slc174, and slc185 single-mutant cells cultured at 32° (Figure S2). We observed no obvious reduction in the steady state levels of either condensin or top2 mRNA in any slc mutant, arguing against an indirect negative effect of slc mutations on the expression of these genes.
We assessed slc mutants for additional phenotypes. We observed no cryosensitivity for growth at 20° (data not shown). However, slc127, slc129, and slc174, as well as most slc mutants of the Cut14 subunit of condensin, exhibited thermosensitivity for growth at 36° (Figure S3A). Condensin takes part in the attachment of kinetochores to microtubules during mitosis, and in the signaling and/or repair of DNA damage during interphase, as indicated by the hypersensitivity to hydroxyurea (HU) and ultraviolet light of cut14-Y1 and cnd2-1 mutants (Aono et al. 2002; Akai et al. 2011; Tada et al. 2011). To test for a possible role of slc genes in these processes, we examined the growth of slc mutants in the presence of increasing amounts of thiabendazole, an inhibitor of microtubule polymerization, or in the presence of HU. The cut14-71, slc127, and slc129 mutants, and to a lesser extent ark1-190, showed hypersensitivity to TBZ (Figure S3B). In addition, slc127, slc129, slc174, and cnd1-175 were hypersensitive to HU (Figure S3C). However, we observed no hypersensitivity to ultraviolet radiation ranging from 50 to 100 J/m2 (not shown).
Identification of slc mutations
The arp9 gene was cloned by functional complementation of the slc127 thermosensitive phenotype using the pUR19-B1 genomic library (Barbet et al. 1992). Sequencing arp9 in the slc127 mutant strain revealed a C37T mutation (numbering starting from the ATG) that created a premature stop codon. The slc127 mutation was therefore renamed arp9-127. We failed to clone the three other slc genes by functional complementation. Those genes were identified by an alternative approach combining next-generation, whole-genome sequencing of slc mutants strain and genetic mapping of slc point mutations. We identified 109 ± 8 SNPs in each slc mutant strains (File S1, File S2, File S3). First, pairwise comparisons of the lists of SNPs using a home-made script allowed us to identify 5 ± 3 SNPs that were unique to each mutant background. Second, each slc mutation was assigned to one chromosome by cosegregation analyses (Anders et al. 2008), further refining the number of candidate SNPs to 3 ± 2 per mutant genome. Finally, candidate single-base mutations were identified by genetic linkage analyses and confirmed by Sanger sequencing (Table 1). slc129 corresponds to snf21, slc174 to cph2, and slc185 to ulp2. Except for snf21, which is an essential gene, we confirmed that deleting arp9, cph2, or ulp2 was synthetically lethal with cut3-477 at 32° (Figure S4).
nut2 is a suppressor of cut3-477
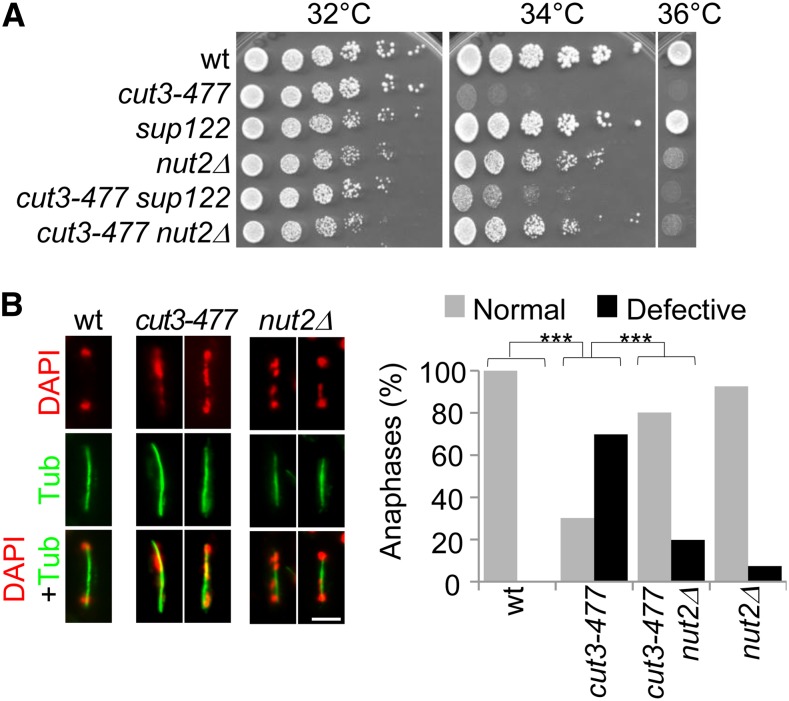
