Summary
In the mouse olfactory bulb, information from sensory neurons is extensively processed by local interneurons before being transmitted to the olfactory cortex by mitral and tufted (M/T) cells. The precise function of these local networks remains elusive because of the vast heterogeneity of interneurons, their diverse physiological properties, and their complex synaptic connectivity. Here we identified the parvalbumin interneurons (PVNs) as a prominent component of the M/T presynaptic landscape by using an improved rabies-based trans-synaptic tracing method for local circuits. In vivo two-photon targeted patch recording revealed that PVNs have exceptionally broad olfactory receptive fields, and exhibit largely excitatory and persistent odor responses. Trans-synaptic tracing indicated that PVNs receive direct input from widely distributed M/T cells. Both the anatomical and functional extent of this M/T→PVN→M/T circuit contrasts with the narrowly confined M/T→granule cell→M/T circuit, suggesting that olfactory information is processed by multiple local circuits operating at distinct spatial scales.
Introduction
Most brain areas contain highly heterogeneous neural circuits that are spatially intermingled and that serve different functions. Heterogeneity is manifested in diverse cell types, synaptic connectivity patterns, and physiological response profiles (Monyer and Markram, 2004). Understanding information processing by neural circuits, therefore, depends on our ability to identify and study local circuits individually, yet within the context of their larger network. Recent advances in imaging, physiology, and molecular genetics have improved the resolution with which we can identify and functionally isolate neural circuits and their individual components in the intact brain (Denk et al., 2012; Luo et al., 2008). Here, we combined an improved rabies-based method for trans-synaptic tracing of local circuits with in vivo electrophysiology to investigate the unique functional attributes of a particular type of local interneurons in the olfactory bulb (OB) and its synaptic connectivity.
In the OB, olfactory information is represented by two consecutive layers of excitatory neurons, the olfactory receptor neurons (ORNs) and mitral and tufted (M/T) cells. ORNs send afferent axons to the OB where they converge into discrete glomeruli, forming a spatially distributed “odor map” (Sakano, 2010). At the second layer, olfactory information is represented by populations of M/T cells, the output neurons of the OB. Although individual M/T cells receive direct input from a single class of ORNs, olfactory representations in the M/T populations is not a mere reflection of their cognate ORN input (Murthy, 2011). In fact, M/T cells receive a large fraction of their synapses from local interneurons, most of which are inhibitory (Egger and Urban, 2006), and OB local interneurons outnumber M/T cells by two orders of magnitude (Shepherd, 2004).
The local inhibitory networks in the OB are highly diverse in terms of cellular markers (Batista-Brito et al., 2008), morphology (Kosaka and Kosaka, 2011), and physiology (Tan et al., 2010). For example, granule cells (GCs) form a local inhibitory circuit with mitral cells (MCs) via dendrodendritic interactions (Jahr and Nicoll, 1980; Shepherd, 2004). GCs mediate recurrent inhibition whereby GCs inhibit, in a reciprocal manner, the exact same MC that excited them. Additionally, GCs can mediate lateral inhibition between different MCs through inhibition of nearby MCs (Margrie et al., 2001; Shepherd et al., 2007). Other inhibitory networks involving periglomerular neurons have been suggested to contribute to the decorrelation of M/T ensembles, a process suggested to be central for odor discrimination (Aungst et al., 2003; Cleland, 2010). However, the mechanisms by which these and other local networks in the OB transform olfactory information remain poorly understood, partly due to the heterogeneity of the physiological properties and connectivity patterns of interneurons. For example, it remains unclear which local interneuron types provide direct input to M/T cells, and what are their spatial distributions with respect to that of M/T cells.
The rabies virus (RV)-mediated mono-synaptic tracing strategy (Wickersham et al., 2007) has been combined with mouse genetics to identify presynaptic partners of defined neuronal subtypes in different regions of the brain and spinal cord. This system utilized a mutant RV, which lacks the rabies gene encoding the envelope glycoprotein (RG) required for viral spread. This mutant RV is pseudotyped with the avian EnvA envelope glycoprotein, which restricts the cells the virus can initially transduce to those that express the EnvA receptor TVA. To reconstitute infectious RV particles in vivo in a defined neuronal population of interest (the “starter cell” population), neurons are first transduced with conditional adeno-associated virus (AAV) vectors that express TVA and RG in a manner that is dependent on the expression of the Cre recombinase (Wall et al., 2010; Watabe-Uchida et al., 2012) or the tetracycline transactivator tTA2 (Miyamichi et al., 2011). The starter cells can thus be restricted to particular populations based on cell-type specific Cre or tTA2 expression. Neurons expressing TVA and RG can then be transduced by EnvA-pseudotyped and RG-deleted RV to produce starter cells, from which their presynaptic partners are infected by reconstituted RV. Here, we improved upon previous tracing methods so as to more precisely trace connectivity within local circuits. We combined our improved trans-synaptic tracing with mouse genetics and in vivo physiology to investigate local interneuron circuits in the mouse OB. Our study suggests that reciprocal connections between M/T cells and parvalbumin neurons (PVNs) underlie a broad feedback control of the OB output.
Results
Optimizing Local Circuit Analysis Using Trans-synaptic Tracing
Two technical limitations need to be overcome in order for RV-mediated trans-synaptic labeling to be applied for the analysis of local circuit connectivity with precision. First, to achieve unambiguous interpretation of local connections, starter cells should be unequivocally labeled to distinguish them from their presynaptic partners. Second, non-specific labeling by RV need to be close to zero to avoid false positives in the identification of presynaptic partners. Currently applied methods suffer from one or both of these limitations (see below). Therefore, we first attempted to optimize the RV method for tracing local circuits.
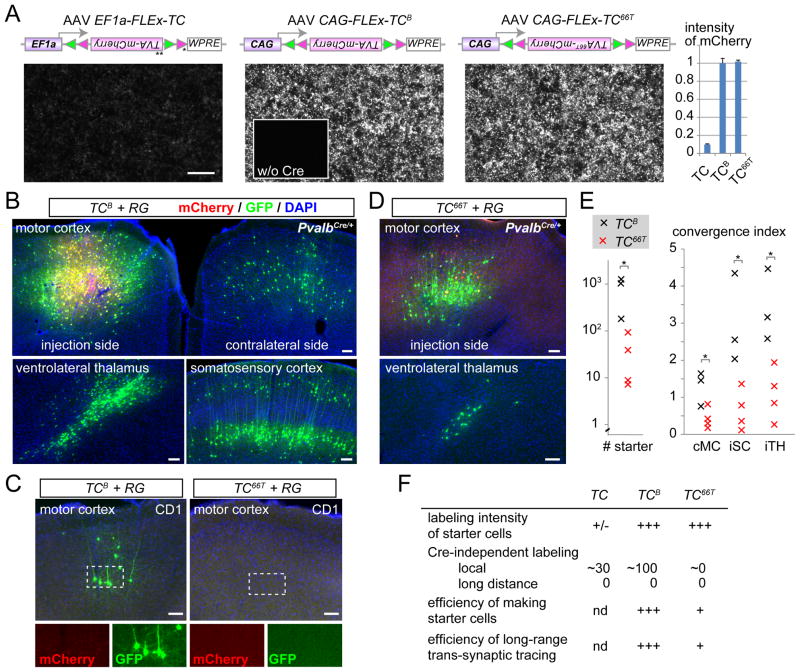
In the system developed by Watabe-Uchida et al. (2012), starter cells are co-transduced by two AAV vectors that express 1) a TVA-mCherry fusion protein in a Cre-dependent manner and 2) RG to complement RV-ΔG-GFP, allowing trans-synaptic tracing from Cre-expressing cells. We found that, depending on the brain region and cell type, starter cells were not consistently detected due to variable expression of mCherry. To improve starter cell identification, we changed the promoter and sequences surrounding the start codon of the TVA-mCherry expression cassette (Figure 1A; see Experimental Procedures). The new construct CAG-FLEx-TCB (for TVA-mCherryBright) produced 10-fold greater mCherry fluorescence in cultured HEK293 cells compared with the original EF1a-FLEx-TC (Figure 1A). We also made similar modifications to the RG-expressing vector, and produced CAG-FLEx-RG in AAV2 (AAV serotype 2). For in vivo test, a 1:1 mixture of CAG-FLEx-TCB and CAG-FLEx-RG was introduced into the motor cortex of wild-type (negative controls) or PvalbCre/+ knock-in (Hippenmeyer et al., 2005) mice. The EnvA-pseudotyped RV (RV-ΔG-GFP+EnvA) was introduced into the same location two weeks later, and animals were sacrificed for analysis 4 days afterwards. In the PvalbCre/+ mice, we found 811±324 (mean±SEM, n=3 animals) mCherry+/GFP+ starter cells clearly labeled with mCherry near the injection site (Figure 1B, E). Consistent with results from a previous study (Miyamichi et al., 2011), thousands of mCherry−/GFP+ cells (GFP+ cells in short hereafter) were found near the injection site. Further away from the injection site, we found GFP+ cells in locations consistent with known long-range presynaptic partners of motor cortex neurons (Figure 1B) (Iriki et al., 1989). In the negative control animals that did not express Cre, however, significant numbers of GFP+ cells (74±30, n=4 animals) were also present near the injection site (Figure 1C, left panel), as was reported by Watabe-Uchida et al. (2012). The Cre-independent labeling was restricted to within ~500 μm of the injection site and was not present in long-range connections, and is likely caused by Cre-independent expression of TVA (see below), as TVA is known to be highly sensitive to EnvA-pseudotyped virus (Wall et al., 2010). Leaky TVA expression also produced local background RV infection when using the tTA/TRE strategy (Miyamichi et al., 2011; Figure S1A). As leaky expression of RG is unlikely sufficient to reconstitute infectious RV particles, this does not compromise long-distance tracing of presynaptic partners. However, local GFP+ cells produced by direct infection of Cre-negative cells by RV-ΔG-GFP+EnvA as a result of leaky TVA expression cannot be distinguished from trans-synaptic spread from starter cells to local presynaptic partners, compromising specificity of local trans-synaptic tracing.
