Abstract
The epigenetic regulator BMI1 is upregulated progressively in a wide variety of human tumors including colorectal cancer. In this study, we assessed the requirement for Bmi1 in intestinal tumorigenesis using an autochthonous mouse model in which Apc was conditionally ablated in the intestinal epithelium. Germline mutation of Bmi1 significantly reduced both the number and size of small intestinal adenomas arising in this model, and it acted in a dose-dependent manner. Moreover, in contrast to wildtype controls, Bmi1−/− mice showed no increase in median tumor size, and a dramatic decrease in tumor number, between 3 and 4 months of age. Thus, Bmi1 is required for both progression and maintenance of small intestinal adenomas. Importantly, Bmi1 deficiency did not disrupt oncogenic events arising from Apc inactivation. Instead, the Arf tumor suppressor, a known target of Bmi1 epigenetic silencing, was upregulated in Bmi1 mutant tumors. This was accompanied by significant upregulation of p53, which was confirmed by sequencing to be wildtype, and also elevated apoptosis within the smallest Bmi1−/− adenomas. By crossing Arf into this cancer model, we showed that Arf is required for the induction of both p53 and apoptosis, and it is a key determinant of the ability of Bmi1 deficiency to suppress intestinal tumorigenesis. Finally, a conditional Bmi1 mutant strain was generated and used to determine the consequences of deleting Bmi1 specifically within the intestinal epithelium. Strikingly, intestinal-specific Bmi1 deletion suppressed small intestinal adenomas in a manner that was indistinguishable from germline Bmi1 deletion. Thus, we conclude that Bmi1 deficiency impairs the progression and maintenance of small intestinal tumors in a cell autonomous and highly Arf-dependent manner.
Keywords: Bmi1, Apc, colorectal cancer, intestine, apoptosis, Arf
Introduction
Colorectal carcinoma (CRC) is the 4th leading cause of cancer deaths worldwide, resulting in over 600,000 fatalities a year (1). Upregulation of Wnt pathway activity, through loss of the Apc tumor suppressor or deregulation of the β-catenin proto-oncogene, is an early event in the development of most colon adenomas (2). Additional mutations and epigenetic changes are associated with tumor development and progression. The oncogene BMI1 is frequently overexpressed in human CRC, and the degree of upregulation correlates with disease progression and is predictive of poor patient survival (3–5). This suggests that BMI1 enables both the development and metastatic progression of CRC. Knockdown of Bmi1 in human CRC cell lines has been shown to suppress their proliferation in vitro and in xenografts (6). However, the role of Bmi1 in the initiation of autochthonous intestinal tumors has not been investigated.
Much of our understanding of Bmi1's in vivo role has come from analysis of germline Bmi1−/− mice (7). These animals have a shortened lifespan and they display cerebellar and hematopoietic abnormalities. These tissue defects reflect a requirement for Bmi1 to maintain the self-renewal and proliferative capacity of adult neuronal and hematopoietic stem/progenitor cells via epigenetic silencing of the Ink4a-Arf and Cdkn1a loci (8–10). Bmi1's ability to silence these tumor suppressors and promote stem cell characteristics has been linked to Bmi1's oncogenic activity in various tumor types (9, 11). In CRC, intestinal stem cells are thought to be the targets of transformation (12), reflecting the importance of Wnt/β-catenin signaling in the maintenance of these cells. Bmi1 is expressed in intestinal stem cell populations, including both the +4 position and LGR5+ cells (13–15), but its requirement for their function has not been demonstrated. Here, we use mouse models to assess the requirement for Bmi1 in the development of autochthonous small intestinal tumors and show that it plays a crucial role.
