Abstract
One of the most important pathways which are frequently affected in colorectal cancer is p53/ (MDM2)/p14ARF pathway. We aim to determine the methylation pattern of p14/ARF in relation to mutation of p53. This correlation was studied to investigate whether their alterations could be considered as a predictor factor of prognosis in colorectal cancer and whether it can be useful in early-stage diagnosis. Statistical analyses show that p14/ARF hypermethylation was correlated with rectum location (p = 0.004), primary TNM stage (p = 0.016), and advanced Astler–Coller stage (p = 0.024). The RT-PCR that revel 31 % of patients did not express p14/ARF mRNA or at very low level. A high concordance between CpG hypermethylation and the low levels (p < 0.005) was shown. In addition, our analyses demonstrate that patients with mutation in the p53 gene have a lack of the protein expression (p < 0.005). This category with negative expression of p53 had a shorter survival rate (p < 0.005). On the one hand, MSP pattern of p14/ARF were correlated with a lack of p53 expression (p = 0.007). We found that p53/p14ARF pathway was frequently deregulated among our patients. In our study, we demonstrate that hypermethylation of p14/ARF occurs early during CRC tumorogenesis. However, we did not find correlation between p14/ARF and survival. These results suggest that p14/ARF methylation pattern may constitute a predictor factor of CRC in early stage but it could not be considered as a prognostic factor. On the other hand and because of the reversibility of the methylation mechanism, it may be appropriate to target the demethylation of p14/ARF to develop new drogues for CRC.
Keywords: Colorectal cancer, p14/ARF methylation, p53 alteration, Prognostic factor, Tunisian population
Introduction
Extensive molecular analyses have revealed that colorectal carcinogenesis is characterized by a multistep process of genetic and epigenetic alterations. P53/MDM2/p14ARF pathway is usually affected in colorectal carcinogenesis. Indeed, these proteins are actively involved in the apoptosis which represents a principal physiological control mechanism. Any alteration affecting one of these molecules could lead to abnormal cell survival and will start the carcinogenesis process. The p53 is a key regulator of cell cycle checkpoints; it plays an important role inducing cell death after DNA damage or under conditions of cellular stress [1].
The prevalence of p53 mutations in colorectal cancer is highly variable among different series and may be estimated from 40 to 60 % of patients [2, 3]. Its expression is maintained at a very low level in the normal cells [4]. However, it has been demonstrated that mutations in p53 gene increase the half life of the protein which is associated with overexpression in the nucleus [5]. Furthermore, the main cellular function of the MDM2 oncoprotein is to control the level of p53 through an autoregulatory feedback loop. In cancers, MDM2 overexpression deregulates this feedback, and the interaction between MDM2 and p53 is blocked [6]. Recently, the p14/ARF protein has been investigated acting as intermediate in the MDM2/P53 pathway regulation.
This protein has been also identified as a tumor suppressor gene promoting the rapid degradation of MDM2 and leading to p53 stabilization and its nuclear accumulation [6, 7]. In fact, p14/ARF bounds and blocks MDM2 to inhibit the nucleocytoplasmic shuttling of p53 and induces its nuclear retention, production, and activation [8–10]. Furthermore, it acts upstream of p53 and answers to a negative feedback regulation, which suggests that p53 mutations or its inactivation by MDM2 amplification are often accompanied by overexpression of p14/ARF [11]. P53-positive tumors are also likely to have sustained epistatic mutations such as MDM2 amplification or p14/ARF loss or inactivation [12]. Nuclear import and export is a feature of both p53 and MDM2, such that nuclear p53 absence is associated with tumors with a poor prognosis [13]. Many analyzes suggest that p14/ARF influences the subcellular localization of MDM2 [14]. Consequently, the localization of these proteins and the relationship between their levels of expression are likely to be important in many carcinogenesis. Previous studies have examined p14/ARF mRNA expression in breast cancers, with evidence suggesting altered expression and an association with p53 [15, 16].
Moreover, the literature describes other process by which p14/ARF gene can be inactivated in many cancers such as deletion, promoter hypermethylation, or mutations [17]. In colorectal cancer (CRC), the p14/ARF inactivation was proved to be the result of promoter hypermethylation [18]. This last seems to be rich in CpG dinucleotides methylation of the cytosine residues at the CpG islands; this region plays an important role in the inactivation of gene expression. The transcription of p14/ARF can be deregulated by the hypermethylation [18]. Transcriptional silencing of the p14/ARF gene through CpG hypermethylation of the DNA promoter is an important event in the genetic regulation of cancers and would be associated with its carcinogenesis process [18]. This epigenetic mechanism occurs in many cancers and was mainly studied in glioma and bladder cancers [17, 19]. Actually, only few recent studies were published concerning hypermethylation and loss of expression of p14/ARF in colorectal cancer [7, 20].
The complexity and the close relationship between p53 and p14/ARF prompted us to describe these mutational profiles and expression in Tunisian colorectal cancer. In our study, we aimed to determine the p14/ARF expression level and its promoter methylation pattern in relation to mutational status of p53. First, we analyzed the relationship between epigenetic profiles and mutation status of p14/ARF and p53 genes with clinicopathological parameters. Next, we investigated whether the promoter methylation and the mRNA expression, respectively of p14/ARF and p53, were a predictor of the disease progression and the prognosis of colorectal cancer patients in Tunisian population.
Patient specimens
We underwent a retrospective study from 1995 to 2011 regarding patients with CRC, diagnosed in the laboratory of Pathology, Mongi Slim Hospital, Tunis. The individuals had neither gastrointestinal diseases nor a history of tumor. In the 167 cases included in this study, samples were taken not only from the tumoral area but also from the margin, corresponding to distant resection and were histologically free from precancerous lesions and cancer. The data collected for all patients included sex, age, tumor localization, TNM stage, and Astler–Coller stage. For DNA and the RNA extraction, representative samples of frozen sample tumoral mucosa (112) and paraffin-embedded tissues (55) were obtained from the files of 167 patients with CRC. The patient group included 83 women and 84 men. The mean age of the Tunisian patients (at the time of tissue collection) was 57 years. On histological exam, the tumor location was divided into 99 colon and 68 rectum. Furthermore, the pathologic classification of tumors was made according to the international TNM staging system: we identified 30 in primary stage (stages I and II) and 137 in advanced stage (stages III and IV). Concerning the Astler–Coller stage, we found 53 cases at early Astler–Coller stage and 114 at the advanced ones.