Figure 1. Optimizing Cell Type-specific Trans-synaptic Tracing.
(A) Schematic of the construct and DNA transfection in cultured HEK293 cells. To increase the expression level of Cre-dependent pEF1-FLEx-TC (Watabe-Uchida et al., 2012) (left), we changed the promoter to CAG, removed an upstream and out-of-frame ATG (*) between the promoter and the open reading frame after Cre mediated recombination, and added a Kozak sequence before the start codon (**) for optimal translation (Kozak, 1987). The resultant cassette (pCAG-FLEx-TCB, middle) was further modified by introducing a point mutation in TVA to construct pCAG-FLEx-TC66T (right). All constructs, when co-transfected with a CAG-Cre plasmid, allowed 293 cells to express mCherry, while mCherry was not expressed in the absence of Cre (middle inset and data not shown). mCherry expression level was markedly increased in CAG-FLEx-TCB and CAG-FLEx-TC66, as quantified in the right graph (mean ± SEM); mCherry intensity of CAG-FLEx-TCB is normalized to 1.
(B) Proof-of-principle demonstration of Cre-dependent trans-synaptic tracing by transducing a mixture of two AAV2 vectors containing CAG-FLEx-RG and CAG-FLEx-TCB into the primary motor cortex (M1) of PvalbCre/+ mice, followed by RV-ΔG-GFP+EnvA. Starter cells (yellow) and AAV-transduced cells that did not receive RV (red) were restricted to the injection site, and trans-synaptically labeled GFP+ neurons (green) were found in presynaptic areas of the M1, including the contralateral M1 (cMC), ipsilateral somatosensory cortex (iSC), and ipsilateral ventrolateral thalamus (iTH).
(C) Negative controls co-transducing AAV2 CAG-FLEx-TCB or CAG-FLEx-TC66T with AAV2 CAG-FLEx-RG into CD1 mice with no Cre expression. mCherry from AAV was rarely detected, but GFP from RV was detected near the injection site when CAG-FLEx-TCB was used. In contrast, no GFP or mCherry was detected when CAG-FLEx-TC66T was used.
(D) A proof-of-principle demonstration of trans-synaptic tracing by transducing PvalbCre/+ mice with AAV2 CAG-FLEx-TC66T and AAV2 CAG-FLEx-RG. Similar to panel (B), starter cells in M1 and local and long-range presynaptic partners were detected.
(E) Quantification of number of starter cells (left) and convergence index for long distance tracing. Each “x” represents an individual experiment with CAG-FLEx-TCB (black) or CAG-FLEx-TC66T (red). cMC, contralateral motor cortex; iSC, ipsilateral somatosensory cortex; iTH, ipsilateral thalamus.
(F) Summary of three variants of trans-synaptic tracing.
Scale bars, 100 μm.
See Figure S1 for additional controls and quantification.
To decrease TVA activity without affecting the expression level of the TVA-mCherry fusion protein, we introduced a point mutation (Glu66 → Thr, or 66T) in the TVA receptor known to reduce EnvA-enveloped viral transduction by ~10 fold in a cell culture assay (Rong et al., 1998). The expression levels of CAG-FLEx-TC66T and CAG-FLEx-TCB were comparable in HEK293 cells, as assayed by mCherry fluorescence intensity (Figure 1A). When CAG-FLEx-TC66T was used instead of CAG-FLEx-TCB along with CAG-FLEx-RG, followed by RV-ΔG-GFP+EnvA, Cre-independent labeling of GFP in the negative control animals was drastically reduced in the motor cortex (0.5±0.5 GFP+ cells, n=4 animals; Figure 1C, right) and in the OB (0.8±0.5 GFP+ cells, n=6 animals) of wild-type CD1 mice that did not express Cre (see Figure S1 for more controls). By contrast, when CAG-FLEx-TC66T and CAG-FLEx-RG were introduced into the motor cortex of PvalbCre/+ mice, we found 36±20 (n=4 animals) mCherry+/GFP+ starter cells clearly labeled with mCherry near the injection site. As expected, hundreds of local GFP+ cells were labeled near the injection site (Figure 1D), although the convergence index (defined as the number of presynaptic GFP+ cells divided by the number of starter cells) for long distance trans-synaptic labeling was significantly lower than those obtained by using CAG-FLEx-TCB (Figure 1E).
In summary, both CAG-FLEx-TCB and CAG-FLEx-TC66T allowed trans-synaptic tracing and unequivocal labeling of starter cells with the mCherry marker. TCB is suitable for efficient long distance tracing but cannot be used for mapping local circuits near the injection site due to high Cre-independent transduction by EnvA-pseudotyped RV. By contrast, TC66T is suitable for analyzing local circuits with near-zero Cre-independent transduction of EnvA-pseudotyped RV, albeit with reduced number of starter cells (Figure 1E, F).
Parvalbumin Neurons Features Prominently in the Presynaptic Landscape of Mitral/Tufted Cells
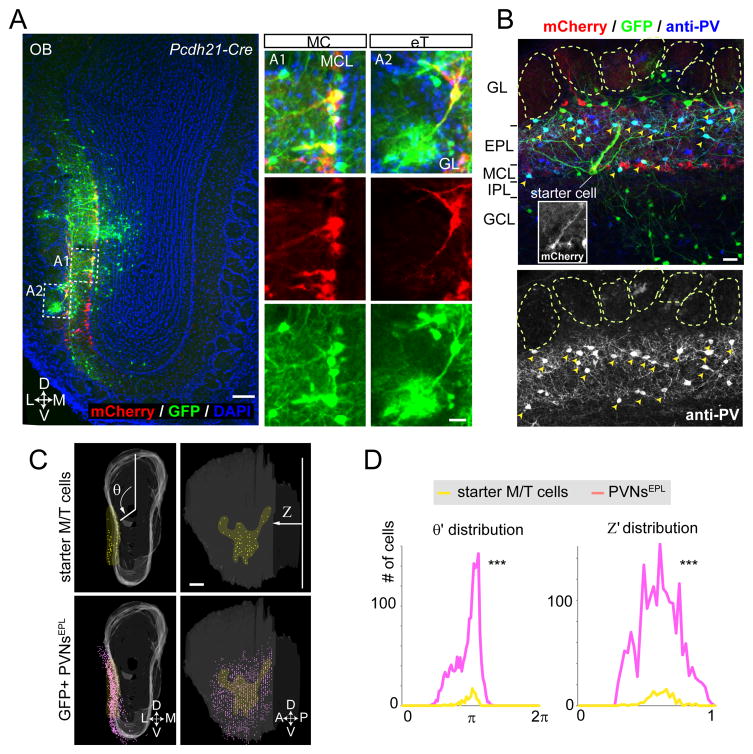
We next applied CAG-FLEx-TC66T-based trans-synaptic tracing to map the presynaptic partners of M/T cells in the OB. We used Pcdh21-Cre mice (Nagai et al., 2005) to restrict starter cells to M/T cells. We targeted M/T cells of the ventrolateral OB (1.6 mm from the dorsal surface). All starter cells (mCherry+/GFP+) in 4 animals were located either in the superficial layer between glomeruli and the external plexiform layer (EPL; 23±12), or in the mitral cell layer (MCL; 23±11) (Figure 2A), consistent with both the distribution of external tufted cells (eTs) or mitral cells (MCs) and with the expression pattern of Pcdh21-Cre. 1997±588 mCherry−/GFP+ (hereafter GFP+ cells), presumed presynaptic partners of the starter M/T cells, were located in different layers in the OB, including 362±80 periglomerular and external tufted cells in the glomerular layer, 946±261 cells in the EPL, 107±41 cells in the mitral cell layer, 324±159 granule cells and 259±64 deep short axons cells (dSACs) in the granule cell layer (Figure 2A–B). In addition, we detected 218±82 presumed cortical feedback projection neurons in the anterior olfactory nucleus (AON) and anterior piriform cortex (APC, Figure S6D–G), two cortical targets of M/T cells.
Figure 2. Parvalbumin Neurons in the External Plexiform Layer Are Prominent Presynaptic Partners of M/T Cells.
(A) Starter cells were limited to the mitral and tufted cell populations by transducing Pcdh21-Cre mice with AAV2 CAG-FLEx-RG and AAV2 CAG-FLEx-TC66T followed by RV-ΔG-GFP+EnvA. In this 60-μm coronal section, starter cells are located in the mitral cell layer (MCL) and the boundary between glomeruli and external plexiform layer (eT, for external tufted cells). L, lateral; M, medial; D, dorsal; V, ventral. Scale bars, 100 μm for the low magnification image; 20 μm for the high magnification images.
(B) A typical example of a 60-μm coronal section containing a starter MC (inset) and GFP+ presynaptic neurons stained by anti-PV antibodies. GL, glomerular layer; EPL, external plexiform layer; MCL, mitral cell layer; IPL, internal plexiform layer; GCL, granule cell layer. Neurons co-labeled with GFP and anti-PV antibodies are indicated by yellow arrowheads. Scale bar, 20 μm.