Results and Discussion
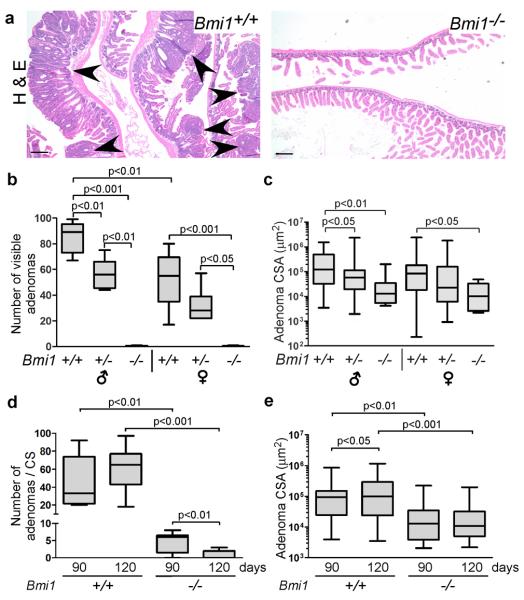
To investigate Bmi1's role in intestinal tumorigenesis, we took advantage of an established mouse model in which a single conditional Apcfl allele (16) is inactivated throughout the intestinal epithelium using the Vil-cre transgene (17). After somatic recombination of the wildtype Apc allele, these mice develop numerous small intestinal adenomas and a lower incidence of colon adenomas (18). The resulting tumor load causes morbidity between 100–140 days of age. Importantly, this precedes the typical maximal lifespan of germline Bmi1−/− adult mice (approximately 150 days), allowing us to intercross these mice and assess how germline Bmi1 mutation affects the tumor phenotype of Apc;Vil-cre mice. We generated cohorts of Apcfl/+;Vil-cre mice that were Bmi1+/+, Bmi1+/− or Bmi1−/−. Males and females were considered separately because intestinal tumorigenesis is gender dependent (18). Any graph showing pooled data from male and female mice (as noted in the figure legend), illustrates a phenotype that is overtly present in both sexes. Initially, we examined the tumor phenotypes of 120 day old mice. We found that visible small intestinal tumors were numerous in Bmi1+/+ animals, but almost completely absent in Bmi1−/− mice (Figure 1a,b). Bmi1 heterozygotes displayed an intermediate phenotype, suggesting a dose-dependent effect (Figure 1b). Analysis of all tumors (visible by eye or microscopic analysis) confirmed that Bmi1 mutation caused a significant, dose-dependent reduction in the size of small adenomas (Figure 1c). Indeed, the median and maximal cross-sectional areas (CSA) of Bmi1−/− adenomas were more than tenfold lower than those of Bmi1+/+ tumors (Figure 1c). Additionally, even when considering lesions of any size, Bmi1−/− mice had significantly fewer small intestinal adenomas (p<0.001) than Bmi1+/+ controls (Figure 1d). Importantly, Bmi1 status did not alter the efficiency of Cre-mediated recombination (Supplemental Figure 1a) or impair key oncogenic events arising from Apc inactivation including accumulation of nuclear β-Catenin and c-Myc (Supplemental Figure 1b). Thus, we conclude that Bmi1 mutation acts in a dose-dependent manner to suppress small intestinal tumor development.
Figure 1. Bmi1 is required for small intestinal adenoma development, progression and maintenance.
a Representative Haematoxylin & Eosin stained sections of small intestine from 120 day old Apcfl/+;Vil-cre mice show adenomas (marked by arrowheads) in Bmi1+/+ but not Bmi1−/− context. Original magnification 4X. Scale bar represents 200 μm. b,c Quantification of (b) the number of visible (diameter > 1mm) adenomas, and (c) the cross-sectional area (CSA) of all adenomas, in 120-day old animals according to Bmi1 status and gender (n= 4–8). d,e Comparison of (d) the total number of small intestinal adenomas per histological cross-section (CS), and (e) adenoma cross-sectional area (CSA), in 90-day old versus 120-day old mice shows that Bmi1 is required for their maintenance and progression. Data were pooled for male and females, using equal numbers of each (n=8–10 for combined genders). For (a–e) mouse small intestine was dissected, fixed in formalin, coiled and subject to histological processing followed by paraffin embedding. Sections were examined using a Nikon Eclipse E600 microscope with a SPOT RT digital camera. SPOT Basic imaging software was utilized for capture and area/length measurements. Data are presented as box plots with marked median values and whiskers spanning the 5th to the 95th percentiles. For statistical analyses (b,c) ANOVA was performed with a Tukey post-test for paired comparisons or (d,e) a student's 2-sided t-test was performed for paired comparisons.
