Abstract
The differentiation and activation of both innate and adaptive immune cells is highly dependent on a coordinated set of transcriptional and post-transcriptional events. Chromatin-modifiers and transcription factors regulate the accessibility and transcription of immune genes, respectively. Immune cells also express miRNA and RNA-binding proteins that provide an additional layer of regulation at the mRNA level. However, long noncoding RNA (lncRNA), which have been primarily studied in the context of genomic imprinting, cancer, and cell differentiation, are now emerging as important regulators of immune cell differentiation and activation. In this review, we provide a brief overview of lncRNA, their known functions in immunity, and discuss their potential to be more broadly involved in other aspects of the immune response.
Introduction
Cells of the immune system are equipped with the capacity to undergo dramatic changes in their transcriptional program in order to rapidly mobilize expression of genes important in host-defense. The differentiation and activation of both innate and adaptive immune cells is highly dependent on a coordinated set of transcriptional and post-transcriptional events. Hundreds of protein-coding genes are induced and repressed during both of these processes. For example, in the innate immune response, the bodies first line of defense against pathogen attack, dendritic cells and macrophages express both common and unique sets of toll-like receptors, intracellular signaling molecules, chemokines and cytokines [1]. Chromatin-modifying complexes determine which regions of the genome are accessible to transcription factors, which, in turn, help regulate transcription [2–4]. Furthermore, RNA-binding proteins and miRNA can regulate the products of transcription in various ways (e.g. protein translation, RNA turnover, and splicing). More recently, lncRNAs have been shown to regulate various biological process and several studies now suggest that they play important roles during the differentiation and activation of immune cells.
Long noncoding RNAs
lncRNAs are arbitrarily defined as noncoding RNAs that have at least 200 nucleotides and are described in several reviews [5–9]. Although the majority of a lncRNA sequence should lack coding potential, it is possible that some lncRNAs encode small peptides [10,11]. Therefore, it may be useful to define lncRNAs as long RNA molecules that perform molecular functions that are distinct from encoding proteins. In humans, lncRNAs are often polyadenylated and their spliced transcripts can consist of one or more exons. In general, most lncRNAs have fewer exons than mRNA and are shorter than most mRNA[12].
