Abstract
Cultivars of rice (Oryza sativa L.), especially of the type with large spikelets, often fail to reach the yield potential as expected due to the poor grain-filling on the later flowering inferior spikelets (in contrast to the earlier-flowering superior spikelets). The present study showed that the size and grain weight of superior spikelets (SS) was greater than those of inferior spikelets (IS), and the carbohydrate supply should not be the major problem for the poor grain-filling because there was adequate amount of sucrose in IS at the initial grain-filling stage. High resolution two-dimensional gel electrophoresis (2-DE) in combination with Coomassie-brilliant blue (CBB) and Pro-Q Diamond phosphoprotein fluorescence stain revealed that 123 proteins in abundance and 43 phosphoproteins generated from phosphorylation were significantly different between SS and IS. These proteins and phosphoproteins were involved in different cellular and metabolic processes with a prominently functional skew toward metabolism and protein synthesis/destination. Expression analyses of the proteins and phosphoproteins associated with different functional categories/subcategories indicated that the starch synthesis, central carbon metabolism, N metabolism and cell growth/division were closely related to the poor grain-filling of IS. Functional and expression pattern studies also suggested that 14-3-3 proteins played important roles in IS poor grain-filling by regulating the activity of starch synthesis enzymes. The proteome and phosphoproteome obtained from this study provided a better understanding of the molecular mechanism of the IS poor grain-filling. They were also expected to be highly useful for improving the grain filling of rice.
Introduction
Rice (Oryza sativa L.) is one of the world’s most important staple crops. It is essential for global food security, especially in the populous Asian and African regions [1]. The grains of rice grow on the spikelets, which can be classified as SS or IS according to their location on a branch and the time of flowering [2], [3]. In general, SS are on the apical primary branches, while IS on the proximal secondary branches on a rice plant. By comparison, SS flower earlier and fill faster with larger and heavier grains than IS [2]–[4]. The poor grain-filling of IS on rice cultivars, especially for the “super” varieties developed recently that bear numerous spikelets per panicle, has become a subject for study, as it not only negatively affects the final yield but also the milling and quality of the rice [5]–[7].
The grain-filling of rice is largely a process of starch accumulation, since the starch contributes 90% of the dry weight of an unpolished mature grain [8]. However, it has been reported that the carbohydrates may not be the only limiting factor [3], [5], [9]. Low activities of the enzymes that convert sucrose to starch, such as sucrose synthase (SuSase), adenosine diphosphateglucose pyrophosphorylase (AGPase), starch synthase (StSase), and starch branching enzyme (SBE), might also contribute to the low filling rate and weight of the grains on IS [10]–[12]. In addition, a low abscisic acid (ABA)/ethylene ratio and cytokinins and indole acetic acid (IAA) contents were also considered important in this regard [4], [13], [14]. Exogenously applied ABA or mild water stress, which resulted in a significant increase of grain ABA content at the early grain-filling stage, significantly stimulated the grain-filling of IS [12], [15].
A complex biological process, filling of a rice grain involves 21,000 genes including 269 that are closely related to various physiological and biochemical pathways [16]. Thus, to understand the process thoroughly, not only the conventional physiological and biochemical means, but also the molecular methods, must be applied. Ishimaru et al. showed that the gene expressions of vacuolar invertase (INV3), SuSase (RSus3), and AGPase (AGPL1 and AGPS2) were much lower in IS than SS [11]. Recently, applying the high-throughput sequencing technology, differences in gene expressions between the rice spikelets was unveiled. A DNA microarray and real-time PCR analysis showed a group of genes relating to the starch metabolism, and with their enhanced expression profiles, the higher transcript levels in SS than IS at the early (EGS) and mid-grain-filling stages (MGS) [17]. The expression profiles of the miRNAs showed that 189 of them were differentially expressed between SS and IS [18]. However, the genome DNA sequence can only show, to a certain degree, the possible existence of corresponding functions of genes. It cannot predict whether or when the genes can be expressed. On the other hand, proteomic profiling can provide valuable insights regarding molecular mechanisms of poor grain-filling of IS. Thus far, there is no expression patterns of the abovementioned proteins available to enable the differentiation between SS and IS at the grain-filling stages. We proposed that the grain-filling disparity stemmed from the differential expressions of relevant proteins in the temporal and space of the spikelets.
Using the proteomic approach, we aimed to differentiate the protein expressions and to determine their relationship with the poor IS grain-filling during the EGS, MGS and late grain-filling stage (LGS). A 2-DE gel-based proteomic methodology was applied. The proteomes of SS and IS were compared. The phosphoprotein regulations of SS and IS at different grain-filling stages were differentiated using Pro-Q Diamond phosphoprotein in gel stain.
Results
Physiological Characteristics of Grain-filling of SS and IS
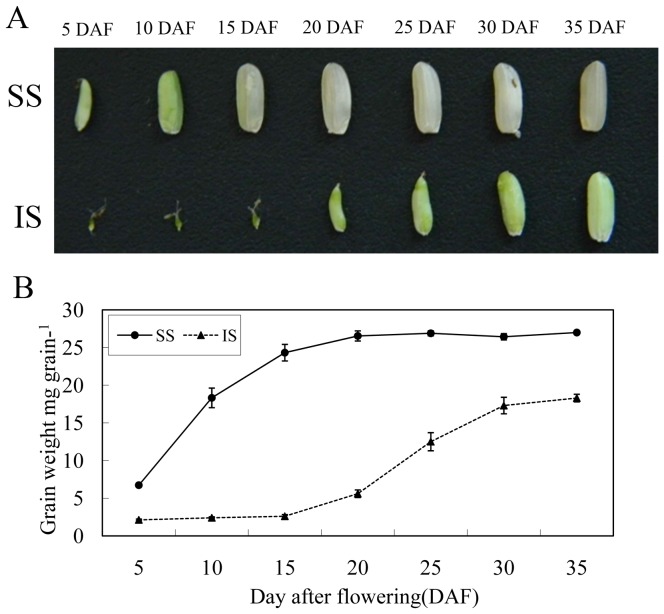
A developmental asynchronism was observed on the grain-fillings between SS and IS. After flowering, the size of SS increased rapidly, but the development of IS appeared stagnant from 5 to 15 DAF (Fig. 1A). The grain weight on SS increased rapidly from 5 to 20 DAF, while that on IS did not show significant increases until 20 to 35 DAF (Fig. 1B). Nonetheless, the end grain weight of IS was lower than that of SS (Fig. 1B).
Figure 1. Superior spikelets (SS) and inferior spikelets (IS) in rice.
A: Developmental changes in representative SS and IS B: Grain weight of SS and IS. Experiments were repeated 6 times. Error bars represent standard errors.
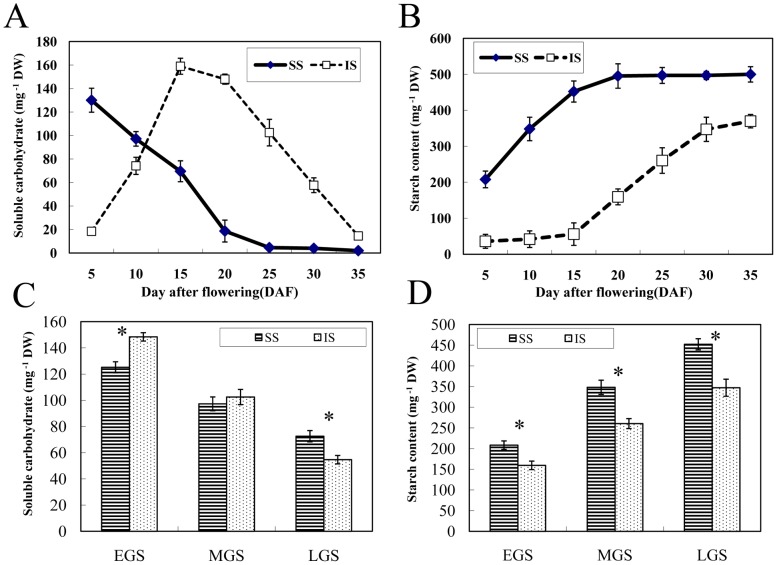
The sucrose content in SS decreased after 5 DAF. In contrast, in IS, it increased rapidly after 5 DAF, peaked at 15 DAF, and decreased thereafter (Fig. 2A). Similar patterns of change on starch content and grain weight on SS and IS were observed (Fig. 1B & 2B). As shown in Fig. 2C and D, higher soluble carbohydrates contents were found on IS than SS in EGS and MGS, but lower in LGS; and, that SS had a higher starch content than IS throughout the periods.
Figure 2. Concentrations of soluble carbohydrate and starch in grains on superior spikelets (SS) and inferior spikelets (IS) in rice of rice.
A: Dynamic change of soluble carbohydrate concentration in grains on SS and IS. B: Dynamic change of starch concentration in grains on SS and IS. C: Difference of soluble carbohydrate concentration between SS and IS at three stages. D: Difference of starch concentration between SS and IS at three stages. Experiments were repeated 6 times. Error bars represent standard errors. EGS, MGS and LGS represent early, mid-, and late grain-filling stage, respectively, of endosperm development stages of SS and IS. Asterisk (*) represents significant difference (P = 0.05) between SS and IS.
2-DE Analysis and Identification of Differentially Expressed Proteins and Phosphoproteins on SS and IS
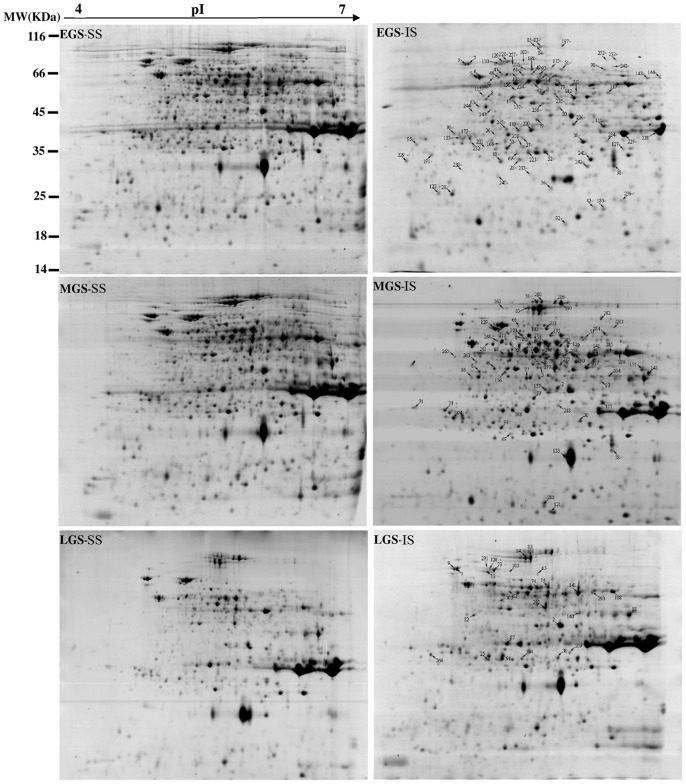
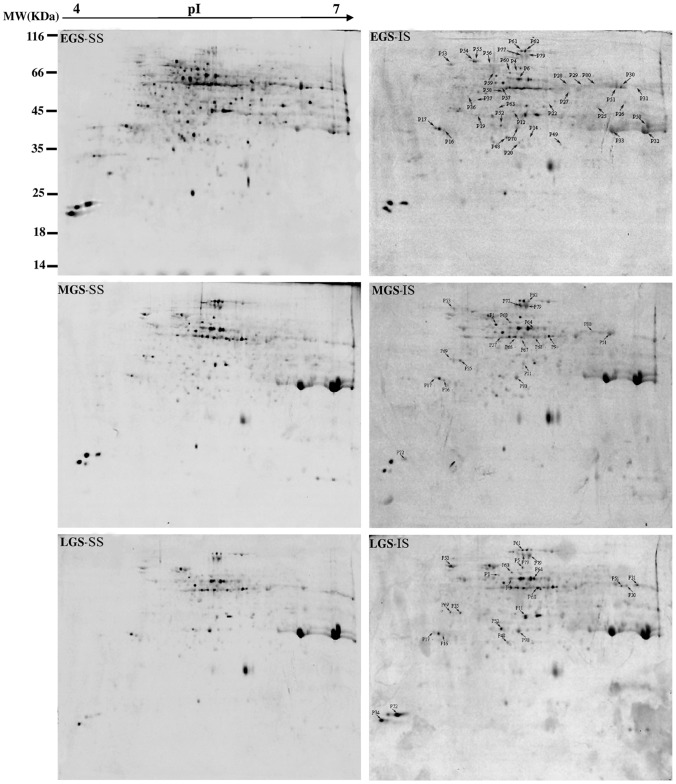
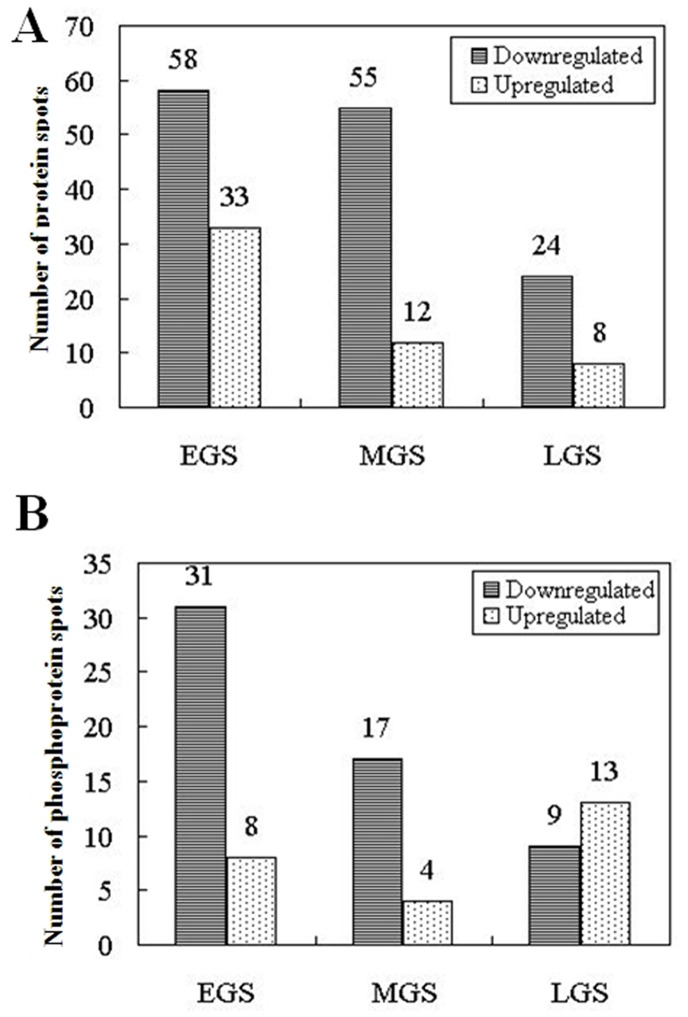
In this study, all proteins from SS and IS were resolved and identified by using the high-resolution 2-DE followed by the CBB (Fig. 3) or Pro-Q Diamond phosphoprotein gel staining (Fig. 4). On the CBB-stained 2-DE gels (Fig. 3), 1,300±75 spots/gel in EGS, 1,350±45 spots/gel in MGS and 1,000±40 spots/gel in LGS could be visualized. It was determined that the intensities of 91 spots in EGS, 67 spots in MGS and 32 spots in LGS differed significantly (≥1.5-fold) between SS and IS. Of these protein spots, 58 (63.7%) were downregulated and 33 (36.3%) upregulated in EGS, 55 (82.1%) downregulated and 12 (17.9%) upregulated in MGS, and 24 (75%) downregulated and 8 (36.3%) upregulated in LGS for IS, as compared to SS (Fig. 5A). The 2-DE maps stained with Pro-Q Diamond resolved 455±40, 425±35 and 390±36 phosphoprotein spots/gel in EGS, MGS, and LGS, respectively (Fig. 4). On IS, a total of 39 spots in EGS, 21 spots in MGS and 22 spots in LGS were found to be differentially phosphorylated. Of these phosphoprotein spots, 31 (79.5%) were downregulated and 8 (20.5%) upregulated in EGS, 17 (81%) downregulated and 4 (19%) upregulated in MGS, and 9 (40.9%) downregulated and 8 (59.1%) upregulated in LGS (Fig. 5B).