We also examined the effect of Bmi1 mutation on colon adenomas, and found that Bmi1−/− animals had fewer visible tumors than Bmi1+/+ controls (Supplemental Figure 2a,b). Unfortunately, because colon tumors arise at low frequency in Apcfl/+;Vil-cre mice it was difficult to establish statistical significance, and this was only achieved for male mice (Supplemental Figure 2b). Given this challenge, we decided to focus on the small intestinal phenotype. First, we wanted to determine the effect of Bmi1 mutation at earlier stages of tumor development, and thus examined 90 day animals. At this younger age, we also observed significantly fewer (p <0.01) and smaller (p<0.01) lesions in Bmi1−/− animals versus Bmi1+/+ controls (Figure 1d,e). Moreover, our comparison of the 90 and 120 day time points showed that these two genotypes had differential effects on tumor progression (Figure 1d,e). In Bmi1+/+ mice, tumor number did not change significantly between 90 and 120 days, but tumor size increased significantly (p<0.05). In contrast, Bmi1−/− mice showed no increase in median tumor size between 90 and 120 days. Furthermore, the incidence of tumors decreased significantly (p<0.01) between these two ages. Taken together, these findings suggest that Bmi1 loss suppresses small intestinal tumor progression and maintenance.
We wanted to determine how Bmi1 loss impedes tumor progression. In many other tumor types, Bmi1's oncogenic activity is at least partially dependent upon its ability to repress the Ink4a/Arf locus (19). Thus, we wanted to explore the potential contribution of the p16Ink4a and p19Arf tumor suppressors. While we could not perform western blot for p16Ink4a and p19Arf in Bmi1−/− adenomas because of their small size, and reliable detection by immunohistochemistry in the intestine has not been demonstrated, we were able to screen wildtype and Bmi1+/− adenomas for these proteins. These two genotypes showed no obvious difference in p16Ink4a levels (Supplemental Figure 3a), but p19Arf was typically elevated in Bmi1+/− tumors, compared to Bmi1+/+ controls (Figure 2a). Given this, it seemed likely that p19Arf was also upregulated in Bmi1−/− tumors. Thus, we examined the small intestinal adenomas for the known tumor suppressive effects of p19Arf. Initially, we assayed for senescence-associated β-galactosidase (SA-βgal). However, we saw no detectable differences between Bmi1+/+ versus Bmi1−/− adenomas at either 90 days (little detectable SA-βgal) or 120 days (low level SA-βgal staining; Supplemental Figure 3b). Next, we assessed the proliferative index of adenomas by quantification of BrdU incorporation at 90 days (Figure 2b). Unexpectedly, the proportion of proliferating cells was significantly higher in Bmi1 mutant adenomas than Bmi1+/+ controls, even when controlling for tumor size (Figure 2b). Thus, neither enhanced senescence nor impaired proliferation can account for the tumor suppressive effects of Bmi1 loss. Finally, we assayed for apoptosis by quantification of cleaved caspase-3 in adenomas at 90 days (Figure 2c). We found that this was increased significantly (p<0.01) in smaller (CSA<104 μm2) Bmi1−/− adenomas, but not in larger Bmi1−/−tumors. We also detected cleaved caspase-3 in non-transformed regions of Bmi1−/− jejunum and ileum, including in Bmi1−/−mice that lacked Vil-cre and therefore Apc inactivation (Supplemental Figure 4a–c). Thus, Bmi1 loss increased the predisposition of emerging intestinal adenomas, and also normal intestinal epithelium, to undergo apoptosis.
