Abstract
Epidemiologic data suggest that maternal microbial infections may cause fetal neurodevelopmental disorders, potentially increasing susceptibility to heavy psychopathologies such as schizophrenia, schizophreniform disorder, autism, pervasive developmental disorders, bipolar disorders, psychosis, epilepsy, language and speech disorders, and cognitive impairment in adult offspring. However, the molecular pathomechanisms underlying such a relationship are not clear. Here we analyze the potential role of the maternal immune response to viral infection in determining fetal brain injuries that increase the risk of neurological disorders in the adult. We use influenza infection as a disease model and human axon guidance pathway, a key process in the formation of neural network during midgestation, as a potential fetal target of immune insults. Specifically, we examined influenza A H1N1 hemagglutinin (HA), an antigenic viral protein, for amino acid sequence similarity to a random library of 188 axon guidance proteins. We obtain the results that (1) contrary to any theoretical expectations, 45 viral pentapeptide matches are distributed throughout a subset of 36 guidance molecules; (2) in 24 guidance proteins, the peptide sharing with HA antigen involves already experimentally validated influenza HA epitopes; and (3) most of the axon guidance vs HA peptide overlap is conserved among influenza A viral strains and subsets. Taken together, our data indicate that immune cross-reactivity between influenza HA and axon guidance molecules is possible and may well represent a pathologic mechanism capable of determining neurodevelopmental disruption in the fetus.
Key words: influenza A H1N1 virus, hemagglutinin, immune cross-reactivity, schizophrenia, autism, bipolar disorder
Introduction
Little information exists about the molecular basis underlying schizophrenia. Abnormalities in neurotransmitters (eg, dopamine, 5-hydroxytryptamine, glutamate, and gamma amino butyric acid systems),1–4 intracellular cytoskeletal assembly,5 and synapse-associated proteins6,7 have been thoroughly studied, but attempts to unequivocally relate schizophrenia to alterations in specific molecules are unsatisfying. The old dopamine hypothesis of schizophrenia1 turned toward a more complex context, involving neurotransmitter interactions and neurocircuits.8 Also, in many cases, genetic mutations and polymorphisms of genes putatively associated to schizophrenia do not appear related to an increased risk for schizophrenia.9–16
Likewise, autism and an ample category of autism-like disorders are of unknown etiology.17–19 Several hypotheses have been advanced to explain risk for autism, pervasive developmental disorders, speech or language impairment, Asperger’s syndrome, and Rett syndrome. Genetic factors20–22 and prenatal exposure to ultrasounds,23 environmental stress,24,25 and microbial infections,26–29 have been invoked as risk factors that can disrupt fetal brain development. However, notwithstanding worldwide extensive and intensive research efforts to solve the mystery of autism and autism-like pathologies, science and medicine are still facing a lack of knowledge of the molecular basis of such neurodisorders.17–19
Actually, it seems that the primum movens in autism and schizophrenia is not related to a single molecule or lesion but might deal with a defective neural connectivity originating during neurodevelopment30–32 as a result of prenatal insult(s).33 In particular, an association between prenatal viral influenza infection and an increased risk for autism and schizophrenia in the adult offspring has been repeatedly studied,33–38 and maternal immune activation during pregnancy, rather than direct infection of the fetus, has been advanced as a possible causative insult.39–42 The time of exposure to influenza virus has been invoked to be a critical factor, and a risk of schizophrenia in humans appears to be higher for influenza exposure from early to midgestation.33,39,43 Accordingly, in an experimental model of prenatal exposure to human influenza virus in mice, Fatemi and colleagues37,44 demonstrated that abnormal changes occur in the offspring following influenza infection at E9 (which corresponds to the middle of the first trimester in humans),37,45 E16 (which corresponds to the middle of the second trimester in humans),46 and E18 (which corresponds to late second trimester in humans).38 The extent of aberrant changes was higher in offspring of E16-infected mice, thus suggesting that infection during middle second trimester leads to heavier effects in the exposed offspring than during middle first or late second trimester.46 On the other hand, in spite of the epidemiologic and experimental evidence that prenatal exposure to influenza infection may be linked to autism, schizophrenia, and other neurological alterations in the childhood,29–46 the mechanicistic role(s) of influenza infection in the etiology of such disorders remain to be elucidated.33
In the last decade, the possibility of accessing complete human and microbial proteomes provided a new platform for dissecting the host-virus relationships at the phenetic peptide level.47–50 Indeed, sharing of peptide modules between microbes and the human host may prelude to a subversion of host cellular processes and immune responses.49–53 Hence, analysis of the shared motifs and of the proteins involved in the overlap may help identify altered cellular functions and associated (immune)pathologies and, possibly, open the way to new preventive/therapeutic approaches54–58 and more specific diagnostic tools.58 In exploring the hexapeptide identity platform between the influenza A H5N1 and Homo sapiens proteomes, it was found that peptide sharing involves human proteins such as reelin, neurexin I-α, myosin-IXa, Bardet-Biedl syndrome 10 protein, Williams syndrome transcription factor, disrupted in schizophrenia 1 protein, amyotrophic lateral sclerosis 2 chromosomal region candidate gene 17 protein, fragile X mental retardation 2 protein, and jouberin.48 That is, the influenza A H5N1 polyprotein-vs-human proteome peptide overlap involves human antigens that, when altered, have been reported to be potentially associated with multiple neurological disorders that can include autism, schizophrenia, epilepsy, amyotrophic lateral sclerosis, and sensorineural deafness.
According to this line of research, to understand how maternal immune activation during pregnancy might cause neurodevelopmental alterations and result in psychopathologies in the offspring, in this study we analyzed the pentapeptide overlap between influenza A H1N1 hemagglutinin (HA), a highly antigenic protein,55,59 and human proteins associated to the axon guidance pathway. In fact, guidance molecules have important roles in neurodegenerative diseases during neural development.60,61 Moreover, the relationship between prenatal infection and risk of neurological disorders seems to be temporally circumscribed to midgestation,33,39,46,62 a time frame coincident with guidance protein expression (~16 week of gestation).63
We searched HA and axon guidance molecules for common pentapeptide motifs because the optimal amino acid (aa) length of a B-cell epitope is 5 aa.64–69 Likewise, scientific literature indicates that 5 residues may represent minimal antigenic determinants in T-cell epitopes,70–75 and numerous pentapeptide epitopes able to bind major histocompatibility complex molecules and induce T-cell proliferation have been described.76–86 Therefore, pentapeptide sharing between the viral protein and axon guidance proteins might be indicative of potential immune cross-reactions.
Here, we report that HA and axon guidance molecules have a vast pentapeptide overlap that quantitatively exceeds the expected values and qualitatively is endowed with an immunological potential. Our findings support the hypothesis of an immune basis for increased risk of schizophrenia, autism, and other psychopathologies in the offspring following influenza prenatal infection.
Methods
HA protein sequence, UniProtKB/Swiss-Prot accession number: Q67010, 565 aa long, from influenza A H1N1 virus (National Center for Biotechnology Information Taxonomic identifier: 382845, isolate A/swine/Cambridge/1939) was analyzed for pentapeptide sharing with human axon guidance proteins as follows. First, a viral pentapeptide library was constructed by dissecting the HA primary sequence into pentapeptides offset by 1 residue, ie, MKARL, KARLL, ARLLV, RLLVL, etc. Then, each of the final 561 pentamers was analyzed for occurrence(s) within a library consisting of primary sequences of human proteins involved in axon guidance pathway. To eliminate possible bias in the analysis, the axon guidance library was constructed at random using UniProtKB Database (http://www.uniprot.org/)87 and utilizing the following 5 key words: axon, guidance, proteins, homo, sapiens. The key word-guided search produced 188 human protein entries (see table 1) that directly or indirectly relate to axon guidance pathway and are of different length from 93 aa (SDF1) to 4640 aa (MYCB2) for a total of 176 648 aa. Axon guidance proteins are reported as UniProtKB/Swiss-Prot entry names throughout the article, unless when discussed in detail. Any viral occurrence in the set of guidance proteins was termed a match. Proteins hosting viral match(es) were recorded by UniProtKB/Swiss-Prot entry name and briefly described.
Table 1.
