1. Introduction
Chromatin, a complex of DNA and associated proteins, governs diverse processes including gene transcription, DNA replication and DNA repair [1]. The fundamental unit of chromatin is the nucleosome, consisting of 147bp of DNA wound approximately 1.6 turns around a histone octamer of one (H3-H4)2 tetramer and two H2A-H2B dimers. In order to form nucleosomes, (H3-H4)2 tetramers are deposited first, followed by the rapid deposition of H2A-H2B. Therefore, it is believed that the assembly of (H3-H4)2 tetramers into nucleosomes is the rate-limiting step of nucleosome assembly. Moreover, assembly of H3-H4 into nucleosomes following DNA replication, DNA repair and gene transcription is likely to be a key step in the inheritance of epigenetic information and maintenance of genome integrity. Understanding how assembly of H3-H4 into nucleosomes is regulated is extremely important and will be the focus of this review.
In general, nucleosome assembly is classified into replication-coupled (RC) nucleosome assembly and replication-independent (RI) nucleosome assembly (Figure 1A and Table 1). Both RC and RI nucleosome assembly processes occur in both yeast and mammalian cells despite the fact that yeast cells have only one form of histone H3, which is most similar to the mammalian H3 variant H3.3. In mammalian cells, H3.3 is mainly assembled into nucleosomes in a RI manner while the canonical histone H3 (H3.1 and H3.2), whose expression peaks during S phase in mammalian cells, is assembled into nucleosomes in a RC manner. H3.1 and H3.2 differ by only one amino acid and therefore, throughout the review, we will refer to H3.1 as the canonical histone H3 in mammalian cells. While H3 molecules are different between yeast and mammalian cells, most of the histone chaperones, a group of proteins that bind histones and promote nucleosome assembly and/or exchange without being final products, are conserved from yeast to human cells [2]. Histone chaperones are key factors in regulating nucleosome assembly. Further regulation comes from post-translational modifications of the histone proteins. Therefore, we will separate our discussion below into the regulation of RC and RI nucleosome assembly, highlighting the roles of histone chaperones and modifications on newly synthesized H3 and H4 in these two processes.
Figure 1.
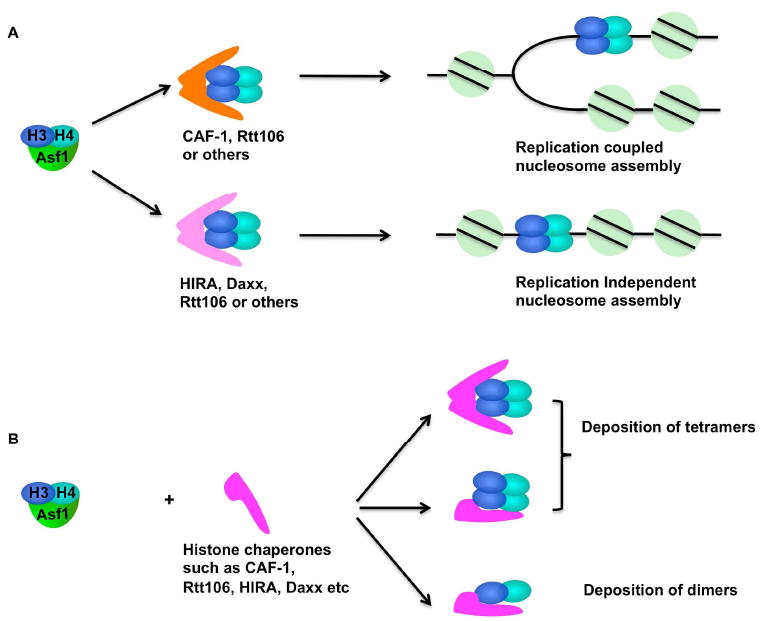
Nucleosome assembly of new H3-H4. (A) There are two major nucleosome assembly pathways: replication coupled (RC) nucleosome assembly and replication independent (RI) nucleosome assembly. Histone chaperone Asf1 binds a H3-H4 dimer, which will be transferred to histone chaperones that are involved in RC or RI nucleosome assembly. (B) Models of formation of histone (H3-H4)2 tetramers, the first building block of a nucleosome. Two H3-H4 dimers from Asf1 can be transferred to a histone chaperone to form one (H3-H4)2 tetramer on a monomeric or dimeric form of the histone chaperon. Alternatively, a H3-H4 heterodimer will be transferred from Asf1 to another histone chaperone, which will deposit two H3-H4 dimers sequentially onto DNA for formation of a (H3-H4)2 tetramer.
Table 1.