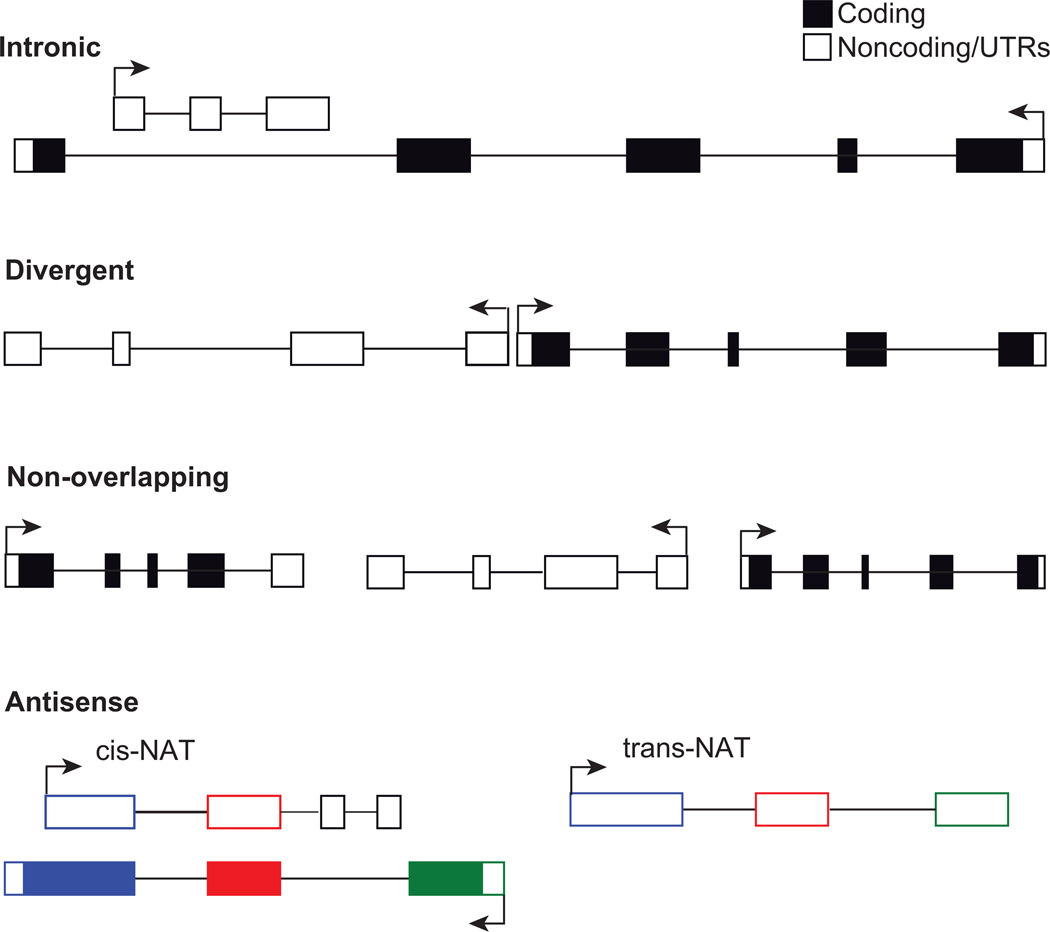
Although several lncRNA were discovered and characterized prior to 2005 [13–15], advances in sequencing and array technologies have led to the discovery of thousands of lncRNA transcripts [16–18]. It is now known that at least 75% of the human genome is transcribed in one cell or another and the majority of these transcripts can be categorized as lncRNA [19]. Consequently, lncRNA genes are located in various chromosomal regions and can be regarded as the major class of RNA genes encoded by the genome. However, the functions of most lncRNA are unknown and it is therefore useful to classify lncRNA based on their location and proximity to protein-coding genes [5](Figure 1). Indeed, the names of many lncRNA are derived from their neighboring protein-coding genes. lncRNA that occur within the introns of protein-coding genes are aptly referred to as intronic lncRNA. Divergently transcribed lncRNA/mRNA pairs have 5’ ends that are proximal to each other and are typically co-regulated [20]. We propose that lncRNA that do not overlap with a protein-coding gene are referred to as non-overlapping lncRNA. Although non-overlapping lncRNA are often referred to as intergenic lncRNA, the latter term is somewhat misleading as the majority of the genome contains bona fide genes that often happen to be lncRNA. When a lncRNA is complementary to another protein-coding or non-coding gene it can be classified as an antisense lncRNA or natural antisense transcript (NAT)[21]. A cis-NAT overlaps with its complementary gene, whereas a trans-NAT does not overlap with its complementary gene. The genesis of trans-NATs can probably be attributed to gene duplication events where the opposite strand has evolved the ability to be transcribed. Indeed, many lncRNAs are likely to be transcribed by so-called pseudogenes that no longer encode protein.
Figure 1.
Classification of lncRNA relative to protein-coding genes. Arrows indicate the direction of transcription. lncRNA contain non-coding exons (unfilled boxes). mRNA exons are composed of coding regions (filled boxes) and UTRs (unfilled boxes). Intronic lncRNA are located within an intron of a protein-coding gene in either direction and do not overlap with exons in the protein-coding gene. Divergent lncRNA/mRNA pairs are have proximal 5’-ends and are transcribed in opposite directions. Non-overlapping lncRNA do not overlap with protein-coding genes and are also referred to as intergenic or interleaved lncRNA. cisNATS are natural antisense