Random Library of Human Proteins Associated to Axon Guidance Pathway Downloaded from UniProtkb Database
| ABL1(1130); ABL2 (1182); ABLM2 (611); ABLM3 (683); ACTB (375); ACTG (375); AGAP2 (1192); ANK1 (1881); ANK2 (3957); AP2A1 (977); AP2A2 (939); AP2B1 (937); AP2M1 (435); AP2S1 (142); ARHGB (1522); ARHGC (1544); B3GN2 (397); BOC (1114); CADH4 (916); CAP1 (475); CAP2 (477); CD166 (583); CD5R1 (307); CDC42 (191); CDK1 (297); CDK5 (292); CHL1 (1208); CLAP1 (1538); CLAP2 (1294); CNTN1 (1018); CO1A1 (1464); CO4A2 (1712); CO4A3 (1670); CO6A3 (3177); COF1 (166); CREB1 (341); CSK21 (391); CSK22 (350); DCC (1447); DLG1 (904); DLG3 (817); DOCK1 (1865); DPYL1 (572); DPYL2 (572); DPYL3 (570); DPYL5 (564); DRAXI (349); DVL1 (695); EFNB1 (346); EGFR (1210); ENAH (591); EPHA7 (998); EPHA8 (1005); EPHB3 (998); ERBB4 (1308); EVL (416); EXT1 (746); EZRI (586); FAK1 (1052); FARP2 (1054); FES (822); FEZ1 (392); FEZ2 (353); FGFR1 (822); FOXD1 (465); FOXD4 (439); FX4L2 (416); FX4L4 (416); FX4L5 (416); FX4L6 (417); FYN (537); FZD3 (666); GFRA1 (465); GFRA3 (400); GLI2 (1586); GLI3 (1580); GRB2 (217); GSK3B (420); HS90A (732); HS90B (724); ITA2 (1181); ITAV (1048); ITB1 (798); ITB3 (788); JIP3 (1336); K319L (1049); KDIS (1771); KGP1 (671); KIF5C (957); KS6A1 (735); KS6A2 (733); KS6A3 (740); LGI1 (557); MET (1390); MK01 (360); MP2K1 (393); MYCB2 (4640); MYH11 (1972); MYH9 (1960); MYO10 (2058); NET1 (604); NFASC (1347); NFIB (420); NOGG (232); NRCAM (1304); NRX1A (1477); NRX3A (1643); PAK1 (545); PAK2 (524); PI51C (668); PLCG1 (1290); PLXA1 (1896); PLXA2 (1894); PLXA3 (1871); PLXA4 (1894); PLXB1 (2135); PLXB2 (1838); PLXB3 (1909); PLXD1 (1925); PO4F2 (409); PRIO (253); PSD95 (724); PTN11 (597); RAC1 (192); RAC2 (192); RAF1 (648); RASH (189); RASK (189); RASN (189); RGMA (450); RHG35 (1513); RHG39 (1083); RHOA (193); RHOB (196); RHOC (193); RHOG (191); RND1 (232); ROBO1 (1651); ROBO2 (1378); ROCK1 (1354); ROCK2 (1388); RRAS (218); RTN4R (473); RYK (604); SCC4 (613); SCN3B (215); SDCB1 (298); SDF1 (93); SEM3E (775); SEM4A (761); SEM4C (833); SEM4D (862); SEM4F (770); SEM4G (838); SEM6A (1030); SEM6D (1073); SH3G2 (352); SHH (462); SHOT1 (631); SIA8B (375); SIAH1 (282); SIAH2 (324); SLIT1 (1534); SLIT2 (1529); SLIT3 (1523); SMO (787); SPON2 (331); SPTA1 (2419); SPTB1 (2137); SPTB2 (2364); SPTN1 (2472); SPTN2 (2390); SPTN4 (2564); SRC (536); SRGP1 (1085); SRGP2 (1071); STIP1 (543); TBB3 (450); TLN1 (2541); TNR16 (427); TRPC4 (977); UBP33 (942); UNC5A (842); UNC5B (945); UNC5D (953); VASP (380); WASL (352); WNT3A (352). |
Note: Proteins are given as UniProtKB/Swiss-Prot entry names and listed in alphabetical order with aa length in parentheses.
The 2 libraries were searched for aa groupings that were common sequences using pentapeptides as probes. As discussed above, a pentapeptide can be a sufficient minimal determinant for epitope-paratope interaction and thus can act as an immune unit and play a crucial role in cellular immunoreactivity and antigen-antibody recognition.64–86,88,89
Multialignment sequence analysis was carried with T-Coffee program90,91 using HA aa primary sequences corresponding to the following HA Swiss-Prot entries: Q9WFX3, 566 aa (strain A/Brevig Mission/1/1918 [H1N1]); P03455, 566 aa (strain A/Swine/New Jersey/ 11/1976 [H1N1]); Q9WCE3, 566 aa (strain A/Duck/Australia/749/1980 [H1N1]); P26142, 122 aa (strain A/Camel/Mongolia/1982 [H1N1]); Q07FI5, 565 aa (strain A/China:Nanchang/11/1996 [H1N1]); Q289M7, 565 aa (strain A/New Zealand:South Canterbury/35/2000 [H1N1]); B4URD6, 565 aa (strain A/Russia:St.Petersburg/ 8/2006 [H1N1]); and A8C8J4, 565 aa (strain A/USA: Texas/UR06-0195/2007 [H1N1]).
The Immune Epitope Database and Analysis Resources (IEDB; www.immuneepitope.org)92 was used to search for influenza A HA-derived B- and/or T-cell epitopes raised in the human host.
Results
Pentapeptide Sharing Between Influenza A H1N1 HA and Human Axon Guidance Proteins
Table 1 lists the 188 human guidance molecules derived from UniProtKB databank and analyzed for exact pentapeptide matching to HA protein (UniProtKB/Swiss-Prot accession: Q67010) from influenza A H1N1 virus, isolate A/swine/Cambridge/1939. The viral protein was chosen based on the following criteria: (1) known to belong to an influenza strain (swine/Cambridge/1939) that had been studied in the context of the relationship between schizophrenia and the occurrence of influenza epidemics93 and (2) of significant antigenic and immunogenic impact.55,59
Pentapeptide matching to influenza A H1N1 HA protein was carried out as follows. A pentapeptide library of 561 pentamers overlapped by 4 residues was created for influenza A H1N1 HA protein; then, a protein library was created by downloading aa sequences of the 188 guidance molecules listed in table 1 from UniProtKB database. Each viral pentapeptide from the first library was used to search for instances of the same pentapeptide in the axon guidance protein library, and the protein(s) sharing the match(es) were recorded.
The final data on the peptide sharing are reported in table 2. It can be seen that 36 out of the 188 guidance proteins listed in table 1, collected as described under Methods, have pentapeptide matches to HA. Exactly, 45 viral pentapeptide matches are distributed throughout 36 human proteins related to human axon guidance pathway. The 36 axon guidance proteins are briefly described under table 294–135 (further details and related references are available at http://www.uniprot.org/). The pentapeptide matching pattern varies, with guidance proteins sharing 2 matches (ANK2, AP2A2, CO6A3, FARP2, FES, GLI2, KS6A1, KS6A2, MYCB2, NFASC, ROBO1, and SEM6A) or even 3 matches (AP2M1 and FEZ2) with HA protein.
Table 2.
Distribution of HA Pentapeptide Matches in Human Axon Guidance Proteins
| Sequencea | Posb | Human Axon Guidance Proteinc |
|---|---|---|
| ARLLV | 3 | SEM4G. Semaphorin-4G. It is a ligand of Plexin-B2, required in cerebellar development94 |
| LLVLLd | 5 | ARHGB. Rho guanine nucleotide exchange factor 11. Modulator of RhoGEF glutamate transport95 |
| CO4A3. Collagen α-3(IV) chain. Goodpasture antigen. Binds to a protein that is highly expressed in neurons of the cerebral cortex, hippocampal formation, the basal ganglia, the olfactory bulb and nuclei of the thalamus, the hypothalamus, and the septal area96 | ||
| TLAAT | 11 | SPTA1. Spectrin α chain, erythrocytic 1. Defects in SPTA1 are the cause of epileptic encephalopathy early infantile type 597 |
| LLEKN | 36 | SPTN4. Spectrin beta chain, nonerythrocytic 4. Involved in nervous system membrane biogenesis and in ion channel clustering98 |
| SVNLL | 46 | AP2M1.e AP-2 complex subunit mu. Involved in axonal growth cone motility99 |
| VNLLE | 47 | CO6A3.f Collagen α-3(VI) chain. Protects against neuronal apoptosis100 |
| LLEDS | 49 | GFRA1. GDNF family receptor α-1. Involved in establishing synaptic contacts and promoting the assembly of presynaptic terminals101 |
| HNGKL | 54 | SEM6A.f Semaphorin-6A. It is as a major contributor to the guidance of corticospinal tract axons at multiple choice points102 |
| GNPEC | 80 | KS6A1.f Ribosomal protein S6 kinase α-1. Involved in signaling pathways responsible for tuberous sclerosis complex 2 phosphorylation. Involved in hippocampal signaling cascades in consolidation of fear memory103–106 |
| SLLPA | 86 | GLI2.f Zinc finger protein GLI2. Gli2 alterations affect Slit function in pioneer longitudinal guidance107 |
| LLPAR | 87 | GLI2.f See previous entry |
| NSETG | 101 | AP2M1.e See previous entry |
| SETGA | 102 | AP2M1.e See previous entry |
| PGDFI | 109 | CLAP2. CLIP-associating protein 2. Regulates microtubule plus-end dynamics at the cell cortex108 |
| EELRE | 116 | FEZ2.e Fasciculation and elongation protein zeta-2. Involved in axonal outgrowth and fasciculation109,110 |
| ELREQ | 117 | FEZ2.e See previous entry |
| LREQL | 118 | FEZ2.e See previous entry |
| PSD95. Postsynaptic density protein 95. Disks large homolog 4. Required for synaptic plasticity associated with NMDA receptor signaling111,112 | ||