Figure 3. Representative 2-DE gels of proteins from superior spikelets (SS) and inferior spikelets (IS) at three grain-filling stages.
Three independent replicates for superior and inferior spikelets at each stage. EGS, MGS and LGS represent early, mid-, and late grain-filling stage, respectively, of endosperm development stages of SS and IS. Proteins prepared from SS and IS at the EGS, MGS and LGS were separated by 2-DE and stained by Coomassie Brilliant Blue. The differentially expressed protein spots between SS and IS at three stages were determined according to the method described in “Materials and methods”. MW (in kilodaltons) and pI of the proteins are shown at left and top, respectively.
Figure 4. Representative 2-DE gels of phosphoproteins from superior spikelets (SS) and inferior spikelets (IS) at three grain-filling stages.
Three independent replicates for SS and IS at each stage. EGS, MGS and LGS represent early, mid-, and late grain-filling stage, respectively, of endosperm development stages of SS and IS. Proteins prepared from the EGS, MGS and LGS were separated by 2-DE and stained by Pro-Q Diamond phosphoprotein. The differentially expressed phosphoprotein spots between SS and IS at three stages were determined according to the method described in “Materials and methods”. MW (in kilodaltons) and pI of the proteins are shown at left and top, respectively.
Figure 5. Patterns of change on differentially expressed protein spots (A) and phosphoprotein spots (B) of inferior spikelets (IS) in comparison to superior spikelets (SS).
EGS, MGS and LGS represent early, mid-, and late grain-filling stage, respectively, of endosperm development stages of SS and IS.
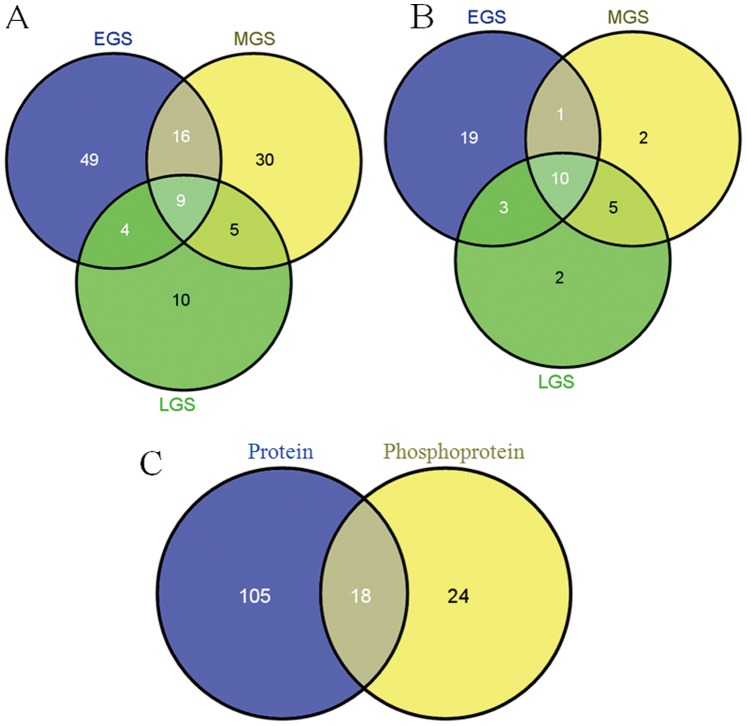
Some of the protein and phosphoprotein spots were further analyzed by MALDI-TOF-MS or LC-ESI-MS/MS. At the end, most of them were successful identified (Table 1 and 2). In all, 81, 61, and 29 unique proteins and 33, 18, and 20 phosphoproteins in EGS, MGS and LGS, respectively, were identified. Among them, 9 proteins and 10 phosphoproteins showed significant differences between SS and IS for all grain-filling stages (Fig. 6A–B). Eighteen proteins, such as 14-3-3, ADPase and Pyruvate orthophosphate dikinase (PPDK), showed significant differences in expression abundance, as well as phosphorylation level, between SS and IS (Fig. 6C), indicating their possible important roles in regulating the grain-filling.
Table 1. List of protein spots with differential expressions between superior and inferior spikelets in 3 grain-filling stages.
| PNa | SNb | Anc | Protein name | Score | Mr(KDa)/pI d | MP.e | CTf | ||||||||||
| EGS | MGS | LGS | |||||||||||||||
| 1- Metabolism | |||||||||||||||||
| 1.1- Sugars conversion | |||||||||||||||||
| 1 | 63g | B9FUS5 | Similar to Xylose isomerase | 243 | 53.8/5.42 | 3 | DR | – | – | ||||||||
| 2 | 271h | Q8H3Q7 | Xylose isomerase | 37 | 53.5/5.66 | 2 | – | UR | – | ||||||||
| 3 | 234h | Q75K72 | Putative beta-1,3-glucanase’ | 37 | 34.7/6.37 | 5 | UR | – | – | ||||||||
| 4 | 213h | Q10DZ9 | Phosphoglucomutase, cytoplasmic 2 | 274 | 62.9/5.63 | 7 | – | DR | – | ||||||||
| 66g | Q10DZ9 | Phosphoglucomutase, cytoplasmic 2 | 243 | 54.7/4.93 | 2 | – | DR | – | |||||||||
| 5 | 273 h | Q9AUQ4 | Phosphoglucomutase | 239 | 62.9/5.63 | 17 | – | DR | – | ||||||||
| 6 | 268h | Q33AE4 | Phosphoglucomutase | 53 | 66.1/6.68 | 2 | – | DR | – | ||||||||
| 7 | 78g | Q93X08 | Similar to UTP–glucose-1-phosphateuridylyltransferase | 108 | 51.8/5.43 | 2 | – | DR | – | ||||||||
| 276h | Q93X08 | UDP-glucose pyrophosphorylase | 355 | 51.7/5.59 | 15 | – | DR | – | |||||||||
| 293h | Q93X08 | UDP-glucose pyrophosphorylase | 143 | 51.7/5.59 | 10 | – | DR | ||||||||||
| 8 | 69g | Q0J8G4 | Similar to Fructokinase | 430 | 35.9/5.02 | 4 | – | DR | – | ||||||||
| 9 | 13g | Q8H8T0 | UDP-L-arabinose mutase 1 | 324 | 40.1/8.21 | 3 | – | DR | – | ||||||||
| 1.2-Starch synthesis | |||||||||||||||||
| 10 | 14g | B8XED2 | ADP-glucose pyrophosphorylase large subunit | 556 | 58.2/5.55 | 4 | DR | – | DR | ||||||||
| 50g | B8XED2 | ADP-glucose pyrophosphorylase large subunit | 355 | 58.2/5.55 | 3 | UR | DR | – | |||||||||
| 49g | B8XED2 | ADP-glucose pyrophosphorylase large subunit | 189 | 58.2/5.55 | 2 | DR | – | – | |||||||||
| 11 | 97g | B8XEC0 | ADP-glucose pyrophosphorylase large subunit | 410 | 58.2/5.71 | 3 | DR | DR | – | ||||||||
| 12 | 141g | P15280 | Similar to Glucose-1-phosphateadenylyltransferase small subunit | 420 | 53.2/5.87 | 3 | – | DR | DR | ||||||||
| 13 | 215h | Q69T99 | Glucose-1-phosphate adenylyltransferase | 500 | 54.8/6.67 | 2 | – | DR | – | ||||||||
| 232h | Q69T99 | Glucose-1-phosphate adenylyltransferase | 52 | 54.8/6.67 | 5 | DR | – | – | |||||||||
| 14 | 98g | A8QXE7 | Granule-bound starch synthase | 347 | 63.4/8.77 | 3 | DR | – | – | ||||||||
| 15 | 226h | Q7X834 | Pullulanase | 954 | 106.4/6.30 | 4 | DR | – | – | ||||||||
| 220h | Q7X834 | Pullulanase | 208 | 106.4/6.30 | 4 | DR | – | – | |||||||||
| 1.3-Pyruvate,orthophosphate dikinase (PPDK) | |||||||||||||||||
| 16 | 83g | Q6AVA8 | Pyruvate,orthophosphate dikinase | 377 | 103.6/5.98 | 3 | DR | – | DR | ||||||||
| 84g | Q6AVA8 | Pyruvate,orthophosphate dikinase | 222 | 103.6/5.98 | 3 | DR | – | DR | |||||||||
| 85g | Q6AVA8 | Pyruvate,orthophosphate dikinase | 367 | 103.6/5.98 | 3 | DR | DR | – | |||||||||
| 1.4- Amino acid | |||||||||||||||||
| 17 | 188g | Q9XEA8 | Cysteine synthase | 547 | 34.4/5.35 | 5 | DR | DR | – | ||||||||
| 18 | 252h | Q0DZE3 | Phenylalanine ammonia-lyase | 231 | 75.5/6.49 | 21 | DR | – | – | ||||||||
| 253h | Q0DZE3 | Phenylalanine ammonia-lyase | 158 | 75.5/6.49 | 16 | DR | – | – | |||||||||
| 19 | 222h | Q93WM3 | Asparaginyl-tRNA synthetase, cytoplasmic 1 | 559 | 62.5/5.90 | 2 | UR | – | – | ||||||||
| 20 | 77g | O49218 | Mthylmalonate semi-aldehyde dehydrogenase | 271 | 57.5/5.99 | 3 | DR | UR | UR | ||||||||
| 21 | 121g | Q8VXC4 | Glycine rich RNA binding protein | 297 | 19.6/6.59 | 2 | – | UR | – | ||||||||
| 22 | 104g | Q84V24 | Aspartate aminotransferase | 282 | 46.0/5.90 | 4 | – | DR | – | ||||||||
| 23 | 284h | Q6ZHC3 | Putative aspartate-tRNA ligase | 100 | 60.8/6.32 | 6 | – | DR | – | ||||||||
| 24 | 46g | Q7XXQ8 | Putative RNA-binding protein | 144 | 41.8/5.08 | 2 | – | DR | – | ||||||||
| 25 | 74g | Q65XK0 | Similar to Ketol-acid reductoisomerase | 288 | 62.7/6.01 | 3 | – | DR | DR | ||||||||
| 26 | 282h | Q2QLY5 | Methionine synthase | 208 | 84.5/6.30 | 13 | – | DR | |||||||||
| 27 | 12g | Q5JNB0 | Similar to Cysteine synthase | 176 | 42.1/6.28 | 2 | – | – | DR | ||||||||
| 28 | 140g | Q651F0 | Similar to 3-dehydroquinate synthase-like protein | 73 | 47.5/8.05 | 2 | – | – | DR | ||||||||
| 1.5- Nucleotides | |||||||||||||||||
| 29 | 111g | P49027 | Guanine nucleotide-binding protein beta subunit-like protein | 394 | 36.7/5.97 | 3 | UR | DR | – | ||||||||
| 30 | 112g | Q2R1V8 | NAD-dependent epimerase/dehydratasefamily protein | 284 | 42.5/5.75 | 3 | DR | – | – | ||||||||
| 31 | 265h | Q5Z6P9 | Putative RAD23 protein | 23 | 43.0/4.77 | 2 | – | UR | – | ||||||||
| 32 | 82g | Q84P58 | Adenosine kinase-like protein | 201 | 40.6/5.57 | 3 | – | – | UR | ||||||||
| 1.6- Lipid | |||||||||||||||||
| 33 | 119g | Q69YA2 | Similar to 3-oxoacyl-[acyl-carrier-protein]synthase I | 99 | 49.5/6.76 | 2 | UR | – | – | ||||||||
| 1.7- Glycolysis | |||||||||||||||||
| 34 | 235h | Q6H6C7 | Phosphoglycerate kinase | 611 | 42.1/5.86 | 19 | DR | – | – | ||||||||
| 236h | Q6H6C7 | Phosphoglycerate kinase | 117 | 42.1/5.86 | 7 | UR | – | – | |||||||||
| 277h | Q6H6C7 | Phosphoglycerate kinase | 448 | 42.1/5.86 | 13 | – | DR | – | |||||||||
| 292h | Q6H6C7 | Phosphoglycerate kinase | 519 | 42.1/5.86 | 22 | – | – | DR | |||||||||
| 35 | 231h | Q75K90 | Phosphoglycerate kinase | 184 | 32.4/9.92 | 9 | DR | – | – | ||||||||
| 36 | 79g | Q53P96 | Fructose-bisphosphate aldolase class-I | 101 | 39.6/6.85 | 2 | DR | UR | – | ||||||||
| 269h | Q53P96 | Fructose-bisphosphate aldolase class-I | 51 | 39.3/7.30 | 6 | – | DR | – | |||||||||
| 1.8- Tricarboxylic acid (TCA) pathway | |||||||||||||||||
| 37 | 144g | Q9ASP4 | Dihydrolipoamide dehydrogenase family protein | 331 | 53.0/5.71 | 3 | UR | – | – | ||||||||
| 38 | 80g | Q7XDC8 | Cytoplasmic malate dehydrogenase | 393 | 35.9/5.75 | 3 | DR | – | – | ||||||||
| Q7XDC8 | Cytoplasmic malate dehydrogenase | 393 | 35.9/5.75 | 3 | – | DR | – | ||||||||||