transcripts that are complementary to an overlapping transcript. Here, the individual exons of the protein-coding gene have unique colors (blue, red, and green) and the colors of the cisNAT indicate corresponding regions of complementarity. transNATs are complementary to a non-overlapping transcript in a different genomic location (inter- or intra-chromosomal) where the colors indicate regions of complementarity. Adapted from Rinn and Chang [5]
Molecular functions of lncRNA
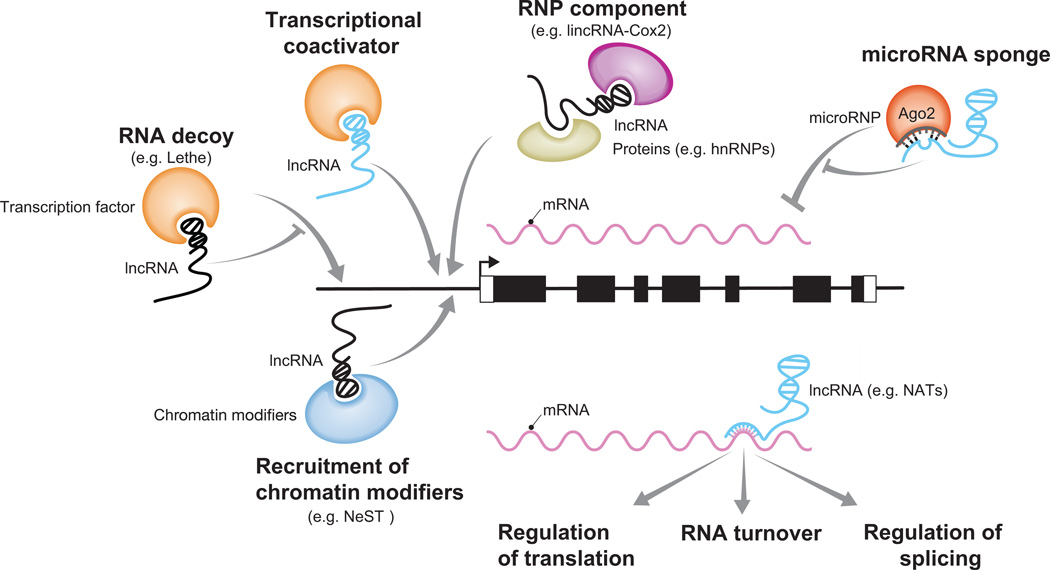
The molecular functions of lncRNA are exceptionally diverse and several comprehensive reviews describe their mechanisms of action in detail [5–8,21]. The precise sequence and structure of a lncRNA probably determines the number and type of molecules (e.g. protein, mRNA, miRNA) that it interacts with. Through these interactions, lncRNA regulate a variety of processes that include transcription, splicing, RNA degradation, and translation (Figure 2).
Figure 2.
Known and proposed functions of lncRNA in immunity. lncRNAs typically regulate transcription of a gene by forming complexes that bind a genomic region upstream of the transcription start site. lncRNA can potentially interact with other transcripts and regulate miRNA pathways, translation, splicing, and RNA turnover. lncRNAs with known molecular functions in immunity are colored black. Potential molecular functions for lncRNA in immunity are colored blue. Adapted from Hu et al [8]. lncRNA: long non-coding RNA, mRNA: messenger RNA, RNP: Ribonucleoprotein, hnRNPs: Heterogenous Ribonucleoproteins, NATs: Natural antisense transcripts.
Many lncRNA have been shown to interact with various chromatin-modifiers that alter the chemical structure of chromatin and determine the accessibility of DNA to transcription factors and RNA polymerases (e.g. Xist [22], Air [23], HOTAIR [24], Nest [25]). Other lncRNA are known to form ribonuceloproteins that regulate the transcription of specific sets of genes (e.g. lincRNA-Cox2 [26] and lincRNA-p21 [27]). lincRNA-p21 and hnRNP-K form a complex that bind specific regions of the genome and repress the transcription of genes in the p53 pathway. lncRNA can also act as decoys for DNA-binding proteins by mimicking the sequence or structure of the target DNA (e.g. gas5 [28], PANDA [29], DHFR minor [30] and possibly Lethe [31]). Like mRNA, lncRNA can also interact with miRNAs by hybridizing to partially complementary sequences [32,33] Additionally, some miRNA-binding lncRNA have the potential to act as molecular sponges that quench the effects of miRNA on their primary mRNA targets [34–39].