| EQLSS | 120 | FGFR1. Fibroblast growth factor receptor 1. Required for normal mesoderm patterning and correct axial organization during normal development of the gonadotropin-releasing hormone neuronal system113 |
| SSVSS | 123 | ROBO1.f Roundabout homolog 1. Receptor for SLIT1 and SLIT2. Acts as molecular guidance cue in axonal navigation at the ventral midline of the neural tube and projection of axons to different regions during neuronal development114,115 |
| SVSSL | 124 | DOCK1. Dedicator of cytokinesis protein 1. Required for projection of commissural axons in the neural tube116 |
| SSLER | 126 | SEM6A.f See previous entry |
| PNHNT | 141 | MYCB2.f E3 ubiquitin-protein ligase MYCBP2. Key regulator for axon guidance, outgrowth, and synapse development117 |
| LTKKG | 167 | FARP2.f FERM, RhoGEF and pleckstrin domain-containing protein 2. Involved in the response of neuronal growth cones to class-3 semaphorins118 |
| TKKGN | 168 | ANK1. Ankyrin-1. Integral membrane protein. Isoform Br21 is expressed in brain119 |
| ANK2.f Ankyrin-2. Brain ankyrin. Structurally defines terminal microdomains of peripheral somatosensory axons120 | ||
| VNNKG | 182 | MYCB2.f See previous entry |
| RFTPE | 225 | ANK2.f See previous entry |
| GDTII | 253 | NFASC.f Neurofascin. Involved in neurite extension, axonal guidance, synaptogenesis, and myelination121 |
| DTIIF | 254 | NFASC.f See previous entry |
| EATGN | 259 | ROBO1.f See previous entry |
| GNLIA | 262 | AP2A1. AP-2 complex subunit α-1. Component of the adaptor protein complex 2. Expressed in forebrain, spinal cord, cerebellum122 |
| AP2A2.f AP-2 complex subunit α-2. Component of the adaptor protein complex 2. Huntingtin-interacting protein 9122,123 | ||
| NLIAP | 263 | FARP2.f See previous entry |
| TSNAS | 283 | DLG1. Disks large homolog 1. Dlg1-PTEN interaction regulates myelin thickness to prevent peripheral nerve overmyelination124 |
| SLPFQ | 305 | KS6A1.f See previous entry |
| KS6A2.f Ribosomal protein S6 kinase α-2. Chronic activation of p90RSK has been shown in human epileptic hippocampus. Involved in hippocampal signaling cascades in consolidation of fear memory104–106 | ||
| KYVRS | 321 | FES.f Tyrosine-protein kinase Fes/Fps. Cooperates to induce neurite extension125 |
| TGLRN | 332 | SLIT1. Slit homolog 1 protein. Molecular guidance cue. Function seems to be mediated by interaction with ROBO receptors114,115,126 |
| GLRNI | 333 | NRX3A. Neurexin-3-α. Neuronal synaptic adhesion molecule127 |
| SIQSR | 339 | ROCK2. Rho-associated protein kinase 2. Plays a role in the regulation of spine and synaptic properties in the hippocampus128,129 |
| SRGLF | 342 | ITA2. Integrin α-2. Integrin-ECM interactions regulate axonal outgrowth and pathfinding130 |
| AELLV | 439 | FES.f See previous entry |
| LLVLLd | 441 | ARHGB. See previous entry |
| CO4A3. See previous entry | ||
| AKEIG | 473 | RAC1. Ras-related C3 botulinum toxin substrate 1. Plays an essential role in neuronal development131 |
| QILAI | 528 | SEM4F. Semaphorin-4F. Expressed during neurodevelopment and in the adult brain. Attached to PSD95 in glutamatergic synapses132 |
| STVAS | 534 | AP2A2.f See previous entry |
| VASSL | 536 | ARHGC. Rho guanine nucleotide exchange factor 12. LARG protein. Mediates the action of repulsive guidance molecule133 |
| KS6A2.f See previous entry | ||
| LVLLV | 540 | CO6A3.f See previous entry |
| SNGSL | 555 | DCC. Netrin receptor DCC. Mediates axon attraction of neuronal growth cones during neurodevelopment upon ligand binding134 |
| EFNB1. Ephrin-B1. Regulates axon guidance by reverse signaling through a PDZ-dependent mechanism135 |
aPentapeptide sequence, with aa given in 1-letter code.
baa position along the HA sequence.
cHuman axon guidance proteins given with UniProtKB/Swiss-Prot entry and recommended name.
dLLVLL occurs twice in HA, at aa pos 5 and 441.
eGuidance protein with three pentapeptide matches to HA.
fGuidance protein with two pentapeptide matches to HA.
Number of Occurrences of HA Pentapeptide Matches in Human Axon Guidance Proteins Is Independent of the Protein Length
Mathematically, the number of times a perfect n-peptide match might occur at random in 2 proteins is directly proportional to the product of the protein aa lengths and inversely proportional to the number of possible aa (20) raised to n. In the present case, for a protein to have a HA pentapeptide match, it must have a minimal length equal to 5664 aa. For the 2 libraries analyzed here (eg, the HA pentapeptide set amounting to 565 aa, and 176 648 aa, respectively), the theoretical number of pentapeptide matches is 31.2.
Hence, it has to be observed that, as an average value, the comprehensive overlapping extent reported in table 2 (equal to 45 pentapeptide matches) is about 1.5-fold higher than the theoretical value.
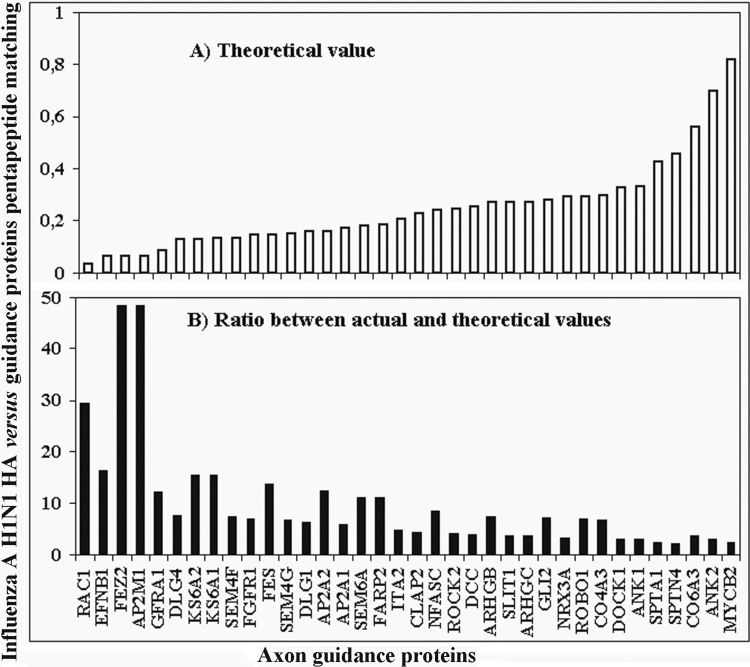
In addition, it is of interest that HA pentapeptide matching varies among the 36 guidance molecules listed in table 2 independently of the protein length (figure 1). In fact, short polypeptides such as FEZ2 and AP2M1 (353 and 435 aa, respectively; see table 1) have up to 3 HA pentapeptide occurrences (table 2). That is, the actual values are respectively higher 48 and 39 times than the theoretical ones. The theoretical number of HA pentapeptide matches for each of the 36 human axon guidance proteins, which are listed in table 2 and range from 192 aa (RAC1) to 4640 aa (MYCB2), is reported in figure 1A. The ratio between theoretical and actual HA pentapeptide matching is shown in figure 1B. On the whole, figure 1 shows that, mostly, the actual number of HA pentapeptide matches in the 36 human axon guidance proteins is independent of the protein length.
Fig. 1.
Actual-vs-theoretical pentapeptide matching between HA and human axon guidance proteins. Axon guidance proteins are listed along the x-axis according to increasing aa length. Actual matching values derived from table 2.
HA-vs-Axon Guidance Peptide Overlap Is Highly Conserved Among Influenza A H1N1 Viral Strains
Next, we investigated whether the pentapeptide commonality with human guidance molecules was a property unique to the HA sequence under study (Q67010, from isolate A/H1N1/swine/Cambridge/1939). To this end, a multialignment sequence analysis was carried out on HA aa sequences from the following influenza A HIN1 strains: Brevig Mission/1/1918 (Q9WFX3); Swine/New Jersey/11/1976 (P03455); strain A/Duck/Australia/749/1980 (Q9WCE3); Camel/Mongolia/1982 (P26142); China:Nanchang/11/1996 (Q07FI5); New Zealand:South Canterbury/35/2000 (Q289M7); Russia:St.Petersburg/8/2006 (B4URD6); and USA:Texas/UR06-0195/2007 (A8C8J4). The multialignment analysis is reported in figure 2. It can be seen that the HA-vs-axon guidance peptide overlap described in table 2 is highly conserved among influenza A H1N1 strains, even at the heptapeptide level (see the EELREQL heptapeptide shared with FEZ2). In other words, the HA peptide sequences shared with guidance molecules and derived from an influenza strain that characterized a H1N1 swine infection in 1939 are highly conserved in influenza A H1N1 strains that temporally succeeded from 1918 to 2007, are present in different world geographical areas, and have been responsible for infections in humans and other hosts (birds, pigs, camels).
Fig. 2.
HA peptide modules common to human axon guidance proteins are highly conserved among influenza A virus H1N1 strains. CLUSTAL W multiple sequence alignment of HA sequences from influenza A virus H1N1 strains was obtained using the T-Coffee program.90,91 HA UniProtKB/Swiss-Prot accessions correspond to: Q67010 (swine/Cambridge/1939); Q9WFX (Brevig Mission/1/1918); P03455 (Swine/NewJersey/11/1976); Q9WCE3 (Duck/Australia/749/1980); P26142 (Camel/Mongolia/1982); Q07FI5 (China:Nanchang/11/1996); Q289M7 (New Zealand: South Canterbury/35/2000); B4URD6 (Russia: St.Petersburg/8/2006); A8C8J4 (USA:Texas/UR06-0195/2007); HA sequence under study (Q67010) is given boxed; aa given in 1-letter code; viral peptide modules common to human axon guidance molecules (see table 2) are given in italics; asterisks highlight the heptapeptide common to viral HAs and human FEZ2.
