Abstract
Until now, models of psychiatric diseases have typically been animal models. Whether they were to be used to further understand the pathophysiology of the disorder, or as drug discovery tools, animal models have been the choice of preference in mimicking psychiatric disorders in an experimental setting. While there have been cellular models, they have generally been lacking in validity. This situation is changing with the advent of patient-specific induced pluripotent stem cells (iPSCs). In this article, we give a methodological evaluation of the current state of the iPS technology with reference to our own work in generating patient-specific iPSCs for the study of autistic spectrum disorder (ASD). In addition, we will give a broader perspective on the validity of this technology and to what extent it can be expected to complement animal models of ASD in the coming years.
Keywords: Induced pluripotent stem cells, Autism, SHANK3
Introduction
Autistic spectrum disorder (ASD) is a complex and heterogeneous early onset neurodevelopmental condition characterized by profound impairments in social and language skills and by stereotypical behaviours. ASD has a substantial and complex underlying genetic component (Devlin and Scherer 2012) with more than 100 genes implicated in the aetiology, none of which alone account for more than 2 % of cases (Betancur 2011). Furthermore, single gene defects, CNVs and syndromic forms account for only 10–20 % of cases (Betancur 2011). This complex genetic heterogeneity overlapping as it does with other conditions such as severe intellectual impairment and epilepsy (Devlin and Scherer 2012) and combined with a lack of other diagnostic biomarkers means that ASD is still clinically defined by behavioural assessments (Lord et al. 1989; Lord et al. 1994). In this respect, considerable work has gone into developing animal models of ASD in terms of defining behavioural assays reflecting the core symptoms of the disorder: social interaction, communication and repetitive behaviours (Crawley 2012). Furthermore, an increasing catalogue of genes associated with ASD has been replicated in transgenic models, and environmental factors implicated in the disorder, such as prenatal exposure to valproate, have been successfully shown to produce ASD-like symptoms in mice according to these behavioural assays (Roullet et al. 2013). However, drug discovery for psychiatric disorders generally has faced serious problems, largely emanating from the failure of the predictive validity of animal models of disease (Markou et al. 2009; van der Worp et al. 2010). Drug discovery in this area has therefore been slow, and although there are some recent additions to the pipeline for syndromic forms of ASD (Krueger and Bear 2011; Hampson et al. 2012), there is currently not a single approved treatment for ASD in Japan or the five major European jurisdictions, and only risperidone and aripiprazole, two atypical antipsychotics, are approved in the US for the management of irritability (Nightingale 2012).
Studies in vitro using either rodent primary neurons or immortalized cell lines over-expressing a molecular target of interest have been and will continue to be valuable in high-throughput screening in the drug discovery process (Eglen and Reisine 2011). However, these systems also have questionable validity either by virtue of simply being non-human or from artefacts of over-expression. A significant advance in the generation of more physiologically relevant cell types came from the successful isolation and culture of human embryonic stem cells (hES) (Thomson et al. 1998). Within the context of psychiatric disorders, this permitted for the first time the possibility of generating human neurons in vitro and protocols have since been developed to generate a number of different neuronal subtypes by directed differentiation with morphogens along known developmental trajectories (Muguruma et al. 2010). However, in addition to ethical concerns with hES generation, modelling diseases with reduced penetrance or complex genetics in hES cells is still not straightforward. Work towards developing embryonic stem (ES) cells from interspecies human/animal hybrid embryos attempted to overcome the latter issue but faced additional ethical concerns and legislative barriers (St John et al. 2008) and are technically cumbersome to deliver.
As a consequence of the groundbreaking work of the Yamanaka lab, somatic cells can now be reprogrammed into pluripotent stem cells from a simple patient biopsy (typically skin, blood or hair) (Takahashi and Yamanaka 2006; Takahashi et al. 2007). This technology therefore allows the delivery of human neurons in vitro, derived precisely from patients suffering from a specific disorder. Where that disorder is neurodevelopmental, with a strong genetic component such as ASD, then this becomes an attractive technology since it allows us to recapitulate in a culture dish precisely those developmental events that are putatively abnormal during the aetiology of the disorder (Marchetto and Gage 2012). These patient-specific induced pluripotent stem cells (iPSCs) present several important advantages over other in vitro systems. The derived cells are human yet do not have the accompanying ethical issues surrounding human ES cells. Furthermore, given that none of the genes implicated in ASD are fully penetrant for the condition, the influence of genetic background is problematic for approaches to modelling ASD in vitro where mutations are engineered into pre-existing pluripotent human cell lines. Patient-specific iPSCs represent a powerful new approach to capture both the primary genetic lesion and the genetic background from human individuals with confirmed diagnoses of ASD. Furthermore, it facilitates the study of larger deletions in complex genes containing internal promoters or polygenic cases which are currently difficult to engineer using genome editing technologies.
In addition to providing a methodological perspective on the iPSC technology, this review will seek to identify where patient-specific iPSCs can be expected to complement animal models in identifying disease mechanisms in ASD as part of the early stages of the drug discovery process. It is clear however that this technology is not limited to this stage of the drug discovery process, and iPSCs and their differentiated derivatives are increasingly being used in the development of assays for the screening of compound libraries to identify disease modifying drugs (Yang et al. 2013) and the development of more sophisticated in vitro toxicology assays (Anson et al. 2011). It has even been suggested that iPSCs could be generated from participants of clinical trials to interrogate how individual genetic variation affects drug responses and conceivably also to test new compounds in ‘in vitro clinical trials’ (Eglen and Reisine 2011).