At least two lncRNA are known to regulate protein translation in one way or another. Several regions of lincRNA-p21 are complementary to the mRNA of CTNNB1 and JUNB, and binding of lincRNA-p21 to these mRNA is associated with their translational repression [40]. The antisense Uchl1 gene overlaps with two exons in the 5’-end of the Uchl1 protein-coding gene[41]. Antisense Uchl1 RNA promotes association of Uchl1 mRNA with active polysomes and enhances protein translation. A lncRNA that is antisense to the Zeb2 protein-coding gene affects Zeb2 protein-levels and may also regulate its translation [42]. Other lncRNAs are involved in different aspects of mRNA turnover, including mRNA decay and mRNA stabilization. Two lncRNAs, termed 1/2-sbsRNAs are partially complementary to the 3’UTRs of some mRNA and have the potential to form a duplex. The lncRNA-mRNA duplexes are proposed to bind Staufen 1, which drives staufen-mediated mRNA decay [43]. BACE1-AS is a lncRNA that is anti-sense to the protein-coding gene that encodes β-secretase-1 (BACE1), an enzyme associated with Alzheimer's disease pathophysiology. BACE1-AS is proposed to protect BACE1 from mRNA degradation through base-paring of the respective transcripts [44]. lncRNA gadd7 interacts with TDP43, a protein that binds and stabilizes Cdk6 mRNA. The interaction between gadd7 and TDP43 leads to increased degradation of Cdk6 mRNA [45]. lncRNAs can also regulate alternative splicing of pre-mRNAs. The lncRNA MALAT1 interacts with serine/arginine splicing factors, which affects the distribution of splicing factors in nuclear speckle domains and alternative splicing of pre-mRNA [46]. Small nucleolar lncRNAs (sno-lncRNAs) interact with FOX-family splicing factors and influence alternative splicing of pre-mRNAs [47].
Expression of lncRNAs in immune cells
To date, most lncRNA have been primarily studied in the context of genomic imprinting, cancer and cell differentiation [5,8]. However, lncRNA are now emerging as important regulators of various other biological processes, including the immune response. CD11c+ dendritic cells express lncRNAs when they are stimulated with lipopolysaccharide, an activator of TLR4 signaling [16]. CD8+ T cells express hundreds of lncRNA genes, many of which are specific to lymphocytes and are dynamically regulated during differentiation or activation [48]. CD4+ T cells are also reported to express many lncRNAs [49]. Hundreds of lncRNAs are expressed in mice exposed to severe acute respiratory virus or influenza virus [50], many of which are regulated downstream of type I IFN signaling. Rhabdomyosarcoma cells are also reported to express lncRNAs when they are infected with enterovirus 71 [51]. Although these studies clearly demonstrate that lncRNAs are expressed in different immunological contexts, their molecular functions are largely unexplored.
lncRNAs in adaptive immunity
In Th2 cells, lincR-Ccr2–5’AS is transcribed in the opposite direction of the chemokine Ccr2 and is located between Ccr3 ad Ccr3 [52]. Silencing of LincR-Ccr2–5’AS led to lower expression of the neighboring Ccr1, Ccr2, Ccr3, and Ccr5 genes. These chemokines are required for trafficking of Th2 cells to the lungs and knockdown of lincR-Ccr2–5’AS also reduced migration of Th2 cells to the lung.
TMEVPG1/ NeST is a lncRNA that was initially proposed to control Theiler’s virus persistence in mice [53]. Following infection with Theiler’s virus, TMEVPG1/ NeST is expressed in immune cells of susceptible SJL/J mice that develop persistent infection, but not in those from the resistant B10.S strain. TMEVPG1/ NeST is located in a region of the genome near the IFN-γ gene. Earlier studies demonstrated that transcription of TMEVPG1/NeST was Th1 selective and dependent on Stat4 and T-bet, transcriptions factors that are involved in Th1 cell differentiation [54]. Gomez et al developed B10.S mice that express either the SJL/J-derived or B10.S-derived NeST RNA as a transgene in both CD4+ and CD8+ T cells [25]. Unlike the parental B10.S strain, which clear Theiler’s virus, the B10.S mice that overexpress SJL/J-derived NeST, phenocopy the SJL/J parental strain by becoming persistently infected and developing demyelinating lesions in the brain. NeST also confers differential susceptibility to Salmonella enterica Typhimurium. B10.S mice succumb to lethal Salmonella infection, while SJL/J or the B10.S mice that overexpress the SJL/J-derived NeST were able to survive this infection. The higher expression of SJL/J derived NeST in these mice, led to the inducible synthesis of IFN-γ in CD8+ T cells causing decreased Salmonella enterica pathogenesis. NeST acts as an enhancer-like lncRNA by enhancing transcription of IFN-γ. NeST RNA binds WDR5, which alters histone 3 lysine 4 trimethylation at the IFN-γ locus. Consequently, both IFN-γ RNA and IFN-γ protein levels are increased in activated CD8+ T cells. Collectively, these findings highlight the importance of lncRNAs in host defense and indicate that like protein coding genes, lncRNAs can confer susceptibility to infectious diseases.
lncRNAs in innate immunity
Recently, several groups have provided increasing levels of evidence that collectively demonstrate the functional importance of lncRNAs in innate immunity [26,31,55,56]. A KIR antisense lncRNA is expressed in progenitor cells or pluripotent cell lines and its overexpression in NK cells leads to decreased expression of the KIR protein-coding gene [55]. KIR antisense lncRNA overlaps with KIR-coding exons 1 and 2, as well as a proximal promoter that is upstream of KIR. Transcription of KIR antisense lncRNA appears to be regulated by myeloid zinc finger 1, which leads to silencing of KIR through an unknown mechanism.