(C) A representative example of 3D-reconstructed OB showing starter M/T cell populations (yellow dots) and GFP+ PVNsEPL (magenta dots). White shadow represents the 2D surface of the MCL in the 3D OB model. θ represents rotation angle from the polar axis (vertical line in the left top panel), and Z represents the relative distance from the most posterior section. A, anterior; P, posterior. Scale bar, 200 μm.
(D) Histograms showing distribution of normalized θ and Z (θ′ and Z′) for pooled data (n=3 mice). Notably, both θ′ and Z′ distribution of PVNsEPL was significantly broader than those of starter cells. Statistical significance was tested by the Levene’s test against the null hypothesis assuming the equal variance between two populations (*** p<0.001).
See Figure S2 for 3D-reconstruction of labeled OB samples; Figure S3 for tracing with a longer survival period after RV infection; Figure S6 for cortical feedback input to M/T cells.
Among the different cell types labeled in the OB, the most prominent population was located in the EPL with a star-like shape, suggesting an interneuron identity (Kosaka et al., 1994). To characterize the cell types of these GFP+ cells, we co-stained the OB sections with antibodies against calretinin, calbindin, parvalbumin (PV), tyrosine hydroxylase and Tbr2. Among the antibodies used, anti-PV antibodies most frequently co-labeled GFP+ neurons in the EPL (Figure 2B). Indeed, 88.1±1.6% of GFP+ cells in the EPL were PV+ neurons (PVNs) (n=3 animals). To test whether this enrichment reflects connection specificity or bias of RV infection, we directly injected RV carrying its own glycoprotein in its envelope (RV-ΔG-GFP+RG) into the OB, which should transduce all cell types capable of receiving RV whether or not they are synaptically connected with M/T cells. We found that only 33.0±1.6 % of GFP+ cells in the EPL were PVNs (n=3 animals, Figure S1D). Thus, the enrichment of PVNs in the M/T trans-synaptic tracing experiment reflects connection specificity: PVNs are prominent presynaptic partners of M/T cells.
To compare the distribution of labeled cells in different layers and of different cell types across samples, we mapped them to a common 3D reference OB using a modified 3D reconstruction protocol (Miyamichi et al., 2011) (see Supplemental Experimental Procedures and Figure S2). As an example, Figure 2C (top panels) shows the lateral view of a representative OB sample (same as Figure 2A), with 31 eTs and 27 MCs starter cells (shown in yellow). A distribution of the 1,234 GFP+ PVNs in the EPL (PVNsEPL) are shown in magenta. Interestingly, GFP+ PVNsEPL were more broadly distributed across the OB surface than their postsynaptic M/T starter cells (see Figure S2D for an additional example). To quantify this observation, we introduced a cylindrical coordinate system into the OB model, where Z represents the position along the anterior-posterior axis and θ represents the angle from the dorsal polar axis (Figure 2C). We normalized individual samples to a common frame, and use θ′ and Z′ to represent normalized values (see Supplemental Experimental Procedures). The histograms in Figure 2D show that PVNsEPL were significantly more widely distributed than their target M/T cells in both axes (Levene’s test for equal variance; p<0.001). Notably, longer survival period for RV to spread and label pre-synapitc neurons did not change tracing efficiency of local circuits or general labeling patterns in the OB (Figure S3). Thus, M/T cells receive input from PVNs that are more broadly distributed spatially than the M/T cells themselves.
Two-photon Targeted Patch of PVNsEPL in the Mouse OB
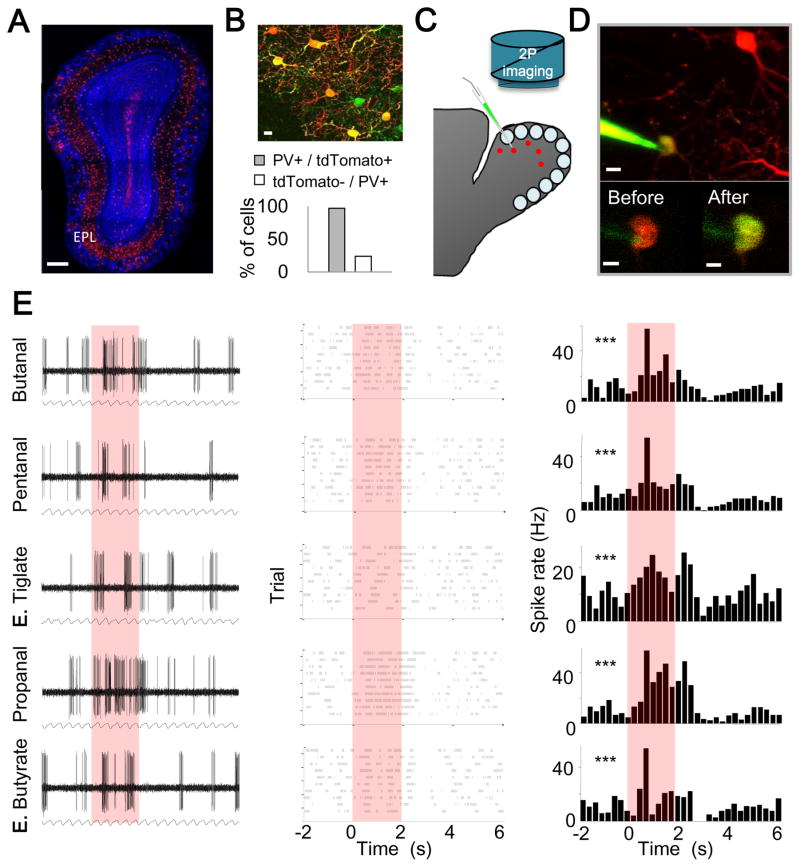
Given that PVNsEPL are prominent presynaptic partners of M/T cells, we next combined mouse genetics with imaging and physiology to determine their odor response properties in vivo. To visualize PVNs, we produced mice carrying PvalbCre/+ and a Cre-dependent tdTomato reporter (Hippenmeyer et al., 2005; Madisen et al., 2010). As expected from known distributions of PVNs in the OB, tdTomato+ cells were found predominantly in the EPL, with small numbers in the GL and GCL (Figure 3A). While not all PV+ EPL cells were tdTomato+, the vast majority of EPL tdTomato+ cells were PV+ as assessed by immunostaining (97.2%, n= 280 cells from 2 mice; Figure 3B). Therefore, we regard tdTomato+ neurons in the EPL of these mice as PVNsEPL. We targeted PVNsEPL by combining in vivo two photon imaging of the dorsal surface of the OB with loose patch recordings (Figure 3C) (Margrie et al., 2003). Unlike in the cortex, where PVNs are known to be fast spiking interneurons, PVNsEPL in the OB did not have a unique spike shape by which we can validate correct targeting (Figure S4). To validate correct targeting, therefore, we filled each cell with electrode dye after recording (Figure 3D). Only neurons that were successfully patched, recorded throughout the session, and double-labeled with tdTomato and the electrode dye were included in our dataset.
Figure 3. Two Photon Targeted Recording of OB PVNs in vivo.
(A) A confocal micrograph of an OB coronal section from a PvalbCre/+; Rosa26Ai9/+ mouse, showing conditional expression of tdTomato in PVNs (red). EPL, external plexiform layer. Scale bar, 250 μm.
(B) Top, a merged imaged of PV immunostaining (green) and tdTomato expression (red). Yellow cells denote double labeling. Scale bar, 15 μm. Bottom, in the EPL, P(PV+/tdTomato+) is 97.2%, and P(tdTomato-/PV+) is 24.0% (n= 280 cells from 2 mice). Based on our estimate that 1/3 of EPL cells are PV+ (Figure S1D), we infer using Bayes’ Rule that P(PV+/tdTomato-) is ~11%.
(C) Schematic of two-photon targeted patch (TPTP) recording from PV+ neurons.
(D) Top, a two-photon micrograph showing an in vivo TPTP recording from a PV+ neuron. Scale bar, 10 μm. Bottom, Two-photon micrograph of the cell body before (left) and after recording (right). Yellow somata denote successful labeling. Scale bar, 5 μm.