The nmt41-cut3 cut3-477 tester strain forms pink colonies at 32° on synthetic medium supplemented with phloxine B and 20 μM thiamine (Figure 1B). During the screening procedure, we picked a mutant colony that remained white on synthetic medium plus phloxine B plus thiamine at 32°, suggesting a possible suppressive effect on cut3-477. Backcrosses confirmed the presence of a suppressor mutation, named sup122, distinct from the cut3 locus and independent of nmt41-cut3, which restored the growth of cut3-477 at restrictive temperature (Figure 4A). The sup122 mutation segregates 2:2 in crosses and is recessive. Genome resequencing of the sup122 mutant strain combined with genetic mapping revealed a point mutation in the nonessential nut2 gene (Table 1 and File S4), and we confirmed that deleting nut2 restored growth of cut3-477 at 34° (Figure 4A). To investigate the mechanism through which the lack of nut2 partly suppressed the thermosensitivity of cut3-477, we assessed chromosome segregation in cut3-477 nut2Δ double-mutant cells at 34° (Figure 4B). The frequency of defective anaphases was markedly reduced in cut3-477 nut2Δ cells compared with cut3-477, although chromosome segregation was altered in ~7% of anaphases in nut2Δ. Thus, nut2 is required for normal chromosome segregation and its deficiency rescues chromosome segregation when condensin is impaired by the cut3-477 mutation.
Figure 4.
The lack of nut2 partly compensates for cut3-477 deficiency. (A) Cells of indicated genotypes were serially diluted and spotted onto complete medium. (B) Cells exponentially growing at 30° were shifted at 34° for one generation, fixed, and processed for immunofluorescence against α-tubulin (Tub) and DNA staining with DAPI. Bar, 5 μm. Chromosome segregation was assessed in late anaphase cells (spindle >5 μm, n > 100). ***P < 0.001, χ2 test.
Discussion
To identify genes that potentially interact with condensin to mediate chromatin interaction and compaction, we performed an unbiased genetic screen for genetic interaction partners of the Cut3/Smc4 ATPase subunit of condensin in fission yeast. This screen identified novel mutations in the condensin subunits Cut14, Cnd1, and Cnd3, and in Aurora-B kinase, which regulates condensin binding to chromatin (Tada et al. 2011), demonstrating its efficiency in identifying bona fide condensation factors. In addition, we isolated mutations in arp9, snf21, cph2, and ulp2, four genes not previously implicated in mitotic chromosome condensation in fission yeast. We show that all these mutations cause chromatin bridges and/or segregation anomalies in anaphase, consistent with defective chromosome condensation. In the course of this screen, we independently identified nut2Δ as a suppressor of the thermosensitive growth and segregation defects caused by the condensin mutation cut3-477. Thus, Arp9, Snf21, Cph2, Ulp2, and Nut2 may play a role in mitotic chromosome condensation.
The presence of single alleles of arp9, snf21, cph2, or ulp2 indicates that the screen is not saturated and therefore that additional genes synthetically lethal with cut3 remain to be identified. Consistent with this, using high-throughput genetic interaction mapping Ryan et al. (2012) reported negative genetic interactions between cut3 and at least 32 non-essential genes, including ulp2. Surprisingly, however, the screen failed to detect arp9 and cph2, although cph2 was present in the deletions library they used for screening.