Similar data of conservativeness were found in examining other influenza A subtypes (data not shown).
The Pentapeptide Overlap Between HA and Guidance Proteins Has an Immunologic Potential
As discussed under Introduction, neuropathologic diseases such as autism, schizophrenia, bipolar disorder with psychotic features, brief psychosis, and psychosis could potentially ensue from maternal immune responses to influenza developed during the pregnancy.29,33–46 Aiming at understanding the molecular mechanism(s) linking anti-influenza maternal immune response(s) and fetal brain insult(s), we proceeded trying to determine the immunologic potential of the HA-vs-axonal proteins pentapeptide overlap described in table 2, and figures 1 and 2.
For this purpose, we investigated the potential cross-reactivity among influenza A epitopes and human axonal guidance molecules using IEDB (www.immuneepitope.org), a public database developed to classify epitope data that had been experimentally validated and controlled in a number of research laboratories worldwide.92 Specifically, the IEDB database was searched for B- and T-cell peptide epitopes derived from influenza A H1N1 HA, targeted by human humoral and/or cellular response(s), and sharing pentapeptide(s) with axon guidance molecules (see table 2). The results obtained following the IEDB search are reported in table 3. It can be seen that 24 out of the 45 pentapeptides shared between HA and axon guidance proteins are, sometimes repeatedly, present in 42 influenza A H1N1 HA-derived epitopes as consecutively overlapping and/or adjacent pentapeptide sequences as exemplified in epitope ID 798, ADYEELREQLSSVSS, and epitope ID 80050, TGLRNIPSIQSRG (with peptide fragments shared between HA and guidance proteins given in italics). Data similar to those reported in table 3 were also found in B- and T-cell epitopes derived from other influenza A subtypes (data not shown).
Table 3.
Peptides Shared Between Influenza H1N1 HA and Guidance Proteins Are Present in B- and/or T-Cell Epitopes
| IEDB IDa | Epitope Sequenceb,c | Immune Recognition Context | Influenza A H1N1 Virus Strain |
|---|---|---|---|
| 798 | ADYEELREQLSSVSS | T | New Caledonia/20/1999 |
| 6801 | CPKYVRSAKL | T | Puerto Rico/8/1934 |
| 26959 | ILAIYSTVASSL | T | California/04/2009 |
| 28063 | IPSIQSRGL | T | Puerto Rico/8/1934 |
| 29691 | IYSTVASSLVL | T | Puerto Rico/8/1934 |
| 35415 | LEDSHNGKL | T | Denver/1957 |
| 61460 | SSVSSFERF | T | camel/Mongolia/1982 |
| 79792 | DTIIFEANGNLIA | T | New Caledonia/20/1999 |
| 79809 | ELLVLLENERTLD | T | New Caledonia/20/1999 |
| 79864 | IIFEANGNLIAPW | T | New Caledonia/20/1999 |
| 80034 | SSLPFQNVHPVTI | T | New Caledonia/20/1999 |
| 80050 | TGLRNIPSIQSRG | T | New Caledonia/20/1999 |
| 113324 | DGFLDIWTYNAELLV | T | New Caledonia/20/1999 |
| 113375 | EQLSSVSSFERFE | T | New Caledonia/20/1999 |
| 113595 | KYVRSAKLRMVT | T | New Caledonia/20/1999 |
| 125913 | ELLVLLENERTLDYHDS | T | California/04/2009 |
| 126340 | ITFEATGNLVVPRYAFA | T | California/04/2009 |
| 128403 | DDGFLDIWTYNAELLVL | T | New Caledonia/20/1999 |
| 129178 | KVKSQLKNNAKEIGNG | T | New Caledonia/20/1999 |
| 129221 | LEKNVTVTHSVNLLEDS | T | New Caledonia/20/1999 |
| 129319 | LRMVTGLRNIPSIQSRG | T | New Caledonia/20/1999 |
| 129320 | LRNIPSIQSRGLFGAIA | T | New Caledonia/20/1999 |
| 129890 | SFWMCSNGSLQCRICI | T | New Caledonia/20/1999 |
| 129938 | SLGAISFWMCSNGSLQ | T | New Caledonia/20/1999 |
| 130394 | YQILAIYSTVASSLVLL | T | New Caledonia/20/1999 |
| 144630 | ELLVLLENERTLDYHDSNVK | T | California/04/2009 |
| 144688 | IDYEELREQLSSVSSFERFE | T | California/04/2009 |
| 144696 | IYQILAIYSTVASSLVLVVSLGA | T | California/04/2009 |
| 144732 | NLYEKVRSQLKNNAKEIGNG | T | California/04/2009 |
| 144810 | VLEKNVTVTHSVNLLEDK | T | California/04/2009 |
| 144813 | VTHSVNLLEDKHNGKLCK | T | California/04/2009 |
| 150978 | EKNVTVTHSVNLLED | B | California/04/2009 |
| 150998 | ILGNPECESLSTASS | B | California/04/2009 |
| 150999 | IPSIQSRGLFGAIAG | B | California/04/2009 |
| 151026 | LREQLSSVSSFERFE | B | California/04/2009 |
| 151036 | NLLEDKHNGKLCKLR | B | California/04/2009 |
| 151056 | SQLKNNAKEIGNGCF | B | California/04/2009 |
| 151063 | TFEATGNLVVPRYAF | B | California/04/2009 |
| 151064 | TPKGAINTSLPFQNI | B | California/04/2009 |
| 151075 | YNAELLVLLENERTL | B | California/04/2009 |
| 151076 | YPGDFIDYEELREQL | B | California/04/2009 |
| 152919 | QLSSVSSFERFEIFPKTSSW | T | California/04/2009 |
Note: IEDB was searched for influenza A HA epitopes that had produced humoral or cellular immune responses in the human host. The search generated 537 epitopes; of which, 120 refer to H1N1 strains, and 42 correspond to H1N1 HA-derived sequences containing peptide fragments (in italics) shared between HA and guidance proteins (see table 2).
aFor epitope further details and references, refer to www.immuneepitope.org.92
baa sequences given in 1-letter code.
cPeptide fragments shared between HA and guidance proteins are given in italics.
In short, table 3 suggests that 24 fundamental axon guidance molecules (ARHGB, ARHGC, CLAP2, CO4A3, CO6A3, DCC, EFNB1, FARP2, FES, FEZ2, FGFR, GFRA1, ITA2, KS6A1, KS6A2, NFASC, NRX3A, PSD95, RAC1, ROBO1, ROCK2, SEM4F, SEM6A, and SLIT1; see table 2 for a brief protein description) are potentially hittable by cross-reactions following human immune responses against influenza A H1N1. In this regard, it is pertinent to note that autoantibodies against neurofascin (UniProtKB/Swiss-Prot entry: NFASC, an axon guidance protein that shares a pentapeptide with the HA epitope IEDB ID 79792, see tables 2 and 3) result in axonal injury.136 Moreover, it has to be noted that table 3 describes only linear epitopic sequences. Actually the issue of conformational epitopes might add to the cross-reactivity potential illustrated in tables 2 and 3. As a matter of fact, a search through IEDB epitopes shows, eg, that the discontinuous HA epitope, S138S139P141N142K171G172S173S174Y175P176K177 S179K180S181V183N211E260T262 (IEDB ID: 1779502) from the A/California/04/2009 H1N1 virus,137 hosts the pentapeptide KGSSY that is present in the axon guidance molecule ROBO1 (aa pos 1525–1529).
Discussion
This study demonstrates that anti-HA humoral and/or cellular immune responses that follow influenza A H1N1 infection in humans have the potential to cross-react with human axon guidance molecules. In the context of our random guidance molecule selection, such a potential immunologic cross-reactivity involves 24 crucial guidance proteins (ARHGB, ARHGC, CLAP2, CO4A3, CO6A3, DCC, EFNB1, FARP2, FES, FEZ2, FGFR, GFRA1, ITA2, KS6A1, KS6A2, NFASC, NRX3A, PSD95, RAC1, ROBO1, ROCK2, SEM4F, SEM6A, and SLIT1; see table 2 for a brief protein description). Qualitatively, the immunogenic potential of influenza A H1N1 HA epitopes is often multitargeting. The sequence EELREQLSSVSS (pos 116–127, table 2) is exemplar in this regard. It contains an heptapeptide in common with FEZ2 (EELREQL), and shares a pentapeptide with PSD95 (LREQL), FGFR1 (EQLSS), and ROBO1 (SSVSS). That is, an anti-influenza immune response targeting a viral epitope containing the EELREQLSSVSS sequence might cross-react with 4 fundamental axon guidance proteins that have been already implicated in the genesis of schizophrenia, autism, and bipolar disorders.
– FEZ2 (or Fasciculation and elongation protein zeta-2) is an ortholog of the C. elegans unc-76 gene,109 and is involved in axonal outgrowth and fasciculation.110 That is, it is necessary for normal axonal bundling and elongation within axon bundles.