| 39 | 151g | Q7F280 | Similar to NADP-isocitrate dehydrogenase | 209 | 46.4/6.34 | 2 | – | DR | – | ||||||||
| 40 | 275h | Q6H7M1 | Putative fumarylacetoacetate hydrolase | 79 | 47.1/5.94 | 5 | – | DR | – | ||||||||
| 41 | 274h | Q6Z702 | Putative 3-isopropylmalate dehydratase large subunit | 104 | 55.5/7.33 | 12 | – | DR | – | ||||||||
| 1.9- Pentose phosphate pathway (PPP) | |||||||||||||||||
| 42 | 219h | Q0J8A4 | Glyceraldehyde-3-phosphate dehydrogenase | 337 | 36.7/6.81 | 4 | – | UR | – | ||||||||
| 43 | 65g | Q84ZY2 | Putative transketolase | 319 | 80.6/6.12 | 3 | – | – | DR | ||||||||
| 1.10- Fermentation | |||||||||||||||||
| 44 | 175g | Q10MW3 | Similar to Pyruvate decarboxylase isozyme 3 | 497 | 65.8/5.53 | 5 | DR | DR | – | ||||||||
| 45 | 18g | Q6ZBH2 | Alcohol dehydrogenase superfamily, zinc-containing protein | 483 | 40.0/6.03 | 3 | – | – | DR | ||||||||
| 1.11- Nitrogen | |||||||||||||||||
| 46 | 286h | Q338N8 | Alanine aminotransferase | 170 | 52.6/6.65 | 11 | – | DR | – | ||||||||
| 47 | 157g | P14655 | Glutamine synthetase,chloroplastic | 265 | 49.8/6.18 | 2 | UR | DR | – | ||||||||
| 48 | 2g | P14656 | Glutamine synthetase cytosolic isozyme | 340 | 39.4/5.51 | 3 | – | DR | DR | ||||||||
| 2-Protein synthesis and destination | |||||||||||||||||
| 2.1- Protein folding and modification | |||||||||||||||||
| 49 | 105g | Q9LWT6 | Similar to 60 kDa chaperonin | 289 | 64.3/5.60 | 2 | DR | – | – | ||||||||
| 47g | Q9LWT6 | Similar to 60 kDa chaperonin | 253 | 64.3/5.60 | 2 | DR | – | – | |||||||||
| 50 | 6g | Q53LQ0 | Protein disulfide isomerase | 226 | 57.0/4.95 | 2 | DR | – | – | ||||||||
| 5g | Q53LQ0 | Protein disulfide isomerase | 259 | 57.0/4.95 | 2 | DR | – | – | |||||||||
| 51 | 7 g | Q84P99 | Heat shock-related protein | 337 | 45.0/5.02 | 2 | DR | – | DR | ||||||||
| 52 | 9g | Q0DJB9 | Similar to Stromal 70 kDa heat shock-related protein | 479 | 48.7/4.57 | 3 | DR | – | DR | ||||||||
| 53 | 129g | Q943K7 | Heat shock protein Hsp70 family protein | 440 | 71.3/5.10 | 4 | DR | DR | – | ||||||||
| 54 | 103 g | Q53P60 | Similar to Heat shock 70 kDa protein | 446 | 73.1/5.49 | 4 | DR | UR | DR | ||||||||
| 55 | 240h | Q2QU06 | 60 kDa chaperonin alpha subunit | 21 | 61.1/5.21 | 2 | UR | – | – | ||||||||
| 56 | 224h | Q5Z907 | T-complex protein 1, epsilon subunit | 535 | 59.1/6.00 | 4 | DR | – | – | ||||||||
| 257h | Q5Z907 | Putative T complex protein | 35 | 59.1/6.02 | 2 | DR | – | – | |||||||||
| 57 | 88g | Q69Y99 | Putative chaperonin 21 precursor | 389 | 26.4/7.71 | 2 | UR | – | – | ||||||||
| 58 | 223h | C7J006 | Cupin region domain containing protein | 406 | 45.3/6.50 | 8 | UR | – | – | ||||||||
| 59 | 218 h | C7IZM0 | Proteinase inhibitor I4, serpin family protein | 264 | 29.2/5.47 | 2 | – | DR | – | ||||||||
| 60 | 261h | Q5QLP0 | Putative heat shock protein 82 | 34 | 70.7/5.15 | 2 | – | DR | – | ||||||||
| 61 | 214h | Q9AUV8 | Phosphorylase | 951 | 106.2/5.94 | 11 | – | DR | – | ||||||||
| 279h | Q9AUV8 | Phosphorylase | 602 | 106.2/5.94 | 24 | – | DR | – | |||||||||
| 280h | Q9AUV8 | Phosphorylase | 434 | 106.2/5.94 | 18 | – | DR | – | |||||||||
| 62 | 270h | Q6Z7L1 | Putative dnaK-type molecular chaperone | 104 | 72.9/5.59 | 8 | – | DR | – | ||||||||
| 63 | 262h | Q0D9G9 | Heat shock protein HtpG 10 | 52 | 92.8/4.98 | 5 | – | DR | – | ||||||||
| 64 | 216h | Q9AYD4 | HSP-70 cofactor 5 | 332 | 36.5/4.72 | 5 | – | DR | – | ||||||||
| 65 | 291h | Q2QV45 | 70 kDa heat shock protein | 79 | 74.0/5.25 | 7 | – | – | DR | ||||||||
| 2.2- Protein synthesis | |||||||||||||||||
| 66 | 247h | Q8H3I3 | Putative 40S ribosomal protein | 52 | 33.0/5.08 | 5 | UR | – | – | ||||||||
| 67 | 94g | P29545 | Elongation factor 1 beta’ | 316 | 23.8/4.86 | 3 | DR | DR | – | ||||||||
| 68 | 71g | Q40680 | Similar to Elongation factor 1 beta 2 | 269 | 24.9/4.36 | 2 | – | DR | – | ||||||||
| 69 | 73g | O24182 | Endosperm lumenal binding protein | 264 | 73.7/5.30 | 4 | – | – | DR | ||||||||
| 128g | O24182 | Endosperm lumenal binding protein | 368 | 73.7/5.30 | 3 | – | – | DR | |||||||||
| 130g | O24182 | Endosperm lumenal binding protein | 264 | 73.7/5.30 | 3 | DR | – | – | |||||||||
| 2.3-Proteolysis | |||||||||||||||||
| 70 | 32g | Q6H852 | Proteasome subunit alpha type 2 | 268 | 25.8/5.39 | 3 | UR | – | – | ||||||||
| 71 | 143g | Q5SNJ4 | Similar to Mitochondrial processing peptidase | 249 | 54.1/6.69 | 2 | UR | – | – | ||||||||
| 2.4-Protein storage | |||||||||||||||||
| 72 | 227h | Q0JJ36 | Glutelin | 488 | 56.2/8.87 | 5 | DR | – | – | ||||||||
| 73 | 228h | Q10JA8 | Glutelin | 385 | 56.0/8.53 | 7 | DR | – | – | ||||||||
| 74 | 233h | Q0DH05 | Alpha-globulin | 66 | 21.0/7.50 | 4 | DR | – | – | ||||||||
| 3-Signal transduction | |||||||||||||||||
| 75 | 70g | Q7XTE8 | Similar to 14-3-3-like protein GF14-6 | 246 | 30.0/4.76 | 3 | UR | UR | UR | ||||||||
| 76 | 38g | Q6YZA9 | Germin-like protein 3 | 354 | 19.6/8.89 | 2 | DR | – | – | ||||||||
| 4-Disease/defense | |||||||||||||||||
| 77 | 28g | Q7XUY5 | Bet v I allergen family protein | 238 | 17.3/4.75 | 2 | UR | – | – | ||||||||
| 78 | 191g | Q7XEL9 | Similar to Chitinase 2 | 328 | 31.2/4.48 | 4 | UR | – | – | ||||||||
| 266h | Q7XEL9 | Similar to Chitinase 2 | 71 | 31.2/4.68 | 5 | – | – | DR | |||||||||
| 79 | 23g | Q7XCK6 | Chitinase | 273 | 28.0/6.28 | 2 | UR | – | – | ||||||||
| 80 | 30g | Q5WMX0 | Similar to ChitinaseIII | 219 | 32.8/6.08 | 2 | UR | DR | UR | ||||||||
| 81 | 42g | Q0DRV6 | Superoxide dismutase | 143 | 15.3/5.71 | 2 | UR | – | – | ||||||||
| 82 | 242h | Q0DJ64 | Superoxide dismutase | 164 | 25.0/7.02 | 9 | UR | – | – | ||||||||
| 83 | 243h | Q0J7H9 | Glyoxalase | 39 | 32.5/5.67 | 5 | UR | – | – | ||||||||
| 84 | 92g | Q9FR35 | Peroxiredoxin-2C | 186 | 17.3/5.58 | 2 | UR | – | – | ||||||||
| 85 | 241h | Q65XA0 | Dehydroascorbate reductase | 281 | 23.6/6.21 | 10 | UR | – | – | ||||||||
| 86 | 256h | Q5QNE8 | Putative 4-methyl-5(B-hydroxyethyl)-thiazol monophosphate biosynthesis enzyme | 32 | 45.1/6.30 | 2 | UR | – | – | ||||||||
| 87 | 197h | Q5Z9Y8 | Late embryogenesis abundant protein | 58 | 27.9/4.40 | 4 | DR | DR | – | ||||||||
| 88 | 115g | Q0JL44 | Sgt1 | 431 | 41.0/4.97 | 4 | UR | – | – | ||||||||
| 89 | 27g | Q8H8U5 | Glutathione S-transferase | 322 | 34.0/8.86 | 3 | UR | – | – | ||||||||
| 90 | 36g | Q9FE01 | L-ascorbate peroxidase | 276 | 27.2/5.21 | 3 | DR | – | – | ||||||||
| 91g | Q9FE01 | L-ascorbate peroxidase | 434 | 27.2/5.21 | 3 | – | UR | UR | |||||||||
| 91 | 145g | Q9SXP2 | Nad-dependent formate dehydrogenase | 345 | 41.4/6.87 | 3 | – | DR | – | ||||||||
| 92 | 248h | Q6H660 | Putative stress-induced protein sti1 | 180 | 64.9/6.38 | 16 | DR | – | – | ||||||||
| 283h | Q6H660 | Putative stress-induced protein sti1 | 227 | 64.9/6.38 | 15 | – | DR | – | |||||||||
| 93 | 136g | Q9AVA6 | Putative selenium binding protein | 306 | 51.3/5.73 | 3 | – | DR | – | ||||||||
| 94 | 288h | Q6ZK46 | Putative early embryogenesis protein | 48 | 58.0/8.44 | 2 | – | UR | – | ||||||||
| 5-Transporters | |||||||||||||||||
| 95 | 62g | Q0JKB4 | Similar to ATP synthase beta chain, mitochondrial precursor | 280 | 59.6/6.10 | 2 | DR | – | DR | ||||||||
| 3g | Q0JKB4 | Similar to ATP synthase beta chain, mitochondrial precursor | 595 | 59.6/6.10 | 3 | UR | DR | – | |||||||||
| 96 | 258h | Q6ZG90 | Putative ATP synthase | 31 | 27.3/6.93 | 6 | DR | – | – | ||||||||
| 97 | 225h | Q93W07 | Vacuolar ATPase B subunit | 488 | 54.0/5.20 | 2 | DR | – | – | ||||||||
| 98 | 229h | Q6Z8K7 | Putative H(+)-transporting ATP synthase | 20 | 26.2/5.06 | 3 | UR | – | – | ||||||||
| 99 | 198h | Q651T8 | Putative vacuolar proton-ATPase | 234 | 68.4/5.30 | 15 | DR | – | – | ||||||||
| 100 | 156g | Q01859 | Mitochondrial F1-ATPase beta subunit | 22 | 59.1/6.30 | 2 | DR | UR | – | ||||||||
| 101 | 190g | P0C521 | ATP synthase F0 subunit 1 | 471 | 55.5/5.85 | 3 | – | DR | – | ||||||||
| 102 | 303 h | Q5N7P8 | ATP synthase subunit beta | 135 | 45.2/5.43 | 2 | – | – | DR | ||||||||
| 6-Cell growth/division | |||||||||||||||||
| 103 | 1g | P0C539 | Actin | 408 | 42.2/5.72 | 3 | DR | – | – | ||||||||
| 104 | 177g | Q8S3Q3 | Similar to Possible apospory-associated like | 97 | 38.1/5.10 | 3 | DR | – | – | ||||||||
| 105 | 122g | P35681 | Similar to Translationally controlled tumor protein | 140 | 19.0/4.51 | 2 | DR | – | – | ||||||||
| 106 | 263h | Q10CU1 | Tubulin beta-7 chain | 25 | 49.8/4.87 | 4 | DR | DR | |||||||||
| 7-Secondary metabolism | |||||||||||||||||