The cut14-71 mutation identified here confers hypersensitivity to TBZ. To the best of our knowledge, this is the first described mutation in condensin that confers pronounced TBZ hypersensitivity. The fact that among eight condensin mutants (seven slc alleles plus cut3-477), solely cut14-71 exhibits this phenotype, suggests that this type of mutation may be rare. Pcs1 recruits condensin at the kinetochore (Tada et al. 2011), and like cut14-71, pcs1Δ is co-lethal with cut3-477 and confers hypersensitivity to TBZ. Thus, cut14-71 may affect the binding and/or the activity of condensin at the kinetochore in particular. cut14-71 changes the glycine 397 of Cut14 into an aspartate. This residue is located at the very beginning of a coiled coil motif and is not conserved throughout evolution. Using the COILS program as a predictive tool (Lupas et al. 1991), we observed no disruption of the coiled-coil organization in the Cut14-71 mutant protein. Furthermore, Cut14 does not seem to directly contact the Pcs1/Mde4 monopolin complex (Tada et al. 2011). Thus, why cut14-71 would have a specific impact on condensin at kinetochores remains enigmatic. The three other slc alleles of cut14 (slc85, slc90, and slc180) all affect conserved residues located within the hinge domain. Notably, slc90 affects a threonine residue conserved from bacteria to human and located near glycine patches clustered at the dimerization interface and essential for dimer formation and DNA binding (Hirano and Hirano 2002). Thus slc85, slc90, and slc180 may weaken the interaction between the hinge domains of Cut3/Smc4 and Cut14/Smc2.
Arp9 is a subunit of the Swi/Snf and RSC (i.e., remodels the structure of chromatin) chromatin remodeling complexes, whereas Snf21 is the ATP-dependent DNA helicase of RSC (Monahan et al. 2008). The fact that both snf21-129 and arp9-127 exhibit similar phenotypes, such as hypersensitivity to TBZ or HU, thermosensitivity, and the greatest frequency of chromatin bridges in anaphase at 32° among slc mutants, is consistent with their coexistence into functionally similar complexes. Arp9 may contribute to the association of RSC with chromatin as a heterodimer with Arp42 (Monahan et al. 2008), and lack of Arp42 or of the Snf5 subunit of Swi/Snf is synthetically sick in combination with cut3-477 (Ryan et al. 2012). These data suggest that chromatin remodeling may play a role in chromosome condensation. ISWI chromatin remodeling complexes have been identified as major components of mitotic chromosomes assembled in Xenopus laevis egg extracts (MacCallum et al. 2002). However, the role of ISWI complexes in the formation of mitotic chromosome in this system appears rather limited. Furthermore, RSC has been implicated in multiple processes such as kinetochore function and recruitment of cohesin along chromosome arms (Hsu et al. 2003; Huang et al. 2004). Given the role of remodeling complexes as transcriptional coactivators, we cannot rule out the possibility that snf21-129 and/or arp9-127 mutations affect chromosome condensation indirectly. The hypersensitivity to HU of snf2-129 and arp9-127 mutants may reflect a defect in DNA replication during S phase, and the chromatin bridges exhibited by these mutants may stem, at least in part, from entry into mitosis with only partially replicated chromosomes. Alternatively, ATP-dependent chromatin remodeling may play an active role in the binding of condensin to “open” chromatin at promoters of active genes (see herein). Further work is required to determine whether or not the link we describe here between chromatin remodeling complexes and condensin is direct.
Cph2 is a zinc finger PHD domain protein associated with the Clr6 histone deacetylase within a complex called Clr6 II, which deacetylates histones mostly within coding regions of the genome (Nicolas et al. 2007). Clr6 II is the functional counterpart of budding yeast RPD3S, and RPD3-dependent deacetylation of histone H4-K5/K12 has been implicated in the recruitment of condensin onto rDNA in budding yeast in response to starvation (Tsang et al. 2007). Clr6 contributes to the regulation of condensin binding to retrotransposons in fission yeast through the deacetylation of H3-K56 (Tanaka et al. 2012), and the lack of Alp13, a nonessential subunit of Clr6 II, has been shown to cause chromosome segregation anomalies similar to those associated with defects in condensation (Nakayama et al. 2003). We found that, like cph2Δ, alp13Δ was colethal with cut3-477 at 32° (see Figure S5). Thus, Cph2 may influence condensin association with chromatin as part of the Clr6 II histone deacetylase complex. Clr6 II may contribute to chromosome condensation by modulating the level of acetylation of nucleosomes, but possibly also of condensin itself, as suggested for the related cohesin complex (Kagami et al. 2011).