– PSD95 (or Postsynaptic density protein 95; alternative names: SAP90, Synapse-associated protein 90; and DLG4, Disks large homolog 4) is highly expressed in postsynaptic density of neurons in the forebrain and in presynaptic region of inhibitory synapses formed by cerebellar basket cells on axon hillocks of Purkinje cells. PSD95 is required for synaptic plasticity associated with NMDA receptor signaling. Indeed, complex and crucial functions are supported by PSD95 as reified by an extremely high number of molecular interactions (see http://www.uniprot.org/uniprot/P78352 for details and references). Of course, these molecular interactions (and related functions) might be perturbed by an immune attack targeting PSD95. For example, PSD95 associates with TMEM16B, a protein with calcium-dependent chloride channel activity,138–141 involved in deciphering and modulating the dynamic neuronal signaling in neurons important for learning and memory.142 Moreover, overexpression or depletion of PSD95 changes the ratio of excitatory to inhibitory synapses in hippocampal neurons.111,112,143–146 Hence, it is feasible to hypothesize (and address research to verify) that an immune attack against PSD95 might disrupt interaction(s) with TMEM16B and alter the mechanisms of learning and memory, thus generating learning disabilities as pathologic consequences.
– FGFR1 (or Fibroblast growth factor receptor 1) is required for normal mesoderm patterning and correct axial organization during normal development of the gonadotropin-releasing hormone neuronal system;147–151
– ROBO1 (or Roundabout homolog 1) is a receptor for SLIT1 and SLIT2 and acts as molecular guidance cue in cellular migration, including axonal navigation at the ventral midline of the neural tube and projection of axons to different regions during neuronal development.114,115,152,153
In essence, an immune response against influenza A infection may cross-react with FEZ2, PSD95, FGFR1, ROBO1 and other crucial guidance proteins, in this way disrupting fundamental molecular interactions in the developing brain. Later, infection(s) with (or vaccination protocols against) different influenza A H1N1 strains or A subtypes might evoke immune responses cross-reactive with the same epitopic targets already damaged during the fetal life. Indeed, as discussed above (see also figure 2), sequence analyses and epitope-coverage information reveal highly conserved protein regions in influenza strains that can be recognized by the human immune system as possible targets for inducing cross-reactions.154 Moreover, cross-reactivity as a pathogenic mechanism linking maternal infections and neuropsychological disturbances in the future adult appears feasible considering the experimental evidence emerged from the many well-known data according to which, eg, (1) short synthetic peptides may define T-cell influenza epitopes155; (2) an exceptionally broad pattern of immunodominance characterizes the primary HLA-DR1-restricted CD4 T-cell response to influenza virus HA in HLA-DR1 transgenic mice156; and (3) a high level of cross-reactivity exists between an influenza virus HA-specific CD4+ T-cell clone derived from a patient with multiple sclerosis and 14 influenza HA variants, 11 viral, 15 human, and 3 myelin-derived peptides.157 Thus, the present data might also represent a clue to investigate the origin and nature of brain reactive autoantibodies that have been repeatedly detected and hypothesized to play a role in neuropsychiatric disorders.158–163 In this framework, our findings indicate that influenza infections and, obviously, anti-influenza vaccination have the potential to cause neuropathologies because of the extensive influenza-vs-human peptide commonality.164 This study might help disentangle the intricated and mysterious context of the risk of autistic and schizophrenic disorders following maternal infections, thus also addressing the issue of influenza vaccination during pregnancy. Indeed, using sequence uniqueness as a criterion for constructing anti-influenza vaccines would eliminate potential autoimmune cross-reactions.48,55,164 More in general, this study represents a methodological approach to dissect the links between microbial infections and neuropathologies at the molecular level.
Funding
Funding from Italian Ministry of University (60%) to DK.
References
- 1. van Praag HM. The possible significance of cerebral dopamine for neurology and psychiatry. Psychiatr Neurol Neurochir. 1967; 70: 361–379 [PubMed] [Google Scholar]
- 2. Seeman P, Ohara K, Ulpian C, et al. Schizophrenia: normal sequence in the dopamine D2 receptor region that couples to G-proteins. DNA polymorphisms in D2. Neuropsychopharmacology. 1993; 8: 137–142 [DOI] [PubMed] [Google Scholar]
- 3. Mukai J, Liu H, Burt RA, et al. Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nat Genet. 2004; 36: 725–731 [DOI] [PubMed] [Google Scholar]
- 4. Charych EI, Liu F, Moss SJ, Brandon NJ. GABA(A) receptors and their associated proteins: implications in the etiology and treatment of schizophrenia and related disorders. Neuropharmacology. 2009; 57: 481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leliveld SR, Bader V, Hendriks P, et al. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J Neurosci. 2008; 28: 3839–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eastwood SL, Cotter D, Harrison PJ. Cerebellar synaptic protein expression in schizophrenia. Neuroscience. 2001; 105: 219–229 [DOI] [PubMed] [Google Scholar]
- 7. Eastwood SL, Harrison PJ. Synaptic pathology in the anterior cingulate cortex in schizophrenia and mood disorders. A review and a Western blot study of synaptophysin, GAP-43 and the complexins. Brain Res Bull. 2001; 55: 569–578 [DOI] [PubMed] [Google Scholar]
- 8. Arguello PA, Gogos JA. A signaling pathway AKTing up in schizophrenia. J Clin Invest. 2008; 118: 2018–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nyegaard M, Demontis D, Thestrup BB, et al. No association of polymorphisms in human endogenous retrovirus K18 and CD48 with schizophrenia. Psychiatr Genet. 2012; 22: 146–148 [DOI] [PubMed] [Google Scholar]
- 10. Sobell JL, Lind TJ, Sigurdson DC, et al. The D5 dopamine receptor gene in schizophrenia: identification of a nonsense change and multiple missense changes but lack of association with disease. Hum Mol Genet. 1995; 4: 507–514 [DOI] [PubMed] [Google Scholar]
- 11. Seeman P, Ulpian C, Chouinard G, et al. Dopamine D4 receptor variant, D4GLYCINE194, in Africans, but not in Caucasians: no association with schizophrenia. Am J Med Genet. 1994; 54: 384–390 [DOI] [PubMed] [Google Scholar]
- 12. Nothdurfter C, Giegling I, Konte B, et al. Lack of association of the 5-HT(3A) receptor with schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2012; 159B: 310–315 [DOI] [PubMed] [Google Scholar]
- 13. Glaser B, Schumacher J, Williams HJ, et al. No association between the putative functional ZDHHC8 single nucleotide polymorphism rs175174 and schizophrenia in large European samples. Biol Psychiatry. 2005; 58: 78–80 [DOI] [PubMed] [Google Scholar]
- 14. Saito S, Ikeda M, Iwata N, et al. No association was found between a functional SNP in ZDHHC8 and schizophrenia in a Japanese case-control population. Neurosci Lett. 2005; 374: 21–24 [DOI] [PubMed] [Google Scholar]
- 15. Cabranes JA, Ancín I, Santos JL, et al. No effect of polymorphisms in the non-duplicated region of CHRNA7 gene on sensory gating P50 ratios in patients with schizophrenia and bipolar disorder. Psychiatry Res. 2012. 10.1016/j.psychres.2012.08.015 [DOI] [PubMed] [Google Scholar]
- 16. Zhang F, Liu C, Chen Y, et al. No association of catechol-O-methyltransferase polymorphisms with schizophrenia in the Han Chinese population. Genet Test Mol Biomarkers. 2012; 16: 1138–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller G. Neurological disorders. The mystery of the missing smile. Science. 2007; 316: 826–827 [DOI] [PubMed] [Google Scholar]
- 18. Mitka M. Rising autism rates still pose a mystery. JAMA. 2010; 303: 602 [DOI] [PubMed] [Google Scholar]
- 19. Gurrieri F. Working up autism: the practical role of medical genetics. Am J Med Genet C Semin Med Genet. 2012; 160C: 104–110 [DOI] [PubMed] [Google Scholar]
- 20. Klei L, Sanders SJ, Murtha MT, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012; 3: 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hedrick A, Lee Y, Wallace GL, et al. Autism risk gene MET variation and cortical thickness in typically developing children and adolescents. Autism Res. 2012. 10.1002/aur.1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kong A, Frigge ML, Masson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012; 488: 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abramowicz JS. Ultrasound and autism: association, link, or coincidence? J Ultrasound Med. 2012; 31: 1261–1269 [DOI] [PubMed] [Google Scholar]
- 24. Dietert RR, Dietert JM, Dewitt JC. Environmental risk factors for autism. Emerg Health Threats J. 2011; 4. 10.3402/ehtj.v4i0.7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kinney DK, Munir KM, Crowley DJ, Miller AM. Prenatal stress and risk for autism. Neurosci Biobehav Rev. 2008; 32: 1519–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ivarsson SA, Bjerre I, Vegfors P, Ahlfors K. Autism as one of several disabilities in two children with congenital cytomegalovirus infection. Neuropediatrics. 1990; 21: 102–103 [DOI] [PubMed] [Google Scholar]