Technical issues in the generation of iPSC lines
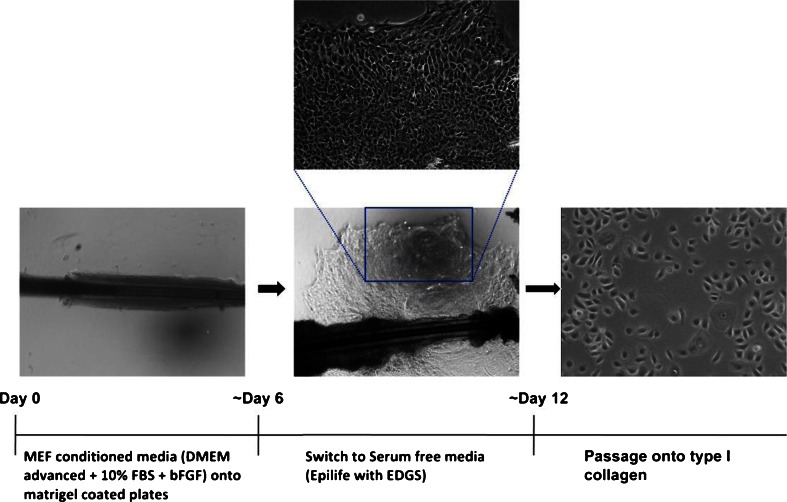
The initial approach to generating iPSCs used fibroblasts as the starting material (Takahashi and Yamanaka 2006; Takahashi et al. 2007). This cell type is accessible by skin biopsy but is invasive and may be considered unsuitable for young children. iPSCs have also been successfully derived from other accessible tissue types including blood and hair keratinocytes (Ye et al. 2009; Aasen et al. 2008). However, although obtaining blood is a routine procedure, it can be difficult to perform on severely autistic children without distress. A promising alternative cell type is keratinocytes derived from the root shaft of scalp hair (Aasen et al. 2008; Aasen and Izpisúa Belmonte 2010). This is a relatively non-invasive procedure and sufficient keratinocytes can be generated from a single hair to generate iPS cell lines (Fig. 1.). Furthermore, we have found that it is possible to maintain viable hair samples for at least 48 h at ambient temperatures in media containing 10 % foetal bovine serum buffered with 15 mM HEPES before plating. This allows for considerable flexibility in sample acquisition and transport.
Fig. 1.
Derivation of keratinocytes from scalp hair. The root shaft of each hair was plated onto matrigel-coated dishes, a presoaked coverslip placed over the top of the dissected hair root, and mouse embryonic fibroblast-conditioned media with 50 μg/ml gentamicin was added to each well. Once the keratinocytes had begun to migrate from the hair shaft onto the matrigel substrate, the media was switched to Epilife with EDGS supplement (Life Technologies), and the gentamicin was removed. For subsequent passaging, keratinocytes were pretreated for 1 h with Y-27632 (10 μM) before being dissociated with Accutase and plating onto type I collagen-coated plates in keratinocyte media (with Y27632 (10 μM) for the first 24 h)
The first group to derive iPSCs from keratinocytes reported that the kinetics and efficiency of reprogramming were substantially increased over that of fibroblasts (Aasen et al. 2008). In our study, switching keratinocytes to ES media 2 or 3 days after transduction with the reprogramming vector gives a very low efficiency of reprogramming (<0.001 %), possibly due to elevated calcium-driving terminal differentiation of the keratinocytes before sufficient reprogramming has occurred (Hennings et al. 1980). We have adopted a protocol similar to that of Zhu et al. (2010) in which keratinocytes, after being transduced with the reprogramming vector, are maintained for an extended period in low calcium serum-free keratinocyte growth media before being adapted to hES media. Using this approach, we observe a similar efficiency of reprogramming to fibroblasts (∼0.05 %), but with slightly faster kinetics (colonies are ready to transfer into feeder-free conditions after 21 days). The lower reprogramming efficiency we observe in keratinocytes compared to the original report (Aasen et al. 2008) has also been observed by others (Carey et al. 2009) and could be due to a number of factors including the titre of virus used and the nature of the reprogramming construct. Rather than individual retroviral vectors for the four reprogramming factors, we use a polycistronic lentiviral construct flanked with loxp sites at an MOI of two to increase the likelihood of deriving lines with single integrands, thus allowing easier removal of the vector, and decreasing the risk of insertional mutagenesis (Papapetrou and Sadelain 2011). However, it should be noted that the iPS field is increasingly moving towards the use of non-integrating systems such as Sendai virus (Nishimura et al. 2011), episomal (Yu et al. 2009) and modified RNA vectors (Warren et al. 2010).