Figure 2. Bmi1 loss promotes Arf-dependent, p53 stabilization and apoptosis within small intestinal adenomas.
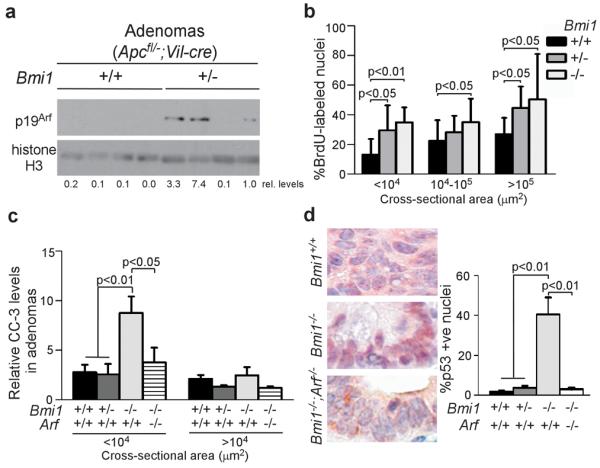
a Western blot of lysates from small intestinal adenomas demonstrates higher p19Arf expression in Bmi1+/− versus Bmi1+/+ adenomas. Lysates were prepared from small intestinal adenomas that were minced, digested for 1 hour at 37°C in dispase (150 u/ml final; Gibco, Japan), triturated and lysed in RIPA buffer (200 mM Tris pH 7.4, 130 mM NaCl, 10% glycerol, 0.1% SDS, 0.5% DOC, 1% Triton) containing protease inhibitors (Roche, Germany). Protein was separated by SDS-PAGE and western blot was performed with anti-p19Arf (top panel; BD Biosciences, San Jose CA, NB200-106) and Histone H3 (lower panel; Cell Signaling, Danvers MA, 4620) antibodies, followed by incubation with a 1:5000 dilution of HRP-conjugated secondary antibodies (Cell Signaling, Danvers, MA). Relative levels of p19Arf were quantified and normalized to levels of Histone H3 after band density measurements using ImageJ software. b Bmi1+/− and Bmi1−/− adenomas are more proliferative than Bmi1+/+ adenomas in 90-day old mice, as determined by quantification of the average percentage of adenoma nuclei staining positive for incorporated BrdU. Data were stratified by adenoma cross-sectional area. BrdU (10 μg/g body weight; Sigma, St. Louis MO) was injected I.P. two hours prior to euthanasia. c Analysis of average number of cleaved caspase-3 positive adenoma cells, normalized to adenoma cross-sectional area, shows that Bmi1 deficiency elevated apoptosis in the smallest adenomas in an Arf-dependent manner. d Adenomas of 90 day old Bmi1−/−mice have an average high percentage of cells with p53 stabilization, which is lost in the absence of Arf. Original magnification was 40X. For b–d: Adenomas from at least 5 mice of each genotype consisting of both males and females were assayed. Immunohistochemistry on intestinal sections was performed after antigen retrieval in 10 mM citric acid pH 6.0 using a decloaking chamber, followed by standard DAB protocol and haematoxylin counterstain. Antibodies were: anti-BrdU (1:100; Beckton Dickson, Franklin Lakes NJ, 347580), p53 (1:500, Santa Cruz, Santa Cruz CA, FL-393) and cleaved caspase-3 (1:1000, Cell Signaling, Danvers MA, 9661). Tissue processing, image capture/measurements and statistical analysis were performed as described for Figure 1b,c and error bars represent standard deviation.