Ulp2 is a sumo-specific protease that plays a dual role in the cycle of sumoylation/desumoylation of proteins. Sumo (small ubiquitin-related modifier) is a reversible posttranslational protein modifier that is synthesized into an inactive proform. Ulp2 cleaves Sumo propeptide into a mature form that can be conjugated to target proteins, and, through its protease activity, also allows the de-sumoylation of target proteins (Geiss-Friedlander and Melchior 2007). Budding yeast ULP2 has been identified as a multicopy suppressor of the smc2-6 allele of condensin, and lack of ULP2 is colethal with smc4-1 (Bachellier-Bassi et al. 2008; Strunnikov et al. 2001). Thus, Ulp2 seems to play a positive role in chromosome condensation that is conserved in budding and fission yeasts. The targets of Ulp2 in this pathway may be multiple, including topoisomerases and/or condensin. Sumoylation of condensin subunits has been observed and is necessary for condensin recruitment to the rDNA repeats during anaphase (D’Amours et al. 2004; Takahashi et al. 2008). However, the underlying mechanism remains elusive. Ulp2 may play a positive role in this pathway because it seems required for the rDNA enrichment of Smc4-GFP (Strunnikov et al. 2001).
Nut2 is a component of the mediator complex, which physically associates with DNA binding transcription factors and RNA polymerase II. Through these contacts, mediator assists the recruitment of RNA polymerase II to genes promoters and integrates regulatory signals for transcription (Conaway and Conaway 2011). Mediator can stimulate or repress transcription and its subunits exhibit gene-specific effects. The origin of the chromosome segregation defects in nut2Δ cells remains undetermined. However, the fact that the lack of Nut2 improves chromosome segregation in cut3-477 cells strongly suggests that the lack of Nut2 liberates a condensation activity that is limiting in a cut3-477 background, and, therefore, that Nut2 plays a negative role in chromosome condensation. The restored growth of the cut3-477 nut2Δ double mutant at 34° compared with cut3-477 most likely stems from the improved chromosome segregation. Nut2 may be required for the transcription of a negative regulator of chromosome condensation. Alternatively, given that transcription and condensin binding are antagonistic (Clemente-Blanco et al. 2009), an overall reduction in transcription caused by the lack of Nut2 may facilitate condensin binding to chromatin.
A recent report indicates that increasing cohesin occupancy on chromosomes by eliminating the destabilizing activity of Wapl leads to a substantial increase in the compaction of chromosomes (Tedeschi et al. 2013). Mediator and cohesin physically interacts and cooccupy active genes in human cells (Kagey et al. 2010). Thus, lack of Nut2 may compensate for condensin deficiency by increasing cohesin occupancy. However, we think this is unlikely because the lack of fission yeast Wapl (wpl1Δ) does not restore growth of cut3-477 condensin mutant (Figure S6). Rather, given the overlap that seems to exist between condensin and cohesin for their loading onto chromosomes (D’Ambrosio et al. 2008), mediator may play a direct role in chromosome condensation through the recruitment of condensin onto chromatin, and Nut2 may negatively regulate this pathway.
One characteristic of condensin I and II complexes that recently emerged from genome-wide mapping experiments in budding and fission yeasts, C. elegans, chicken DT40 cells, and mouse embryonic stem cells is that both complexes are enriched at promoters of active genes (D’Ambrosio et al. 2008; Dowen et al. 2013; Kim et al. 2013; Kranz et al. 2013). The bacterial condensin of Bacillus subtilis also binds in the vicinity of highly expressed genes (Gruber and Errington 2009). Therefore, transcription somehow influences the binding of condensin complexes to chromosomes. Remarkably, four of the five “non-condensin” genes identified in this screen—arp9, snf21, cph2, and nut2—are implicated in transcription in eukaryotes. Swi/snf and RSC chromatin remodelers alter nucleosome positioning at gene promoters, thereby exposing DNA to the transcriptional machinery. Clr6 II/RPD3S deacetylates histones within the coding region of genes thereby promoting chromatin reformation in the wake of RNA polymerase II and prohibiting antisense transcription (Nicolas et al. 2007; Venkatesh et al. 2012). Furthermore, if we consider all known genetic interaction partners with cut3 referenced in the pombase database (Wood et al. 2012), 28% (11/39) are linked to transcription. Although we cannot exclude the possibility of a role in pathways related to topo II and/or RCA, it is tempting to speculate that this enrichment in transcription cofactors reflects a link between condensin binding to chromosomes and high transcription rate. Local chromatin features established with transcription may play a role in condensin association with chromosome in eukaryotes. Overall, this genetic screen reinforces the idea of a tight functional interplay between condensin association with chromatin and transcriptional processes.