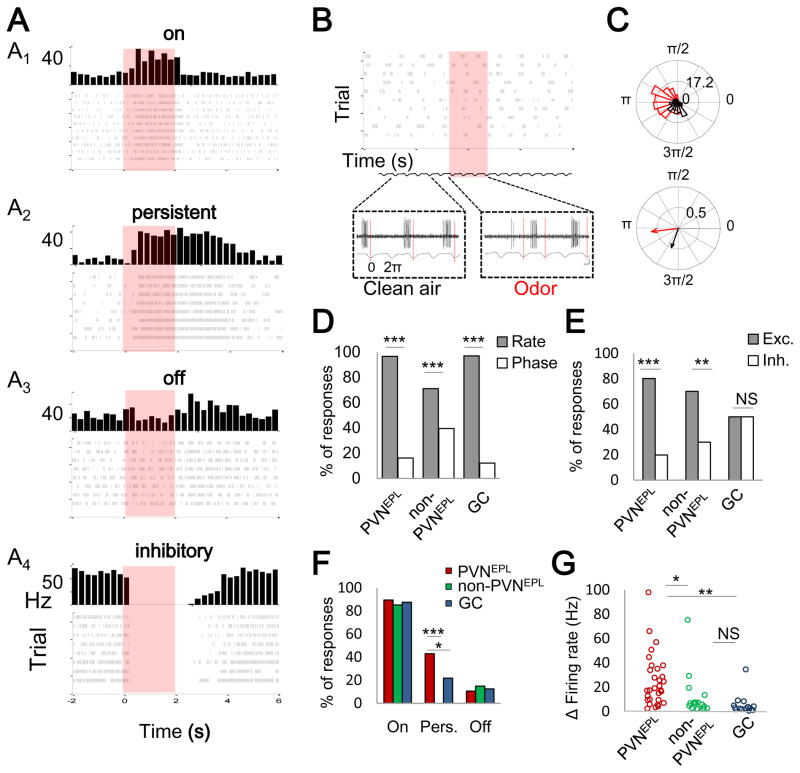
(E) Representative example of a loose patch recording from a PVNEPL presented with 5 odors. Each row represents the responses to a single odor; red box indicates odor presentation (2 sec). Left, raw voltage traces showing spontaneous and odor evoked activity during one trial. The respiration trace is shown below. Middle, raster plots showing spontaneous and odor evoked activity across all trials. Right, peri-stimulus time histogram (PSTH) showing average spike rates across trials (binning 250 msec). Asterisks mark significant responses (paired T-test; ***p<0.001).
See Figure 4 for spontaneous electriphysiological properties of PVNEPL.
PVNsEPL Are Broadly Tuned to Odors
We monitored both the spontaneous and odor-evoked activity of PVNsEPL in anesthetized, freely breathing mice (n=30 cells from 15 mice). Spontaneous firing rates of PVNsEPL were highly heterogeneous, spanning between 0.4 and 42 Hz (mean 13.57±1.81 Hz). Furthermore, 90% of PVNsEPL were spontaneously tuned to the phase of the respiratory cycle, with the activity in most cells locked to the inhalation period (Binomial proportion test; p<0.001, Figure S4). For odor stimulation, we used a panel of 5 monomolecular odors known to activate the dorsal OB. A representative example of one such recording in response to a 2-sec odor stimulus is shown in Figure 3E. Here, the PVNEPL responded significantly to each of the five odors (paired T-test; P< 0.001 for all odors).
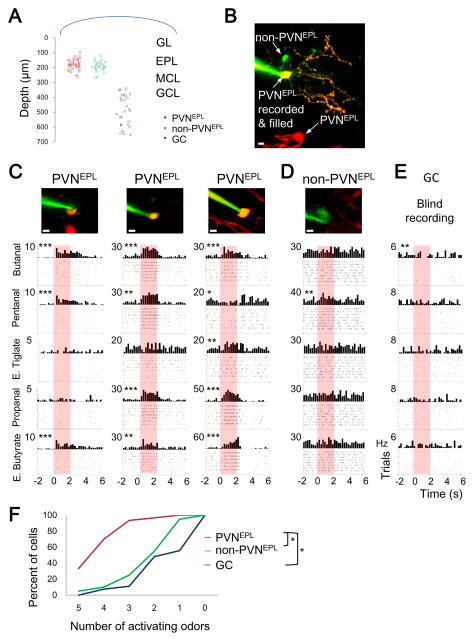
We recorded the odor response profiles of PVNsEPL as well as two additional neuronal populations in the same mice. One additional group was comprised of tdTomato-negative cells in the EPL (n=20 cells, from 11 mice). Although little is known about these cells, previous studies showed that they include either one or a combination of somatostatin, CRH, calretinin, or TH-expressing interneurons, as well as tufted cells (Hamilton et al., 2005; Huang et al., 2013). Based on our estimation that PV+ cells comprise 33% of the EPL cells (Figure S1D), and as only 24% of PV+ neurons did not express tdTomato (Figure 3B), we infer that tdTomato-negative cells expressing PV+ comprise only ~11% of the EPL. Thus, we collectively refer to tdTomato-negative cells in the EPL as non-PVNsEPL. The second interneuron group we used for comparison was the granule cells (GCs), the largest interneuron population in the OB (n=27 cells, from 4 mice). Recordings from PVNsEPL and non-PVNsEPL were made strictly from the EPL (mean recording depth 186.76±42.13μm) using two-photon targeted patch, and GCs were targeted blindly by limiting the pipette depth to the GCL (>340μm; Figure 4A). Additionally, non-PVNsEPL were filled with dye after recording to verify that the recording was from an EPL cell body, rather than an en-passant neurite (Figure 4B). This preparation allowed us to perform the first in vivo functional investigation of sparse, genetically defined subpopulations in the OB.
Figure 4. PVNs Are Broadly Tuned to Odors.
(A) Distribution of the depth of recording of all neurons in our dataset.
(B) A two-photon micrograph of a non-recorded PVNEPL (tdTomato only, red), PVNEPL that was recorded from (TdTomato and dye, yellow) and a non-PVNEPL (dye only, green). Scale bar 10μm.
(C–E) Neuronal responses from (C) three PVNEPL, (D) one non-PVNEPL (E) and one granule cell (GC). PSTHs and raster plots showing odor evoked responses (or lack thereof) to 5 odors. Asterisks mark significant responses *, P<0.05; **, P<0.01; ***, P<0.001 (paired T-test). The two photon micrograph of the recorded neuron is shown above. Scale bar 10 μm.
(F) Cumulative distribution plot of odor selectivity in response to 5 odors for PVNEPL (n=30 neurons, red), non-PVNEPL (n=20 neurons, green) and GCs (n=27 neurons, blue). Tukey’s HSD; *, p<0.05.
Recording the odor-evoked activity of these three groups allowed us to compare their odor response profiles under similar conditions. We first examined the odor selectivity of each group by testing each cell’s spiking response to the panel of 5 odors. Neurons that responded to all 5 odors were considered non-selective or broadly tuned (see Experimental Procedures). Our first striking observation was that PVNsEPL were particularly broadly tuned. For example, Figure 4C shows three PVNsEPL responding to 3, 4, or all 5 odorants (Figure 4C, asterisks). In contrast, non-PVNsEPL and GCs normally responded to only 1–2 odors (Figure 4D–E, asterisks). In total, while 93% (28/30 neurons) of the PVNsEPL responded to 3 or more odors, only 25% (5/20 neurons) of the non-PVNsEPL and 11% (5/27 neurons) of the GCs responded similarly (Figure 4F). Moreover, 33% (10/30 neurons) of the PVNsEPL responded to all odors presented whereas none of the GC and only 5% (1/20 neurons) of the non-PVNsEPL showed such broad tuning (Figure 4F). Statistical analysis validated that the PVNsEPL were significantly more broadly tuned to odors compared to the two other populations we tested (Tukey’s HSD, p<0.05, Figure 4F).
Unique Odor Response Profile of PVNsEPL
Having found that PVNsEPL are particularly broadly tuned to odors, we next analyzed whether they have other unique physiological properties. We first subdivided odor responses into changes in firing rates and in phase tuning (see Experimental Procedures). While PVNsEPL showed both types of odor-evoked responses (e.g. Figure 5A for rate changes; Figure 5B–C for phase tuning changes), the vast majority of odor responses were characterized by changes in firing rate (>95%, Binomial test; p<0.001, Figure 5D). Odor-evoked changes in PVNsEPL firing rate were primarily excitatory (Figure 5A1–A3, Figure 5E), but some neurons showed inhibitory responses (Figure 5A4). Interestingly, most PVNsEPL (>80%) and non-PVNsEPL exhibited either exclusively excitatory or exclusively inhibitory odor evoked responses. By contrast, GCs showed similar levels of excitatory and inhibitory responses, even within a given cell (Figure 5E, Figure S5). These data suggest that PVNsEPL receive predominantly excitatory (or dis-inhibitory) inputs.
Figure 5. Odor Response Profiles of PVNs Are Predominantly Excitatory.
(A) Examples (PSTHs and raster plots) of four types of responses in PVNEPL neurons: on (A1); persistent (A2); off (A3); and inhibitory (A4). Each panel is a different cell and odor pair.
(B) An example of an odor evoked response by phase tuning in a PVNEPL neuron. Top, raster plot for a single odor (8 trials). Bottom, raw spiking traces with regard to the respiration before and during odor presentation.
(C) Phase-analysis of the cell shown in panel (B). Top, binned data of spike respiration phase tuning across all trials before and during odor presentation (black and red, respectively). Bottom, direction and magnitude of the mean resultant phase tuning vector before and during odor stimulation.
(D) Histogram of odor evoked changes in firing rate and odor evoked changes in phase tuning for all cell groups (Binomial proportion test; p<0.001).
(E) Histogram of odor evoked excitatory vs. inhibitory responses for each group (Binomial proportion test; p<0.01)
(F) Histogram of the percentage of on, persistent (pers.) and off odor evoked responses for each group (Binomial proportion test; p<0.05).
(G) Response magnitude of odor evoked changes in firing rate for each group (Tukey’s HSD, P<0.05). In panel (D–G), *, P<0.05; **-P<0.01; ***, P<0.001. NS, not significant.
See Figure S5 for distribution of cells that exhibit excitatory, inhibitory, or both excitatory and inhibitory responses among PVNsEPL, non-PVNsEPL, and GCs.