- 27. De Miranda J, Yaddanapudi K, Hornig M, Villar G, Serge R, Lipkin WI. Induction of Toll-like receptor 3-mediated immunity during gestation inhibits cortical neurogenesis and causes behavioral disturbances. MBio. 2010; 1:e00176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meyer U, Feldon J, Dammann O. Schizophrenia and autism: both shared and disorder-specific pathogenesis via perinatal inflammation? Pediatr Res. 2011; 69: 26R–33R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Patterson PH. Maternal infection and immune involvement in autism. Trends Mol Med. 2011; 17: 389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bunney BG, Potkin SG, Bunney WE., Jr New morphological and neuropathological findings in schizophrenia: a neurodevelopmental perspective. Clin Neurosci. 1995; 3: 81–88 [PubMed] [Google Scholar]
- 31. Arnold SE. Neurodevelopmental abnormalities in schizophrenia: insights from neuropathology. Dev Psychopathol. 1999; 11: 439–456 [DOI] [PubMed] [Google Scholar]
- 32. Bombin I, Mayoral M, Castro-Fornieles J, et al. Neuro psychological evidence for abnormal neurodevelopment associated with early-onset psychoses. Psychol Med. 2012; 25: 1–12 [DOI] [PubMed] [Google Scholar]
- 33. Brown AS, Derkits EJ. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. Am J Psychiatry. 2010; 167: 261–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tochigi M, Okazaki Y, Kato N, Sasaki T. What causes seasonality of birth in schizophrenia? Neurosci Res. 2004; 48: 1–11 [DOI] [PubMed] [Google Scholar]
- 35. Yolken RH, Torrey EF. Viruses, schizophrenia, and bipolar disorder. Clin Microbiol Rev. 1995; 8: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fatemi SH, Earle J, Kanodia R, et al. Prenatal viral infection leads to pyramidal cell atrophy and macrocephaly in adulthood: implications for genesis of autism and schizophrenia. Cell Mol Neurobiol. 2002; 22: 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fatemi SH, Pearce DA, Brooks AI, Sidwell RW. Prenatal viral infection in mouse causes differential expression of genes in brains of mouse progeny: a potential animal model for schizophrenia and autism. Synapse. 2005; 57: 91–99 [DOI] [PubMed] [Google Scholar]
- 38. Fatemi SH, Reutiman TJ, Folsom TD, et al. Maternal infection leads to abnormal gene regulation and brain atrophy in mouse offspring: implications for genesis of neurodevelopmental disorders. Schizophr Res. 2008; 99: 56–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown AS, Begg MD, Gravenstein S, et al. Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch Gen Psychiatry. 2004; 61: 774–780 [DOI] [PubMed] [Google Scholar]
- 40. Smith SE, Li J, Garbett K, Mirnics K, Patterson PH. Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci. 2007; 27: 10695–10702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fatemi SH, Folsom TD, Rooney RJ, et al. The viral theory of schizophrenia revisited: abnormal placental gene expression and structural changes with lack of evidence for H1N1 viral presence in placentae of infected mice or brains of exposed offspring. Neuropharmacology. 2012;62:1290–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Short SJ, Lubach GR, Karasin AI, et al. Maternal influenza infection during pregnancy impacts postnatal brain development in the rhesus monkey. Biol Psychiatry. 2010; 67: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brown AS. The environment and susceptibility to schizophrenia. Prog Neurobiol. 2011; 93: 23–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meyer U, Feldon J, Fatemi SH. In-vivo rodent models for the experimental investigation of prenatal immune activation effects in neurodevelopmental brain disorders. Neurosci Biobehav Rev. 2009; 33: 1061–1079 [DOI] [PubMed] [Google Scholar]
- 45. Shi L, Fatemi SH, Sidwell RW, Patterson PH. Maternal influenza infection causes marked behavioral and pharmacological changes in the offspring. J Neurosci. 2003; 23: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fatemi SH, Folsom TD, Reutiman TJ, et al. Abnormal expression of myelination genes and alterations in white matter fractional anisotropy following prenatal viral influenza infection at E16 in mice. Schizophr Res. 2009;112:46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kanduc D, Stufano A, Lucchese G, Kusalik A. Massive peptide sharing between viral and human proteomes. Peptides. 2008; 29: 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kanduc D. Describing the hexapeptide identity platform between the influenza A H5N1 and Homo sapiens proteomes. Biologics. 2010; 4: 245–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lucchese G. Confronting JC virus and Homo sapiens biological signatures. Front Biosci. 2013; 18: 716–721 [DOI] [PubMed] [Google Scholar]
- 50. Lucchese G. A peptide talk between JC virus and the human host: from silent infection to autoimmunity. Immunopharmacol Immunotoxicol. 2012; 34: 1067–1074 [DOI] [PubMed] [Google Scholar]
- 51. Capone G, Novello G, Bavaro SL, et al. A qualitative description of the peptide sharing between poliovirus and Homo sapiens. Immunopharmacol Immunotoxicol. 2012; 34: 779–785 [DOI] [PubMed] [Google Scholar]
- 52. Novello G, Capone G, Fasano C, Bavaro SL, Polito AN, Kanduc D. A quantitative description of the peptide sharing between poliovirus and Homo sapiens. Immunopharmacol Immunotoxicol. 2012; 34: 373–378 [DOI] [PubMed] [Google Scholar]
- 53. Capone G, Pagoni M, Delfino AP, Kanduc D. Evidence for a vast peptide overlap between West Nile virus and human proteomes. J Basic Microbiol. 2012. 10.1002/jobm.201200204 [DOI] [PubMed] [Google Scholar]
- 54. Lucchese G, Stufano A, Kanduc D. Searching for an effective, safe and universal anti-HIV vaccine: Finding the answer in just one short peptide. Self Nonself. 2011; 2: 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lucchese G, Stufano A, Kanduc D. Proteome-guided search for influenza A B-cell epitopes. FEMS Immunol Med Microbiol. 2009; 57: 88–92 [DOI] [PubMed] [Google Scholar]
- 56. Kanduc D. Epitopic peptides with low similarity to the host proteome: towards biological therapies without side effects. Expert Opin Biol Ther. 2009; 9: 45–53 [DOI] [PubMed] [Google Scholar]
- 57. Stufano A, Capone G, Pesetti B, Polimeno L, Kanduc D. Clustering of rare peptide segments in the HCV immunome. Self Nonself. 2010; 1: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Capone G, Lucchese G, Calabrò M, Kanduc D. West Nile virus diagnosis and vaccination: using unique viral peptide sequences to evoke specific immune responses. Immunopharmacol Immunotoxicol. 2013; 35: 64–70 [DOI] [PubMed] [Google Scholar]
- 59. Wei CJ, Yassine HM, McTamney PM, et al. Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Sci Transl Med. 2012; 4: 147ra114 [DOI] [PubMed] [Google Scholar]
- 60. Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996; 274: 1123–1133 [DOI] [PubMed] [Google Scholar]
- 61. Lin L, Lesnick TG, Maraganore DM, Isacson O. Axon guidance and synaptic maintenance: preclinical markers for neurodegenerative disease and therapeutics. Trends Neurosci. 2009; 32: 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McAlonan GM, Li Q, Cheung C. The timing and specificity of prenatal immune risk factors for autism modeled in the mouse and relevance to schizophrenia. Neurosignals. 2010; 18: 129–139 [DOI] [PubMed] [Google Scholar]
- 63. Andersen SL. Trajectories of brain development: point of vulnerability or window of opportunity? Neurosci Biobehav Rev. 2003; 27: 3–18 [DOI] [PubMed] [Google Scholar]
- 64. Lucchese G, Stufano A, Trost B, Kusalik A, Kanduc D. Peptidology: short amino acid modules in cell biology and immunology. Amino Acids. 2007; 33: 703–707 [DOI] [PubMed] [Google Scholar]
- 65. Kanduc D. Protein information content resides in rare peptide segments. Peptides. 2010; 31: 983–988 [DOI] [PubMed] [Google Scholar]
- 66. Kanduc D. Homology, similarity, and identity in peptide epitope immunodefinition. J Pept Sci. 2012; 18: 487–494 [DOI] [PubMed] [Google Scholar]
- 67. Lucchese G, Sinha AA, Kanduc D. How a single amino acid change may alter the immunological information of a peptide. Front Biosci. 2012; 4: 1843–1852 [DOI] [PubMed] [Google Scholar]
- 68. Lucchese G, Calabro M, Kanduc D. Circumscribing the conformational peptide epitope landscape. Curr Pharm Des. 2012; 18: 832–839 [DOI] [PubMed] [Google Scholar]
- 69. Willers J, Capone G, Lucchese A. Peptides: an arrival point in cancer vaccinology. Front Biosci. 2012; 4: 1381–1392 [DOI] [PubMed] [Google Scholar]
- 70. Rothbard JB. Peptides and the cellular immune response. Ann Inst Pasteur. 1986; 137: 518–526 [Google Scholar]
- 71. Rothbard JB, Taylor WR. A sequence pattern common to T cell epitopes. EMBO J. 1988; 7: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rothbard JB, Pemberton RM, Bodmer HC, Askonas BA, Taylor WR. Identification of residues necessary for clonally specific recognition of a cytotoxic T cell determinant. EMBO J. 1989; 8: 2321–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mathews JH, Allan JE, Roehrig JT, Brubaker JR, Uren MF, Hunt AR. T-helper cell and associated antibody response to synthetic peptides of the E glycoprotein of Murray Valley encephalitis virus. J Virol. 1991; 65: 5141–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rothbard JB, Gefter ML. Interactions between immunogenic peptides and MHC proteins. Annu Rev Immunol. 1991; 9: 527–565 [DOI] [PubMed] [Google Scholar]