| 107 | 166g | Q9XGP7 | Similar to Caffeoyl-CoA O-methyltransferase 2 | 514 | 27.9/5.11 | 3 | DR | – | – | ||||||||
| 108 | 87g | Q9XJ19 | Similar to Caffeoyl-CoA 3-O-methyltransferase | 396 | 29.0/5.21 | 3 | – | – | UR | ||||||||
| 109 | 127g | Q75LL5 | Putative strictosidine synthase | 17 | 54.2/8.75 | 2 | UR | – | – | ||||||||
| 110 | 95g | Q9SXV0 | Similar to Cytochrome C oxidase subunit 6b | 52 | 19.1/4.27 | 2 | UR | – | – | ||||||||
| 111 | 61g | Q10Q21 | Similar to Cytochrome C reductase-processing peptidase subunit I, MPP subunit I, P55 | 160 | 58.9/5.49 | 2 | DR | – | – | ||||||||
| 51g | Q10Q21 | Similar to Cytochrome C reductase-processing peptidase subunit I, MPP subunit I, P55 | 387 | 58.9/5.49 | 3 | DR | DR | ||||||||||
| 112 | 25g | Q84T92 | Chalcone isomerase | 153 | 24.0/5.15 | 2 | – | – | UR | ||||||||
| 9 -Photosynthesis | |||||||||||||||||
| 113 | 26g | Q6Z3V7 | Putative Photosystem I reaction center subunit IV | 178 | 15.5/9.64 | 2 | DR | – | – | ||||||||
| 114 | 8g | P93431 | Ribulose-1,5-bisphosphate carboxylase/oxygenase activase | 498 | 48.1/5.85 | 3 | UR | UR | – | ||||||||
| 86g | P93431 | Ribulose-1,5-bisphosphate carboxylase/oxygenase activase | 352 | 48.1/5.85 | 3 | – | DR | – | |||||||||
| 115 | 48g | Q7X9A7 | Putative rubisco subunit binding-protein alpha subunit precursor | 717 | 61.5/5.36 | 5 | DR | – | – | ||||||||
| 116 | 68g | Q2QW49 | Ribulose bisphosphate carboxylase large chain precursor | 300 | 56.5/9.04 | 3 | – | DR | – | ||||||||
| 108g | Q2QW49 | Ribulose bisphosphate carboxylase large chain precursor | 389 | 56.5/9.04 | 4 | – | – | DR | |||||||||
| 117 | 133g | Q40701 | 23 kDa polypeptide of photosystem II | 336 | 27.1/9.54 | 3 | – | DR | – | ||||||||
| 118 | 285h | Q8S6F2 | Putative rbcL; RuBisCO large subunit from chromosome 10 chloroplast insertion | 361 | 52.9/6.92 | 16 | – | DR | – | ||||||||
| 119 | 304h | Q5QLG3 | Phosphoribulokinase/uridine kinase-like | 89 | 46.1/6.90 | 3 | – | – | UR | ||||||||
| 10- Unclassified | |||||||||||||||||
| 120 | 244h | Q8H684 | OSEYA1 | 52 | 34.1/5.11 | 4 | DR | – | – | ||||||||
| 121 | 230 h | Q0DUA3 | Os03g0197300 protein | 28 | 68.2/5.71 | 3 | DR | – | – | ||||||||
| 122 | 180g | Q53RR0 | Mov34/MPN/PAD-1 family, putative | 16 | 139.3/9.93 | 2 | UR | – | – | ||||||||
| 123 | 123g | B8BKE8 | Hypothetical protein OsI_36050 | 23 | 17.1/11.15 | 2 | DR | – | – | ||||||||
Note:
proteins number,
protein spots number correspond to those on 2-DE gels shown in Fig. 3;
accession number;
theoretical MW (kDa) and pI.
match peptides;
changes on protein spots in superior spikelets compared to inferior spikelets; UR: protein upregulated in inferior spikelets as compared to superior spikelets; DR: protein downregulated in inferior spikelets as compared to superior spikelets; EGS: early grain-filling stage, MGS: mid-grain-filling stage, LGS: late grain-filling stage;
protein spot identified by MALDI-TOF-MS;
protein spot identified by LC-ESI-MS/MS.
Table 2. List of phosphoprotein spots with differential expressions between superior and inferior spikelets in 3 grain-filling stages.
| PNa | SNb | Anc | Protein name | Score | Mr(KDa)/pI e | MP.f | CTg | ||
| EGS | MGS | LGS | |||||||
| 1 Metabolism | |||||||||
| 1.1 Sugars conversion | |||||||||
| 1 | P 1 | Q33AE4 | Phosphoglucomutase | 106 | 66.1/6.70 | 9 | – | DR | DR |
| 2 | P5 | Q9AUQ4 | Phosphoglucomutase | 276 | 62.9/5.60 | 22 | – | – | UR |
| 3 | P 9 | Q93X08 | UDP-glucose pyrophosphorylase | 134 | 51.7/5.60 | 2 | – | DR | – |
| 4 | P26 | Q6ZBH2 | Putative sorbitol dehydrogenase | 263 | 39.3/6.47 | 18 | DR | – | – |
| 5 | P63 | Q8S9Z2 | Putative dTDP-glucose 4,6-dehydratase | 39 | 44.3/6.25 | 5 | DR | – | – |
| 6 | P61 | Q9AUV8 | Phosphorylase | 352 | 106.2/5.94 | 17 | UR | – | DR |
| P62 | Q9AUV8 | Phosphorylase | 258 | 106.2/5.94 | 18 | UR | DR | – | |
| 1.2 Starch synthesis | |||||||||
| 7 | P6 | Q5VNT5 | Glucose-1-phosphate adenylyltransferase | 353 | 57.0/5.70 | 15 | DR | – | – |
| P68 | Q5VNT5 | Glucose-1-phosphate adenylyltransferase | 278 | 57.0/5.70 | 19 | – | DR | DR | |
| 8 | P80 | A8QXE7 | Granule-bound starch synthase | 347 | 63.4/8.77 | 3 | DR | DR | – |
| 1.3 Pyruvate, phosphate dikinase (PPDK) | |||||||||
| 9 | P77 | Q6AVA8 | Pyruvate, phosphate dikinase | 794 | 93.6/5.50 | 26 | DR | – | DR |
| P79 | Q6AVA8 | Pyruvate, phosphate dikinase | 794 | 93.6/5.50 | 26 | DR | – | DR | |
| 1.4 Amino acid metabolism | |||||||||
| 10 | P19 | Q5JNB0 | Cysteine synthase | 33 | 41.8/6.70 | 3 | DR | – | – |
| 11 | P28 | Q0INQ6 | Serine hydroxymethyltransferase | 52 | 50.7/7.69 | 5 | DR | – | – |
| 12 | P30 | Q338N8 | Alanine aminotransferase | 147 | 52.6/6.70 | 10 | DR | – | UR |
| P51 | Q338N8 | Alanine aminotransferase | 291 | 52.6/6.65 | 21 | DR | DR | UR | |
| 13 | P37 | Q0DPW7 | 3-isopropylmalate dehydrogenase | 168 | 41.2/5.40 | 15 | DR | – | – |
| 14 | P22 | Q6H7M1 | Putative fumarylacetoacetate hydrolase | 79 | 47.1/5.94 | 8 | DR | – | – |
| 1.5 Nucleotides | |||||||||
| 15 | P35 | Q6K1R5 | Putative adenosine kinase | 154 | 37.0/5.16 | 14 | – | DR | UR |
| 1.6 Lipid | |||||||||
| 16 | p52 | Q6Z0I4 | Putative enoyl-ACP reductase | 60 | 39.1/8.68 | 7 | DR | – | DR |
| P70 | Q6Z0I4 | Putative enoyl-ACP reductase | 123 | 39.1/8.70 | 10 | – | UR | UR | |
| 1.7 Glycolysis | |||||||||
| 17 | P4 | Q5QMK7 | Phosphoglycerate mutase | 423 | 60.8/5.68 | 20 | DR | – | – |
| P60 | Q5QMK7 | Phosphoglycerate mutase | 310 | 60.8/5.68 | 19 | DR | DR | UR | |
| 18 | P25 | Q6H6C7 | Phosphoglycerate kinase | 28 | 42.1/5.86 | 3 | DR | – | – |
| 19 | P72 | A6MZY0 | Phosphoglycerate kinase | 29 | 18.0/4.70 | 2 | – | DR | UR |
| 20 | P67 | Q10P35 | Enolase 2, putative, expressed | 436 | 47.9/5.50 | 14 | – | DR | – |
| P74 | Q10P35 | Enolase 2, putative, | 26 | 47.9/5.50 | 2 | – | – | UR | |
| 2 Protein synthesis and destination | |||||||||
| 2.1 Protein synthesis | |||||||||
| 21 | P58 | Q6ZI53 | Elongation factor Tu | 156 | 50.4/6.64 | 10 | DR | – | – |
| 22 | P56 | Q6Z058 | Putative Luminal binding protein 5 | 154 | 73.5/5.22 | 6 | DR | – | – |
| 23 | P31 | Q5W6H1 | Putative DNA-binding protein GBP16 | 154 | 43.2/7.08 | 12 | DR | – | UR |
| 2.2 Protein folding and modification | |||||||||
| 24 | p55 | Q0J0U8 | Heat shock protein | 148 | 80.1/5.06 | 17 | UR | – | – |
| 25 | p54 | Q5QLP0 | Putative heat shock protein 82 | 118 | 70.7/5.15 | 13 | UR | – | – |
| 26 | P3 | Q8H903 | 60 kDa chaperonin | 343 | 60.8/5.87 | 23 | – | – | UR |
| 27 | P53 | Q2QV45 | 70 kDa heat shock protein | 682 | 74.0/5.25 | 34 | UR | DR | UR |
| 28 | P57 | Q6Z7B0 | Dnak-type molecular chaperone Bip | 522 | 73.3/5.19 | 22 | DR | UR | – |
| P69 | Q6Z7B0 | Dnak-type molecular chaperone | 427 | 73.3/5.20 | 13 | – | DR | UR | |
| 2.3 Storage protein | |||||||||
| 29 | P32 | Q0JJ36 | Glutelin | 488 | 56.2/8.87 | 5 | UR | – | – |
| p50 | Q0JJ36 | Glutelin | 320 | 56.2/8.87 | 7 | UR | – | – | |
| 30 | P33 | Q10JA8 | Glutelin | 385 | 56.0/8.53 | 7 | UR | – | – |
| P36 | Q10JA8 | Glutelin | 30 | 56.0/8.53 | 2 | DR | – | – | |
| 31 | P49 | Q6YTX6 | Seed protein | 15 | 27.4/7.64 | 4 | DR | – | – |
| 3 Signal transduction | |||||||||
| 32 | P16 | Q10E23 | 14-3-3 protein | 279 | 29.2/4.90 | 8 | DR | DR | DR |
| 33 | p17 | Q7XTE8 | Similar to 14-3-3-like protein GF14-6 | 246 | 30.0/4.76 | 3 | DR | DR | DR |
| 4 Transporters | |||||||||
| 34 | P29 | Q8S7T5 | ATP synthase subunit alpha | 220 | 55.1/6.25 | 14 | DR | – | – |
| 35 | P59 | Q651T8 | Vacuolar proton-ATPase | 175 | 68.4/5.34 | 15 | DR | – | – |
| 36 | P14 | Q6H7I9 | ATP-dependent Clp protease proteolytic subunit | 27 | 31.9/7.20 | 2 | DR | – | – |
| 37 | P66 | Q8S6F3 | ATP synthase subunit beta | 430 | 54.0/5.50 | 20 | – | DR | |
| 5 Cell growth/division | |||||||||
| 38 | P27 | Q0PVB0 | Alpha-tubulin | 71 | 49.6/5.06 | 5 | DR | – | – |
| 39 | P11 | Q6ZK46 | Early embryogenesis protein | 171 | 58.0/8.40 | 5 | – | DR | UR |
| 6 Secondary metabolism | |||||||||
| 40 | P12 | Q19BJ6 | Flavone O-methyltransferase | 125 | 39.7/5.66 | 11 | DR | – | – |
| P64 | Q19BJ6 | Flavone O-methyltransferase | 41 | 39.7/5.70 | 5 | – | DR | DR | |
| 7 Photosynthesis | |||||||||
| 41 | P20 | Q6Z8F4 | Phosphoribulokinase | 110 | 44.8/6.02 | 11 | DR | – | – |
| 8 Unknown | |||||||||
| 42 | P48 | Q0JDG9 | Os04g0404400 protein | 15 | 31.2/5.02 | 2 | DR | – | UR |
Note:
phosphoproteins number,
phosphoprotein spots number correspond to those on 2-DE gels shown in Fig. 4;
score;
theoretical MW (kDa) and pI.