To further examine the physiological characteristics of PVNsEPL, we classified excitatory odor responses into three categories: “on”, “persistent”, and “off” responses. Examples of all three response types from different PVNsEPL are shown in Figure 5A1–A3. All three groups showed predominantly “on” responses (89%, 85%, 87%, for PVNEPL, non-PVNEPL, and GC, respectively, Figure 5F). Interestingly however, and unique to PVNsEPL, over 40% of excitatory responses persisted after odor stimulation had ended (mean persistent response to a 2-sec odor stimulation was 7.13±0.38 sec.). Finally, the magnitude of PVNsEPL responses (quantified by the absolute maximal changes in firing rates) was significantly higher than that of both non-PVNsEPL and GCs (Tukey’s HSD, p<0.05, Figure 5G). Taken together, these data suggest that PVNsEPL exert robust and persistent inhibition upon M/T cells to a wide range of stimuli.
PVNs Integrate Input from Broadly Distributed Mitral/Tufted Cells
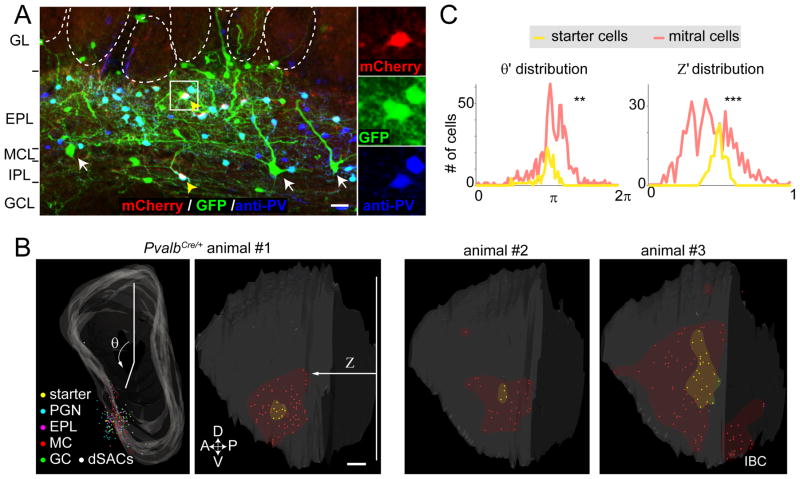
To investigate the underlying basis for the broad odor tuning of PVNsEPL, we applied the CAG-FLEx-TC66T-based trans-synaptic tracing strategy using PvalbCre/+ mice (Hippenmeyer et al., 2005) to determine the presynaptic landscape of the PVNs themselves (n=7 mice). Almost all (>97%) starter cells (mCherry+/GFP+) were clearly labeled with PV antibodies (Figure 6A). As expected from previous anatomical studies, in addition to the PVNsEPL, starter cells were often found in the deeper internal plexiform and granule cell layers (IPL/GCL, Figure 6A; hereafter referred to as PVNsIPL). On average, we detected 11.7±3.7 starter PVNs, and various types of GFP+ neurons in the OB (271.6±83.9 cells per animal) and in the AON/APC (76.9±48.5 cells, Figure S6A–C), which are presumed presynaptic partners of PVNs. GFP+ neurons in the OB included periglomerular and external tufted cells in the glomerular layer (29.4±6.9 cells), EPL neurons (90.1±39.2 cells), MCs (43.0±10.8 cells), granule cells (5.9±1.8 cells) and dSACs (106±33.9 cells). In two samples, we obtained exclusively PVNsEPL as starter cells. In both samples MCs were detected as presynaptic partners, indicating MC→PVNEPL direct connections. The convergence index was 7.8 and 6.3, indicating that multiple MC inputs converge onto each PVNEPL. As noted previously (Marshel et al., 2010; Miyamichi et al., 2011), convergence index derived from RV-based trans-synaptic tracing is likely an underestimate of the real connectivity.
Figure 6. The Presynaptic Landscape of PVNs Includes Widely Distributed MCs.
(A) A representative 60-μm coronal section with starter cells co-stained with anti-PV antibodies. In this section, two PV+ starter cells were detected, one each in the EPL and IPL (yellow arrowheads). White arrows represent mitral cells. Abbreviations as in Figure 2B. Scale bar, 20 μm.
(B) Three representative examples of 3D-reconstructed OBs showing starter PVNs (yellow dots) and GFP+ presynaptic neurons in the frontal (animal #1, left) and lateral (the rest) views. Different cell type/layer is represented by differently colored dots. Area of starter cells and MCs are highlighted by yellow and red shadows. θ and Z are as Figure 2. Note that some samples (for example, animal #3) contain labeled MCs in the mirror imaged area of the injection site (IBC, intra-bulbar connections) (Schoenfeld et al., 1985). Scale bar, 200 μm.
(C) Histograms showing distribution of normalized θ and Z (θ′ and Z′) for pooled data (n=7 mice). Notably, both θ′ and Z′ distribution of presynaptically labeled MCs are significantly broader than those of starter cells. **, p<0.01 and ***, p<0.001 by the Levene’s test against the null hypothesis assuming the equal variance between two populations.
See Figure S6 for cortical feedback input to PVNs.
We next analyzed the spatial distribution of neurons presynaptic to PVNs using 3D-reconstructed OBs. Figure 6B shows a representative example (animal #1) with starter PVNs exclusively in the EPL restricted to the region near the injection site. Presynaptic partners to these PVNEPL starter cells were more widely distributed spatially than the PVNEPL starter cells themselves. Notably, the presynaptic MCs (shown in red) were located most broadly, both along the dorsal-ventral and anterior-posterior axes. Other examples (that contained both PVNEPL and PVNIPL as starter cells) showed a similar trend. The histograms of θ′ and Z′ (Figure 6C, n=7 pooled data) showed that presynaptic MCs were significantly more broadly distributed compared to the starter PVNs along both the dorsal-ventral axis (θ′) and the anterior-posterior axis (Z′) (Levene’s test for equal variance; p<0.01). The combined data of presynaptic partner tracing from the M/T (Figure 2) or the PV (Figure 6) cells as starter cells revealed reciprocal connectivity in the form of MC→PVNEPL→MC. In this feedback loop, individual M/T cells on average labeled 16.7 input PVNsEPL, each of which labeled ~7 input MC cells. Thus, even taking into consideration that the convergence index from RV-based trans-synaptic tracing reflects a lower bound, individual M/T cells di-synaptically communicate with >100 M/T cells that are broadly spread in the OB via PVNs as intermediate neurons (Figure 8).
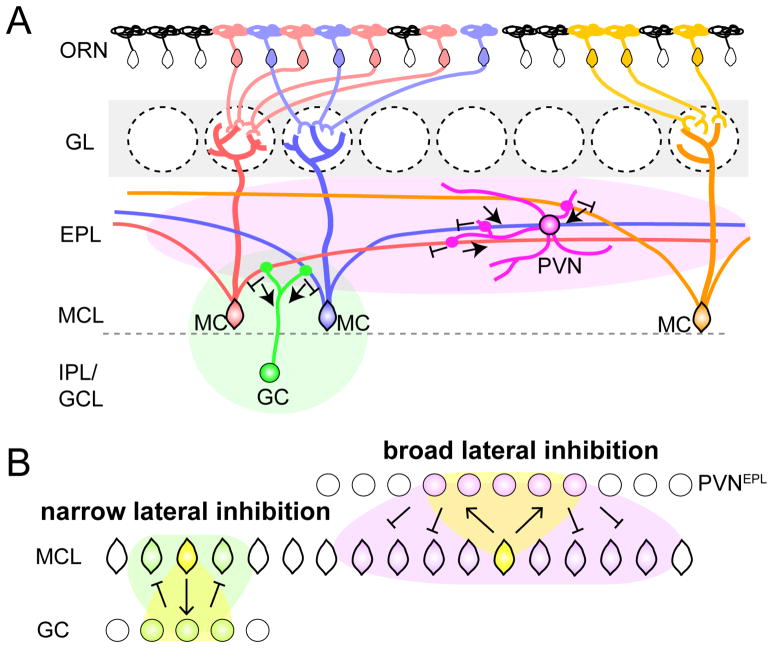
Figure 8. Feedback Circuits by PVNsEPL Underlying Broad Odor Processing.
(A) Schematic summary of the MC→PVNEPL→MC circuit in the OB. Each PVNEPL receives input from, and sends output to, broadly distributed MCs via MCs’ long secondary dendrites in the EPL for broad lateral inhibition of MC output. In contrast, the MC→GC→MC feedback circuit connects nearby MCs for local lateral inhibition. The green and magenta ovals symbolize areas influenced by PVN and GC.
(B) Schematic illustration of the difference in the spatial spread of the two lateral inhibitory feedback loops.