Validation of iPSC pluripotency and quality control
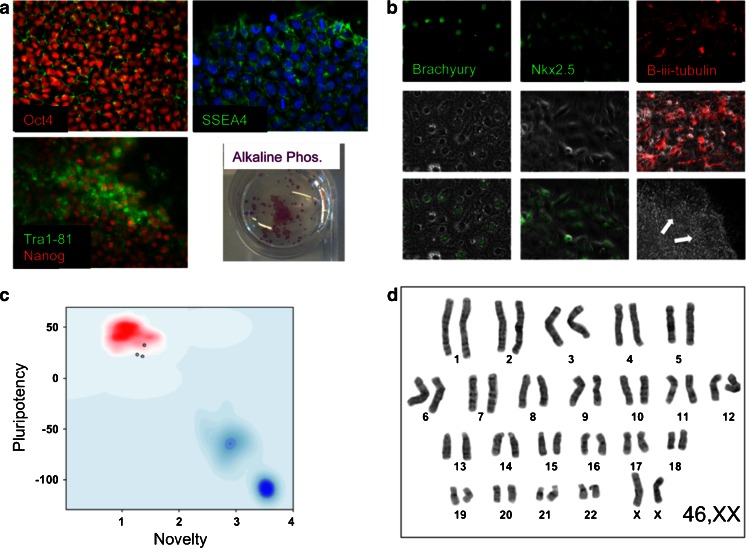
There is no universally agreed set of criteria in the field for demonstration of pluripotency in hiPSCs, although attempts have begun to work towards such a goal (Maherali and Hochedlinger 2008; Daley et al. 2009). Nevertheless, various assays to assess pluripotency have been developed and are essential for the quality control of newly derived iPS lines. These include immunocytochemical and quantitative PCR (qPCR) analysis of markers either associated with or necessary for the maintenance of the pluripotent state. We routinely assess lines for Oct4, Nanog, Tra1-81 and SSEA4 (Fig. 2a). The expression of alkaline phosphatase is also associated with pluripotency (Andrews et al. 1984) and is assessed by a colorimetric assay of enzymatic activity on lightly fixed cells (Fig. 2a (iv)).
Fig. 2.
Representative images for the validation of iPSC pluripotency and quality control. a Undifferentiated iPSCs were probed with primary antibodies to Oct4, Nanog, SSEA4 and Tra1-81 and assessed for alkaline phosphatase enzymatic activity. b To assess the capacity of the iPS lines to spontaneously differentiate into the three embryonic lineages, EBs were generated in vitro for 2 days and then plated onto gelatin-coated plates. The cultures were grown for a further 12 days before fixing and staining for mesodermal, endodermal and neuroectodermal markers Brachyury, Nkx2.5 and β-iii-tubulin, respectively. c For the assessment of pluripotency using ‘PluriTest’, total RNA was extracted from iPS cells and genome wide gene expression analysis was performed on Illumina HT12v4 beadchips at the Genome Centre, Queen Mary, University of London. For Pluritest analysis, intensity data files were uploaded at http://www.pluritest.org and analysis performed according to Muller et al. (2011). d Karyotypes were generated from 70–80 % confluent cells treated with 100 ng/ml colcemid (Life Technologies) for 3 h and subjected to hypotonic lysis in 0.075 M potassium chloride for 10 min at 37 °C. Samples were then fixed in methanol/glacial acetic acid (ratio 3:1) and stained with Giemsa on glass slides for analysis. Twenty metaphase cells were counted for each line and four cells analysed in detail
The teratoma assay has generally been considered the gold standard for demonstrating pluripotency in human ES or iPSCs (Maherali and Hochedlinger 2008; Daley et al. 2009). However, a number of serious shortcomings have been identified with this assay including an absence of agreed objective minimal criteria for assessing and reporting teratomas (Müller et al. 2010) and the reported failure of this assay to robustly distinguish between partially and more fully reprogrammed iPSCs (Chan et al. 2009). Alternative methods that are more quantitative and higher throughput are being developed, such as the array based bioinformatics approach developed by the Loring lab (Müller et al. 2011). We use this method (Fig. 2b) complemented by an in vitro assessment of spontaneous differentiation into the three germ layers from embryoid bodies (Fig. 2c).
In addition to pluripotency, it is also essential to demonstrate genomic stability in the iPS line. This is routinely performed by generating a G-banded karyotype (Fig. 2d). Although a more detailed appraisal of structural abnormalities within chromosomes can be attained by the use of comparative genomic hybridization arrays, this approach cannot completely substitute for karyotyping as the former will not detect balanced translocations or inversions.
We also routinely assess each new iPS line for the capacity to fully neutralise in adherent culture to ensure consistency in down-stream assays. The addition of SMAD inhibitors to iPSCs plated at high density in a neural maintenance media should form a uniform layer of Pax6 + ve neuroepithelial cells (Fig. 3a) (Chambers et al. 2009; Shi et al. 2012). Upon passaging these neuroepithelial cells, a uniform field of neural rosettes should form (Fig. 3b). In the absence of additional patterning factors, these rosettes should then generate neuronal subtypes of the dorsal telencephalon (Fig. 3c).
Fig. 3.
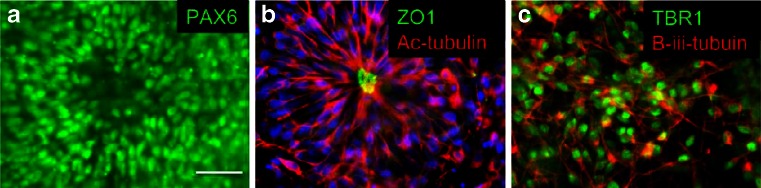
Neuralisation of iPSCs was differentiated at high density in monolayer culture in the presence of SMAD inhibitors. a Uniform PAX6 staining of early neuroepithelial cells. b Rosette formation by neural progenitors after passaging of neuroepithelial cells. The apical lumen of the rosette is stained with the tight junction protein ZO-1, whilst acetylated tubulin forms a radial structure from the lumen outwards. c Early passaging and differentiation of these neural progenitors with the NOTCH inhibitor DAPT give rise to large numbers of TBR1/β-iii-tubulin + ve putative layer VI cortical neurons
Selection of ASD subgroups for the generation of iPSCs and anticipated cellular phenotypes
Just as with animal models, capturing the heterogeneity of disorders such as autism in cellular models presents a challenge. There are several aspects to heterogeneity: clinical, genetic and phenotypic. From an iPS standpoint, the genetic heterogeneity is the most readily addressed and will probably be the starting point for analysis in a number of laboratories. Research groups will gravitate towards specific genetic lesions, hoping that the phenotypes that emerge will have general relevance, but the genetic heterogeneity of ASD and its varying severity between individuals will ultimately require the derivation and comparison of patient specific iPSCs from large numbers of individuals.