- 75. Sant’Angelo DB, Robinson E, Janeway CA, Jr, Denzin LK. Recognition of core and flanking amino acids of MHC class II-bound peptides by the T cell receptor. Eur J Immunol. 2002; 32: 2510–2520 [DOI] [PubMed] [Google Scholar]
- 76. Reddehase MJ, Rothbard JB, Koszinowski UH. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature. 1989; 337: 651–653 [DOI] [PubMed] [Google Scholar]
- 77. Geluk A, Van Meijgaarden KE, Janson AA, et al. Functional analysis of DR17(DR3)-restricted mycobacterial T cell epitopes reveals DR17-binding motif and enables the design of allele-specific competitor peptides. J Immunol. 1992; 149: 2864–2871 [PubMed] [Google Scholar]
- 78. Hemmer B, Kondo T, Gran B, et al. Minimal peptide length requirements for CD4(+) T cell clones–implications for molecular mimicry and T cell survival. Int Immunol. 2000; 12: 375–383 [DOI] [PubMed] [Google Scholar]
- 79. Liebers V, Raulf M, Mazur G, Modrow S, Baur X. Epitope mapping with peptides of Chi t I component III and immunomodulation of the Chi t immune response. J Allergy Clin Immunol. 1993; 92: 334–339 [DOI] [PubMed] [Google Scholar]
- 80. Hashiguchi S, Hino K, Taniguchi Y, et al. Immunodominance of seven regions of a major allergen, Cry j 2, of Japanese cedar pollen for T-cell immunity. Allergy. 1996; 51: 621–632 [DOI] [PubMed] [Google Scholar]
- 81. Moran E, Simmons C, Vinh Chau N, et al. Preservation of a critical epitope core region is associated with the high degree of flaviviral cross-reactivity exhibited by a dengue-specific CD4+ T cell clone. Eur J Immunol. 2008; 38: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Godkin AJ, Thomas HC, Openshaw PJ. Evolution of epitope-specific memory CD4(+) T cells after clearance of hepatitis C virus. J Immunol. 2002; 169: 2210–2214 [DOI] [PubMed] [Google Scholar]
- 83. Miyazaki I, Cheung RK, Gaedigk R, et al. T cell activation and anergy to islet cell antigen in type I diabetes. J Immunol. 1995; 154: 1461–1469 [PubMed] [Google Scholar]
- 84. Tindle RW, Croft S, Herd K, et al. A vaccine conjugate of ‘ISCAR’ immunocarrier and peptide epitopes of the E7 cervical cancer-associated protein of human papillomavirus type 16 elicits specific Th1- and Th2-type responses in immunized mice in the absence of oil-based adjuvants. Clin Exp Immunol. 1995; 101: 265–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tiwari R, Geliebter J, Lucchese A, Mittelman A, Kanduc D. Computational peptide dissection of Melan-a/MART-1 oncoprotein antigenicity. Peptides. 2004; 25: 1865–1871 [DOI] [PubMed] [Google Scholar]
- 86. Lucchese G, Stufano A, Kanduc D. Proposing low-similarity peptide vaccines against Mycobacterium tuberculosis. J Biomed Biotechnol. 2010; 2010: 832341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. UniProt Consortium The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009; 37: D169–D174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Novello G, Capone G, Kanduc D. Reviewing the role of peptide rarity in bacterial toxin immunomics. Front Biosci. 2012; 4: 216–225 [DOI] [PubMed] [Google Scholar]
- 89. Lucchese G, Stufano A, Calabro M, Kanduc D. Charting the peptide crossreactome between HIV-1 and the human proteome. Front Biosci. 2011; 3: 1385–1400 [DOI] [PubMed] [Google Scholar]
- 90. Di Tommaso P, Moretti S, Xenarios I, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011; 39: W13–W17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000; 302: 205–217 [DOI] [PubMed] [Google Scholar]
- 92. Peters B, Sidney J, Bourne P, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005; 3: e91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sham PC, O’Callaghan E, Takei N, Murray GK, Hare EH, Murray RM. Schizophrenia following pre-natal exposure to influenza epidemics between 1939 and 1960. Br J Psychiatry. 1992; 160: 461–466 [DOI] [PubMed] [Google Scholar]
- 94. Maier V, Jolicoeur C, Rayburn H, et al. Semaphorin 4C and 4G are ligands of Plexin-B2 required in cerebellar development. Mol Cell Neurosci. 2011; 46: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Davidkova G, McCullumsmith RE, Meador-Woodruff JH. Expression of ARHGEF11 mRNA in schizophrenic thalamus. Ann N Y Acad Sci. 2003; 1003: 375–377 [DOI] [PubMed] [Google Scholar]
- 96. Mencarelli C, Hammels C, Van Den Broeck J, et al. The expression of the Goodpasture antigen-binding protein (ceramide transporter) in adult rat brain. J Chem Neuroanat. 2009; 38: 97–105 [DOI] [PubMed] [Google Scholar]
- 97. Hamdan FF, Saitsu H, Nishiyama K, et al. Identification of a novel in-frame de novo mutation in SPTAN1 in intellectual disability and pontocerebellar atrophy. Eur J Hum Genet. 2012; 20: 796–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Berghs S, Aggujaro D, Dirkx R, Jr, et al. betaIV spectrin, a new spectrin localized at axon initial segments and nodes of ranvier in the central and peripheral nervous system. J Cell Biol. 2000; 151: 985–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kamiguchi H, Long KE, Pendergast M, et al. The neural cell adhesion molecule L1 interacts with the AP-2 adaptor and is endocytosed via the clathrin-mediated pathway. J Neurosci. 1998; 18: 5311–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cheng IH, Lin YC, Hwang E, et al. Collagen VI protects against neuronal apoptosis elicited by ultraviolet irradiation via an Akt/phosphatidylinositol 3-kinase signaling pathway. Neuroscience. 2011; 183: 178–188 [DOI] [PubMed] [Google Scholar]
- 101. Ledda F. Ligand-induced cell adhesion as a new mechanism to promote synapse formation. Cell Adh Migr. 2007; 1: 137–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Rünker AE, Little GE, Suto F, Fujisawa H, Mitchell KJ. Semaphorin-6A controls guidance of corticospinal tract axons at multiple choice points. Neural Dev. 2008; 3: 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ballif BA, Roux PP, Gerber SA, MacKeigan JP, Blenis J, Gygi SP. Quantitative phosphorylation profiling of the ERK/p90 ribosomal S6 kinase-signaling cassette and its targets, the tuberous sclerosis tumor suppressors. Proc Natl Acad Sci USA. 2005; 102: 667–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Park SA, Kim TS, Choi KS, Park HJ, Heo K, Lee BI. Chronic activation of CREB and p90RSK in human epileptic hippocampus. Exp Mol Med. 2003; 35: 365–370 [DOI] [PubMed] [Google Scholar]
- 105. Sananbenesi F, Fischer A, Schrick C, Spiess J, Radulovic J. Mitogen-activated protein kinase signaling in the hippocampus and its modulation by corticotropin-releasing factor receptor 2: a possible link between stress and fear memory. J Neurosci. 2003; 23: 11436–11443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ahi J, Radulovic J, Spiess J. The role of hippocampal signaling cascades in consolidation of fear memory. Behav Brain Res. 2004; 149: 17–31 [DOI] [PubMed] [Google Scholar]
- 107. Farmer WT, Altick AL, Nural HF, et al. Pioneer longitudinal axons navigate using floor plate and Slit/Robo signals. Development. 2008; 135: 3643–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mimori-Kiyosue Y, Grigoriev I, Lansbergen G, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005; 168: 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bloom L, Horvitz HR. The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc Natl Acad Sci USA. 1997; 94: 3414–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Alborghetti MR, Furlan AS, Kobarg J. FEZ2 has acquired additional protein interaction partners relative to FEZ1: functional and evolutionary implications. PLoS ONE. 2011; 6: e17426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cheng MC, Lu CL, Luu SU, et al. Genetic and functional analysis of the DLG4 gene encoding the post-synaptic density protein 95 in schizophrenia. PLoS ONE. 2010; 5: e15107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kristiansen LV, Beneyto M, Haroutunian V, Meador-Woodruff JH. Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia. Mol Psychiatry. 2006; 11: 737–47, 705 [DOI] [PubMed] [Google Scholar]
- 113. Chung WC, Tsai PS. Role of fibroblast growth factor signaling in gonadotropin-releasing hormone neuronal system development. Front Horm Res. 2010; 39: 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Brose K, Bland KS, Wang KH, et al. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999; 96: 795–806 [DOI] [PubMed] [Google Scholar]
- 115. Kim M, Roesener AP, Mendonca PR, Mastick GS. Robo1 and Robo2 have distinct roles in pioneer longitudinal axon guidance. Dev Biol. 2011; 358: 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Li X, Gao X, Liu G, Xiong W, Wu J, Rao Y. Netrin signal transduction and the guanine nucleotide exchange factor DOCK180 in attractive signaling. Nat Neurosci. 2008; 11: 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Po MD, Hwang C, Zhen M. PHRs: bridging axon guidance, outgrowth and synapse development. Curr Opin Neurobiol. 2010; 20: 100–107 [DOI] [PubMed] [Google Scholar]
- 118. Toyofuku T, Yoshida J, Sugimoto T, et al. FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nat Neurosci. 2005; 8: 1712–1719 [DOI] [PubMed] [Google Scholar]
- 119. Gallagher PG, Forget BG. An alternate promoter directs expression of a truncated, muscle-specific isoform of the human ankyrin 1 gene. J Biol Chem. 1998; 273: 1339–1348 [DOI] [PubMed] [Google Scholar]