Mitral Cell–Granule Cell Reciprocal Connections Are Narrowly Organized
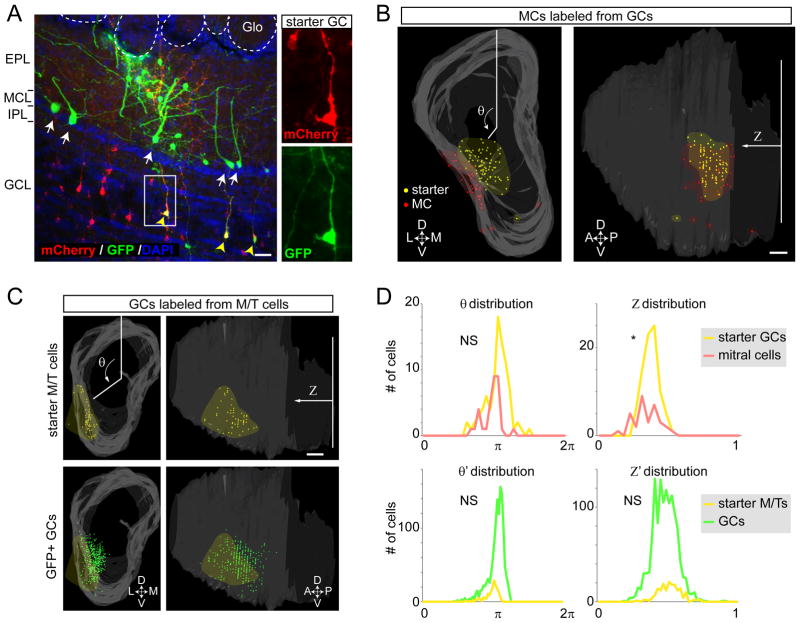
Granule cells (GCs) are known to receive excitatory input from MC’s secondary dendrites in the EPL, and send back inhibitory output to the dendrites of parent and nearby MCs (Shepherd, 2004). This MC→GC→MC feedback loop is thought to mediate recurrent and lateral inhibition. However, in vivo evidence to support these models is still limited, and the spatial distribution of this lateral connectivity is unknown. To investigate the circuit organization of MC–GC connections in comparison to the MC→PVNEPL→MC feedback loop, we used trans-synaptic tracing. It is challenging to restrict starter cells to GCs, however, because GC-specific Cre driver lines have not been reported. Instead, we used a pan-GABAergic driver, GAD2Cre/+ (Taniguchi et al., 2011), and injected Cre-dependent AAVs (CAG-FLEx-TC66T and CAG-FLEx-RG) into the GCL of the OB, taking advantage of the fact that the vast majority GCL neurons are GCs. We obtained two samples (out of 8 OBs injected and analyzed) that contained starter cells only in the GCL, without ‘contamination’ of periglomerular cells, EPL neurons, or dSACs. In one sample (Figure 7A), 82 starter GCs produced 272 presynaptic GFP+ neurons in the OB. In contrast to the neuronal populations presynaptic to M/T cells or PVNs, no neurons in the glomerular layer or external tufted cells were labeled from starter GCs. From the pooled data of both samples, a single starter GC, on average, labeled 0.54 MCs, 0.87 EPL neurons, 0.57 dSACs, and 0.6 pyramidal cells in the AON/APC (Figure S6G). To quantitatively analyze the spatial distribution of presynaptic MCs relative to the starter GCs, we reconstructed the OB as shown above (Figure 7B; 7D, top panels). MCs that were presynaptic to GCs formed clusters close to the starter cells without significant lateral spread. This distribution of presynaptic MC was in sharp contrast to the widespread distribution M/T cells presynaptic to PVNs (Figure 6B, C).
Figure 7. Mitral Cell-Granule Cell Reciprocal Connections Are Narrowly Organized.
(A) A representative 60-μm coronal section with starter cells in the granule cell population (yellow arrowheads). Trans-synaptically labeled MCs (white arrows) and EPL neurons were labeled with GFP. Abbreviations as in Figure 2B. Scale bar, 20 μm
(B) A representative example of 3D-reconstructed OB showing starter GCs (yellow dots) and GFP+ presynaptic MCs (red dots) in the frontal (left panel) and lateral (right panel) views. Area of starter cells and MCs are highlighted by yellow and red shadows. θ and Z are as Figure 2. Scale bar, 200 μm.
(C) A representative example of 3D-reconstructed OB showing starter mitral/tufted (M/T) cells (yellow dots) and GFP+ presynaptic GCs (green dots) in the frontal (left panels) and lateral (right panels) views. Areas of starter M/T cells are highlighted by yellow shadows. Scale bar, 200 μm.
(D) Top: histograms showing distribution of θ and Z for the sample shown in the panel B. The variance of θ for the starter granule cells and presynaptic MCs are not significantly different (NS), while the variance of Z is significantly different (p<0.05). Bottom panels: histograms showing distribution of normalized and pooled θ and Z (θ′ and Z′) for the n=4 samples of condition shown in the panel C. The variance of θ′ and Z′ for the starter mitral/tufted (M/T) cells and presynaptic granule cell (GCs) are not significantly different (NS) by the Levene’s test.
Finally, we analyzed the spatial organization of GC→MC connections from the trans-synaptic tracing data using Pcdh21-Cre (Figure 2). The distribution of presynaptic GCs (Figure 7C, green) was only slightly greater than the area of the starter MCs (Figure 7C, yellow). Pooled and normalized distributions (θ′ and Z′) supported this observation quantitatively (Figure 7D, bottom panels). Thus, the projection neurons of the OB, the mitral/tufted cells, are connected to two local feedback loops involving PVNsEPL and GCs, with striking differences in their physiological properties (Figures 4–5) and anatomical organization (Figures 2, 6–7).
Discussion
Here, we used cell type-specific trans-synaptic tracing and in vivo physiology to characterize the circuit composed of the reciprocal connections between PVNEPL and M/T cells. We discuss below the anatomical and physiological properties of this circuit in comparison with the MC→GC→MC circuit, and suggest that M/T→PVNEPL→M/T connections form a spatially broad lateral inhibitory circuit in the OB (Figure 8). We also discuss the technical advances and limitations of applying trans-synaptic tracing tools to untangle the complexity of local neuronal circuits.
In the OB, odor information from sensory neurons is processed by multiple local networks of interneurons. Local networks thus shape the activity of M/T cells that convey odor information to higher brain regions (Dhawale et al., 2010; Kikuta et al., 2013; Tan et al., 2010). The presynaptic landscape of M/T cells described here provides a rudimentary “roadmap”, upon which physiological and functional studies can be integrated for understanding the complex interactions underlying transformations of olfactory information. From this landscape, the PVNEPL subpopulation emerged as a prominent source of M/T cells input (Figure 2). PVNs have recently attracted much attention elsewhere in the brain (Isaacson and Scanziani, 2011; Tiesinga and Sejnowski, 2009). In the cortex, for example, these cells provide classic feedforward inhibition onto pyramidal cells, controlling both the timing and spatial spread of cortical excitation (Cruikshank et al., 2007; Gabernet et al., 2005; Gibson et al., 1999; Inoue and Imoto, 2006; Poo and Isaacson, 2009). Cortical PVNs have also been implicated in forming gamma oscillations (Sohal et al., 2009), regulating the activation of quiescent hippocampal stem cells (Song et al., 2012), and the onset of developmental critical periods in an experience-dependent manner (Kuhlman et al., 2011). In the OB, the role of PVNs has been inferred largely from anatomical studies, where their connectivity onto M/T cells has been established (Kosaka and Kosaka, 2008, 2011). Here we highlight two prominent features of PVNsEPL: their relatively broad odor tuning and spatially widespread connection patterns. Notably, both of these features are rather unique in comparison to other neurons in the OB (Davison and Katz, 2007; Fantana et al., 2008; Kato et al., 2012; Kikuta et al., 2013). What may be the role of broadly tuned interneurons exerting widespread local inhibition in the OB?
Previous studies examining the inhibition performed in the EPL have largely focused on the MC→GC→MC circuit. This reciprocal connectivity has been proposed to mediate nearest-neighbor lateral inhibition, as well as recurrent inhibition (Margrie et al., 2001; Mori et al., 1999; Yokoi et al., 1995). More recently, this lateral inhibitory circuit has been suggested to function in a spatially distributed manner, among distinct glomerular units that are not limited to the nearest-neighbor glomeruli (Debarbieux et al., 2003; Fantana et al., 2008). Our anatomical and physiological data suggest that the GC feedback loop is narrowly tuned both spatially and functionally, supporting their role in local columnar computation. By contrast, the spatially and functionally broad MC→PVNEPL→MC feedback loop suggests a role in more global computations within the EPL (Figure 8). For example, one computation commonly thought to be performed in the OB is decorrelation of similar odor representations (Cleland, 2010). Although the dominant models of bulbar decorrelation focus on interglomerular mechanisms at the glomerular layer (Aungst et al., 2003; Whitesell et al., 2013) or GCs lateral inhibition, broadly tuned inhibition may also provide a complementary mechanism to promote decorrelation (Poo and Isaacson, 2009; Sohya et al., 2007). Specifically in the OB, PVNsEPL would exert robust inhibition onto M/T cells, effectively elevating their response threshold. Additionally, both theoretical models and experimental observations suggest a role for broadly tuned inhibition in input normalization. In the OB and Drosophila antennal lobe (analogous to vertebrate OB), broadly tuned inhibition, scaled by the total input activity, may enable the limited dynamic range of projection neurons to encode information over a wide range of odorant concentration (Isaacson and Scanziani, 2011; Olsen et al., 2010). The inhibition exerted by PVNEPL is broadly distributed across the input field. Thus, these cells may promote the relative, rather than absolute, odor representation by M/T cells (see Kato et al., the accompanying manuscript).
The broad odor tuning of PVNs is consistent with the broad distribution of M/T→PVN excitatory inputs, while the narrow responsiveness of GCs can be explained by the narrowly distributed connectivity. How do multiple local feedback circuits operate at distinct spatial scales? One hypothesis is that GCs form reciprocal synapses more preferentially on proximal dendrites (closer to the cell bodies), while PVNsEPL do not have such bias. Indeed, a previous pseudorabies virus-mediated multi-synaptic circuit mapping identified cluster-like organization of the GC-M/T connections (Willhite et al., 2006), consistent with the narrow spatial distribution. Our data suggests that GCs connect predominately near the parent mitral cell soma.