Methods
DNA and RNA extraction
Twenty milligrams of genomic DNA was extracted from paraffin embedded and frozen samples of tumoral mucosa. They were treated using the Wizard SV Genomic DNA Purification System according to the manufacturer’s instructions (Promega, Madison, WI). The concentration of the DNA was measured with a spectrophotometer. Total RNA was extracted with TRIZOL reagent (Invitrogen) according to the manufacturer’s instructions. After purification, RNA was dissolved in DEPC-treated water. The cDNA was synthesized by M-MLV Reverse Transcriptase (Invitrogen) and stored at −20 °C until used.
Sodium bisulfite modification of DNA(cDNA synthesis) and methylation-specific PCR of p14/ARF
Two micrograms of genomic DNA from each sample were bisulfite-modified using the EZ DNA methylation kit (ZYMO Research, Orange, CA) according to the manufacturer’s instructions. After treatment, the resulting bisulfite-modified DNA was eluted in 10 μL of the kit elution buffer and stored at −20 °C. Two microliters of the bisulfite-modified DNA were used for each PCR reaction. Two microliters of bisulfite-modified DNA from each sample were amplified independently using the U- and M-specific primers in a 25-μL total volume reaction (Table 1). Each PCR reaction contained a final concentration of 0.4 mmol of each primer (SGS, Köping, Sweden), 0.5-mmol dNTPs, 1× PCR buffer (Promega), 1.5-mM MgCl2 (Promega), and 0.04 units of Taq polymerase (Promega). The PCR products were checked on-chip electrophoresis.
Table 1.
Detail of the primers and cycling parameters
| Sequence (5′-3′) | Product length (pb) | Annealing temperature (°C) |
|---|---|---|
| p14/ARF | ||
| U | ||
| SpUF : TTTTTGGTGTTAAAGGGTGGTGTAGT | 122 | 58° |
| SpUR: CACAAAAACCCTCACTCACAACAA | ||
| M | ||
| SpMF: GTGTTAAAGGGCGGCGTAGC | 132 | 58° |
| SpMR: AAAACCCTCACTCGCGACGA | ||
| P53 | ||
| SPF:CCCAAGCAATGGATGAT | 379 | 56° |
| SPR:TGACAGGAAGCCAAAGG | ||
| Β-actin | ||
| SPF: CCAGGCACCAGGGCGTGATGGTGGGCATGG | 581 | 56° |
| SPR: AGCAGCCGTGGCCATCTCTTGCTCGAAGTC | ||
| P14/ARF | ||
| Sp F: GTGGCCCTCGTGCTGATG | 207 | 58° |
| Sp R: AGCACCACCAGCGTGTCC | ||
| p53 exons | ||
| Exon 5 | ||
| Sp5F: TGTTCACTTGTGCCCTGACT | 266 | 54° |
| Sp5R: CAGCCCTGTCGTCTCTCCAG | ||
| Exon 6 | ||
| Sp6F: GCCTCTGATTCCTCACTGAT | 160 | 53° |
| Sp6R: TTAACCCCTCCTCCCAGAGA | ||
| Exon 7 | ||
| Sp7F: ACTGGCCTCATCTTGGGCCT | 180 | 56° |
| Sp7R: TGTGCAGGGTGGCAAGTGGC | ||
| Exon 8 | ||
| Sp8F: TAAATGGGACAGGTAGGACC | 230 | 54° |
| Sp8R: TCCACCGCTTCTTGTCCTGC | ||
RT-PCR for detection of p53 and p 14/ARF mRNA expression
Total RNA was reverse-transcribed by M-MLV reverse transcriptase (Invitrogen) from which the c DNA was obtained from PCR reaction. PCR primers for p53 and β-actin were outlet in Table 1. RT-PCR of p53 was conducted with an initial step for 5 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at T m (degrees Celsius). cDNA integrity was confirmed by β-actin-specific PCR analyzes. The RT-PCR-products were checked on chips electrophoresis. The amplified band was 379 bp for p53, 207 bp for p14/ARF, and 581 bp for β-actin.
Analyzes of p14/ARF and p53 amplification product by on-chip electrophoresis
The PCR products of p14/ARF and p53 migration have been performed by chips-electrophoresis, for that, we used DNA 1000 LabChips kits, prepared with gel-dye mix, pressurized, then a marker solution and DNA 1500 ladder were added. For this process, 1 μL of each PCR reaction was added into one out of 11 sample wells of a prepared chip. After vortexing, the chip was placed in the BioRad Experion bioanalyzer. The electrophoresis of samples lasted approximately 30 or 40 min. Fragment analyzes was conducted using BioRad Experion software, and an overlay of two electropherogram was used to compare PCR patterns derived from tumor and normal mucosa. Differences in the peak patterns of the overlaid electropherogram were evaluated and two were used for each patient.
Mutational analyzes of p53: PCR amplification and SSCP analyzes
Exons 5 to 8 of p53 gene were amplified by polymerase chain reaction (PCR). The amplification products of p53 were resolved on on-chip electrophoresis. Primer sequences outlet in Table 1. An independent PCR assays was performed for each exon. The standard PCR reaction contained 0.5 μg genomic DNA, 200 μmol/L dNTPs, 1× PCR buffer (Promega), 1.5 mmol/L MgCl2, 1 U of Taq DNA polymerase (Promega), 0.3 μmol/L of each primer (biomatik), H2O, and 5 % dimethyl sulfoxide.
A 2-μL volume of p53 PCR product was denatured in 5 μL of formamide, incubated for 10 min at 95 °C. The overall mutation rate of p53 was identified after chips electrophoresis by the presence of one or two extra bands migration above or below the normal single-stranded products. Occasionally, mutated bands were detected between the single and double-strand bands that may be caused by formation of normal-mutated heterodimers. Samples with mobility shift were confirmed by sequencing using Sanger methods.