Tumor necrosis factor induces the expression of hundreds of lncRNAs in murine fibroblasts [31]. Among these, Lethe, a pseudogene lncRNA, is transcribed upon activation of NFκB, a transcription factor important in inflammation. In turn, Lethe binds directly to RelA, a subunit of the NFκB heterodimeric transcription factor and inhibits NFκB DNA binding activity. These findings suggest that Lethe functions as a post-induction feedback regulator of TNF signaling in order to dampen the inflammatory response.
HOTAIRM1 is a lncRNA that is specifically expressed in myeloid cells [56]. It is antisense to the HOXA genes and is transcribed in NB4 promyelocytic leukemia cells that are activated by all-trans retinoic acid (ATRA) through the retinoic acid receptor. During ATRA-induced granulocyte maturation of NB4 cells, HOTAIRM1 is required for the expression of HOXA1, HOXA4, CD11b, and CD18. These results suggest that HOTAIRM plays an important role during granulocyte maturation.
A synthetic bacterial lipoprotein (Pam3CSK4), that engages TLR2 induces the expression of 62 lncRNAs in mouse bone marrow-derived macrophages [26]. The most significantly induced lncRNAs tended to be located near protein-coding immune genes, suggesting co-regulation of these neighboring genes. lincRNA-Cox2 was amongst the most highly induced of these lncRNAs and is proximal to the prostaglandin-endoperoxide synthase 2 (Ptgs2/Cox2) gene. TLR ligands as well as other inflammatory triggers induced co-expression of both lincRNA-Cox2 and its neighboring Ptgs2 (Cox2) gene. lincRNA-Cox2 was shown to both positively and negatively regulate the transcription of distinct classes of immune genes. In the unstimulated state, lincRNA-Cox2 repressed the expression of 787 genes, including chemokines (Ccl5, Cx3cl1), chemokine receptors (Ccrl) and interferon-stimulated genes (ISGs) (Irf7, Oas1a, Oas1l, Oas2, Ifi204 and Isg15). However, stimulation of TLR2 with Pam3CSK4, induced the expression of 713 distinct genes that were dependent on lincRNA-Cox2, including Tlr1, Il6 and Il23a. Transcriptional repression of the target genes (e.g. Ccl5) was primarily mediated by interactions of lincRNA-Cox2 with heterogeneous nuclear ribonucleoprotein A/B and A2/B1. However, these two hnRNP proteins were not involved in coordinating the TLR induced transcription of IL6, suggesting that lincRNA-Cox2 forms additional regulatory complexes.
lncRNAs and modulation of host-pathogen interactions
It is likely that some lncRNAs participate in host-pathogen interactions that alter the resulting immune response. Many viruses encode miRNA that target host mRNA and are important for viral replication and immune evasion [57]. It is therefore conceivable that host cells express lncRNA that quench the effects of these viral miRNA during an immune response. While some lncRNAs (e.g. lincRNA-Cox2) repress anti-viral gene expression, it is also conceivable that viruses or other pathogens utilize these host lncRNAs to thwart induction of anti-viral genes.
lncRNAs have been described in plasmodium falciparum and several viruses [58–60]. The viral lncRNA are often smaller than 200 nucleotides and regulate a variety of processes, including viral latency, apoptosis, and immunity. Kaposi’s sarcoma-associated herpesvirus (KSHV) expresses a polyadenylated nuclear RNA (lncRNA-PAN) that interacts with IRF4 and is proposed to inhibit transcription of IL-4, IL-18, and IFNγ [61]. In T cells, Herpesvirus expresses the non-coding RNAs HSUR1 and HSUR2, which interact with the host miRNA miR-27. This interaction leads to degradation of the miRNA and altered expression of its target genes[62].