In this study, we improved the RV-based Cre-dependent trans-synaptic tracing system (Wall et al., 2010; Watabe-Uchida et al., 2012) (Figure 1), enabling analysis of local connectivity in any circuit with starter cells defined by a specific Cre line. We provided new evidence in support of synaptic specificity of RV spread. For example, ~88% of RV labeled neurons in the EPL from the starter M/T cells were PVNs, despite the fact that PVNs constitute only ~33% of all RV-infectable cells in the EPL (Figure S1D). Therefore, selective RV spread from M/T cells to PVNsEPL indicates both strong PVNEPL→M/T connections and preferential RV spread through synaptic connections. External tufted cells were not labeled from starter GCs, despite the fact that their secondary dendrites (Igarashi et al., 2012) are intermingled with the ascending dendrites of GCs, also supporting the synapse specificity of RV spread. Our study also extended the utility of RV tracing to mapping connections made by atypical dendro-dendritic connections between M/T and PVNsEPL or GCs (Shepherd et al., 2007), at least qualitatively. However, trans-synaptic tracing is still a method under development and its limitation are not fully understood. For example, a major caveat is that only a subset of presynaptic partners of starter cells are labeled, leading to significant underestimation of the full connectivity matrix. Therefore the quantitative data regarding convergence ratio should only be interpreted in a relative rather than absolute manner. Another limitation of the current study is that we cannot distinguish mitral from tufted cells, as a mitral cell-specific Cre line has yet to be established. As their electrophysiological properties (Nagayama et al., 2004) and axonal projection patterns (Igarashi et al., 2012) are different, it will be interesting to compare the presynaptic landscape of these two distinct classes of projection neurons in the OB as genetic tools become available in the future. Nevertheless, our approach of integrating cell type-specific tracing with in vivo physiology facilitates studies of local circuits individually, yet retaining the context of their larger networks. We expect it to be widely applicable to dissect the function of neural circuits in many other parts of the brain.
Supplementary Information is linked to the online version of the paper.
Experimental Procedures
Animals
All animal procedures followed animal care guidelines approved by Stanford University’s Administrative Panel on Laboratory Animal Care (A-PLAC) and the Hebrew University Animal Care and Use Committee. The Pcdh21-Cre (Nagai et al., 2005) mouse was obtained from RIKEN Bioresource Center (Japan). PvalbCre (Hippenmeyer et al., 2005), GAD2Cre (Taniguchi et al., 2011) and Cre-dependent tdTomato reporter strain ROSA26Ai9 (Madisen et al., 2010) were obtained from the Jackson laboratories.
Surgical and Viral Procedures
To genetically label PV positive neurons (Figure 3–5), a total of n=27 PvalbCre/+; ROSA26Ai9/+ double heterozygous mice were used, both male and female, 8–12 weeks old. Twenty five mice were used for in-vivo electrophysiology experiments and two for histology. For electrophysiology, mice were anesthetized with an intraperitoneal injection of ketamine and medetomidine (100 mg/kg and 0.83 mg/kg, respectively) dna a subcutaneous injection of Carprofen (0.004 mg/g). Additionally, dextrose-saline was injected to prevent dehydration. Experiments lasted up to 11 hours (usually ~6 hrs). The depth of anesthesia was assessed by monitoring the pinch withdrawal reflex and rectal temperature was continuously monitored and maintained at 36°C ±1°C. For imaging and recording, a craniotomy (~1×2 mm) was performed over the OB of one hemisphere, while taking care to ensure that the dura remained intact.
For transsynaptic labeling (Figure 1, 2, 6, and 7), 0.3 μl of 1:1 mixture of AAV CAG-FLEx-RG and AAV CAG-FLEx-TC (TCB for the experiments in Fig. 1 and TC66T for the experiments in Fig. 2, Fig., 6, and Fig. 7) was injected into the brain at postnatal day (PD) 21 by using a stereotactic apparatus (KOPF). During surgery, animals were anesthetized with 65mg/kg ketamine and 13mg/kg xylazine (Lloid Laboratories). For motor cortex injections, the needle was placed 1.5 mm anterior and 1.5 mm lateral from the Bregma, and 0.4 mm from the brain surface. For OB injections, needle was placed 4.2 mm anterior and 0.8 mm lateral from the Bregma, and 1.6 mm from the OB surface. After recovery, animals were housed in regular 12 hour-dark/light cycle with food and water ad libitum. Two weeks later, 0.3 μl of pseudotyped RV (ΔG-GFP+EnvA) was injected into the same brain location under anesthesia. After recovery, animals were housed in a biosafety room for 4 days (or 8 days only for Figure S3) to allow RV to infect, trans-synaptically spread and express sufficient amount of GFP to label presynaptic cells.
Electrophysiology
Imaging was performed on an Ultima two-photon microscope from Prairie Technologies (Middleton, WI), using a 16× water-immersion objective lens (0.8 NA;CF175, Nikon) at 915nm excitation (DeepSee femtosec laser, Spectraphysics). Electrodes (5–10 MΩ) were filled with an internal solution containing (in mM): 140 K-gluconate, 10 KCl, 10 HEPES, 10 Na2-Phosphocreatine, 4 MgATP, 0.4 Na2GTP, 0.5 EGTA, pH adjusted to 7.25 with KOH. 200 μM Alexafluor 488 (Life technologies, Invitrogen) was added in targeted recordings, and dye from the electrode was electroporated into the cell using current pulses. Only tdTomato+ cells that were distinctly filled during this procedure were included in our PVNEPL and non-PVNEPL dataset.
Odor stimuli were delivered directly to the nose via a custom made 5-channel olfactometer with completely isolated odor channels to circumvent contamination between odors. The odors (butyraldehyde, valeraldehyde, ethyl tiglate, propanol and ethyl butyrate) were delivered for 2 secs (4–10 trials/odor) in a pseudo-random order, with an inter-stimulus interval of 18 secs at a final concentration of 50 ppm or 500 ppm. No significant differences between the average data of these concentrations were found so data were pooled. The animal’s respiration was monitored throughout the experiment by a low pressure sensor (1-INCH-D1-4V-MINI, by ‘All sensors’). which was used to identify the inhalation onset during the respiratory cycle and to trigger odor delivery. All data anaylsis was carried out by custom-written Matlab programs.
Additional procedures on Constructs and Virus Preparations, Two-photon Targeted Recording from PV-Positive Neurons, Respiration-Triggered Odor Delivery, Electrophysiological Data Analysis, Immunohistochemistry and Microscopy, and 3D-Reconstruction and Quantitative Analysis, can be found in Supplemental Exeprimental Procedures.
Supplementary Material
Highlights.
An improved method for trans-synaptic tracing of local circuits
Parvalbumin interneurons (PVNs) are dominant presynaptic partners of mitral cells
PVNs are widely responsive to odors with unique physiological signatures
A mitral cell → PVN → mitral cell circuit provides broad feedback control of OB output
Acknowledgments
We thank the Stanford Viral Core for help in generating AAVs, N. Uchida for reagents, C. Manalac for technical assistance, L. Cohen for assitaning with electrophysiology, members of the Luo and Mizrahi Labs for discussion and critical reading of the manuscript, and T. Komiyama and J. Isaacson for communicating unpublished results. We thank D. Feldman and M. Feldman for financial support during the sabbatical of A.M. in the Luo Lab. K.M. is a Research Specialist, and L.L. is an Investigator, of the Howard Hughes Medical Institute. B.C.W. is an NSF Graduate Research Fellow and a Stanford Graduate Fellow. This work was also supported by a HHMI Collaborative Innovation Award, an NIH grant (R01-NS050835) to L.L., and a European Research Council Grant (number 203994) to A.M.