Immunohistochemistry of p53 protein
Serial sections of 4-mm thickness were cut from formalin-fixed paraffin-embedded samples, incubated in an oven at 37 °C overnight, deparaffinized, and rehydrated. The slides were immersed in citrate buffer (pH = 6.0) in a microwave for 2–5 min to unmask the epitopes and then kept at room temperature for 20 min, followed by a Tris-wash for 5 min. The sections on the slides were incubated with peroxidase block to inhibit endogenous peroxidase activity. After washing twice in Tris, the sections were incubated in p53 (1:50, Vision Biosystems) at room temperature for 1 h. They were then incubated with postprimary block for 30 min. Expressions were assessed after incubation of the sections at room temperature with the peroxidase-labeled DAKO Envision System for 30 min, using DAB as a chromogene for 20 min. After washing with distilled water, the sections were then counterstained with hematoxylin. The reaction was considered as positive when a positive nucleus staining of p53.
Statistical analyzes
The relationships between p14/ARF, p53 gene status and the different clinicopathological variables were assessed using χ 2 test. The odds ratio was obtained by unconditional logistic regression analyzes. Survival curves were computed according to the Kaplan–Meier method. All p values cited were two-sided and p values of <0.05 were judged as statistically significant. SPSS software, version 17.0, was used for analyzes,
Results
Analyzes of methylation status and mRNA expression of p14/ARF: correlation with clinicopathological data
Of 167 patients with CRC, 120 (71.8 %) cases were unmethylated (U), 33 (19.7 %) methylated (M), and 14 (8.5 %) methylated and unmethylated (MU) at the same time (Fig. 1). Statistical analyzes shows that the MSP pattern was correlated with location (p = 0.04), Astler–Coller stage (p = 0.024) and with TNM stage (p = 0.016). In fact, we found that 65 % of U phenotype was seen in the colon compared with M and MU phenotype distribution which was equivalent between colon and rectum. For the prognosis factor, the MSP pattern demonstrates that the M and MU bands were correlated with the primary TNM stage: Stages I and II (Table 2) and with advanced Astler–Coller stage (stages C and D; p = 0.024). However, we do not find any statistical association between p14/ARF MSP pattern and the other clinicopathological criteria.
Fig. 1.
MSP analysis of p14/ARF for different patients with colorectal cancer; U and M indicates the U and the M fragments, respectively. Lanes L, ladder at 1,500 bp; NC, negative control of PCR (without primers); 1, positive controls of U reaction; 2, positive controls of M reaction. For patient 1, lanes 3 and 4 reveal the presence of U band and the absence of M band, respectively. For patient 2, lanes 5 and 6 reveal the presence of the U and M band respectively in the same tumor. Patient 3 reveals the absence of U band and the presence of M band (M) in lanes 7 and 8, respectively. Inset, comparison report of density of UM and MU pattern of p14/ARF
Table 2.
Number of CRC samples with p14/AFR methylation pattern, p53 mRNA expression, p53 mutation status, and P53 immunostaning analysis in relation to clinicopathological parameters n (percent)
| Variables | P14/ARF MSP pattern | P53 mutation status | P53 mRNA expression | P53 immunostaning | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U | M | U+M | p | + | − | p | + | − | p | + | − | p | |
| 120 (%) | 33 (%) | 14 (%) | 29 (%) | 138 (%) | 58 (%) | 109 (%) | 57 (%) | 110 (%) | |||||
| Sexe | |||||||||||||
| Male | 63 (52.5) | 17 (51.5) | 4 (28.6) | 0.11 | 12 (41.4) | 72 (52.1) | 0.19 | 33 (56.9) | 51 (46.8) | 0.14 | 31 (54.4) | 70 (63.7) | 0.21 |
| Female | 57 (47.5) | 16 (48.5) | 10 (71.4) | 17 (58.6) | 66 (47.9) | 25 (43.1) | 58 (53.2) | 24 (45.6) | 41 (36.3) | ||||
| Age (year) | |||||||||||||
| <65 | 78 (65) | 15 (45.5) | 8 (57.1) | 0.21 | 17 (58.6) | 84 (60.1) | 0.49 | 31 (53.4) | 70 (64.2) | 0.11 | 33 (57.9) | 51 (46.4) | 0.11 |
| ≥65 | 42 (35) | 18 (54.5) | 6 (42.9) | 12 (41.4) | 54 (39.9) | 27 (46.6) | 39 (35.8) | 24 (42.1) | 57 (53.4) | ||||
| Site | |||||||||||||
| Colon | 78 (65) | 16 (48.5) | 5 (35.7) | 0.040 | 18 (62) | 81 (58.7) | 0.45 | 40 (69) | 59 (54.1) | 0.044 | 37 (64.9) | 56 (50.9) | 0.11 |
| Rectum | 42 (45) | 17 (51.5) | 9 (64.3) | 11 (38) | 57 (41.3) | 18 (31) | 50 (45.9) | 20 (35.1) | 54 (49.1) | ||||
| TNM stage | |||||||||||||
| Stage I+II | 23 (19.2) | 7 (21.2) | 0 (0) | 0.016 | 4 (13.8) | 26 (18.7) | 0.38 | 11 (19) | 19 (17.4) | 0.48 | 15 (26.3) | 15 (13.6) | 0.037 |
| Stage III+IV | 97 (80.8) | 26 (78.7) | 14 (100) | 25 (86.2) | 112 (81.3) | 47 (81) | 90 (82.6) | 63 (73.7) | 74 (86.4) | ||||
| Astler–Coller stage | |||||||||||||
| Stage A+B | 45 (37.5) | 7 (21.2) | 1 (7.1) | 0.24 | 21 (72.4) | 94 (68.1) | 0.41 | 44 (75.8) | 71 (65.1) | 0.1 | 49 (86) | 87 (79) | 0.18 |
| Stage C+D | 75 (62.5) | 26 (68.8) | 13 (92.9) | 8 (27.6) | 44 (31.9) | 14 (24.2) | 38 (34.9) | 8 (14) | 23 (21) | ||||
We also examined the expression of p14/ARF using RT-PCR. Among 167 early lesions with available cDNA, 101/120 colorectal adenomas which are U at p14/ARF expressed high levels of p14/ARF mRNA, whereas 23/52 adenomas with p14/ARF M and 10/52 with MU pattern do not expresses p14/ARF mRNA or very little p14/ARF mRNA level, demonstrating an exact correlation of transcriptional loss with p14/ARF hypermethylation (p < 0.005; Fig. 2; Table 3). No correlation was found with clinicopathological features.
Fig. 2.