Match peptides;
changes of phosphoprotein spots in inferior spikelets as compared to superior spikelets, UR: phosphoprotein upregulated in inferior spikelets as compared to superior spikelets; DR: phosphoprotein downregulated in inferior spikelets as compared to superior spikelets; EGS: early grain-filling stage, MGS: mid-grain-filling stage, LGS: late grain-filling stage.
Figure 6. Differentially expressed protein and phosphoprotein for superior and inferior spikelets.
A: Code numbers of proteins at three stages. B: Code numbers of phosphoproteins at three stages. C: Comparative proteins and phosphopoteins.
Function Classification of Differentially Expressed Proteins and Phosphoproteins for SS and IS
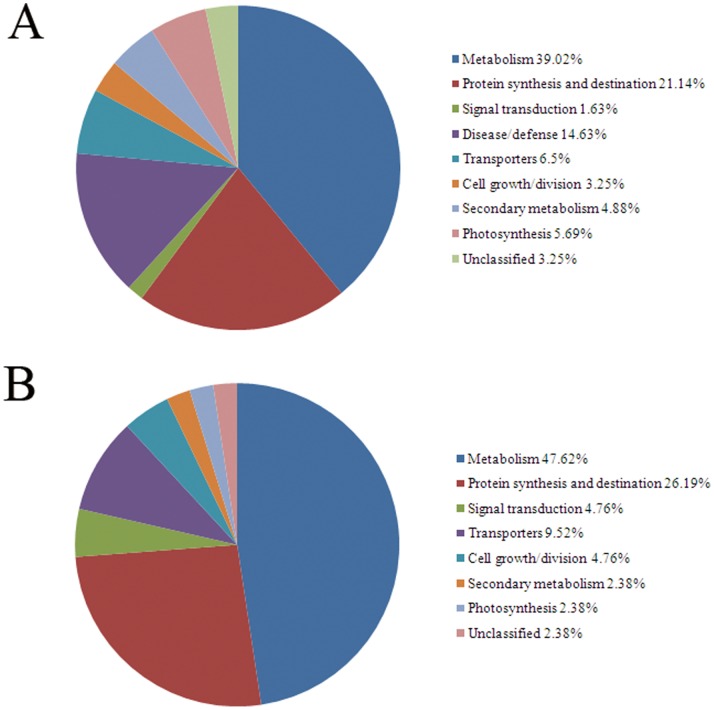
According to genetic ontology, the identified proteins and phosphoproteins were classified into 8 functional categories, i.e., metabolism, protein synthesis/destination, signal transduction, disease/defense response, transporter, cell growth/division, secondary metabolism and photosynthesis (Fig. 7, Table 1 and 2). Those proteins and phosphoproteins without appropriate genetic ontology terms and/or could not be classified into the 9 categories were marked as “unknown” in this report.
Figure 7. Function classifications of identified proteins (A) and phosphoproteins (B).
Of the 123 identified proteins, 39.02% were associated with the metabolism, and 21.14% the protein synthesis/destination (Fig. 7A). For the 43 phosphoproteins, 46.62% were grouped with the metabolism and 26.19% the protein synthesis/destination (Fig. 7B). The metabolism and protein synthesis/destination functions seemed crucial in differentiating the grain-fillings between SS and IS.
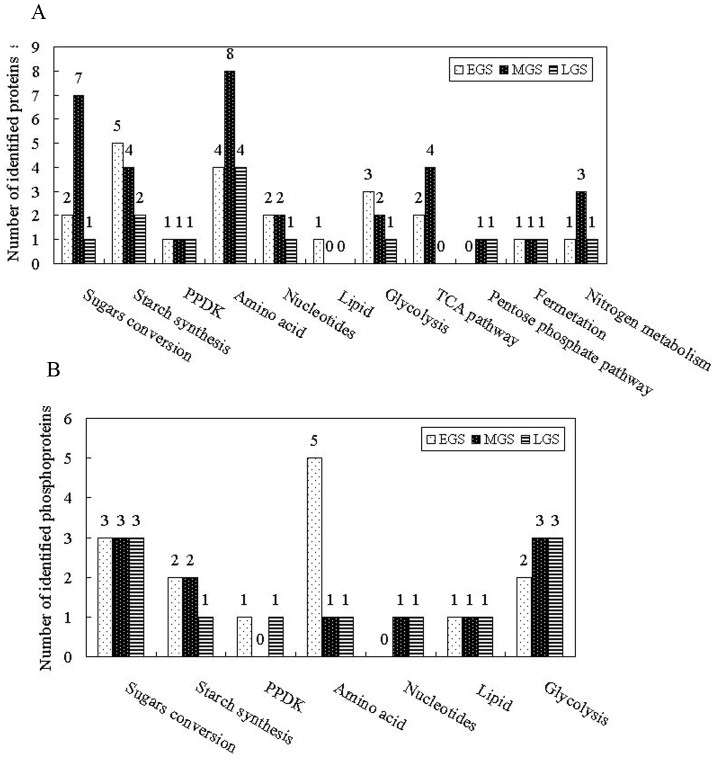
To analyze the metabolic processes of SS and IS at different grain-filling stages, the proteins involved were further divided into 11 subcategories including sugar conversion, starch synthesis, PPDK, amino acid metabolism, nucleotide metabolism, lipid metabolism, glycolysis, tricarboxylic acid (TCA) pathway, pentose phosphate pathway (PPP), fermentation and nitrogen metabolism (Fig. 8A, Table 1). And, the phosphoproteins were divided into 7 subcategories that included sugar conversion, starch synthesis, PPDK, amino acid metabolism, nucleotide metabolism, lipid metabolism and glycolysis (Fig. 8B, Table 2). Plant PPDK was initially discovered in C4 leaves, and recently found to be abundant in developing wheat [19], maize [20] and rice endosperm [21] but the function of PPDK in seed development remains to be elucidated. Therefore, PPDK proteins were organized as an independent category.
Figure 8. Function classifications of identified metabolism-related proteins (A) and phosphoproteins (B).
Western Blotting of Differentially Expressed Proteins
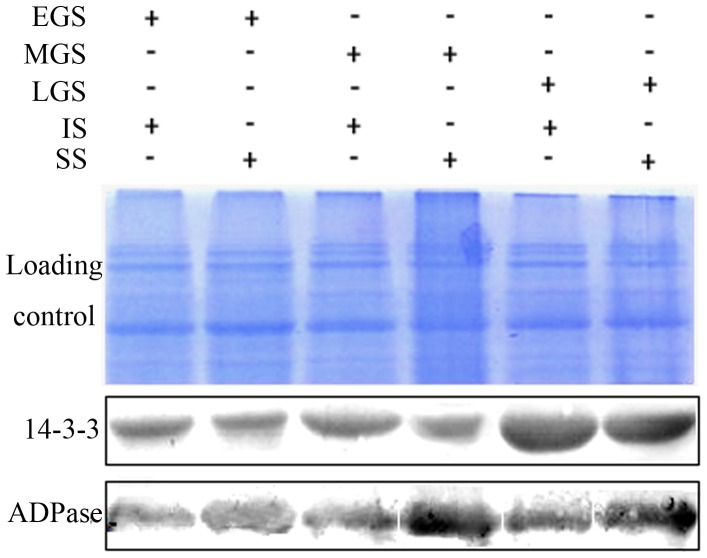
Western blotting was conducted to confirm the differentially expressed proteins detected by 2-DE gels using the mouse antisera as the primary and the HRP-antimouse IgG as the secondary antibodies against 14-3-3 and ADPase (Fig. 9). The 14-3-3 spots showed unregulated and ADPase spots downregulated on IS as compared to SS for all three grain-filling stages.
Figure 9. Western blotting for comparing 14-3-3 protein and ADPase expressions of superior spikelets (SS) and inferior spikelets (IS) at three grain-filling stages.
Gels were divided into two parts at a molecular mass of approximately 66-3-3 protein and ADPase. EGS, MGS and LGS represent early, mid-, and late grain-filling stage, respectively, of endosperm development stages of SS and IS.
Discussion
Physiological Differences between SS and IS
Both grain size and weight of SS were greater than those of IS. It is commonly believed that the poor IS grain-filling was due to a limited carbohydrate supply [22], [23]. However, the present study showed that the soluble carbohydrates in grains on IS was higher than SS during EGS and MGS, indicating that the carbohydrate supply should be more abundant in IS than SS. Then, in LGS, the carbohydrate content became lower on IS than SS. It might be caused by a nutritional scarcity and/or plant senescence. Apparently, the difference of SS and IS grain-filling must be a highly complex process, and to clarify it would require a further exploration on its molecular mechanism. Thus, from the standpoint of protein expression and phosphorylation, we took the proteomic approach to investigate the differences in them between SS and IS during the grain-filling stages of the rice.
Different Proteomic Characteristics of SS and IS Displayed at Three Grain-filling Stages
A comprehensive proteomic analysis with 2-DE was performed to show a total of 156 protein spots with differential abundances by more than 1.5-fold between SS and IS at three grain-filling stages. All of them (represented by 123 unique proteins) were successfully identified by MALDI- TOF- MS or LC-ESI-MS/MS. To verify that some of the differentially expressed proteins were indeed regulated by a post-translational mechanism, such as protein phosphorylation, the 2-DE gels of SS and IS at those grain-filling stages were stained with Pro-Q Diamond dye. The proteomics coupled with the Pro-Q Diamond staining allowed a positive identification of phosphorylated proteins as well as the changes occurred, as Pro-Q Diamond binds specifically to the phosphate moiety of phosphoproteins with high sensitivity and linearity [24], [25]. As a result, 54 phosphoprotein spots (represented by 43 unique phosphoproteins) with significant differences, as assessed by their fluorescence intensities, were identified to be induced by the phosphorylation (≥1.5-fold).
Among the identified proteins and phosphoproteins, 18 proteins showed significant differences in both expression abundance and phosphorylation. Of the 18 proteins, 11 differed defferently at a same grain-filling stage. For example, the expression abundance of 14-3-3 in IS was higher than that in SS at all stages. Contrarily, the phosphorylation was lower in IS. Therefore, the protein expression and the phosphorylation were indeed two different and independent properties in the grain-filling process. Base on the proteomic analysis, the number of differentially expressed proteins or phosphoproteins in EGS was larger than that in MGS or LGS. It indicated that the difference between SS and IS occurred in EGS was most crucial for the grain-filling. In all three grain-filling stages, merely 16 proteins (13%) and 10 phosphoproteins (23.3%) showed significant differences between SS and IS. It suggested that the molecular mechanisms varied at different grain-filling stages as well.
The differentially expressed proteins and phosphoproteins were further annotated by a GO analysis. According to the molecular functions and biochemistry, they were likely to be involved in the biological metabolic processes such as metabolism, protein synthesis and destination, cell growth/division, disease/defense, transporters and signal transduction. A MicroRNA analysis between SS and IS identified 189 differentially expressed microRNAs involved in similar biological metabolic processes [26]. And, our study also found the largest number of the identified proteins or phosphoproteins to be associated with metabolism. This suggested that metabolism could be the most critical function that differentiated the grain-fillings between SS and IS.