Footnotes
Author contributions K.M., Y.S., L.L. and A.M. designed the experiments. K.M. performed and analyzed cell type-specific trans-synaptic labeling experiments. Y.S. performed and analyzed in vivo electrophysiology. M.S. assisted 3D reconstruction and data analysis, B.C.W. provided DNA constructs. K.M., Y.S., L.L. and A.M. wrote the manuscript, with contribution from B.C.W.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aungst JL, Heyward PM, Puche AC, Karnup SV, Hayar A, Szabo G, Shipley MT. Centre-surround inhibition among olfactory bulb glomeruli. Nature. 2003;426:623–629. doi: 10.1038/nature02185. [DOI] [PubMed] [Google Scholar]
- Batista-Brito R, Close J, Machold R, Fishell G. The distinct temporal origins of olfactory bulb interneuron subtypes. J Neurosci. 2008;28:3966–3975. doi: 10.1523/JNEUROSCI.5625-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland TA. Early transformations in odor representation. Trends Neurosci. 2010;33:130–139. doi: 10.1016/j.tins.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruikshank SJ, Lewis TJ, Connors BW. Synaptic basis for intense thalamocortical activation of feedforward inhibitory cells in neocortex. Nat Neurosci. 2007;10:462–468. doi: 10.1038/nn1861. [DOI] [PubMed] [Google Scholar]
- Davison IG, Katz LC. Sparse and selective odor coding by mitral/tufted neurons in the main olfactory bulb. J Neurosci. 2007;27:2091–2101. doi: 10.1523/JNEUROSCI.3779-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarbieux F, Audinat E, Charpak S. Action potential propagation in dendrites of rat mitral cells in vivo. J Neurosci. 2003;23:5553–5560. doi: 10.1523/JNEUROSCI.23-13-05553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Briggman KL, Helmstaedter M. Structural neurobiology: missing link to a mechanistic understanding of neural computation. Nat Rev Neurosci. 2012;13:351–358. doi: 10.1038/nrn3169. [DOI] [PubMed] [Google Scholar]
- Dhawale AK, Hagiwara A, Bhalla US, Murthy VN, Albeanu DF. Non-redundant odor coding by sister mitral cells revealed by light addressable glomeruli in the mouse. Nat Neurosci. 2010;13:1404–1412. doi: 10.1038/nn.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger V, Urban NN. Dynamic connectivity in the mitral cell-granule cell microcircuit. Semin Cell Dev Biol. 2006;17:424–432. doi: 10.1016/j.semcdb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Fantana AL, Soucy ER, Meister M. Rat olfactory bulb mitral cells receive sparse glomerular inputs. Neuron. 2008;59:802–814. doi: 10.1016/j.neuron.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Gabernet L, Jadhav SP, Feldman DE, Carandini M, Scanziani M. Somatosensory integration controlled by dynamic thalamocortical feed-forward inhibition. Neuron. 2005;48:315–327. doi: 10.1016/j.neuron.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Gibson JR, Beierlein M, Connors BW. Two networks of electrically coupled inhibitory neurons in neocortex. Nature. 1999;402:75–79. doi: 10.1038/47035. [DOI] [PubMed] [Google Scholar]
- Hamilton KA, Heinbockel T, Ennis M, Szabo G, Erdelyi F, Hayar A. Properties of external plexiform layer interneurons in mouse olfactory bulb slices. Neuroscience. 2005;133:819–829. doi: 10.1016/j.neuroscience.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Garcia I, Jen HI, Arenkiel BR. Reciprocal connectivity between mitral cells and external plexiform layer interneurons in the mouse olfactory bulb. Front Neural Circuits. 2013;7:32. doi: 10.3389/fncir.2013.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi KM, Ieki N, An M, Yamaguchi Y, Nagayama S, Kobayakawa K, Kobayakawa R, Tanifuji M, Sakano H, Chen WR, et al. Parallel mitral and tufted cell pathways route distinct odor information to different targets in the olfactory cortex. J Neurosci. 2012;32:7970–7985. doi: 10.1523/JNEUROSCI.0154-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Imoto K. Feedforward inhibitory connections from multiple thalamic cells to multiple regular-spiking cells in layer 4 of the somatosensory cortex. J Neurophysiol. 2006;96:1746–1754. doi: 10.1152/jn.00301.2006. [DOI] [PubMed] [Google Scholar]
- Iriki A, Pavlides C, Keller A, Asanuma H. Long-term potentiation in the motor cortex. Science. 1989;245:1385–1387. doi: 10.1126/science.2551038. [DOI] [PubMed] [Google Scholar]
- Isaacson JS, Scanziani M. How inhibition shapes cortical activity. Neuron. 2011;72:231–243. doi: 10.1016/j.neuron.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahr CE, Nicoll RA. Dendrodendritic inhibition: demonstration with intracellular recording. Science. 1980;207:1473–1475. doi: 10.1126/science.7361098. [DOI] [PubMed] [Google Scholar]
- Kato HK, Chu MW, Isaacson JS, Komiyama T. Dynamic sensory representations in the olfactory bulb: modulation by wakefulness and experience. Neuron. 2012;76:962–975. doi: 10.1016/j.neuron.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta S, Fletcher ML, Homma R, Yamasoba T, Nagayama S. Odorant response properties of individual neurons in an olfactory glomerular module. Neuron. 2013;77:1122–1135. doi: 10.1016/j.neuron.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka K, Heizmann CW, Kosaka T. Calcium-binding protein parvalbumin-immunoreactive neurons in the rat olfactory bulb. 1. Distribution and structural features in adult rat. Exp Brain Res. 1994;99:191–204. doi: 10.1007/BF00239586. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Kosaka K. Heterogeneity of parvalbumin-containing neurons in the mouse main olfactory bulb, with special reference to short-axon cells and betaIV-spectrin positive dendritic segments. Neurosci Res. 2008;60:56–72. doi: 10.1016/j.neures.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Kosaka K. “Interneurons” in the olfactory bulb revisited. Neurosci Res. 2011;69:93–99. doi: 10.1016/j.neures.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Tring E, Trachtenberg JT. Fast-spiking interneurons have an initial orientation bias that is lost with vision. Nat Neurosci. 2011;14:1121–1123. doi: 10.1038/nn.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margrie TW, Meyer AH, Caputi A, Monyer H, Hasan MT, Schaefer AT, Denk W, Brecht M. Targeted whole-cell recordings in the mammalian brain in vivo. Neuron. 2003;39:911–918. doi: 10.1016/j.neuron.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Margrie TW, Sakmann B, Urban NN. Action potential propagation in mitral cell lateral dendrites is decremental and controls recurrent and lateral inhibition in the mammalian olfactory bulb. Proc Natl Acad Sci U S A. 2001;98:319–324. doi: 10.1073/pnas.011523098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Amat F, Moussavi F, Wang C, Wickersham I, Wall NR, Taniguchi H, Tasic B, Huang ZJ, He Z, et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature. 2011;472:191–196. doi: 10.1038/nature09714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Markram H. Interneuron Diversity series: Molecular and genetic tools to study GABAergic interneuron diversity and function. Trends Neurosci. 2004;27:90–97. doi: 10.1016/j.tins.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Mori K, Nagao H, Yoshihara Y. The olfactory bulb: coding and processing of odor molecule information. Science. 1999;286:711–715. doi: 10.1126/science.286.5440.711. [DOI] [PubMed] [Google Scholar]
- Murthy VN. Olfactory maps in the brain. Annu Rev Neurosci. 2011;34:233–258. doi: 10.1146/annurev-neuro-061010-113738. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Sano H, Yokoi M. Transgenic expression of Cre recombinase in mitral/tufted cells of the olfactory bulb. Genesis. 2005;43:12–16. doi: 10.1002/gene.20146. [DOI] [PubMed] [Google Scholar]
- Nagayama S, Takahashi YK, Yoshihara Y, Mori K. Mitral and tufted cells differ in the decoding manner of odor maps in the rat olfactory bulb. J Neurophysiol. 2004;91:2532–2540. doi: 10.1152/jn.01266.2003. [DOI] [PubMed] [Google Scholar]
- Olsen SR, Bhandawat V, Wilson RI. Divisive normalization in olfactory population codes. Neuron. 2010;66:287–299. doi: 10.1016/j.neuron.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo C, Isaacson JS. Odor representations in olfactory cortex: “sparse” coding, global inhibition, and oscillations. Neuron. 2009;62:850–861. doi: 10.1016/j.neuron.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L, Gendron K, Strohl B, Shenoy R, Wool-Lewis RJ, Bates P. Characterization of determinants for envelope binding and infection in tva, the subgroup A avian sarcoma and leukosis virus receptor. J Virol. 1998;72:4552–4559. doi: 10.1128/jvi.72.6.4552-4559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H. Neural map formation in the mouse olfactory system. Neuron. 2010;67:530–542. doi: 10.1016/j.neuron.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Schoenfeld TA, Marchand JE, Macrides F. Topographic organization of tufted cell axonal projections in the hamster main olfactory bulb: an intrabulbar associational system. J Comp Neurol. 1985;235:503–518. doi: 10.1002/cne.902350408. [DOI] [PubMed] [Google Scholar]
- Shepherd GM. The synaptic organization of the brain. 5. Oxford ; New York: Oxford University Press; 2004. [Google Scholar]
- Shepherd GM, Chen WR, Willhite D, Migliore M, Greer CA. The olfactory granule cell: from classical enigma to central role in olfactory processing. Brain Res Rev. 2007;55:373–382. doi: 10.1016/j.brainresrev.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohya K, Kameyama K, Yanagawa Y, Obata K, Tsumoto T. GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J Neurosci. 2007;27:2145–2149. doi: 10.1523/JNEUROSCI.4641-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhong C, Bonaguidi MA, Sun GJ, Hsu D, Gu Y, Meletis K, Huang ZJ, Ge S, Enikolopov G, et al. Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature. 2012;489:150–154. doi: 10.1038/nature11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Savigner A, Ma M, Luo M. Odor information processing by the olfactory bulb analyzed in gene-targeted mice. Neuron. 2010;65:912–926. doi: 10.1016/j.neuron.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiesinga P, Sejnowski TJ. Cortical enlightenment: are attentional gamma oscillations driven by ING or PING? Neuron. 2009;63:727–732. doi: 10.1016/j.neuron.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc Natl Acad Sci U S A. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Whitesell JD, Sorensen KA, Jarvie BC, Hentges ST, Schoppa NE. Interglomerular lateral inhibition targeted on external tufted cells in the olfactory bulb. J Neurosci. 2013;33:1552–1563. doi: 10.1523/JNEUROSCI.3410-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willhite DC, Nguyen KT, Masurkar AV, Greer CA, Shepherd GM, Chen WR. Viral tracing identifies distributed columnar organization in the olfactory bulb. Proc Natl Acad Sci U S A. 2006;103:12592–12597. doi: 10.1073/pnas.0602032103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi M, Mori K, Nakanishi S. Refinement of odor molecule tuning by dendrodendritic synaptic inhibition in the olfactory bulb. Proc Natl Acad Sci U S A. 1995;92:3371–3375. doi: 10.1073/pnas.92.8.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.