- 120. Engelhardt M, Vorwald S, Sobotzik JM, Bennett V, Schultz C. Ankyrin-B structurally defines terminal microdomains of peripheral somatosensory axons. Brain Struct Funct. 2012. 10.1007/s00429-012-0443-0 [DOI] [PubMed] [Google Scholar]
- 121. Kriebel M, Wuchter J, Trinks S, Volkmer H. Neurofascin: a switch between neuronal plasticity and stability. Int J Biochem Cell Biol. 2012; 44: 694–697 [DOI] [PubMed] [Google Scholar]
- 122. Lau AW, Chou MM. The adaptor complex AP-2 regulates post-endocytic trafficking through the non-clathrin Arf6-dependent endocytic pathway. J Cell Sci. 2008; 121: 4008–4017 [DOI] [PubMed] [Google Scholar]
- 123. Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998; 7: 1463–1474 [DOI] [PubMed] [Google Scholar]
- 124. Cotter L, Ozçelik M, Jacob C, et al. Dlg1-PTEN interaction regulates myelin thickness to prevent damaging peripheral nerve overmyelination. Science. 2010; 328: 1415–1418 [DOI] [PubMed] [Google Scholar]
- 125. Laurent CE, Smithgall TE. The c-Fes tyrosine kinase cooperates with the breakpoint cluster region protein (Bcr) to induce neurite extension in a Rac- and Cdc42-dependent manner. Exp Cell Res. 2004; 299: 188–198 [DOI] [PubMed] [Google Scholar]
- 126. Wong K, Park HT, Wu JY, Rao Y. Slit proteins: molecular guidance cues for cells ranging from neurons to leukocytes. Curr Opin Genet Dev. 2002; 12: 583–591 [DOI] [PubMed] [Google Scholar]
- 127. Tanaka H, Miyazaki N, Matoba K, Nogi T, Iwasaki K, Takagi J. Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin. Cell Rep. 2012; 2: 101–110 [DOI] [PubMed] [Google Scholar]
- 128. Zhou Z, Meng Y, Asrar S, Todorovski Z, Jia Z. A critical role of Rho-kinase ROCK2 in the regulation of spine and synaptic function. Neuropharmacology. 2009; 56: 81–89 [DOI] [PubMed] [Google Scholar]
- 129. Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003; 4: 446–456 [DOI] [PubMed] [Google Scholar]
- 130. Myers JP, Santiago-Medina M, Gomez TM. Regulation of axonal outgrowth and pathfinding by integrin-ECM interactions. Dev Neurobiol. 2011; 71: 901–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Corbetta S, Gualdoni S, Ciceri G, et al. Essential role of Rac1 and Rac3 GTPases in neuronal development. FASEB J. 2009; 23: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Armendáriz BG, Bribian A, Pérez-Martínez E, et al. Expression of Semaphorin 4F in neurons and brain oligodendrocytes and the regulation of oligodendrocyte precursor migration in the optic nerve. Mol Cell Neurosci. 2012; 49: 54–67 [DOI] [PubMed] [Google Scholar]
- 133. Hata K, Kaibuchi K, Inagaki S, Yamashita T. Unc5B associates with LARG to mediate the action of repulsive guidance molecule. J Cell Biol. 2009; 184: 737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Chen Q, Sun X, Zhou XH, et al. N-terminal horseshoe conformation of DCC is functionally required for axon guidance and may be shared by other neural receptors. J Cell Sci. 2012. 10.1242/jcs.111278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Bush JO, Soriano P. Ephrin-B1 regulates axon guidance by reverse signaling through a PDZ-dependent mechanism. Genes Dev. 2009; 23: 1586–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Mathey EK, Derfuss T, Storch MK, et al. Neurofascin as a novel target for autoantibody-mediated axonal injury. J Exp Med. 2007; 204: 2363–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010; 328: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Stephan AB, Shum EY, Hirsh S, Cygnar KD, Reisert J, Zhao H. ANO2 is the cilial calcium-activated chloride channel that may mediate olfactory amplification. Proc Natl Acad Sci USA. 2009; 106: 11776–11781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Pifferi S, Cenedese V, Menini A. Anoctamin 2/TMEM16B: a calcium-activated chloride channel in olfactory transduction. Exp Physiol. 2012;97:193–199 [DOI] [PubMed] [Google Scholar]
- 140. Billig GM, Pál B, Fidzinski P, Jentsch TJ. Ca2+-activated Cl− currents are dispensable for olfaction. Nat Neurosci. 2011; 14: 763–769 [DOI] [PubMed] [Google Scholar]
- 141. Stöhr H, Heisig JB, Benz PM, et al. TMEM16B, a novel protein with calcium-dependent chloride channel activity, associates with a presynaptic protein complex in photoreceptor terminals. J Neurosci. 2009; 29: 6809–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Huang WC, Xiao S, Huang F, Harfe BD, Jan YN, Jan LY. Calcium-activated chloride channels (CaCCs) regulate action potential and synaptic response in hippocampal neurons. Neuron. 2012; 74: 179–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Piserchio A, Pellegrini M, Mehta S, et al. The PDZ1 domain of SAP90. Characterization of structure and binding. J Biol Chem. 2002; 277: 6967–6973 [DOI] [PubMed] [Google Scholar]
- 144. Kristiansen LV, Patel SA, Haroutunian V, Meador-Woodruff JH. Expression of the NR2B-NMDA receptor subunit and its Tbr-1/CINAP regulatory proteins in postmortem brain suggest altered receptor processing in schizophrenia. Synapse. 2010; 64: 495–502 [DOI] [PubMed] [Google Scholar]
- 145. Funk AJ, Rumbaugh G, Harotunian V, McCullumsmith RE, Meador-Woodruff JH. Decreased expression of NMDA receptor-associated proteins in frontal cortex of elderly patients with schizophrenia. Neuroreport. 2009; 20: 1019–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Beneyto M, Meador-Woodruff JH. Lamina-specific abnormalities of NMDA receptor-associated postsynaptic protein transcripts in the prefrontal cortex in schizophrenia and bipolar disorder. Neuropsychopharmacology. 2008; 33: 2175–2186 [DOI] [PubMed] [Google Scholar]
- 147. Tabarés-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol Psychiatry. 2009; 14: 563–589 [DOI] [PubMed] [Google Scholar]
- 148. Carter CJ. Schizophrenia susceptibility genes directly implicated in the life cycles of pathogens: cytomegalovirus, influenza, herpes simplex, rubella, and Toxoplasma gondii. Schizophr Bull. 2009; 35: 1163–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Tochigi M, Iwamoto K, Bundo M, Sasaki T, Kato N, Kato T. Gene expression profiling of major depression and suicide in the prefrontal cortex of postmortem brains. Neurosci Res. 2008; 60: 184–191 [DOI] [PubMed] [Google Scholar]
- 150. Gaughran F, Payne J, Sedgwick PM, Cotter D, Berry M. Hippocampal FGF-2 and FGFR1 mRNA expression in major depression, schizophrenia and bipolar disorder. Brain Res Bull. 2006; 70: 221–227 [DOI] [PubMed] [Google Scholar]
- 151. Klejbor I, Myers JM, Hausknecht K, et al. Fibroblast growth factor receptor signaling affects development and function of dopamine neurons - inhibition results in a schizophrenia-like syndrome in transgenic mice. J Neurochem. 2006; 97: 1243–1258 [DOI] [PubMed] [Google Scholar]
- 152. Anitha A, Nakamura K, Yamada K, et al. Genetic analyses of roundabout (ROBO) axon guidance receptors in autism. Am J Med Genet B Neuropsychiatr Genet. 2008; 147B: 1019–1027 [DOI] [PubMed] [Google Scholar]
- 153. Potkin SG, Turner JA, Guffanti G, et al. FBIRN A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr Bull. 2009; 35: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Squires RB, Noronha J, Hunt V, et al. Influenza research database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respi Viruses. 2012; 6: 404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Townsend AR, Rothbard J, Gotch FM, Bahadur G, Wraith D, McMichael AJ. The epitopes of influenza nucleoprotein recognized by cytotoxic T lymphocytes can be defined with short synthetic peptides. Cell. 1986; 44: 959–968 [DOI] [PubMed] [Google Scholar]
- 156. Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol. 2007; 81: 7608–7619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Markovic-Plese S, Hemmer B, Zhao Y, Simon R, Pinilla C, Martin R. High level of cross-reactivity in influenza virus hemagglutinin-specific CD4+ T-cell response: implications for the initiation of autoimmune response in multiple sclerosis. J Neuroimmunol. 2005; 169: 31–38 [DOI] [PubMed] [Google Scholar]
- 158. Braunschweig D, Ashwood P, Krakowiak P, et al. Autism: maternally derived antibodies specific for fetal brain proteins. Neurotoxicology. 2008; 29: 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Goines P, Haapanen L, Boyce R, et al. Autoantibodies to cerebellum in children with autism associate with behavior. Brain Behav Immun. 2011; 25: 514–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Williams S, Sakic B, Hoffman SA. Circulating brain-reactive autoantibodies and behavioral deficits in the MRL model of CNS lupus. J Neuroimmunol. 2010; 218: 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Ching KH, Burbelo PD, Carlson PJ, Drevets WC, Iadarola MJ. High levels of Anti-GAD65 and Anti-Ro52 autoantibodies in a patient with major depressive disorder showing psychomotor disturbance. J Neuroimmunol. 2010; 222: 87–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Gonzalez-Gronow M, Cuchacovich M, Francos R, et al. Antibodies against the voltage-dependent anion channel (VDAC) and its protective ligand hexokinase-I in children with autism. J Neuroimmunol. 2010; 227: 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Croen LA, Braunschweig D, Haapanen L, et al. Maternal mid-pregnancy autoantibodies to fetal brain protein: the early markers for autism study. Biol Psychiatry. 2008; 64: 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kanduc D. Peptide cross-reactivity: the original sin of vaccines. Front Biosci. 2012; 4: 1393–1401 [DOI] [PubMed] [Google Scholar]