Low Activity of Starch Synthesis-related Enzymes and Poor IS Grain-filling
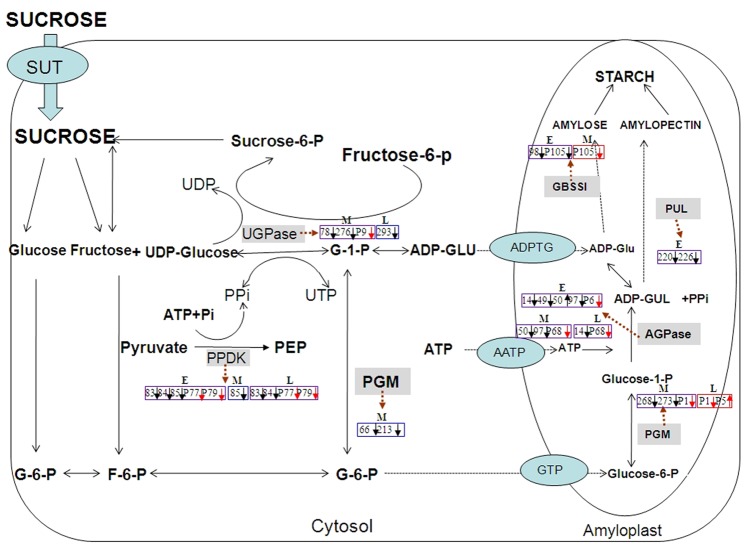
As previous reports indicated, low activities of the starch synthesis enzymes, such as SuSase, AGPase, StSase, and SBE, closely related to the poor grain-filling of IS [10]–[12], [27]. Recently, the DNA microarray analysis showed that there were more enhanced expression profiles and higher transcript levels on a group of genes related to the starch metabolism on SS than IS [17]. Our comparative proteomic study indicated that most of such proteins were downregulated on IS at different grain-filling stages. It seemed to confirm, at protein expression level, that the low activity of the starch synthesizing enzymes was the main reason that caused the poor grain-filling of IS. On the basis of our proteomic data and other published results in literature [28], we proposed a model for the defference regulatory networks of starch synthesis between SS and IS filling (Fig. 10). A difference in the enzymatic activity and carbohydrate content between SS and IS was indeed evident in our study which led to the following conclusion. During EGS, the lower abundance of starch synthesis proteins (i.e., AGPase, GBSS and Pul) on IS, i.e., AGPase, granule-bound starch synthase (GBSS) and pullulanase (Pul), slowed the starch accumulation, as compared to SS. And, in MGS, the downregulated expression of sugar converting proteins, i.e., phosphoglucomutase (PGM) and UDP-glucose pyrophosphorylase (UDPase), and starch synthesizing proteins (AGPase) further slowed the grain-filling. Finally, in LGS, IS had a low protein abundance of UDPase and AGPase accompanied by a low carbohydrate content leading to the low starch accumulation in IS.
Figure 10. Difference on starch biosynthesis pathways between superior spikelets (SS) and inferior spikelets (IS).
Model of starch biosynthetic pathway according to Reference [28]. Blue pane represents differentially expressed proteins, red pane differed phosphorylation, and purple pane either differentially expressed proteins or differed phosphorylation. E, M and L represent early, mid-, and late grain-filling stage, respectively, of endosperm development stages of SS and IS. Black arrow represents proteins downregulated or upregulated in IS as compared to SS. Red arrow represents phosphoproteins downregulated or upregulated in IS as compared to SS. PGM: Phosphoglucomutase; UGPase: UDP-glucose pyrophosphorylase; AGPase: ADPglucose pyrophosphorylase; ADPGT: ADPglucose transporter; AATP: plastidic ATP transporter; GTP: Glucose-6-Phosphate transporter; GBSS: Granule bound starch synthase; Pul: Pullulanase; PPDK: Pyruvate, orthophosphate dikinas.
The starch synthesis enzyme activity has been shown to be regulated by the protein phosphorylation [29], [30]. In the present study, 5 starch synthesis proteins, i.e., PGM, UDPase, phosphorylase, AGPase and GBSS, were stained with Pro-Q Diamond dye to reveal the differences between SS and IS on phosphorylation. These proteins have been identified as phosphoproteins previously. PGM activity can be significantly increased by the signaling kinase p21-activated kinase 1 (PAK1)-mediated phosphorylation of PGM selectively on threonine 466 [31]. A resent study found that the serine phosphorylation of Wx protein played an important role in regulating GBSS activity at the posttranslational level [32]. Consequently, phosphorylation could induce changes in the starch synthesis enzyme activity. Moreover, protein phosphorylation is considered a prerequisite for the formation of the starch synthesizing protein complex (SSPC)in cereal endosperm [29], [33], [34]. And, in wheat and maize amyloplast, the starch synthesis enzymes, such as AGPase, SBE, and StSase, can assemble into a SSPC to function in the starch biosynthesis [20], [35]. The formation of SSPC in cereal endosperm explains how the mutations, which affect specific starch biosynthesis enzymes, have pleiotropic effects on other enzymes in the starch synthesis pathway. Different levels of phosphorylation in starch synthesis enzymes might lead to varied expressions of SSPC in SS and IS as well.
There are also other SSPC enzymes, such as PPDK found in maize and the wheat amyloplast SSPC [20] that lack defined roles in starch synthesis. For example, we identified 3 distinctive PPDK isoforms in SS and IS. It catalyzes the reversible conversion of pyruvate, ATP and Pi into phosphoenolpyruvate (PEP), AMP, and PPi. A recent proteomic study showed the differentially expressed PPDK isoforms in developing wheat [19], maize [20] and rice endosperm [21], indicating its close relationship with the seed development. A protein expression analysis on maize endosperms showed that PPDK might play an important role in the starch-protein balance through the inorganic pyrophosphate-dependent restriction of ADP-glucose synthesis [19]. Hence, it was speculated that the different abundance of protein expressions and phosphorylation’s of PPDK might contribute to the varied activities of the starch synthesis enzymes in SS and IS. The hypothesis has yet been proven, and PPDK’s role in the grain-filling remains to be further studied.
Slowed Glycolysis, TCA Cycle and Alcohol Fermentation vs. Reduced Energy Supply and Building Materials for IS
The central carbon metabolism (glycolysis and TCA cycle) provides energy, cofactor regeneration, and building materials for the interconversions and synthesis of metabolites [36], [37]. The change in the concentration of the metabolites is thus used as an indicator for the biological process. Likewise, the dynamic changes of the protein expression during the grain development of rice reflected the progress of the glycolysis and TCA cycle, which peaked in EGS or MGS. The rice grains were filled from EGS to MGS going through the sink establishment in EGS by endosperm cell enlargement, and thereafter, the starch synthesis. Completion of the prominent glycolysis and TCA cycle in EGS and MGS require an adequate supply of ATP and the synthesis of cellular components essential for the cell enlargement and starch synthesis [21]. In our study, the expression abundance of most of the identified proteins relating to the glycolysis and TCA cycle was found to be lower in the grains on IS than SS. The key glycolysis enzymes were downregulated during EGS and MGS, and those associated with the TCA cycle in MGS. The results suggested that the slowed glycolysis and TCA cycle in IS during EGS and MGS reduced the progress of the endosperm cell enlargement and starch synthesis, which were detrimental to the sink establishment in the grains.
Three glycolysis proteins, phosphoglycerate mutase (PGAM), phosphoglycerate kinase (PGK) and enolase, were detected by means of Pro-Q Diamond staining. PGK was also identified in oilseed rape by the same staining method, and its phosphopeptide by LC-MS/MS/MS [25]. PGAM phosphorylation was detected in arabidopsis seeds [38]. And, the enolase was phosphorylated on a single tyrosine [39]. These enzymes encountered varied phosphorylation on SS and IS, indicating that the glycolysis in IS could be artificially altered through phosphorylation manipulation. Our earlier phosphoproteomic work [15] showed that the addition of ABA could indeed change the phosphorylation of PGK and enolase in IS to promote the grain-filling.
During LGS, the two key enzymes in alcohol fermentation, ADH and transketolase, were downregulated in IS. The fermentation is a two-step process that includes the branching from the glycolysis pathway at pyruvate with concomitant oxidization of NADH to NAD+, and then, the generating of ATP anaerobically [40]. In a typical development of seeds or bulky organs (such as wheat seeds and potato tubers), the internal oxygen concentration is greatly reduced [41]–[43]. The upregulation of alcohol fermentation pathway is very important for maintaining an appropriate ATP level for the starch synthesis under low oxygen tension [21]. Thus, downregulation of the alcohol fermentation in IS at LGS might also cause a reduction on ATP limiting the normal synthesis of starches.
Consequently, it appeared that the changes occurred on the expression and phosphorylation of the proteins associated with glycolysis, TCA cycle and/or alcohol fermentation could bring about the poorer-than-expected grain-filling on IS.
Difference on N Metabolisms between SS and IS
Adequate supply of N is crucial for rice grain-filling, as it directly affects the carbon assimilation and biomass accumulation in the grains [44], [45]. In the present study, alanine aminotransferase (AlaAT) and glutamine synthetase (GS) associated with N metabolism were identified with 3 protein spots. A previous report showed AlaAT gene expressed at high levels in the developing rice grains, and suggested its involvement in N metabolism during the maturation of the rice grains [46]. GS activity in rice grains was found to be positively related to the grain yield [47]. The expressions of GS and AlaAT were both downregulated on IS in MGS and LGS, possibly resulted from an insufficient N supply in the paddy soil that led to an accelerated aging of the rice plants at the later stages. Our earlier proteomic studies showed that an appropriate N fertilization at the late growth stage could significantly promote the GS protein accumulation on IS [45], as suggested by the abovementioned hypothesis. Thus, the low expressions of AlaAT and GS during MGS and LGS could be additional factors affecting the grain-filling of IS.
Low Expression Abundance of Cell Growth/Division Proteins and Grain-filling of IS
Abundant evidences have shown that the low endosperm cell division rate and cell count can cause poor grain-filling on IS [2], [4], [13]. Our study identified 4 proteins and 2 phosphoproteins relating to the IS cell growth/division. The translationally controlled tumor protein (TCTP) was identified and downregulated on IS in EGS. TCTP has been implicated in important cellular processes, such as cell growth, cell cycle progression and malignant transformation, as well as the protection of cells against various stress conditions and apoptosis [3]. TCTP has also been shown to interact with elongation factors, eEF1A [48] and tubulin [49]. The eEF1A functions in the coordinated regulation of the multiple cellular processes including growth, division, and transformation [50]. Tubulin has previously been associated with the cell division during embryogenesis and in a number of environmental and developmental signals [51], [52]. Recently, arabidopsis TCTP (AtTCTP) was found to function in the ABA-mediated stomatal movement, in addition to regulation of the plant growth [49]. In general, TCTP is considered to play a role in (1) cell cycle control, (2) growth factor, (3) ABA response, and (4) interaction with elongation factors, eEF1A and tubulin. Interestingly, two putative TCTP interaction proteins, elongation factor 1 beta and tubulin, were also downregulated and had a very similar downregulation profile like TCTP in EGS on IS. It suggested a possible involvement of this pathway in the IS cell division as well. In addition, tubulin can be stained by Pro-Q Diamond dye to differentiate phosphorylation between SS and IS. And, the phosphorylation sites of tubulin were determined during the development of oilseed rape seeds, and its phosphorylation was considered to be important for the cell division [25].
GLP have been found in various organs (i.e., leaves, roots, and floral tissues) in plants, and under different physiological conditions (e.g., seed germination, stress, and pathogen attack) [47], [53]. In plum fruits, two GLPs genes were identified, and their transcripts were upregulated with the increase of endogenous auxin, indicating that GLPs could be part of the auxin signal network that mediates the cell division and expansion [54]. Auxin is an important signal in the cereal endosperm development [55], and, low auxin leads to a low cell division on rice IS [13]. In the present study, the GLP expression was lower on IS in EGS, indicating a possible difference existed in the auxin signal pathways between SS and IS that led to a lowered cell division of IS.
Previous reports showed that the application of ABA and auxin (IAA) in EGS significantly increased the endosperm cell division rate and cell count on IS [4]. Collectively, it was concluded that, in addition to regulating the cell count of IS, TCTP with eEF1A and tubulin functioned in the ABA-mediated cell development, and that GLP in the auxin signal pathway.
14-3-3 Protein and Grain-filling of IS
In our study, 14-3-3 proteins from SS and IS differed significantly in regard to both protein expression abundance and phosphorylation. The proteins are a family of conserved regulatory molecules expressed in all eukaryotic cells [56]–[58]. They are capable of binding a multitude of functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors [59]. A recent proteomic study on soybean and oilseeds also found the proteins highly expressed during the seed development with an involvement in the signaling and metabolic pathways [25], [60]. Furthermore, 14-3-3 are phosphopeptide-binding proteins with its phosphorylation detected in oilseed rape [25]. However, no in vivo phosphorylation of the proteins in rice grains has been identified thus far.
Stained with Pro-Q Diamond dye, 14-3-3 protein showed phosphorylation levels different between SS and IS. Based on the following analysis, it appeared that 14-3-3 protein played a critically important role in the poor IS grain-filling. Firstly, in a proteomic interaction analysis on wheat and barley endosperms, the largest category of the 14-3-3 binding proteins was associated with the carbohydrate metabolism that included plastidic enzymes for starch synthesis and modification [61]. And, some of the binding proteins, such as GBSS, PPDK and ADP glucose pyrophosphorylase large subunit, were also detected in our study showing a lower expression abundance on IS than SS. Secondly, the previous studies reported an inhibitive ability of 14-3-3 protein on the activity of starch synthesis enzymes [61]–[63], while Sehnke et al. found the reduction of the 14-3-3 proteins correlated to the starch granule formation with an increase in the starch content on their study of arabidopsis leaf starch [62]. Previously, we also observed the appropriate addition of ABA to be favorable for starch synthesis on IS. In the proteomic study that followed, a downregulation of the 14-3-3 protein under the ABA treatment was clearly demonstrated [15]. Therefore, it seemed apparent that a highly expressed 14-3-3 protein was detrimental to the starch formation and accumulation. And, the protein expression of 14-3-3 on IS was shown by the 2-DE and western blotting results to be higher than SS in all three grain-filling stages. Lastly, 14-3-3 protein participates in various signal transduction and regulatory processes by interacting with diverse target proteins in a sequence-specific and phosphorylation-dependent manner [64], [65]. Since the activity regulation of a certain enzymes can be a two-step process of phosphorylation and complex formation with 14-3-3 protein [56], [66], our finding of the differentiated 14-3-3 phosphorylation between SS and IS suggested another potential factor in the complex mechanism.