Conclusions
To date, more than a hundred human lncRNAs with experimentally determined biological functions have been annotated in lncrna db [63]. We propose that lncRNA have the potential to perform a variety of molecular functions in the immune system (Figure 2). However, there are multiple challenges and concerns associated with the functional characterization of lncRNAs [9]. It is possible that some lncRNA are the product of promiscuous transcription or only perform minor molecular functions [64]. Many human lncRNA do not have obvious homologs in mouse [12], and it will be important to determine whether species-specific lncRNA are responsible for major differences between the mouse and human immune systems. It is paramount that the functions of lncRNA in immunity are carefully assessed in genetically manipulated mice. Just like protein-coding genes, genetic manipulation of some lncRNA will produce a dramatic phenotype [25,65], while others will lack an obvious phenotype [66,67]. Many lncRNA appear to be expressed at low levels in immune cells [26] and it will be important to determine their functional relevance. While it is tempting to dismiss lncRNA with low expression values as transcriptional noise, it is likely that current methods to detect transcription are not sufficiently sensitive. Furthermore, it should be noted that relatively low levels of NeST (~0.5 molecules/cell) produce a dramatic phenotype [25]. As many lncRNA are specifically expressed in certain cell types or subcellular compartments [68], it will also be important to assess the expression of lncRNA in different immune cells and their respective compartments. In summary, we propose that lncRNA will be involved various aspects of the immune response that will often be specific to a particular immune cell or subcellular location. A better understanding of the biology of these genes could uncover new targets for therapeutic intervention in both infectious and inflammatory diseases.
Highlights.
lncRNAs are the largest class of non-coding RNA genes in mammalian genomes.
lncRNAs are expressed in a variety of immune cells.
Recent studies demonstrate that lncRNA perform major functions in innate and immune cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Chaussabel D. Unique gene expression profiles of human macrophages and dendritic cells to phylogenetically distinct parasites. Blood. 2003;102:672–681. doi: 10.1182/blood-2002-10-3232. [DOI] [PubMed] [Google Scholar]
- 2.Nusinzon I. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proceedings of the National Academy of Sciences. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 4.Doyle SL, O'Neill LAJ. Toll-like receptors: From the discovery of NFκB to new insights into transcriptional regulations in innate immunity. Biochemical Pharmacology. 2006;72:1102–1113. doi: 10.1016/j.bcp.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 5. Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. •• A comprehensive review of lncRNA
- 6.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon J-H, Abdelmohsen K, Gorospe M. Post-transcriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2012 doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. •• A carefully considered review of lncRNAs in mammalian cell differentiation
- 9.Ulitsky I, Bartel DP. lincRNAs: Genomics, Evolution, and Mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell. 2013 doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pachnis V, Brannan CI, Tilghman SM. The structure and expression of a novel gene activated in early mouse embryogenesis. The EMBO Journal. 1988;7:673–681. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Molecular and Cellular Biology. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 16.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SPA, Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 18.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 19.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proceedings of the National Academy of Sciences. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magistri M, Faghihi MA, Laurent GS, III, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends in Genetics. 2012 doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb Proteins Targeted by a Short Repeat RNA to the Mouse X Chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 24.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gomez JA, Wapinski OL, Yang YW, Bureau J-F, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST Long ncRNA Controls Microbial Susceptibility and Epigenetic Activation of the Interferon-γ Locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. •• Using mouse genetics, this study demonstrates that a lncRNA (NeST) can dramatically alter the susceptibility and adaptive immune response of a mouse to two different pathogens. Furthermore, a relatively small number of lncRNA molecules are responsible for these altered phenotypes.
- 26. Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, et al. A Long Noncoding RNA Mediates Both Activation and Repression of Immune Response Genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. •• This study demonstrates that hundreds of immune genes are both positively and negatively regulated by lincRNA-Cox2 during an innate immune response. Many of the target genes are repressed at the transcriptional level and are mediated by a RNP complex that consists of lincRNA-Cox2 and hnRNP A/B or hnRNP A2/B1.
- 27.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Science Signaling. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 31. Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge R-M, Chang HY, Gingeras T. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife. 2013;2 doi: 10.7554/eLife.00762. • This study demonstrates that a lncRNA can modulate a transcription factor that is essential to the immune response.