In our own studies, we have chosen in the first instance to focus on a subset of ASD individuals at the extreme end of the spectrum who have a clear genetic lesion in a gene that has a strong association with the disorder. This limits the degree to which it may be possible to make more general comments about ASD based on any initial findings but should increase the signal-to-noise ratio that will undoubtedly be problematic in looking for phenotypes in the wider ASD population. However, any phenotype observed can then be investigated in additional ASD subgroups.
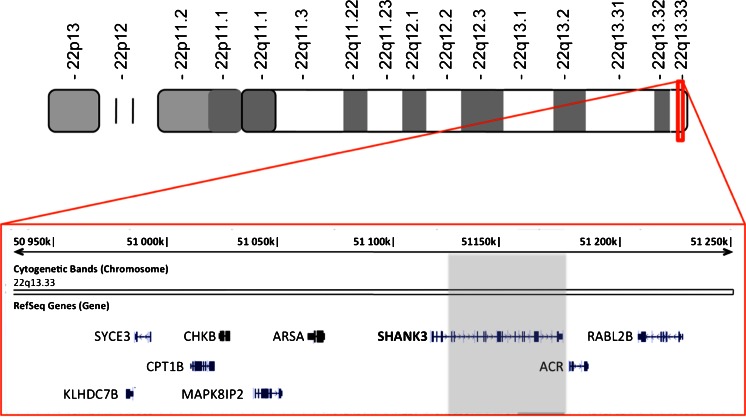
We have generated iPSCs from individuals with deletions in the gene SHANK3 who have been diagnosed with ASD. This includes an individual with an interstitial deletion extending from the third intron of SHANK3 to beyond the end of the gene (Fig. 4).
Fig. 4.
Diagram of the extent of the genetic lesion in the gene SHANK3 in an autistic individual. A 4-year-old boy with absent speech, developmental delay and a confirmed diagnosis of autism was found to have a deletion of the gene SHANK3 on chromosome 22q by comparative genomic hybridization array. The deletion extends from the third intron of SHANK3 to beyond the end of the gene
This deletion encompasses all of the major protein interaction domains of the gene product and leaves the individual effectively hemizygous for SHANK3. Deletions in SHANK3 are strongly associated with ASD (Jeffries et al. 2005; Durand et al. 2007) and have been shown to be critical in the neurological disorder Phelan–McDermid syndrome which itself has a strong association with ASD (>50 %) (Bonaglia et al. 2001; Phelan and McDermid 2012). SHANK3 plays a central role in the post-synaptic density (PSD) of glutamatergic synapses acting as a scaffolding protein for receptors and the actin cytoskeleton (Verpelli et al. 2012). Within the PSD, SHANK3 forms a tetramer with Homer which in turn generates a higher order lattice structure that links to the actin cytoskeleton and plays an important role in stabilizing the structure of the dendritic spine (Hayashi et al. 2009). Studies in which SHANK3 gene dosage have been reduced in primary neuronal cultures by siRNA have shown reductions in dendritic spine density (Roussignol et al. 2005; Verpelli et al. 2011). Conversely, overexpression of SHANK3 in aspiny cerebellar neurons was shown to be sufficient to induce dendritic spine formation (Roussignol et al. 2005). Overexpression of SHANK3 constructs containing truncating mutations found in individuals with ASD has also been found to have a dominant-negative effect on spine induction and maturation in primary neuronal cultures (Durand et al. 2012). In addition to effects on the morphology and number of spines, more subtle effects have been observed on the functional connectivity of dendritic spines where SHANK3 gene-dosage is reduced. Most strikingly perhaps is a specific reduction reported in mGluR5 and in activity-dependent long-term depression (Verpelli et al. 2011). These insights gained into the role of SHANK3 in synapse formation and stability have come in the most part from studies in primary rodent cells. Such studies could undoubtedly be undertaken in human iPSCs, but in addition to providing a human context for such work, iPSCs will also allow for the study of developmental aspects of neurobiology that are not possible in terminally differentiated primary neurons and are impractical in animal models. For example, it is not clear that a simple reduction in gene dosage of the full length transcript is responsible in all cases of ASD associated with mutations in SHANK3. The SHANK3 gene has a complex transcriptional profile with both alternatively spliced exons and intragenic promoters (Maunakea et al. 2010; Wilson et al. 2003). The heterogeneity in the clinical presentation of individuals with mutations and microdeletions in different regions of the SHANK3 gene is possibly due to differential effects of isoform dosage. Indeed, variable phenotypes are observed in the five different lines of mice that have been reported, carrying different truncating mutations in the SHANK3 gene as models of ASD (Jiang and Ehlers 2013, for review). Clarifying the role of so many isoforms in neuronal development, synapse formation, stabilization and plasticity within the context of animal models of ASD will clearly be a formidable task. The capacity to generate differentiated human neurons from iPSCs following a default developmental trajectory offers a much more tractable approach to examining the role of SHANK3 isoforms in neuronal development, synaptogenesis and plasticity. The most relevant findings in vitro can then be examined in animal models to determine the behavioural consequences and responses to pharmacological interventions based on those isoform differences.