p19Arf is known to promote apoptosis through stabilization of p53 (20). Thus, we screened for p53 levels in adenomas by immunohistochemical staining (Figure 2d). The mean percentage of p53-positive nuclei was significantly higher in Bmi1−/− adenomas than in either Bmi1+/+ or Bmi1+/− adenomas (p<0.01). Indeed, 64% of Bmi1−/− adenomas, but 0% of Bmi1+/+ adenomas, had more than 25% p53-positive nuclei. Notably, we observed p53 stabilization in both smaller (CSA<104 μm2) and larger Bmi1−/− tumors (data not shown), even though only the small lesions had elevated apoptosis. This raised the question of whether the stabilized p53 was wildtype or mutant, particularly in the larger Bmi1−/− tumors. To address this question, we used laser capture to isolate DNA from Bmi1−/− and wildtype small intestinal tumors, and screened for p53 mutations by PCR and sequencing of exons 5–9 (Supplemental Figure 5). Unexpectedly, all of these sequences were wildtype (data not shown). Thus, we conclude that p53 is activated in a high fraction of Bmi1−/− adenoma cells but elevated apoptosis is somehow limited to the smaller tumors.
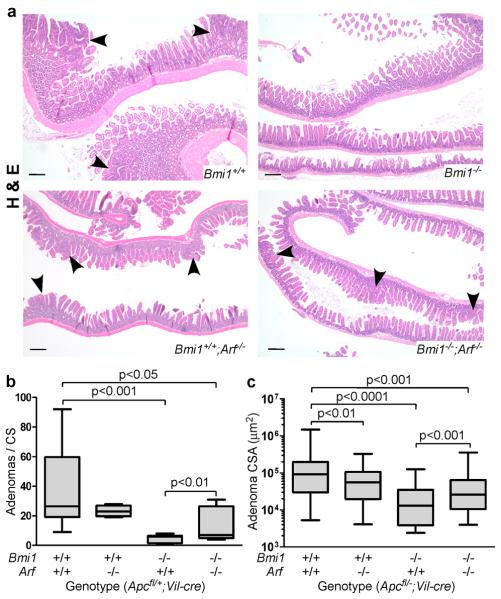
We wanted to assess the degree to which p19Arf contributes to the phenotypes of Bmi1 mutant tumors. Thus, we crossed an Arf null strain (21) into our mouse model to generate Apcfl/+;Vil-cre mice that were wildtype, Arf−/−, Bmi1−/− or Bmi1−/−;Arf−/−. Since the introduction of mutant Arf caused some animals to develop tumor-associated morbidity (cachexia and hunching) by 90 days, we selected this age for analysis (Figure 3). Previous studies showed that Ink4a/Arf−/− mice have a lower small intestinal tumor burden than wildtype controls (22). Consistent with this report, we found that Arf−/− mice had a similar tumor incidence as wildtype controls (Figure 3a,b) but the median size of these Arf−/− tumors was significantly reduced (p<0.01; Figure 3a,c). In stark contrast, Bmi1−/−;Arf−/− mice had a significant increase in both the number (p<0.01; Figure 3a,b) and size (p<0.001; Figure 3a,c) of small intestinal adenomas, compared to Bmi1−/− animals. Indeed, the tumor phenotypes of these Bmi1−/−;Arf−/− animals now approached that of the Arf−/− animals (Figure 3); there was no significance difference in total tumor number (Figure 3b) or tumor size (Figure 3c) between these two genotypes, but Bmi1−/−;Arf−/− mice did have significantly fewer visible lesions than Arf−/− animals (p<0.01, data not shown) suggesting that the rescue is not complete. Because we cannot follow the cohort beyond 90 days, we were unable to determine whether Arf loss altered the influence of Bmi1-deficiency on the progression and/or maintenance of tumors between 90 to 120 days. Importantly, analysis of the 90 day tumors showed that Arf loss significantly suppressed the elevation of cleaved caspase 3 (p<0.05, Figure 2c), and the stabilization of p53 (Figure 2d; p<0.01), within the Bmi1−/− adenomas. Thus, we conclude that Arf plays a major role in the ability of Bmi1 deficiency to suppress small intestinal tumors through activation of p53 and induction of apoptosis.