Supplementary Material
Acknowledgments
We thank J. P. Javerzat in whose lab the tester strain nmt41-cut3 cut3-477 was created, the Yeast Genetic Resource Center for strains, K. Gull for Tat1 antibody and F. Palladino, G. Yvert, and V. Vanoosthuyse for comments on the manuscript. X. R. was supported by a postdoctoral fellowship from the CNRS and L. F. by a PhD studentship from la Ligue Contre le Cancer. This work was supported by an ATIP grant from CNRS to P.B.
Footnotes
Communicating editor: C. S. Hoffman
Literature Cited
- Akai Y., Kurokawa Y., Nakazawa N., Tonami-Murakami Y., Suzuki Y., et al. , 2011. Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open Biol. 1: 110023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders A., Watt S., Bahler J., Sawin K. E., 2008. Improved tools for efficient mapping of fission yeast genes: identification of microtubule nucleation modifier mod22–1 as an allele of chromatin- remodelling factor gene swr1. Yeast 25: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono N., Sutani T., Tomonaga T., Mochida S., Yanagida M., 2002. Cnd2 has dual roles in mitotic condensation and interphase. Nature 417: 197–202 [DOI] [PubMed] [Google Scholar]
- Bachellier-Bassi S., Gadal O., Bourout G., Nehrbass U., 2008. Cell cycle-dependent kinetochore localization of condensin complex in Saccharomyces cerevisiae. J. Struct. Biol. 162: 248–259 [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, et al. , 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Barbet N., Muriel W. J., Carr A. M., 1992. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene 114: 59–66 [DOI] [PubMed] [Google Scholar]
- Basi G., Schmid E., Maundrell K., 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123: 131–136 [DOI] [PubMed] [Google Scholar]
- Baxter J., Sen N., Martinez V. L., De Carandini M. E., Schvartzman J. B., et al. , 2011. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science 331: 1328–1332 [DOI] [PubMed] [Google Scholar]
- Baxter J., Aragon L., 2012. A model for chromosome condensation based on the interplay between condensin and topoisomerase II. Trends Genet. 28: 110–117 [DOI] [PubMed] [Google Scholar]
- Bernard P., Drogat J., Dheur S., Genier S., Javerzat J. P., 2010. Splicing factor Spf30 assists exosome-mediated gene silencing in fission yeast. Mol. Cell. Biol. 30: 1145–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Blanco A., Mayan-Santos M., Schneider D. A., Machin F., Jarmuz A., et al. , 2009. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 458: 219–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Blanco A., Sen N., Mayan-Santos M., Sacristan M. P., Graham B., et al. , 2011. Cdc14 phosphatase promotes segregation of telomeres through repression of RNA polymerase II transcription. Nat. Cell Biol. 13: 1450–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R. C., Conaway J. W., 2011. Function and regulation of the Mediator complex. Curr. Opin. Genet. Dev. 21: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S., Haering C. H., 2011. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 21: 552–559 [DOI] [PubMed] [Google Scholar]
- Cuylen S., Metz J., Haering C. H., 2011. Condensin structures chromosomal DNA through topological links. Nat. Struct. Mol. Biol. 18: 894–901 [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C., Schmidt C. K., Katou Y., Kelly G., Itoh T., et al. , 2008. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 22: 2215–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D., Stegmeier F., Amon A., 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117: 455–469 [DOI] [PubMed] [Google Scholar]
- Dowen J. M., Bilodeau S., Orlando D. A., Hübner M. R., Abraham B. J., et al. , 2013. Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem Cell Rev. I: 371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F., 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J. M., Ellenberg J., 2006. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 16: 333–344 [DOI] [PubMed] [Google Scholar]
- Gruber S., Errington J., 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137: 685–696 [DOI] [PubMed] [Google Scholar]
- Haeusler R. A., Pratt-Hyatt M., Good P. D., Gipson T. A., Engelke D. R., 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 22: 2204–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M., Hirano T., 2002. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 21: 5733–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., 2012. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 26: 1659–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J. M., Huang J., Meluh P. B., Laurent B. C., 2003. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol. Cell. Biol. 23: 3202–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hsu J. M., Laurent B. C., 2004. The RSC nucleosome-remodeling complex is required for Cohesin’s association with chromosome arms. Mol. Cell 13: 739–750 [DOI] [PubMed] [Google Scholar]
- Hudson D. F., Marshall K. M., Earnshaw W. C., 2009. Condensin: architect of mitotic chromosomes. Chromosome Res. 17: 131–144 [DOI] [PubMed] [Google Scholar]