Selection of cell types for analysis
Our current understanding of the function of many of the genes associated with autism directs us towards the glutamatergic synapse (Ebert and Greenberg 2013), but of course, there is no guarantee that the ‘autistic phenotype’ entirely resides there. Many of the ‘synaptic’ genes that are implicated in ASD have broad expression patterns. SHANK proteins, for example, are expressed in the thymus and gut endothelium, as well as the brain (Redecker et al. 2006; Huett et al. 2009). It may be no coincidence that autistic patients typically have gastrointestinal comorbidities and that the disorder is thought by many to have an immunological component (Buie et al. 2010; McAllister and Patterson 2012). Even if we accept that within the brain we are indeed looking for a glutamatergic synaptic phenotype, the question arises—which neuronal subtypes are most affected and from which regions of the brain? A relatively consistent finding in ASD neuropathology is reduced minicolumn spacing and neuronal density in the prefrontal cortex (Casanova et al. 2002; Buxhoeveden et al. 2006; Casanova et al. 2010). A number of robust protocols exist for the differentiation of human iPSCs and ES cells into rostrodorsal cortical neurons either from embryoid bodies (Pankratz et al. 2007) or in adherent culture using SMAD pathway inhibitors (Chambers et al. 2009; Shi et al. 2012). Interestingly, the sequential production of infragranular to supergranular cortical neuronal subtypes is recapitulated in these in vitro systems (Eiraku et al. 2008; Shi et al. 2012).
Another of the more consistent findings reported in post-mortem studies of ASD brains is cell loss in the Purkinje layer of the cerebellum (Ritvo et al. 1986; Bailey et al. 1998; Palmen et al. 2004; Whitney et al. 2008). However, it is also clear from these studies that this is not an invariable feature of autism and it is yet unclear if this represents etiologically distinct subgroups of ASD. The effects on Purkinje neurons of knocking-out SHANK3 in animal models of ASD are unlikely to be informative in this respect as it have been shown that, although in the mouse cerebellum, the expression of SHANK3 is highly expressed in the granule cell layer it is barely detectable in the Purkinje cell layer (Verpelli et al. 2012). Interestingly, however, we have found that the expression pattern of SHANK3 in human brain displays the opposite pattern such that the expression is high in Purkinje cells and undetectable in the granule cell layer (Fig. 5).
Fig. 5.

SHANK3 staining in normal human cerebellum 7 μM paraffin- embedded sections from human cerebellum were probed with a SHANK3 antibody (Sigma-Aldrich, UK), visualized with 3,3′-diaminobenzidine tetrahydrochloride and counterstained with Harris’s hematoxylin
This is a clear example of where animal models could lead to misleading findings, and a human neuronal model would prove valuable. Indeed, a striking overlap in the synaptic pathophysiology in Purkinje cells observed between syndromic (Fragile X) and non-syndromic (Neuroligin-3 knock-out) mouse models of ASD is unlikely to be recapitulated in a SHANK3 knock-out mouse model for this reason (Baudouin et al. 2012). However, although at present, there are reports of deriving both cerebellar granule and Purkinje cell neurons from mouse ES cells (Su et al. 2006; Muguruma et al. 2010), it is unknown if similar induction protocols will efficiently generate these neuronal subtypes from human iPSCs. Indeed, some other potential sites for autism pathology have not yet been addressed in iPSCs, neurons of the amygdala, for example (Baron-Cohen 2000), but there seems no reason in principle why such protocols could not be developed.
A more fundamental limitation might lie around the issue of cell autonomous versus system phenotypes. We have no right at the moment to assume that autism has a simple cellular phenotype. The clinical features of autism—social behaviour, language and repetitive behaviours—do not reside in a single cell type. They are emergent properties of brain systems. Even the anatomical features of the disease will not necessarily be reducible to a simple cellular phenotype. Alterations in brain volume, for example, have been documented in a number of imaging and post-mortem studies of ASD. There has been a consistent finding of brain overgrowth in ASD beginning around the age of 1 year, levelling off by 3 years, and possibly followed by a decline in later life (Redcay and Courchesne 2005). Interestingly, however, this early overgrowth is not uniform across the brain and is most apparent in the frontal cortex, amygdala and cerebellum (Carper et al. 2002; Sparks et al. 2002; Courchesne et al. 2001). The neuropathological findings that are evident upon closer cytological examination in these regions could conceivably be reducible to cellular phenotypes, but not necessarily so.
However, of the studies published so far using iPSCs to discover cellular phenotypes associated with psychiatric disease, the data are encouraging. For example, Brennand et al. (2011) report alterations in connectivity associated with neurons derived from schizophrenia. More relevant to ASD, Pasca et al. (2011) described a phenotype associated with Timothy syndrome (TS), a syndromic form of autism, linked to mutations in the CACNA1C gene. Neurons derived from iPSCs carrying this mutation predictably have a calcium channel deficit, but more interestingly, they also have a histogenic deficit; they generate fewer infra-granular callosal neurons expressing SATB2 and a concomitant increase in CTIP2 expressing neurons. Interestingly, the authors also report that their findings were only partly validated in a TS transgenic mouse model, such that infragranular SATB2 neurons were reduced but numbers of CTIP2 expressing neurons were unchanged. They note that this could reflect an in vitro differentiation artefact or more interestingly could represent a species difference between mouse and human.