Figure 3. The tumor suppressive effects of Bmi1 deficiency are Arf-dependent.
a Representative Haematoxylin & Eosin stained sections of small intestine from 90 day old Apcfl/+;Vil-cre mice show adenomas (marked by arrowheads) in a Bmi1+/+, Bmi1+/+;Arf−/−, Bmi1−/−;Arf−/− but not Bmi1−/− context. Original magnification 4X. Scale bar represents 200 μm. b,c Quantification of the effect of Bmi1 and Arf status on (b) the total number of adenomas per cross-section (CS) and (c) adenoma size, as measured by cross-sectional area (CSA), in 90 day old Apcfl/+;Vil-cre mice (n≥5 for each genotype consisting of both males and females) shows that Arf loss suppresses Bmi1+/+ tumors while promoting Bmi1−/− tumors. Tissue processing and image capture/measurements were performed as described for Figure 1. For statistical analysis, ANOVA was performed, followed by an Unpaired Fisher's LSD test for paired comparisons.
We had previously noted the Bmi1 deficiency also causes apoptosis within the normal intestinal epithelium (Supplemental Figure 4). Unexpectedly, our analysis of the Bmi1−/−;Arf−/− compound mutant mice showed that Arf inactivation had no significant impact on this normal tissue apoptosis. Thus, Bmi1 loss promotes apoptosis through distinct mechanisms in normal, versus transformed, intestinal epithelium. We wished to establish whether the Bmi1 mutant tumor suppression was intrinsic to the intestinal epithelium. To address this, we engineered a conditional Bmi1 (Bmi1fl) mutant mouse strain that allowed Cre-dependent deletion of Bmi1 core functions (encoded by exons 4–8), and confirmed that this conditional Bmi1 mutant allele recapitulates all of the developmental defects characteristic of germline Bmi1 mutants (Supplemental Figure 6).
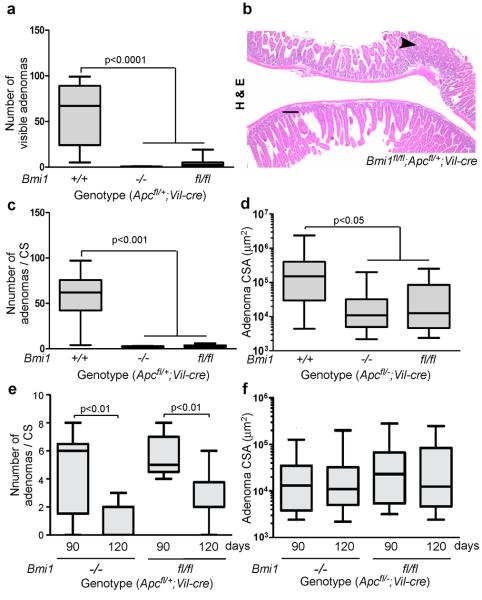
We used this conditional allele to generated cohorts of Bmi1fl/fl;Apcfl/+;Vil-cre mice and Apcfl/+;Vil-cre controls, and compared their small intestinal tumor to each other, and to that of germline Bmi1−/−;Apcfl/+;Vil-cre mice. Initially, we examined animals at 120 days. We found that the intestine-specific Bmi1 deletion significantly reduced the number of visible lesions (p<0.0001, Figure 4a,b), the number of total lesions (p<0.001, Figure 4c) and the size of lesions (p<0.05, Figure 4d) in the small intestine. Importantly, the conditional Bmi1 mutant phenotypes were all indistinguishable from those yielded by germline Bmi1 deletion (Figure 4a–d). We also examined the tumor phenotypes in 90 day old mice to address the issue of progression and maintenance. At this time point, mice the intestinal-specific deletion of Bmi1 also resulted in significantly fewer (p <0.05) and smaller (p<0.01) lesions that the Bmi1+/+ controls (data not shown). Moreover, exactly as seen in germline Bmi1−/−;Apcfl/+;Vil-cre mutants (Figure 4e,f), Bmi1fl/fl;Apcfl/+;Vil-cre mice had significantly more adenomas at 90 versus 120 days (p<0.01, Figure 4e), and there was no increase in tumor size between these two time points (Figure 4f). Thus, the ability of Bmi1 deficiency to suppress the progression, and maintenance, of small intestinal tumors reflects an intrinsic defect of the transformed epithelium. To complement this, we also examined the level of apoptosis and the induction of p53 within the small intestinal adenomas. As anticipated, given the observed tumor suppression, conditional Bmi1 deletion promoted