Chips electrophoresis of p53 and p14/ARF mRNA expression in colorectal cancer tissues. Lanes L, ladder at 1,500 bp; NM, mRNA expression of p53 and p14/ARF in normal mucosa normalized with B-actin; 1–6 reveal the presence of β-actin in all cases; 1, 3, 4, and 6 showed expression of p53 in 372 bp, and therefore, 2 and 5 revealed lack of p53 mRNA expression in 372 bp position; 1, 2, 3, 5 showed expression of p14/ARF, and therefore, 4 and 6 revealed lack of p/14ARF mRNA expression in the 207-bp position
Table 3.
Relationship among p14/ARF methylation, p53 mRNA expression, and SSCP analysis in colorectal cancer patients
| P14/ARF | ||||
|---|---|---|---|---|
| U (%) | M (%) | U+M (%) | p | |
| n = 120 | n = 33 | n = 14 | ||
| P14/ARF | ||||
| RT + (n = 115) | 101 (84.2) | 10 (30.3) | 4 (28.6) | <0.005 |
| RT − (n = 52) | 19 (15.8) | 23 (69.7) | 10 (71.4) | |
| p53 | ||||
| RT + (n = 58) | 50 (41.7) | 7 (21.2) | 1 (7.1) | 0.007 |
| RT − (n = 109) | 70 (58.3) | 26 (78.8) | 13 (92.2) | |
| P53 mutation status | ||||
| M (n = 29) | 21 (17.5) | 7 (21.2) | 1 (7.1) | 0.5 |
| WT (n = 138) | 99 (82.5) | 26 (78.8) | 13 (92.9) | |
M mutation, WT wild type
Analyzes of mRNA expression, mutational status and immunostaning of p53 in CRC: correlation with clinincopathological data
The samples were considered negative when they were positive for β-actin and negative for p53 (Fig. 2). In our set, 109 cases (65.3 %) showed positive expression for p53 while 58 (34.7 %) cases were negative (Table 3).
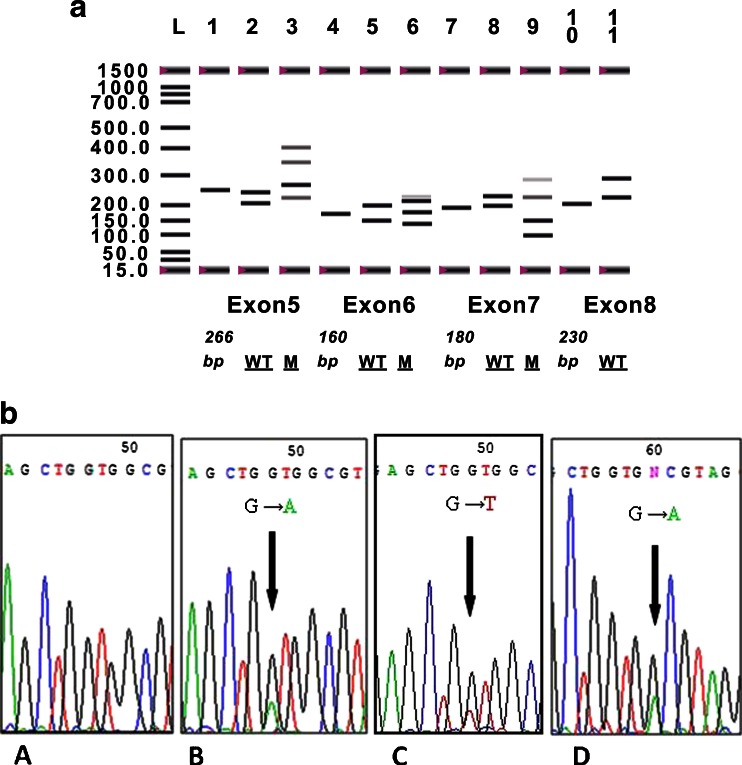
The p53 exons 5, 6, 7 and 8 were successfully amplified in all cases, which gave expected PCR fragment of 266, 160, 180, and 230 bp, respectively. After SSCP analyzes, among 167 cases of CRC, 17.4 % (29/167) was found harboring altered p53. Furthermore, 11 cases showed alterations in exon 5, 5 in exon 6, and 14 in exon 7, whereas no mutation was found in exon 8 (Fig. 3). Out of these 29 cases, 19 (65.5 %) were transition and 10 (34.5 %) transversion. All mutated samples in SSCP analyses were confirmed by sequencing (Fig. 3a, b; Table 4). No significant involvement has been detected through our statistical analyses between p53, SSCP analysis, or p53 mRNA expression and clinicopathological variables. The comparison between p53 mRNA expression and p53 alteration revealed an association among the variables (p < 0.005). In fact, we found that 75.9 % (22) of patients who had mutation in the p53 gene have a lack of the protein expression (Table 5).
Fig. 3.
a Single-strand conformational polymorphism analysis of exons 5, 6, 7, and 8 of the p53 gene. Lanes L, ladder at 1,500 bp; 1, exon 5 tumoral sample with 266-bp band; 2, exon 5 WT; 3, exon 5 sample with mutated band; 4, tumoral sample of exon 6 with 160-bp band; 5, exon 6 sample with Wild Type; 6, exon 6 sample with mutated band; 7, tumoral sample of exon 7 with 180-bp band; 8, exon 7 sample with wild type; 9, exon 7 sample with mutated band; 10, tumoral sample of exon 8 with 230-bp band; and 11, exon 8 sample with wild type. M mutated, WT wild type. b Partial electropherograms representing the normal and mutant profiles of exons 5, 6, and 7. A, Insertion of T. B, Insertion of C. C, transition of G to A leading to a frame-shift mutation
Table 4.