Apart from the insight these results provide into Timothy syndrome, it suggests further that phenotypes associated with more complex ‘system’ properties of the brain might be accessible to study in iPSCs. Further encouragement in this direction is provided by the remarkable capacity of embryonic and induced pluripotent stem cells to generate in vitro highly ordered three-dimensional tissue structures driven by largely intrinsic processes (Sasai 2013). Eiraku et al. have demonstrated in a manner consistent with the default model of neural induction that in embryoid bodies (EBs) of a uniform size and in a minimal media without inhibitors or instructive morphogens, a multilayered cortical-like structure is formed (Eiraku et al. 2008; Nasu et al. 2012). These structures progress through an initial neuroepithelial-like stage with a layer of Pax6 + ve cells displaying apico-basal polarity, and sequentially generate neural progenitor cells such as Tbr2 + ve/Pax6-ve transient amplifying cells, followed by Cajal–Rezius cells and the deep-layer 6 and 5 cortical neurons and finally smaller numbers of more superficial layer neurons. However, it is important to note that, although clearly distinguishable layers are reported to be formed, the usual inside–out pattern of cortical layers is not established in these structures and more superficial layer neurons are under-represented. Despite these current shortcomings, it is clear that the potential exists to not only study molecular interactions and cell fate in iPSCs but that it will become increasingly possible to study four-dimensional histogenesis in this system in a manner that could inform and complement developmental studies in vivo.
In summary, we envisage that in vitro models derived from iPSCs will complement rather than replace animal models of ASD and other psychiatric disorders in the future. It is anticipated that any disease-associated phenotypes observed in iPSC-derived assays of ASD will serve to inform experiments in animal models which will continue to be indispensible in establishing the crucial link between cellular and molecular events and behaviour. Although it remains to be seen whether the iPSC approach will reveal details of the aetiology of ASD beyond those that emerge from animal models of this disorder, the relative simplicity and tractability of this approach are appealing, while the fact that it is human and truly disease-related give iPSCs a face validity that has been missing from other cellular models. The approach we have outlined here in relation to our own studies of SHANK3 can be, and are being, reproduced in many laboratories with different genetic and sporadic sources of cells. This first generation of iPSC projects will undoubtedly have their limitations but may well begin to cast light on aspects of disease aetiology as yet undiscovered.
Acknowledgments
We acknowledge the financial support of the agency for Innovation by Science and Technology (IWT), Ellipsgebouw, Brussels (Belgium). IWT O&O project nr: IWT 090574, project title: the generation of human iPS cells for drug discovery iPS.
We acknowledge the financial support of the National Institute for Health Research (NIHR) Biomedical Research Centre for Mental Health at South London and Maudsley NHS Foundation Trust and Institute of Psychiatry, King’s College London. This article presents independent research funded by the NIHR. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115300, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies’ in kind contribution.
We would like to thank Dr. Claire Troakes of the MRC Neurodegenerative Diseases Brain Bank at the Institute of Psychiatry, King’s College London for providing the human brain sections.
Abbreviations
- ASD
Autistic spectrum disorder
- iPSCs
Induced pluripotent stem cells
- hES
Human embryonic stem cells
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilić J, Pekarik V, Tiscornia G, Edel M, Boué S, Izpisúa Belmonte JC. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26(11):1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Aasen T, Izpisúa Belmonte JC. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat Protoc. 2010;5(2):371–382. doi: 10.1038/nprot.2009.241. [DOI] [PubMed] [Google Scholar]
- Andrews PW, Meyer LJ, Bednarz KL, Harris H. Two monoclonal antibodies recognizing determinants on human embryonal carcinoma cells react specifically with the liver isozyme of human alkaline phosphatase. Hybridoma. 1984;3(1):33–39. doi: 10.1089/hyb.1984.3.33. [DOI] [PubMed] [Google Scholar]
- Anson BD, Kolaja KL, Kamp TJ. Opportunities for use of human iPS cells in predictive toxicology. Clin Pharmacol Ther. 2011;89(5):754–758. doi: 10.1038/clpt.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, Luthert P, Dean A, Harding B, Janota I, Montgomery M, Rutter M, Lantos P. A clinicopathological study of autism. Brain. 1998;121(Pt 5):889–905. doi: 10.1093/brain/121.5.889. [DOI] [PubMed] [Google Scholar]
- Baron-Cohen (2000) The amygdala theory of autism. Neurosci Biobehav Rev 1–10 [DOI] [PubMed]
- Baudouin SJ, Gaudias J, Gerharz S, Hatstatt L, Zhou K, Punnakkal P, Tanaka KF, Spooren W, Hen R, De Zeeuw CI, Vogt K, Scheiffele P. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science. 2012;338(6103):128–132. doi: 10.1126/science.1224159. [DOI] [PubMed] [Google Scholar]
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Borgatti R, Felisari G, Gagliardi C, Selicorni A, Zuffardi O. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am J Hum Genet. 2001;69(2):261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S et al (2011) Modelling schizophrenia using human induced pluripotent stem cells. Nature 1–7 [DOI] [PMC free article] [PubMed]