both the stabilization of p53, and elevated levels of apoptosis, in a similar manner to germline Bmi1 loss (data not show). Finally, we screened for apoptosis in the normal epithelium. In this setting, the intestinal specific deletion of Bmi1 gave no induction of cleaved caspase 3 (data not shown), in stark contrast to the significantly increased cleaved caspase 3 levels resulting from germline Bmi1 mutation (p<0.01, Supplementary Figure 4). This further distinguishes the normal epithelial apoptosis from that of the adenomas, by showing that it is both Arf independent and non-cell autonomous.
Figure 4. The oncogenic effect of Bmi1 in small intestinal tumorigenesis reflects its action within the intestinal epithelium.
a–d Comparison of (a) the number of visible adenomas (diameter>1 mm), (b,c) the total number of adenomas per cross-section (CS) and (d) adenoma size, as measured by CSA, shows a similar degree of tumor suppression in the small intestines of 120 day Apcfl/+;Vil-cre mice that are Bmi1fl/fl versus Bmi1−/− (both genders; n=8 or greater). For (b), original magnification was 4X and the scale bar represents 200 μm. e,f The relative (e) number and (f) size of small intestinal adenomas in Bmi1−/− or Bmi1fl/fl Apcfl/+;Vil-cre mice at 90 days versus 120 days (both genders; n=5–10) phenocopies those of Bmi1−/−;Apcfl/+;Vil-cre mice. The Bmi1 conditional allele (Bmi1fl) was generated using standard gene targeting to insert loxP sites in introns 3 and 8 and validated as shown in Supplemental Figure 6. Tissue processing, image capture/measurements and statistical analyses were performed as described for Figure 1.
We initiated this study because of the known presence of high Bmi1 expression in human CRC (4, 5). Our goal was to assess the requirement for Bmi1 in the context of autochthonous intestinal tumors. Our data clearly show that Bmi1 loss reduces both the number and size of small intestinal adenomas. Additionally, as Bmi1 animals age, we actually see a reduction in the tumor burden that reflects an apparent cap on tumor size, and a clear reduction in the total number of tumors. Thus, taken together, these data support a key role for Bmi1 in tumor development, progression and maintenance. This tumor suppression correlates with upregulation of Arf, stabilization of p53 and a significant increase in the level of apoptosis in smaller adenomas. Our analysis of Arf null mice showed that these molecular changes are linked, and that they make a major contribution to anti-tumorigenic effect of Bmi1 loss in small intestinal adenomas. Finally, our data show that the Arf-dependent induction of apoptosis, and the consequent tumor suppression, reflects a cell autonomous requirement for Bmi1 within the intestinal epithelium.
Supplementary Material
Acknowledgements
The authors dedicate this paper to the memory of Officer Sean Collier, for his caring service to the MIT community and for his sacrifice. This work was supported by an NCI/NIH grant to J.A.L. who is a Ludwig Scholar at MIT, and fellowships to M.A.M. (CIHR), R.F. (Ludwig) and K.I.H. (NSF). We thank the Koch Institute Swanson Biotechnology Center for key technical support, particularly personnel in the ES Cell & Transgenic, Histology and Microscopy facilities. We also thank Sylvie Robine (Institut Curie) for the Vil-cre mouse and members of the Lees and Jacks labs for thoughtful comments.