As the data shown, it seemed likely the differently expressed abundance and phosphorylation of 14-3-3 to be the root cause for the low activity of starch synthesis enzymes. And in turn, the resulting poor grain-filling of IS was observed. Nonetheless, the underlying mechanism of 14-3-3 protein in regulating the starch synthesis still remains to be fully uncovered.
This study identified 123 proteins proteomically different between SS and IS of the rice. The proteins’ possible roles in regulating the grain-filling were categorized based on the biological protein-interaction or reverse genetic functions of the genes. The classification illustrated the vast complexity of the grain-filling process, which might include metabolism, proteins synthesis and destination, signal transduction, transporters and cell growth/division.
For the first time, the Pro-Q Diamond Fluorescence Staining Method was applied to enable positive differentiation of the 43 phosphoproteins between SS and IS. Nonetheless, due to availability of methodologies, the current study could not identify all the phosphoproteins in the grains. Methods with a highly efficient phosphoprotein enrichment capability, such as TiO2 and Ti4+-MAC, could be of help in obtaining an improved profiling for the grain-filling of rice.
Materials and Methods
Plant Material and Sampling
The experiments were carried out at the Experimentation Station of Fujian Agriculture and Forestry University, Fuzhou, Fujian, China (119.280E, 26.080 N) during the rice growing season. The large-panicle rice cultivars, Jinhui No. 809 (Oryza sativa Indica), was used. The seeds were immersed in a water bath at room temperature for 24 h, and subsequently germinated under moist conditions at 37°C for 48 h. The germinated seeds grew in a paddy field. The seedlings at the 5-leaf stage were transplanted with a spacing of 0.15×0.15 m and one seedling per hill. Each 4×4 m planting plot received a fertilization of 225 kg N ha−1, 112.5 kg P2O5 ha−1, and 180 kg K2O ha−1. The phosphorus fertilizer was used as the basal, and the potassium for the top dressing. The field soil was sandy loam with available N, phosphorus, and potassium of 200.1, 147.3 and 208.2 mg kg−1, respectively.
Five hundred panicles that headed on a same day were tagged. The flowering date and position of each spikelet on the tagged panicles were recorded. The duration of anthesis from the first spikelets to the last on a panicle was 7 d. SS and IS were distinguished according to the previous report [2]. Thirty tagged panicles were sampled every 5 days from 0 to 35 day after flowering (DAF, the day was accounted from the first day of flowering). Spikelet samples for protein extraction were obtained 5, 10, 15, 20, 25, and 30 DAF. They were frozen in liquid N immediately after collection, and stored at −80°C.
Determinations of Grain Weight, Soluble Carbohydrate and Sucrose Content
Ten tagged panicles were sampled every 5 d from heading to maturity. SS and IS samples were separated, blanched in an oven at 105°C for 15 min, and dried at 80°C to a constant weight. Thirty grains were used for soluble carbohydrate and sucrose determinations. The samples were dried at 70°C to a constant weight, and dehulled for further analysis. Soluble carbohydrate and starch contents of SS and IS were determined as described by Yoshida et al [67].
Protein Extraction, 2-DE and Statistical Analyses
Proteins in the developing SS were extracted after harvested at 5, 10 and 15 DAF, while at 20, 25 and 30 DAF for IS, when their endosperm development were in EGS, MGS and LGS, respectively. Total protein was extracted in the same manner as reported previously [15]. Briefly, 1.0 g of spikelets was mixed with 0.5 g polyvinyl pyrrolidone, ground to fine powder in a mortar, and subsequently immersed in liquid N. The sample was suspended in a pre-cooled, 10% trichloroacetic acid in acetone containing 0.07% β-mercaptoethanol solution, and kept at −20°C overnight. The thawed sample was then centrifuged at 15,000 rpm for 30 min at 0–4°C. After decanting the supernatant, the precipitate was washed with a pre-cooled, 100% acetone containing 0.07% β-mercaptoethanol solution followed by centrifugation at 15,000 rpm for 30 min at 0–4°C. These procedures were repeated approximately every 8 h until the supernatant was achromatic. Then, the precipitate was dried under vacuum to yield sample pellet. The dried pellet was dissolved in a lysis buffer containing 8 mol/L urea, 4% CHAPS, 40 mmol/L Tris and 65 mmol/L DTT. The mix was homogenized by using ultrasound for 20–25 min, followed by centrifugation at 15,000 rpm for 15 min at 0–4°C. The supernatant was collected and stored at −80°C for proteomic analysis. The protein content was determined by using a previously described method [68].
Isoelectric focusing was carried out with 1.3 mg protein per strip using 24 cm long Immobiline Dry-strips pH 4–7 Linear (GE Biosciences). The protein were loaded onto an IPG strip and focused using an Ettan IPGphor 3 System (GE Healthcare) with a ceramic manifold as follows: gradient to 500 V for 1 h; gradient to 1000 V for 2 h; gradient to 8 KV for 3 h; step and hold at 8 KV for 3 h. The strips were then equilibrated upon gentle agitation on a stirrer twice for 15 min in an equilibration buffer (0.1 M Tris-HCl pH 8.8, 6 M urea, 30% (v/v) glycerol, and 2% (w/v) SDS) containing, firstly, 65 mM DTT, and, then, 2.5% (w/v) iodoacetamide. SDS-PAGE was performed in 26×20 cm, 10.5% (v/v) polyacrylamide gels using an Ettan Dalt 6 Multiple Apparatus (GE Biosciences) at 15°C. Fifteen mA per gel was used during running time until the dye front reached the end of gel. Gels were then stained with colloidal Coomassie Blue G-250 using at least 300 ml of staining solution per gel [69]. The gels were also stained with Pro-Q Diamond phosphoprotein gel stain (Molecular probes, Invitrogen) by fixing in 50% ethanol/10% acetic acid overnight, washing three times (15 min each) in deionized water, incubating in Pro-Q Diamond stain for 120 min in the dark, destaining with three washes of 20% acetonitrile in 50 mM sodium acetate (pH 4.0) and washing twice in deionized water. Images were acquired with Typhoon Variable Model Images Trio+ (GE Healthcare) using 532 nm excitation and a 580 nm band pass emission filter. After fluorescent images were acquired, the gels were stained with Coomassie Blue G-250 again. Gels were then scanned with an ImageScanner (GE Healthcare), and analyzed with Imagemaster 5.0 (GE Healthcare). Protein spots displaying ≥1.5-fold increase or decrease in abundance on average (p<0.05) were selected for protein identification.
In-gel Protein Digestion
In-gel protein digestion was performed with modifications as described previously [70]. In brief, the protein spots from same 2D-Gels were finely excised and transferred to siliconized 0.5 mL Eppendorf tubes. Each gel piece was rinsed twice with deionized water, destained in 25 mM ammonium bicarbonate in water/acetonitrile (50/50) solution (a 1∶1 solution of 30 mM potassium ferricyanide and 100 mM sodium thiosulfate), and then equilibrated in 50 mM ammonium bicarbonate to pH 8. After dehydrated with acetonitrile in a Speed-Vac Centrifuge (Thermo Fisher Scientific, Waltham, MA), the gel spots were rehydrated in a minimal volume of trypsin (Promega, USA) solution (12.5 µg/mL in 25 mM NH4HCO3) and incubated at 37°C overnight. The supernatants were transferred to a 200 µl microcentrifuge tube, and the gels were extracted once with the buffer (67% acetonitrile containing 2.5% trifluoroacetic acid).
Mass Spectrometric Analysis
MALDI-TOF-MS analysis
The peptide extract was combined with the supernatant of the gel spots and dried thoroughly in a Speed-Vac. The extract from the protein digestion (tryptic peptides) was re-suspended in 5 µl of 0.1% trifluoroacetic acid. In 1∶1 ratio, the peptide samples were mixed with a saturated solution of α-cyano-4-hydroxy-trans-cinnamic acid in 30% acetonitrile containing 0.1% trifluoroacetic acid. One µl of the aliquots were, then, spotted onto the stainless steel sample target plates. Protein spots of interest were cut from the gels and sequenced by Tandem mass spectrometry (MS/MS) with a fuzzy logic feedback control, a Reflex III MALDI-TOF System (Bruker) equipped with a delayed ion extraction. Both MS and MS/MS analyses were carried out with air as the collision gas using 1-kV collision energy.
LC-ESI-MS/MS analysis
For the protein samples that were not successfully identified by MALDI-TOF- MS, the extracted tryptic fragments were further characterized by using a LTQ-XL Mass Spectrometer (Thermo Scientific, USA) coupled with an Accera System (Thermo Fisher Scientific). A biobasic C18 column (100×0.18 mm; particle size: 5 um, Thermo Scientific, USA) was used to separate the digested proteins. Solvent A was 0.1% HCO2H mixed in water, and Solvent B was 0.1% HCO2H mixed in CH3CN. The gradient was held at 2% Solvent B for 2 min, and increased linearly up to 90% Solvent B over the course of 60 min. The peptides were eluted from a C18 column at a flow rate of 160 µl/min and then electrosprayed directly into an LTQ mass spectrometer using a spray voltage of 3.5 kV and at the capillary temperature of 275°C. Acquisitions were performed in data-dependent MS/MS scanning mode.
Database Searching
MS data were analyzed, and peak lists generated using Flexanalysis version 2.0 (Bruker). MS peaks were selected between 800 and 3,000, and filtered with a signal to noise ratio greater than 15 to exclude masses derived from trypsin autolysis. Search parameters allowed for one missed tryptic cleavage site, the carbamidomethylation of cysteine, possible oxidation of methionine, and a precursor ion mass tolerance of 100 ppm. Peptide masses were searched using Protein Prospector Mascot (http://www.matrixscience.com). O. sativa database was used for protein identification. The probability score at 95% confidence level as calculated using the software was the criterion for correct identification.
The raw data obtained from LC-ESI-MS/MS were used to search on the O. sativa database from Uniprot (http://www.uniprot.org/) using Proteome Discover 1.2 (Thermo Fisher Scientific). The carbamidomethylation of cysteine was set as a fixed modification, while the oxidation of methionine, a variable modification. Trypsin was the proteolytic enzyme and one missed cleavage was allowed. The search parameters included a precursor ion mass tolerance of 2.5 Da, a fragment mass tolerance of 0.5 Da, and a maximum missed cleavage of 2.
Western Blotting Analysis
Western blotting on the 1-DE gels was performed as described previously. Rabbit antisera to 14-3-3 and ADPase were obtained from Huada Proteomic Research Center, Beijing, China. In brief, the 1-DE gels were transferred to NC membranes for 3 h at 50 V in a transfer buffer (48 mM Tris, 39 mM glycine, 1.3 mM SDS and 20% methanol) at 4°C. The membranes were blocked with 5% BSA, and then, incubated with rabbit antisera to 14-3-3 or ADPase as the primary antibodies. Horseradish peroxidase (HRP) conjugated goat anti-rabbit antibodies were used as the secondary antibodies. Antibody-tagged protein spots were detected by 3, 3′-diaminobenzidine (DAB).
Acknowledgments
We would like to thank Prof Jinzhi Huang, visiting Scholar from the USA for his careful review of the manuscript, as well as valuable comments and suggestions.