Sequencing of P53 in 29 patients with colorectal cancer
| Exon/codon | WT codon | Mutant codon | WT AA | Mutant AA | Effect | ID | Cancer | |
|---|---|---|---|---|---|---|---|---|
| E5/181 | CGC | TGC | Transition | ARG | CYS | MS | 17 | CRC |
| E5/175 | CGC | CAC | Transition | ARG | HIS | MS | 20 | CRC |
| E5/173 | GTG | ATG | Transition | VAL | MET | MS | 37 | CRC |
| E5/154 | GGC | GGT | Transition | ARG | TRP | MS | 39 | CRC |
| E5/166 | TCA | ATC | Transversion | SER | ILE | FM | 64 | CRC |
| E5/177 | CCC | CTC | Transition | PRO | LEU | MS | 112 | CRC |
| E5/159 | GCC | GAC | Transition | ALA | ASP | MS | 113 | CRC |
| E5/176 | TGC | TGG | transition | CYS | TRP | MS | 128 | CRC |
| E5/126 | CAG | TAG | Transition | GLU | STOP | TP | 132 | CRC |
| E5/171 | GAG | CAG | Transversion | GLU | GLN | MS | 147 | CRC |
| E5/175 | CGC | CAC | Transition | ARG | HIS | MS | 148 | CRC |
| E6/196 | CGA | TGA | Transition | ARG | STOP | TP | 74 | CRC |
| E6/205 | TAT | CAT | Transition | TYR | HIS | MS | 22 | CRC |
| E6/204 | CAG | TAG | Transition | LYS | STOP | TP | 68 | CRC |
| E6/213 | CGA | CTA | Transversion | ARG | LEU | MS | 121 | CRC |
| E6/209 | AGA | AAA | Transition | AGR | LYS | MS | 6 | CRC |
| E7 /229 | TGT | TGA | Transversion | CYS | STOP | TP | 2 | CRC |
| E7/248 | CGG | TGG | Transition | ARG | TRP | MS | 19 | CRC |
| E7/245 | GGC | AGC | Transversion | GLY | SER | MS | 28 | CRC |
| E7/234 | TAC | CAC | Transition | TYR | HIS | MS | 72 | CRC |
| E7/248 | CGG | CAG | Transition | ARG | GLN | MS | 58 | CRC |
| E7/250 | CCC | CTC | Transition | PRO | LEU | MS | 162 | CRC |
| E7/254 | ATC | ACC | Transition | ILE | THE | MS | 10 | CRC |
| E7/238 | TGT | TAT | Transition | CYS | TYR | MS | 11 | CRC |
| E7/242 | GAG | TAG | Transversion | GLU | STOP | TP | 82 | CRC |
| E7/239 | GGC | AGC | Transversion | GLY | SER | FM | 83 | CRC |
| E7/244 | GGC | AGC | Transversion | GLY | SER | FM | 15 | CRC |
| E7/283 | TGT | TGA | Transversion | CYS | STOP | TP | 67 | CRC |
| E7/248 | CGG | CCG | Transversion | ARG | PRO | MS | 122 | CRC |
| E7/248 | CGG | CAG | Transition | ARG | GLN | MS | 92 | CRC |
Table 5.
Relationship among p53 m RNA expression and p53 mutation in colorectal cancer patients n (percent)
| P53 mRNA expression | p | ||
|---|---|---|---|
| Negative expression | Positive expression | ||
| p53 mutation status | |||
| SSCP+M | 22 (75.9) | 7 (24.1) | <0.005 |
| SSCP–WT | 36 (26.1) | 102 (73.9) | |
SSCP+ positive mobility shift of p53 (p53 mutated profile), SSCP− negative mobility shift of p53 (p53 wild type)
For the immunoreactions, there were no significant differences with respect to clinicopathological characteristics between weak, moderate, or strong staining. Therefore, the three latter groups were classified as positive in our following analyzes. In total, 57 (34.1 %) of the 167 tumors showed positive immunoreactivity for p53. The immunoreactivity of p53 was invariably confined to the nucleus, and normal mucosa samples were negative for p53 expression (Fig. 4a) and positive in the tumoral area (Fig. 4b). According to clinicopathological variables, no significant involvement was detected through our statistical analyses except for the TNM stage. However, p53 was associated with TNM advanced stage, and we found a loss of expression of p53 (86.4 %) in stages III and IV (p = 0.037; Table 2). We found significant association between p53 expression and mutational status analyses (p < 0.005; Table 6). Indeed, we observed that the patients with negative expression of p53 demonstrate the absence of mobility shift in the DNA by SSCP analysis at 28.26 % (39/138), and the patient with overexpression of p53 demonstrate the presence of mobility shift in the DNA by SSCP 3at 1.1 % (9/29).
Fig. 4.
p53 Nuclear immunostaning. a Negative expression of p53 in healthy mucosa. b Positive expression of p53 in colorectal cancer. Inset, high magnification of p53 expression in tumor
Table 6.
Relationship between p53 protein expression detected by IHC and p53 gene mutation status of 167 CRC patients n (percent)
| P53 | IHC − 10 % | IHC + 10 % | p |
|---|---|---|---|
| M | 20 (68.9) | 9 (31.1) | <0.005 |
| WT | 39 (28.26) | 99 (71.74) |
M mutated, WT wild type
Relationship between the p14/ARF methylation and p53 analyzes
Statistical results did not show any association between the methylation pattern of p14/ARF and the mutational status of p53. Interestingly, combined analyses of p53 mRNA expression and p14/ARF methylation pattern showed a significant association. In fact, we found that the majority of cases with M (26 cases) and UM (13 cases) patterns of p14/ARF were correlated with a lack expression of p53 (p = 0.007; Table 3).
The relationship between the alteration in p14/ARF and p53 and patients survival
The Kaplan–Meier survival curve for p14/ARF showed that there was no correlation between its methylation pattern and the specific disease survival (p = 0.41; Fig. 5a). For the p53 mRNA expression, we found that the patients with negative expression of p53 had shorter survival than patients with positive expression of p53 (p = 0.000; Fig. 5b).
Fig. 5.
Association between p14/ARF hypermethylation, p53 m RNA expression and patient survival. a No significant survival between p14/ARF methylation pattern and patients survival (p = 0.41) and b patients with positive mRNA expression of p53 had shorter survival compared with patients who had a negative expression of p53 (p = 0.000)
Discussion
The p14/ARF is considered as a tumor suppressor protein, its inactivation by hypermethylation has been extensively described in many cancers [4, 11]. The frequency of its promoter hypermethylation varies in different tumor types [21]. This last alteration was particularly intriguing in view to the interplay between p14/ARF and its impact in the p53 pathway during tumorogenesis.
Methylation status of p14/ARF in colorectal cancer remains unclear and is until now not studied in Tunisian population. Therefore, to elucidate the implication of p14/ARF through the colorectal carcinogenesis, the MSP pattern, mRNA expression, and different clinicopathological data were studied.
In this study, the methylation of the p14/ARF gene occurred in 28.2 %. This level among colorectal cancer patients varies between 22 and 50 %. This difference may be due to the diversity of populations [22–24].