- Buie, T., Campbell, D. B., Fuchs, G. J., Furuta, G. T., Levy, J., VandeWater, J., et al. (2010). Evaluation, Diagnosis, and Treatment of Gastrointestinal Disorders in Individuals With ASDs: A Consensus Report. PEDIATRICS, 125(Supplement), S1 S18. doi:10.1542/peds.2009-1878C [DOI] [PubMed]
- Buxhoeveden DP, Semendeferi K, Buckwalter J, Schenker N, Switzer R, Courchesne E. Reduced minicolumns in the frontal cortex of patients with autism. Neuropathol Appl Neurobiol. 2006;32(5):483–491. doi: 10.1111/j.1365-2990.2006.00745.x. [DOI] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A. 2009;106(1):157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carper RA, Moses P, Tigue ZD, Courchesne E. Cerebral lobes in autism: early hyperplasia and abnormal age effects. NeuroImage. 2002;16(4):1038–1051. doi: 10.1006/nimg.2002.1099. [DOI] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Switala AE, Roy E. Minicolumnar pathology in autism. Neurology. 2002;58(3):428–432. doi: 10.1212/WNL.58.3.428. [DOI] [PubMed] [Google Scholar]
- Casanova MF, El-Baz A, Vanbogaert E, Narahari P, Switala A. A topographic study of minicolumnar core width by lamina comparison between autistic subjects and controls: possible minicolumnar disruption due to an anatomical element in-common to multiple laminae. Brain Pathol. 2010;20(2):451–458. doi: 10.1111/j.1750-3639.2009.00319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, Daley GQ, Schlaeger TM. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27(11):1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Karns CM, Davis HR, Ziccardi R, Carper RA, Tigue ZD, Chisum HJ, Moses P, Pierce K, Lord C, Lincoln AJ, Pizzo S, Schreibman L, Haas RH, Akshoomoff NA, Courchesne RY. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57(2):245–254. doi: 10.1212/WNL.57.2.245. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Translational animal models of autism and neurodevelopmental disorders. Dialogues Clin Neurosci. 2012;14(3):293–305. doi: 10.31887/DCNS.2012.14.3/jcrawley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Lensch MW, Jaenisch R, Meissner A, Plath K, Yamanaka S. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell. 2009;4(3):200–201. doi: 10.1016/j.stem.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22(3):229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Rogé B, Héron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39(1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2012;17(1):71–84. doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eglen R, Reisine T. Primary cells and stem cells in drug discovery: emerging tools for high-throughput screening. Assay Drug Dev Technol. 2011;9(2):108–124. doi: 10.1089/adt.2010.0305. [DOI] [PubMed] [Google Scholar]
- Ebert DH, Greenberg ME. Activity-dependent neuronal signalling and autism spectrum disorder. Nature. 2013;493(7432):327–337. doi: 10.1038/nature11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3(5):519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Hampson DR, Gholizadeh S, Pacey LK. Pathways to drug development for autism spectrum disorders. Clin Pharmacol Ther. 2012;91(2):189–200. doi: 10.1038/clpt.2011.245. [DOI] [PubMed] [Google Scholar]
- Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu RM, Li H, Sala C, Hayashi Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137(1):159–171. doi: 10.1016/j.cell.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennings H, Michael D, Cheng C, Steinert P, Holbrook K, Yuspa SH. Calcium regulation of growth and differentiation of mouse epidermal cells in culture. Cell. 1980;19(1):245–254. doi: 10.1016/0092-8674(80)90406-7. [DOI] [PubMed] [Google Scholar]
- Huett A, Leong JM, Podolsky DK, Xavier RJ. The cytoskeletal scaffold Shank3 is recruited to pathogen-induced actin rearrangements. Exp Cell Res. 2009;315(12):2001–2011. doi: 10.1016/j.yexcr.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries AR, Curran S, Elmslie F, Sharma A, Wenger S, Hummel M, Powell J. Molecular and phenotypic characterization of ring chromosome 22. Am J Med Genet A. 2005;137(2):139–147. doi: 10.1002/ajmg.a.30780. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Ehlers MD. Modelling autism by SHANK gene mutations in mice. Neuron. 2013;78(1):8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu Rev Med. 2011;62:411–429. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, Schopler E. Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord. 1989;19(2):185–212. doi: 10.1007/BF02211841. [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24(5):659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;3(6):595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Gage FH. Modelling brain disease in a dish: really? Cell Stem Cell. 2012;10(6):642–645. doi: 10.1016/j.stem.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Markou A, Chiamulera C, Geyer MA, Tricklebank M, Steckler T. Removing obstacles in neuroscience drug discovery: the future path for animal models. Neuropsychopharmacology. 2009;34(1):74–89. doi: 10.1038/npp.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK, Patterson PH. Introduction to special issue on neuroimmunology in brain development and disease. Dev Neurobiol. 2012;72(10):1269–1271. doi: 10.1002/dneu.22054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muguruma K, Nishiyama A, Ono Y, Miyawaki H, Mizuhara E, Hori S, Kakizuka A, Obata K, Yanagawa Y, Hirano T, Sasai Y. Ontogeny-recapitulating generation and tissue integration of ES cell-derived Purkinje cells. Nat Neurosci. 2010;13(10):1171–1180. doi: 10.1038/nn.2638. [DOI] [PubMed] [Google Scholar]