Funding Statement
This work was sponsored by the National Natural Science Foundation of China (No. 31271670), Provincial Natural Science Foundation of Fujian, China (No. 2012J01075, 2013J01092), and Department of Education Science and Technology Project of Fujian, China (No. JK2011014). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fageria N (2007) Yield physiology of rice. J Plant Nutr 30: 843–879. [Google Scholar]
- 2. Ishimaru T, Matsuda T, Ohsugi R, Yamagishi T (2003) Morphological development of rice caryopses located at the different positions in a panicle from early to middle stage of grain filling. Funct Plant Biol 30: 1139–1149. [DOI] [PubMed] [Google Scholar]
- 3. Mohapatra P, Patel R, Sahu S (1993) Time of flowering affects grain quality and spikelet partitioning within the rice panicle. Funct Plant Biol 20: 231–241. [Google Scholar]
- 4. Yang JC, Zhang JH, Wang ZQ, Liu K, Wang P (2006) Post-anthesis development of inferior and superior spikelets in rice in relation to abscisic acid and ethylene. J Exp Bot 57: 149–160. [DOI] [PubMed] [Google Scholar]
- 5. Yang JC, Zhang JH (2010) Grain-filling problem in ‘super’ rice. J Exp Bot 61: 1–4. [DOI] [PubMed] [Google Scholar]
- 6. Peng SB, Cassman KG, Virmani S, Sheehy J, Khush G (1999) Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop Sci 39: 1552–1559. [Google Scholar]
- 7. Yang JP, Zhang S, Wang Z, Visperas Z, Zhu RM (2002) Grain and dry matter yields and partitioning of assimilates in japonica/indica hybrid rice. Crop Sci 42: 766–772. [Google Scholar]
- 8. Yoshida S (1972) Physiological aspects of grain yield. Annual Rev Plant Physiol 23: 437–464. [Google Scholar]
- 9. Tao LX, Wang X, Huang XL (2003) Effects of endogenous IAA on grain-filling of hybrid rice. Chin J Rice Sci 17: 149–155. [Google Scholar]
- 10. Kato T, Shinmura D, Taniguchi A (2007) Activities of enzymes for sucrose-starch conversion in developing endosperm of rice and their association with grain filling in extra-heavy panicle types. Plant Prod Sci 10: 442–450. [Google Scholar]
- 11. Ishimaru T, Hirose T, Matsuda T, Goto A, Takahashi K, et al. (2005) Expression patterns of genes encoding carbohydrate-metabolizing enzymes and their relationship to grain filling in rice (Oryza sativa L.): Comparison of caryopses located at different positions in a panicle. Plant Cell Physiol 46: 620–624. [DOI] [PubMed] [Google Scholar]
- 12. Tang T, Xie H, Wang YX, Lu B, Liang JS (2009) The effect of sucrose and abscisic acid interaction on sucrose synthase and its relationship to grain filling of rice (Oryza sativa L.). J Exp Bot 60: 2641–2652. [DOI] [PubMed] [Google Scholar]
- 13. Yang JC, Zhang JH, Wang ZQ, Zhu QS (2003) Hormones in the grains in relation to sink strength and postanthesis development of spikelets in rice. Plant Growth Regul 41: 185–195. [Google Scholar]
- 14. Zhang H, Tan GL, Yang LN, Yang JC, Zhang JH, et al. (2009) Hormones in the grains and roots in relation to post-anthesis development of inferior and superior spikelets in japonica/indica hybrid rice. Plant Physiol Bioch 47: 195–204. [DOI] [PubMed] [Google Scholar]
- 15. Zhang ZX, Chen J, Lin S, Li Z, Cheng RH, Fang CX, Chen HF, Lin WX (2012) Proteomic and phosphoproteomic determination of ABA’s effects on grain-filling of Oryza sativa L. inferior spikelets. Plant Sci 185: 259–273. [DOI] [PubMed] [Google Scholar]
- 16. Zhu T, Budworth P, Chen W, Provart N, Chang HS, et al. (2003) Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol J 1: 59–70. [DOI] [PubMed] [Google Scholar]
- 17. Zhu GH, Ye NH, Yang JC, Peng XX, Zhang JH (2011) Regulation of expression of starch synthesis genes by ethylene and ABA in relation to the development of rice inferior and superior spikelets. J Exp Bot 11: 3907–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Teo CJ, Thuc LV, Yeoh KA, Chan WS, Chivukula SV, et al. (2011) Profiling the differentially expressed genes in two rice varieties during rapid grain-filling stages. Acta Physiol Plant 33: 2259–2268. [Google Scholar]
- 19. Mechin V, Thevenot C, Le Guilloux M, Prioul JL, Damerval C (2007) Developmental analysis of maize endosperm proteome suggests a pivotal role for pyruvate orthophosphate dikinase. Plant Physiol 143: 1203–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, et al. (2009) Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol 149: 1541–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu SB, Li T, Deng ZY, Chong K, Xue Y, et al. (2008) Dynamic Proteomic Analysis Reveals a Switch Between Central Carbon Metabolism and Alcoholic Fermentation in Oryza sativa Filling Grains1. Plant Physiol 48: 908–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murty P, Murty K (1982) Spikelet sterility in relation to nitrogen and carbohydrate contents in rice. Indian J Plant Physiol 25: 40–48. [Google Scholar]
- 23. Zhu QS, Cao XZ, Luo YQ (1988) Growth analysis on the process of grain filling in rice. Acta Agronomica Sin 3: 182–192. [Google Scholar]
- 24. Chitteti BR, Peng Z (2007) Proteome and phosphoproteome dynamic change during cell dedifferentiation in Arabidopsis. Proteomics 7: 1473–1500. [DOI] [PubMed] [Google Scholar]
- 25. Agrawal GK, Thelen JJ (2006) Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol Cell Proteomics 5: 2044–2059. [DOI] [PubMed] [Google Scholar]
- 26. Peng T, Lv Q, Zhang J, Li JZ, Du YX, et al. (2011) Differential expression of the microRNAs in superior and inferior spikelets in rice (Oryza sativa L.). J Exp Bot 62: 4943–4954. [DOI] [PubMed] [Google Scholar]
- 27. Mohapatra PK, Sarkar RK, Kuanar SR (2009) Starch synthesizing enzymes and sink strength of grains of contrasting rice cultivars. Plant Sci 176: 256–263. [Google Scholar]
- 28. Thitisaksakul M, Jiménez RC, Arias MC, Beckles DM (2012) Effects of environmental factors on cereal starch biosynthesis and composition. J Cereal Sci 2012 56: 67–80. [Google Scholar]
- 29. Tetlow IJ, Wait R, Lu Z, Akkasaeng R, Bowsher CG, et al. (2004) Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 16: 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cohen P (1985) The role of protein phosphorylation in the hormonal control of enzyme activity. Eur J Biochem 151: 439–448. [DOI] [PubMed] [Google Scholar]
- 31. Gururaj A, Barnes CJ, Vadlamudi RK, Kumar R (2004) Regulation of phosphoglucomutase 1 phosphorylation and activity by a signaling kinase. Oncogene 23: 8118–8127. [DOI] [PubMed] [Google Scholar]
- 32. Teng B, Zeng R, Wang Y, Liu Z, Zhang Z, et al. (2012) Detection of allelic variation at the Wx locus with single-segment substitution lines in rice (Oryza sativa L.). Mol Breeding 30: 583–595. [Google Scholar]
- 33. Colleoni C, Myers AM, James MG (2003) One-and two-dimensional native PAGE activity gel analyses of maize endosperm proteins reveal functional interactions between specific starch metabolizing enzymes. J Appl Glycosci 50: 207–212. [Google Scholar]
- 34. Dinges JR, Colleoni C, James MG, Myers AM (2003) Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell 15: 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tetlow IJ, Beisel KG, Cameron S, Makhmoudova A, Liu F, et al. (2008) Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol 146: 1878–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Farré EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, et al. (2001) Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids, and sugar alcohols in potato tubers using a nonaqueous fractionation method. Plant Physiol 127: 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fernie AR, Carrari F, Sweetlove LJ (2004) Respiratory metabolism: glycolysis, the TCA cycle and mitochondrial electron transport. Curr Opin Plant Biol 7: 254–261. [DOI] [PubMed] [Google Scholar]
- 38. Meyer LJ, Gao J, Xu D, Thelen JJ (2012) Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol 159: 517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cooper JA, Esch FS, Taylor S, Hunter T (1984) Phosphorylation sites in enolase and lactate dehydrogenase utilized by tyrosine protein kinases in vivo and in vitro. J Biol Chem 259: 7835–7841. [PubMed] [Google Scholar]
- 40. Tadege M, Dupuis I, Kuhlemeier C (1999) Ethanolic fermentation: new functions for an old pathway. Trends plant Sci 4: 320–325. [DOI] [PubMed] [Google Scholar]
- 41. Geigenberger P (2003) Response of plant metabolism to too little oxygen. Curr opin plant biol 6: 247–256. [DOI] [PubMed] [Google Scholar]
- 42. Rolletschek H, Weschke W, Weber H, Wobus U, Borisjuk L (2004) Energy state and its control on seed development: starch accumulation is associated with high ATP and steep oxygen gradients within barley grains. J Exp Bot 55: 1351–1359. [DOI] [PubMed] [Google Scholar]
- 43. Van Dongen JT, Roeb GW, Dautzenberg M, Froehlich A, Vigeolas H, et al. (2004) Phloem import and storage metabolism are highly coordinated by the low oxygen concentrations within developing wheat seeds. Plant Physiol 135: 1809–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramasamy S, Ten Berge H, Purushothaman S (1997) Yield formation in rice in response to drainage and nitrogen application. Field Crop Res 51: 65–82. [Google Scholar]
- 45. Zhang ZX, Chen J, Li Z, Chen HF, Fang CX, et al. (2012) Differential proteomic expressions between superior and inferior spikelets of rice in response to varied nitrogen treatments. Aust J Crop Sci 6: 316–325. [Google Scholar]
- 46. Kikuchi H, Hirose S, Toki S, Akama K, Takaiwa F (1999) Molecular characterization of a gene for alanine aminotransferase from rice (Oryza sativa.L). Plant Mol Biol 39: 149–159. [DOI] [PubMed] [Google Scholar]
- 47. Wojtaszek P, Stobiecki M, Bolwell GP (1997) Changes in the composition of exocellular proteins of suspension-cultured Lupinus albus cells in response to fungal elicitors or CuCl2 . J Exp Bot 48: 2015–2021. [Google Scholar]
- 48. Cans C, Passer BJ, Shalak V, Nancy-Portebois V, Crible V, et al. (2003) Translationally controlled tumor protein acts as a guanine nucleotide dissociation inhibitor on the translation elongation factor eEF1A. P Nat Acad Sci USA 100: 13892–13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim YM, Han YJ, Hwang OJ, Lee SS, Shin AY, et al. (2012) Overexpression of Arabidopsis translationally controlled tumor protein gene AtTCTP enhances drought tolerance with rapid ABA-induced stomatal closure. Mol Cells 33: 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Negrutskii BS, El’skaya AV (1998) Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucleic Acid Res 60: 47–78. [DOI] [PubMed] [Google Scholar]
- 51. Mayer U, Jürgens G (2002) Microtubule cytoskeleton: a track record. Curr Opin plant biology 5: 494–501. [DOI] [PubMed] [Google Scholar]
- 52. Vassileva VN, Fujii Y, Ridge RW (2005) Microtubule dynamics in plants. Plant biotechnol-NAR 22: 171–178. [Google Scholar]
- 53. Membré N, Berna A, Neutelings G, David A, David H, et al. (1997) cDNA sequence, genomic organization and differential expression of three Arabidopsis genes for germin/oxalate oxidase-like proteins. Plant Mol Biol 35: 459–469. [DOI] [PubMed] [Google Scholar]
- 54. El-Sharkawy I, Mila I, Bouzayen M, Jayasankar S (2010) Regulation of two germin-like protein genes during plum fruit development. J Exp Bot 61: 1761–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lur HS, Setter TL (1993) Role of auxin in maize endosperm development (timing of nuclear DNA endoreduplication, zein expression, and cytokinin). Plant Physiol 103: 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chung HJ, Sehnke PC, Ferl RJ (1999) The 14-3-3 proteins: cellular regulators of plant metabolism. Trends in plant Sci 4: 367–371. [DOI] [PubMed] [Google Scholar]
- 57. Ferl RJ (1996) 14-3-3 proteins and signal transduction. Annu Rev Pharmacol 47: 49–73. [DOI] [PubMed] [Google Scholar]
- 58. Roberts MR (2003) 14-3-3 proteins find new partners in plant cell signalling. Trends Plant Sci 8: 218–223. [DOI] [PubMed] [Google Scholar]
- 59. Fu H, Subramanian RR, Masters SC (2000) 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol 40: 617–647. [DOI] [PubMed] [Google Scholar]
- 60. Schoonheim PJ, Sinnige MP, Casaretto JA, Veiga H, Bunney TD, et al. (2007) 14-3-3 adaptor proteins are intermediates in ABA signal transduction during barley seed germination. Plant J 49: 289–301. [DOI] [PubMed] [Google Scholar]
- 61. Alexander RD, Morris PC (2006) A proteomic analysis of 14-3-3 binding proteins from developing barley grains. Proteomics 6: 1886–1896. [DOI] [PubMed] [Google Scholar]
- 62. Sehnke PC, Chung HJ, Wu K, Ferl RJ (2001) Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. P Natl Acad Sci USA 98: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Song,JM, Dai S, Li HS L, Liu AF, Chen DG, et al. (2009) Expression of a wheat endosperm 14-3-3 protein and its interactions with starch biosynthetic enzymes in amyloplasts. Acta Agronomica Sin 35: 1445–1450. [Google Scholar]
- 64. Mackintosh C (2004) Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J 381: 329–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Comparot S, Lingiah G, Martin T (2003) Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. J Exp Bot 54: 595–604. [DOI] [PubMed] [Google Scholar]
- 66. Sehnke PC, Ferl RJ (1996) Plant metabolism: enzyme regulation by 14-3-3 proteins. Curr Biol 6: 1403–1405. [DOI] [PubMed] [Google Scholar]
- 67.Yoshida S, Forno D, Cock J, Gomez K (1976) Determination of sugar and starch in plant tissue. Laboratory manual for physiological studies of rice. Annual Rev Plant Physiol, Philippines: 46–49.
- 68. Garrels J (1983) Quantitative two-dimensional gel electrophoresis of proteins. Method Enzymol 100: 411–423. [DOI] [PubMed] [Google Scholar]
- 69. Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, et al. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25: 1327–1333. [DOI] [PubMed] [Google Scholar]
- 70. Lin XM, Wu LN, Li H, Wang S, Peng XX (2008) Downregulation of Tsx and OmpW and upregulation of OmpX are required for iron homeostasis in Escherichia coli. J Proteome Res 7: 1235–1243. [DOI] [PubMed] [Google Scholar]