The MSP pattern of p14/ARF are habitually represented by two entities: U and M patterns. Interestingly, in a recent paper, we have published [18], a new result was found showing the presence of U and M bands in the same sample that indicates the hemi-methylated pattern (MU) [18]. This proportion represents 8.5 % of our set. These bands were analyzed in terms of intensity, and we found that the majority of cases showed a greater intensity of the methylation band. This result confirms that it is a physiological and progressive transition from the U pattern towards the promoter hypermethylation. Comparing p14/ARF MSP pattern with clinicopathological parameters showed an association between p14/ARF hemi-methylated pattern and CRC rectum site (p = 0.04). Indeed, our analysis revealed a similar distribution of p14/ARF methylation between the colon (48.5 %) and the rectum (51.5 %). Moreover, hemi-methylated pattern was generally observed in the rectum (64.3 %). However, Burri et al. demonstrate that methylation of p14/ARF gene was significantly more frequent in right-sided than in left-sided tumors [25]. In this frame, Lee et al. showed no statistical significance between the MSP pattern and colorectal cancer site [23].
Based on the prognostic parameters, we did not detect any association between the hypermethylation and the survival rate. This result was similar to many previous studies of the literature [21, 26].
Conversely, we found that methylation of p14/ARF promoter gene was associated with the stage of the disease. In fact, in the primary TNM stages of our tumors, stages I and II, the M and hemi-methylated status were seen at 57.6 and 100 %, respectively (p = 0.016). Furthermore, the infiltrative growth of the tumor (p = 0.024; Astler–Coller C and D) was associated with M and hemi-methylated pattern of the promoter gene. These results showed that the inactivation of p14/ARF were associated with early stages of the tumor which were also characterized by small diameter and absence or rare metastatic lymph nodes. Despite their primary stages, these tumors were correlated with infiltrative growth process. These results proved that these patterns are very aggressive.
Dominguez et al. [27] also reported a significant correlation between methylation of p14/ARF gene and poor prognosis in breast, colon, and bladder carcinomas. Thus, our result and those of the literature indicate that methylation process constitutes the major mechanism of p14/ARF inactivation and could be used as a biomarker for CRC [18].
P14/ARF is a candidate for hypermethylation with loss and inactivation of its protein. It contains a documented CpG island which can be silenced by this genetic and epigenetic alteration. However, few works have evaluated the methylation of p14/ARF in association with its expression. For further comprehension of this loss, we associate this fact to its regulation by mRNA expression. Therefore, we conducted a specific analysis of p14/ARF mRNA expression by RT-PCR and evaluated its impact in the genesis and prognosis in our cohort. Interestingly and according to our results, a high concordance was shown between CpG hypermethylation and the low levels of the p14/ARF mRNA pattern (p < 0.005). Consequently, our data confirmed by others in recent literature, suggests that epigenetic regulation by promoter hypermethylation is the predominant mechanism involved in the deregulation of p14/ARF and may contribute to silencing of p14/ARF mRNA expression in CRC patients [28–30]. Moreover, no association was found between p14/ARF mRNA expression and patient survival.
According to these results, we can conclude that the inactivation of tumor suppressor genes by aberrant hypermethylation is a fundamental process involved in the progression of many malignant tumors, including gastrointestinal cancer [31, 32].
After rigorous validation on RT-PCR and immunohistochemistry investigation, a molecular signature associated with p53 mutant phenotypes in this subset of CRC was identified. Approximately the same values were found between the p53 transcriptional (mRNA) and translational (protein) profile with the p53 mutation status. In fact, we note that mutant forms of p53 have two distinct expressions phenotypes namely positive or negative. This could be the result of different mutations in the p53 gene: activation or inactivation mutation. With regard to the p53 wild type, the lack of expression (39 and 36 cases) is probably associated with the involvement of epi-mutation (p53 promoter hypermethylation), knowing that these data have been recently reported in the literature.
Therefore, we found that p53 mutation and its abnormal expression may affect the occurrence and the development of CRC in synergy. It was reported that in 50 % of human cancers, the p53 gene is mutated. The gain of oncogenicity or the loss of tumor suppressor function of p53 is due to two alterations, such as its inactivation through missens mutations or its overexpression by transcription of p53 mutant form. This fact is considered as metastatic signatures in CRC [33]. It contributes to tumor aggressiveness and results in poor survival [34–37].
The present study demonstrates an association between p53-negative mRNA expression and poor survival in our cohort. The genomic instability associated with p53 mRNA overexpression is the cause of developing a risk phenotype, aggressive progression, and early death as reported in previous works [32, 38].
The basic functions of p14/ARF inactivation, is predicted to reduce the p53 aberrant protein resulting from the mutation of the p53 [32]. Many researches showed that high frequency of p14/ARF promoter methylation had been previously reported to occur in tumors without TP53 mutations [4, 38]. An inverse correlation between TP53 mutations and epigenetic inactivation of p14/ARF in CRC do not always hold true [39].
Conversely, p53/p14/ARF axis was considered as the major pathway involved in the regulation of cell proliferation, apoptosis, and DNA repair [40, 41]. Although these two proteins are mechanically dependant, this complex was (p53/p14/ARF) frequently deregulated through the strong association between p53 expression and p14/ARF methylation. In fact, we showed that M (78.8 %) and hemi-methylated (92.2 %) patterns were observed in tumor samples with a lack of p53 expression. This result is logical as p14/ARF methylation causes loss of p14/ARF functions and induces its absence in the nucleolus. As a result, it cannot bind to MDM2, resulting in its liberation. In this case, MDM2 acts as an oncogene, degrade p53 by ubiquitinylation, and blocks the normal cell cycle.
In the literature, controversial results between p53 expression and p14/ARF inactivation has been observed not only in CRC but also in gastric and lung carcinomas [5, 42–45]. Eischen et al. [46] reported that control of p53 by p14/ARF occurs under specific stressful conditions and their effects on p53 functions may be dependent of the p53/p14ARF pathway in some tumor types. If one gene is abnormal, the p53/p14/ARF pathway function is blocked.
In conclusion, we found that p53/p14/ARF pathway was frequently deregulated in our patients. Herein, we demonstrate that hypermethylation of p14/ARF occurs early during CRC tumorogenesis. However, we did not find any correlations between p14/ARF and survival. These results suggest that p14/ARF methylation pattern may constitute a predictor factor of CRC in early stage but it cannot be considered as a prognostic factor.