- Müller FJ, Goldmann J, Löser P, Loring JF. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell. 2010;6(5):412–414. doi: 10.1016/j.stem.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Müller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, Danner S, Goldmann JE, Herbst A, Schmidt NO, Aldenhoff JB, Laurent LC, Loring JF. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8(4):315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasu M, Takata N, Danjo T, Sakaguchi H, Kadoshima T, Futaki S, Sekiguchi K, Eiraku M, Sasai Y. Robust formation and maintenance of continuous stratified cortical neuroepithelium by laminin-containing matrix in mouse ES cell culture. PLoS One. 2012;7(12):e53024. doi: 10.1371/journal.pone.0053024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale S. Autism spectrum disorders. Nat Rev Drug Discov. 2012;11(10):745–746. doi: 10.1038/nrd3771. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Sano M, Ohtaka M, Furuta B, Umemura Y, Nakajima Y, et al. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J Biol Chem. 2011;286(6):4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmen SJ, van Engeland H, Hof PR, Schmitz C. Neuropathological findings in autism. Brain. 2004;127(Pt 12):2572–2583. doi: 10.1093/brain/awh287. [DOI] [PubMed] [Google Scholar]
- Pankratz MT, Li XJ, Lavaute TM, Lyons EA, Chen X, Zhang SC. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25(6):1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou EP, Sadelain M. Generation of transgene-free human induced pluripotent stem cells with an excisable single polycistronic vector. Nat Protoc. 2011;6(9):1251–1273. doi: 10.1038/nprot.2011.374. [DOI] [PubMed] [Google Scholar]
- Paşca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Paşca AM, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Publ Group. 2011;17(12):1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K, McDermid HE (2012) The 22q13.3 deletion syndrome (Phelan–McDermid syndrome). Mol Syndromol 2(3–5):186–201 [DOI] [PMC free article] [PubMed]
- Redcay E, Courchesne E. When is the brain enlarged in autism? A meta-analysis of all brain size reports. Biol Psychiatry. 2005;58(1):1–9. doi: 10.1016/j.biopsych.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Redecker P, Bockmann J, Böckers TM. Expression of postsynaptic density proteins of the ProSAP/Shank family in the thymus. Histochem Cell Biol. 2006;126(6):679–685. doi: 10.1007/s00418-006-0199-9. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Scheibel AB, Duong T, Robinson H, Guthrie D, Ritvo A. Lower Purkinje cell counts in the cerebella of four autistic subjects: initial findings of the UCLA-NSAC Autopsy Research Report. Am J Psychiatry. 1986;143(7):862–866. doi: 10.1176/ajp.143.7.862. [DOI] [PubMed] [Google Scholar]
- Roullet FI, Lai JK, Foster JA. In utero exposure to valproic acid and autism - A current review of clinical and animal studies. Neurotoxicol Teratol. 2013 Feb 8 [Epub ahead of print] [DOI] [PubMed]
- Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, Bockaert J, Fagni L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005;25(14):3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Cytosystems dynamics in self-organization of tissue architecture. Nature. 2013;493(7432):318–326. doi: 10.1038/nature11859. [DOI] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15(3):477–486. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks BF, Friedman SD, Shaw DW, Aylward EH, Echelard D, Artru AA, Maravilla KR, Giedd JN, Munson J, Dawson G, Dager SR. Brain structural abnormalities in young children with autism spectrum disorder. Neurology. 2002;59(2):184–192. doi: 10.1212/WNL.59.2.184. [DOI] [PubMed] [Google Scholar]
- St John JC, Armstrong L, Minger SL, Campbell KH. Law should recognize value of interspecies embryos. Nature. 2008;451(7179):627. doi: 10.1038/451627a. [DOI] [PubMed] [Google Scholar]
- Su HL, Muguruma K, Matsuo-Takasaki M, Kengaku M, Watanabe K, Sasai Y. Generation of cerebellar neuron precursors from embryonic stem cells. Dev Biol. 2006;290(2):287–296. doi: 10.1016/j.ydbio.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O’Collins V, Macleod MR. Can animal models of disease reliably inform human studies? PLoS Med. 2010;30:7(3). doi: 10.1371/journal.pmed.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpelli C, Schmeisser MJ, Sala C, Boeckers TM. Scaffold proteins at the postsynaptic density. Adv Exp Med Biol. 2012;970:29–61. doi: 10.1007/978-3-7091-0932-8_2. [DOI] [PubMed] [Google Scholar]
- Verpelli C, Dvoretskova E, Vicidomini C, Rossi F, Chiappalone M, Schoen M, Di Stefano B, Mantegazza R, Broccoli V, Böckers TM, Dityatev A, Sala C. Importance of Shank3 protein in regulating metabotropic glutamate receptor 5 (mGluR5) expression and signaling at synapses. J Biol Chem. 2011;286(40):34839–34850. doi: 10.1074/jbc.M111.258384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ER, Kemper TL, Bauman ML, Rosene DL, Blatt GJ. Cerebellar Purkinje cells are reduced in a subpopulation of autistic brains: a stereological experiment using calbindin-D28k. Cerebellum. 2008;7(3):406–416. doi: 10.1007/s12311-008-0043-y. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40(8):575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YM. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell. 2013;S1934-5909(13):00139–2. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang YY, Dang CV, Spivak JL, Moliterno AR, Cheng L. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114(27):5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324(5928):797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7(6):651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]