- Iwasaki O., Tanaka A., Tanizawa H., Grewal S. I., Noma K., 2010. Centromeric localization of dispersed Pol III genes in fission yeast. Mol. Biol. Cell 21: 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami A., Sakuno T., Yamagishi Y., Ishiguro T., Tsukahara T., et al. , 2011. Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Rep. 12: 1189–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., et al. , 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Zhang T., Wong N. C., Davidson N., Maksimovic J., et al. , 2013. Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat Commun. 4: 2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Hirano M., Kobayashi R., Hirano T., 1998. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science 282: 487–490 [DOI] [PubMed] [Google Scholar]
- Kranz A. L., Jiao C. Y., Winterkorn L. H., Albritton S. E., Kramer M., et al. , 2013. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol. 14: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Tanasa B., Tyurina O. V., Zhou T. Y., Gassmann R., et al. , 2010. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466: 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano T., 2005. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 19: 1269–1287 [DOI] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J., 1991. Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- MacCallum D. E., Losada A., Kobayashi R., Hirano T., 2002. ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol. Biol. Cell 13: 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan B. J., Villen J., Marguerat S., Bahler J., Gygi S. P., et al. , 2008. Fission yeast SWI/SNF and RSC complexes show compositional and functional differences from budding yeast. Nat. Struct. Mol. Biol. 15: 873–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno M. B., Duran A., Ribas J. C., 2000. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast 16: 861–872 [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Nakayama J., Xiao G., Noma K., Malikzay A., Bjerling P., et al. , 2003. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 22: 2776–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa N., Nakamura T., Kokubu A., Ebe M., Nagao K., et al. , 2008. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J. Cell Biol. 180: 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E., Yamada T., Cam H. P., Fitzgerald P. C., Kobayashi R., et al. , 2007. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 14: 372–380 [DOI] [PubMed] [Google Scholar]
- Ohta S., Bukowski-Wills J. C., Sanchez-Pulido L., Alves Fde L., Wood L., et al. , 2010. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142: 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza I., Haering C. H., Rutkowska A., 2013. Condensin: crafting the chromosome landscape. Chromosoma 122: 175–190 [DOI] [PubMed] [Google Scholar]
- Ryan C. J., Roguev A., Patrick K., Xu J., Jahari H., et al. , 2012. Hierarchical modularity and the evolution of genetic interactomes across species. Mol. Cell 46: 691–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y., Sutani T., Yamashita Y., Saitoh S., Takeuchi M., et al. , 1994. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 13: 4938–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J., Douziech M., Bazile F., Pascariu M., Bonneil E., et al. , 2009. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol. Cell 34: 416–426 [DOI] [PubMed] [Google Scholar]
- Strunnikov A. V., Aravind L., Koonin E. V., 2001. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 158: 95–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T., Yuasa T., Tomonaga T., Dohmae N., Takio K., et al. , 1999. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 13: 2271–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K., Susumu H., Sakuno T., Watanabe Y., 2011. Condensin association with histone H2A shapes mitotic chromosomes. Nature 474: 477–483 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Dulev S., Liu X., Hiller N. J., Zhao X., et al. , 2008. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet. 4: e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Tanizawa H., Sriswasdi S., Iwasaki O., Chatterjee A. G., et al. , 2012. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol. Cell 48: 532–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A., Wutz G., Huet S., Jaritz M., Wuensche A., et al. , 2013. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501: 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang C. K., Li H., Zheng X. S., 2007. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 26: 448–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Hudson D. F., Ribeiro S. A., Trinkle-Mulcahy L., Spence J. M., et al. , 2006. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol. 8: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S., Smolle M., Li H., Gogol M. M., Saint M., et al. , 2012. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489: 452–455 [DOI] [PubMed] [Google Scholar]
- Wood V., Harris M. A., McDowall M. D., Rutherford K., Vaughan B. W., et al. , 2012. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 40: D695–D699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T. H., Baines A. J., et al. , 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93: 491–500 [DOI] [PubMed] [Google Scholar]
- Xing H., Vanderford N. L., Sarge K. D., 2008. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat. Cell Biol. 10: 1318–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.