Finally, simultaneous assessment of p14/ARF methylation and abnormal expression of p53 may work as a biological indicator for early diagnosis of colorectal cancer, which may provide a theoretical basis for genetic intervention in clinical practice. With regard to our results, several revolutionary prospects can be opened in modern oncology: (1) Intervention of p14ARF methylation may be considered a powerful biomarkers in early colorectal cancer diagnosis. (2) Knowing that the methylation process is a reversible phenomenon, demethylation could be considered as targeted therapy.
Acknowledgment
The authors acknowledge the support from the Ministry of Health and Scientific Research.
Conflicts of interest
None
References
- 1.Schuler M, Green DR. Mechanisms of p53-dependent apoptosis. Biochem Soc Trans. 2001;29:684–688. doi: 10.1042/BST0290684. [DOI] [PubMed] [Google Scholar]
- 2.Paluszkiewicz P, Berbeæ H, Pawlowska-Wakowicz B, Cybulski M, Paszkowska A. p53 protein accumulation in colorectal cancer tissue has prognostic value only in left sided colon tumors. Cancer Detect Prev. 2004;28:252–259. doi: 10.1016/j.cdp.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Soussi T, Beroud C. Significance of p53 mutations in human cancer: a critical analysis of mutations at CpG dinucleotides. Hum Mutat. 2003;21:192–200. doi: 10.1002/humu.10189. [DOI] [PubMed] [Google Scholar]
- 4.Vishwa jet A, Yukio T, Kouki I. Methylation of p14arf gene in meningiomas and its correlation to the p53 and mutation. Mod Pathol. 2004;17:705–710. doi: 10.1038/modpathol.3800111. [DOI] [PubMed] [Google Scholar]
- 5.Sajjad K, Arif A. Correlation of p53 over-expression and alteration in p53 gene detected by polymerase chain reaction-single strand conformation polymorphism in adenocarcinoma of gastric cancer patients from India. World J Gastroenterol. 2009;11:1381–1387. doi: 10.3748/wjg.15.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vishwa JA, Yukio T, Kouki I. Methylation of p14ARF gene in meningiomas and its correlation to the p53 expression and mutation. Mod Pathol. 2004;17:705–710. doi: 10.1038/modpathol.3800111. [DOI] [PubMed] [Google Scholar]
- 7.Manel E, Silvia T, Minoru T, Gabriel C, Miguel AP, Stephen BB, et al. Herman2 hypermethylation-associated inactivation of p14ARF is independent of p16INK4a methylation and p53 mutational status 1. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- 8.Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, et al. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llanos S, Clark PA, Rowe J, Peters G. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol. 2001;3:445–452. doi: 10.1038/35074506. [DOI] [PubMed] [Google Scholar]
- 10.Xirodimas D, Saville MK, Edling C, Lane DP, Laín S. Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene. 2001;20:4972–4983. doi: 10.1038/sj.onc.1204656. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Aguilera A, Sanchez-Beato M, Garcia JF, Prieto I, Pollan M, Piris MA. p14ARF nuclear overexpression in aggressive B cell lymphomas is a sensor of malfunction of the common tumor suppressor pathways. Blood. 2002;99:1411–1418. doi: 10.1182/blood.V99.4.1411. [DOI] [PubMed] [Google Scholar]
- 12.Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 13.Lu W, Pochampally R, Chen L, Traidej M, Wang Y, Chen J. Nuclear exclusion of p53 in a subset of tumors requires MDM2 function. Oncogene. 2000;19:232–240. doi: 10.1038/sj.onc.1203262. [DOI] [PubMed] [Google Scholar]
- 14.Esteller M, Cordon-Cardo C, Corn PG, Meltzer SJ, Pohar KS, Watkins DN, et al. p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. Cancer Res. 2001;61:2816–2821. [PubMed] [Google Scholar]
- 15.Silva J, Domínguez G, Silva JM, García JM, Gallego I, Corbacho C, et al. Analysis of genetic and epigenetic processes that influence p14ARF expression in breast cancer. Oncogene. 2001;20:4586–4590. doi: 10.1038/sj.onc.1204617. [DOI] [PubMed] [Google Scholar]
- 16.Domínguez G, Silva J, Silva JM, Garcia JM, Larrondo FJ, Vargas J, et al. Different expression of P14ARF defines two groups of breast carcinomas in terms of TP73 expression and TP53 mutational status. Genes Chromosomes Cancer. 2001;31:99–106. doi: 10.1002/gcc.1123. [DOI] [PubMed] [Google Scholar]
- 17.He J, Qiao J-b, Zhu H. p14ARF promoter region methylation as a marker for gliomas diagnosis. Med Oncol. 2011;28:1218–1224. doi: 10.1007/s12032-010-9651-8. [DOI] [PubMed] [Google Scholar]
- 18.Lilia KBM, Amira A, Mariem K, Ines C, Olfa El A, Abdel Majid BH, et al. Loss of galectin-3 expression in mucinous colorectal carcinomas is associated with 50CpG island methylation in Tunisian patients. Appl Immunohistochem Mol Morphol. 2011;19:258–265. doi: 10.1097/PAI.0b013e3181f869bb. [DOI] [PubMed] [Google Scholar]
- 19.Ken K, Hideki E, Takenari G, Hiroyuki K, Kenryu N, Motoshi K, et al. p16INK4a and p14ARF methylation as a potential biomarker for human bladder cancer. Biochem Biophys Res Commun. 2006;33:9790–9796. doi: 10.1016/j.bbrc.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura M, Watanabe T, Klangby U, Asker C, Wiman K, Yonekawa Y, et al. Ohgaki H.p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol. 2001;11(2):159–168. doi: 10.1111/j.1750-3639.2001.tb00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito S, Takefumi O, Hiroshi S, Masayuki W, Yoshihiro K, Masaru M, et al. Promoter hypermethylation and quantitative expression analysis of CDKN2A (p14 ARF and p16 INK4a) gene in esophageal squamous cell carcinoma. Anticancer Res. 2007;27:3345–3354. [PubMed] [Google Scholar]