Footnotes
Conflict of Interests There are no competing financial interests in relation to the work described.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010 Dec 15;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER. Molecular genetics of colorectal cancer. Annual review of pathology. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 3.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. The Journal of clinical investigation. 2005 Jun;115(6):1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tateishi K, Ohta M, Kanai F, Guleng B, Tanaka Y, Asaoka Y, et al. Dysregulated expression of stem cell factor Bmi1 in precancerous lesions of the gastrointestinal tract. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006 Dec 1;12(23):6960–6966. doi: 10.1158/1078-0432.CCR-06-0449. [DOI] [PubMed] [Google Scholar]
- 5.Li DW, Tang HM, Fan JW, Yan DW, Zhou CZ, Li SX, et al. Expression level of Bmi-1 oncoprotein is associated with progression and prognosis in colon cancer. J Cancer Res Clin Oncol. 2010 Jul;136(7):997–1006. doi: 10.1007/s00432-009-0745-7. [DOI] [PubMed] [Google Scholar]
- 6.Yu T, Chen X, Zhang W, Colon D, Shi J, Napier D, et al. Regulation of the potential marker for intestinal cells, Bmi1, by beta-catenin and the zinc finger protein KLF4: implications for colon cancer. The Journal of biological chemistry. 2012 Feb 3;287(6):3760–3768. doi: 10.1074/jbc.M111.316349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994 Apr 1;8(7):757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 8.Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell stem cell. 2007 Jun 7;1(1):87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Gieni RS, Hendzel MJ. Polycomb group protein gene silencing, non-coding RNA, stem cells, and cancer. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2009 Oct;87(5):711–746. doi: 10.1139/O09-057. [DOI] [PubMed] [Google Scholar]
- 10.Subkhankulova T, Zhang X, Leung C, Marino S. Bmi1 directly represses p21Waf1/Cip1 in Shh-induced proliferation of cerebellar granule cell progenitors. Molecular and cellular neurosciences. 2010 Oct;45(2):151–162. doi: 10.1016/j.mcn.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009 Jul 15;29(28):8884–8896. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009 Jan 29;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 13.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nature genetics. 2008 Jul;40(7):915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009 Mar 6;136(5):903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 15.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011 Oct 13;478(7368):255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colnot S, Niwa-Kawakita M, Hamard G, Godard C, Le Plenier S, Houbron C, et al. Colorectal cancers in a new mouse model of familial adenomatous polyposis: influence of genetic and environmental modifiers. Lab Invest. 2004 Dec;84(12):1619–1630. doi: 10.1038/labinvest.3700180. [DOI] [PubMed] [Google Scholar]
- 17.el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004 Jul;39(3):186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 18.Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007 Oct 15;67(20):9721–9730. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- 19.Grinstein E, Wernet P. Cellular signaling in normal and cancerous stem cells. Cell Signal. 2007 Dec;19(12):2428–2433. doi: 10.1016/j.cellsig.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 20.Sherr CJ, Bertwistle D, DENB W, Kuo ML, Sugimoto M, Tago K, et al. p53-Dependent and - independent functions of the Arf tumor suppressor. Cold Spring Harbor symposia on quantitative biology. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 21.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997 Nov 28;91(5):649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 22.Gibson SL, Dai CY, Lee HW, DePinho RA, Gee MS, Lee WM, et al. Inhibition of colon tumor progression and angiogenesis by the Ink4a/Arf locus. Cancer research. 2003 Feb 15;63(4):742–746. [PubMed] [Google Scholar]
- 23.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999 Jan;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 24.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995 Sep 26;92(20):9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000 Feb;26(2):113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.