- 32.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013 doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sumazin P, Yang X, Chiu H-S, Chung W-J, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, et al. An Extensive MicroRNA-Mediated Network of RNA-RNA Interactions Regulates Established Oncogenic Pathways in Glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubio-Somoza I, Weigel D, Franco-Zorilla J-M, García JA, Paz-Ares J. ceRNAs: miRNA target mimic mimics. Cell. 2011;147:1431–1432. doi: 10.1016/j.cell.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al. In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147:382–395. doi: 10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-Independent Regulation of the Tumor Suppressor PTEN by Competing Endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biology. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon J-H, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 Suppresses Target mRNA Translation. Molecular Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012 doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 42.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes & Development. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St-Laurent G, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Li D, Zhang W, Guo M, Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay [Internet] The EMBO Journal. 2012 doi: 10.1038/emboj.2012.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The Nuclear-Retained Noncoding RNA MALAT1 Regulates Alternative Splicing by Modulating SR Splicing Factor Phosphorylation. Molecular Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin Q-F, Yang L, Zhang Y, Xiang J-F, Wu Y-W, Carmichael GG, Chen LL. Long Noncoding RNAs with snoRNA Ends. Molecular Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 48. Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen W, Mattick JS. Genome-wide identification of long noncoding RNAs in CD8+ T cells. The Journal of Immunology. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. • This study suggests that lymphoid cells express specific repertoires of lncRNA that are likely to regulate T-cell differentiation and activation.
- 49.Pagani M, Rossetti G, Panzeri I, de Candia P, Bonnal RJP, Rossi RL, Geginat J, Abrignani S. Role of microRNAs and long-non-coding RNAs in CD4 +T-cell differentiation. Immunol Rev. 2013;253:82–96. doi: 10.1111/imr.12055. [DOI] [PubMed] [Google Scholar]
- 50.Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, et al. Unique Signatures of Long Noncoding RNA Expression in Response to Virus Infection and Altered Innate Immune Signaling. mBio. 2010;1:e00206–10–e00206–18. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin Z, Guan D, Fan Q, Su J, Zheng W, Ma W, Ke C. lncRNA expression signatures in response to enterovirus 71 infection. Biochemical and Biophysical Research Communications. 2013;430:629–633. doi: 10.1016/j.bbrc.2012.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013 doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vigneau S, Rohrlich P-S, Brahic M, Bureau J-F. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. Journal of Virology. 2003;77:5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting Edge: Influence of Tmevpg1, a Long Intergenic Noncoding RNA, on the Expression of Ifng by Th1 Cells. The Journal of Immunology. 2012;189:2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright PW, Huehn A, Cichocki F, Li H, Sharma N, Dang H, Lenvik TR, Woll P, Kaufman D, Miller JS, et al. Identification of a KIR antisense lncRNA expressed by progenitor cells. Genes Immun. 2013 doi: 10.1038/gene.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. • This study demonstrates that lncRNA play an important role during granulocyte maturation and suggest that lncRNAs are probably important in the differentiation of other immune cells.
- 57.Skalsky RL, Cullen BR. Viruses, microRNAs, and Host Interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Broadbent KM, Park D, Wolf AR, Van Tyne D, Sims JS, Ribacke U, Volkman S, Duraisingh M, Wirth D, Sabeti PC, et al. A global transcriptional analysis of Plasmodium falciparum malaria reveals a novel family of telomere-associated lncRNAs. Genome Biology. 2011;12:R56. doi: 10.1186/gb-2011-12-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scaria V, Pasha A. Long Non-Coding RNAs in Infection Biology. Front. Gene. 2012;3:308. doi: 10.3389/fgene.2012.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Q, Jeang K-T. Long noncoding RNAs and viral infections. BioMedicine. 2013;3:34–42. doi: 10.1016/j.biomed.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rossetto CC, Pari GS. Kaposi's sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. Journal of Virology. 2011;85:13290–13297. doi: 10.1128/JVI.05886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cazalla D, Yario T, Steitz JA. Down-Regulation of a Host MicroRNA by a Herpesvirus saimiri Noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Research. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bird A. Genome Biology: Not Drowning but Waving. Cell. 2013;154:951–952. doi: 10.1016/j.cell.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved Function of lincRNAs in Vertebrate Embryonic Development despite Rapid Sequence Evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. doi: 10.1261/rna.033217.112. [no date], [no volume]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]