- 22.Christine N, Christine S, Roger D, Alex K, Karin D. Hypermethylation of the 5′ CpG island of the p14 ARF flanking exon 1 β in human colorectal cancer displaying a restricted pattern of p53 overexpression concomitant with increased MDM2 expression. Clin Epigenetics. 2012;4:9. doi: 10.1186/1868-7083-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Lee M, Sup Han W, Kyoung Kim O, Hee Sung S, Sun Cho M, Lee SN, et al. Prognostic value of p16INK4a and p14ARF gene hyperméthylation in human colon cancer. Pathol Res Pract. 2006;202:415–424. doi: 10.1016/j.prp.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 24.Kominami K, Nagasaka T, Cullings HM, Hoshizima N, Sasamoto H, Young J, et al. Methylation in p14ARF is frequently observed in colorectal cancer with low-level microsatellite instability. J Int Med Res. 2009;37:1038–1045. doi: 10.1177/147323000903700408. [DOI] [PubMed] [Google Scholar]
- 25.Burri N, Shaw P, Bouzourene H, Sordat I, Sordat B, Gillet M, et al. Methylation silencing and mutations of the p14ARF and p16INK4 genes in colon cancer. Lab Invest. 2001;81:217–229. doi: 10.1038/labinvest.3780230. [DOI] [PubMed] [Google Scholar]
- 26.Kondo I, Iida S, Takagi Y, Sugihara K. MDM2 mRNA expression in the p53 pathway may predict the potential of invasion and liver metastasis in colorectal cancer. Dis Colon Rectum. 2008;51:1395–1402. doi: 10.1007/s10350-008-9382-5. [DOI] [PubMed] [Google Scholar]
- 27.Dominguez G, Silva J, Garcia JM, Silva JM, Rodriguez R, Munoz C, et al. Prevalence of aberrant methylation of p14ARF over p16 INK4a in some human primary tumors. Mutat Res. 2003;530:9–17. doi: 10.1016/S0027-5107(03)00133-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C, Guo X, Zhang L, Lu Z, Ma N, Cheng Y, et al. Methylation-related silencing of p14ARF gene correlates with telomerase activity and mRNA expression of human telomerase reverse transcriptase in hepatocellular carcinoma. J Surg Oncol. 2008;98(6):462–468. doi: 10.1002/jso.21131. [DOI] [PubMed] [Google Scholar]
- 29.Ito T, Nishida N, Fukuda Y, Nishimura T, Komeda T, Nakao K. Alteration of the p14(ARF) gene and p53 status in human hepatocellular carcinomas. J Gastroenterol. 2004;39(4):355–361. doi: 10.1007/s00535-003-1302-9. [DOI] [PubMed] [Google Scholar]
- 30.Xiaofang L, Kun T, Shaoping Y, Zaiqiu W, Hailong S. Correlation between promoter methylation of p14ARF, TMS1/ASC, and DAPK, and p53 mutation with prognosis in cholangiocarcinoma. World J Surg Oncol. 2013 (in press). [DOI] [PMC free article] [PubMed]
- 31.Chaar I, Amara S, Khiari M, Ounissi D, Dhraif M, Ben Hamida AEM, et al. Relationship between MDM2 and p53 alterations in colorectal cancer and their involvement and prognostic value in the Tunisian population. Appl Immunohistochem Mol Morphol. 2013 (in press). [DOI] [PubMed]
- 32.Xiong C, Li J, Yan L, Huang Y. Methylation of p14ARF and abnormal expression of p53 and mdm2 in colorectal cancer: role and correlation. Sci Res Essays. 2010;5:1669–1675. [Google Scholar]
- 33.Molleví DG, Serrano T, Ginestà MM, Valls J, Torras J, Navarro M, et al. Mutations in TP53 are a prognostic factor in colorectal hepatic metastases undergoing surgical resection. Carcinogenesis. 2007;28:1241–1246. doi: 10.1093/carcin/bgm012. [DOI] [PubMed] [Google Scholar]
- 34.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Joerger AC, Fersht AR. Structure-function-rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 36.Levine AJ, Finlay CA, Hinds PW. P53 is a tumor suppressor gene. Cell. 2004;116:S67–S69. doi: 10.1016/S0092-8674(04)00036-4. [DOI] [PubMed] [Google Scholar]
- 37.Liu DP, Song H, Xu Y. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene. 2009;29:949–956. doi: 10.1038/onc.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chittenden TW, Howe EA, Culhane AC, Sultana R, Taylor JM, Holmes C, et al. Functional classification analysis of somatically mutated genes in human breast and colorectal cancers. Genomics. 2008;91:508–511. doi: 10.1016/j.ygeno.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen L, Kondo Y, Hamilton SR, Rashid A, Issa JP. P14 methylation in human colon cancer is associated with microsatellite instability and wild-type p53. Gastroenterology. 2003;124:626–633. doi: 10.1053/gast.2003.50102. [DOI] [PubMed] [Google Scholar]
- 40.Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, et al. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- 41.Bartek J, Lukas J. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 2001;490:117–122. doi: 10.1016/S0014-5793(01)02114-7. [DOI] [PubMed] [Google Scholar]
- 42.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 43.Ishida K, Zhu BL, Maeda H. Novel approach to quantitative reverse transcription PCR assay of mRNA component in autopsy materials using the TaqMan fluorogenic detection system: dynamics of pulmonary surfactant apoprotein. Forensic Sci Int. 2000;113:127–131. doi: 10.1016/S0379-0738(00)00252-8. [DOI] [PubMed] [Google Scholar]
- 44.Ito T, Nishida N, Fukuda Y, Nishimura T, Komeda T. INK4a-ARF alterations and p53 mutations in hepatocellular carcinomas. Oncogene. 2001;20:7104–7109. doi: 10.1038/sj.onc.1204902. [DOI] [PubMed] [Google Scholar]
- 45.Iida S, Akiyama Y, Nakajima T, Ichikawa W, Nihei Z, Sugihara K. Alterations and hypermethylation of the p14(ARF) gene in gastric cancer. Int J Cancer. 2000;87:654–658. doi: 10.1002/1097-0215(20000901)87:5<654::AID-IJC6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 46.Eischen CM, Alt JR, Wang P. Loss of one allele of ARF rescues Mdm2 haploinsufficiency effects on apoptosis and lymphoma development. Oncogene. 2004;23:8931–8940. doi: 10.1038/sj.onc.1208052. [DOI] [PubMed] [Google Scholar]