A summary of factors involved in RC and RI nucleosome assembly.
| Nucleosome Assembly Pathway | Replication-Coupled (RC) | Replication-Independent (RI) |
|---|---|---|
|
| ||
| Biological Process | DNA replication | Transcription/others |
|
| ||
| Function | Epigenetic inheritance | Histone turnover |
| Gene expression | ||
|
| ||
| Histone Chaperone | Asf1 | Asf1 |
| CAF-1 | HIRA(HIR1) | |
| Rtt106 (Yeast) | Daxx | |
| Rtt106 (Yeast) | ||
|
| ||
| Histone H3 | H3.1 | H3.3 |
|
| ||
| Histone H3 Modifications1 | H3K56Ac | H3K56Ac |
| H3 N-terminal tail Ac | ||
|
| ||
| Histone H4 Modifications | H4K5,12Ac | H4S47ph |
We mentioned solely those modifications that have been shown to have a role in regulating the indicated nucleosome assembly pathways.
2. Replication coupled (RC) nucleosome assembly
Nucleosomes are barriers for DNA replication and therefore, nucleosomes ahead of the replication fork must be temporarily disassembled or remodeled in order for the DNA replication machinery to gain access to the DNA. Immediately following DNA replication, replicated DNA is assembled into nucleosomes using both parental and newly synthesized histone proteins. Numerous studies indicate that the assembly of replicated DNA into nucleosomes is coupled to the on-going DNA replication [3, 4]. It is hypothesized that coupling nucleosome assembly to DNA replication ensures proper inheritance of chromatin structure, propagation of epigenetic marks on histones to daughter cells and maintenance of genome integrity. Supporting this hypothesis, mutations in genes involved in DNA RC nucleosome assembly result in increased sensitivity to DNA damaging agents and compromised maintenance and inheritance of heterochromatin states in yeast and mammalian cells [5-9].
At the molecular level, two separate pathways are likely involved in RC nucleosome assembly. First, parental histones in front of the replication fork are transferred onto the replicated DNA. While how this process is coupled to ongoing DNA replication remains elusive, recent evidence indicates that (H3-H4)2 tetramers are transferred as a single unit for nucleosome formation. While it has been known for a while that parental histones do not mix with newly synthesized histones to form nucleosomes during S phase of the cell cycle [10-12], several studies proposed that parental (H3-H4)2 tetramers may split into two dimers for nucleosome formation [13, 14]. Recently, using stable isotope labeling of amino acids in cell culture (SILAC) combined with quantitative mass spectrometry (MS), it has been shown that parental (H3.1-H4)2 tetramers do not mix with newly synthesized (H3.1-H4)2 tetramers, whereas parental H2A-H2B and newly-synthesized H2A-H2B can be found within one nucleosome following DNA replication [15]. These results not only clarify a major question in the field, but also reinforce the idea that the assembly of (H3-H4)2 tetramers, both parental and newly synthesized, is likely to be a key step in the inheritance of chromatin states and high order chromatin structure. Since only one daughter cell receives the parental (H3-H4)2 tetramer at any given DNA position, epigenetic marks on H3-H4 in the individual nucleosome cannot be maintained. Instead, epigenetic marks on H3-H4 can only be maintained in a group of nucleosomes in a functional domain.
Second, newly synthesized H3-H4 can be deposited onto replicating DNA to form nucleosomes in a histone chaperone dependent manner. Below, we introduce the histone chaperones involved in RC nucleosome assembly and then discuss how newly synthesized (H3-H4)2 tetramers are deposited onto replicating DNA during S phase of the cell cycle.
2.1. Histone chaperones involved in de novo nucleosome assembly of H3-H4
The classic histone chaperone regulating RC nucleosome assembly is chromatin assembly factor 1 (CAF-1), identified in human cells using a SV40 replication-coupled nucleosome assembly assay [3, 16, 17]. CAF-1 is a three subunit (p150, p60 and RbAp48 in humans) protein complex known to bind histones H3-H4 containing modifications indicative of newly synthesized H3-H4 [18, 19]. Thus, CAF-1 is likely involved in promoting de novo nucleosome assembly of newly synthesized H3-H4. Whether CAF-1 has any role in the transfer of parental H3-H4 during S phase of the cell cycle is not known. In human cells, CAF-1 preferentially co-purifies with the canonical histone, H3.1, in comparison to H3.3, indicative of CAF-1’s role in the deposition of H3.1-H4 during S phase of the cell cycle [13]. However, how CAF-1 recognizes H3.1-H4 preferentially over H3.3-H4 remains to be determined.
Anti-silencing factor 1 (Asf1) was discovered as a histone chaperone that promotes CAF-1 dependent nucleosome assembly [20, 21]. Asf1 purified from yeast to human cells cannot promote RC nucleosome assembly by itself using an in vitro SV40 DNA RC nucleosome assembly assay [22, 23], consistent with the idea that Asf1’s role in RC nucleosome assembly is not direct deposition of H3-H4 onto replicating DNA. Several studies indicate that Asf1 is likely involved in regulating the modification and/or nuclear import of newly synthesized H3-H4. For example, in budding yeast, Asf1 is essential for acetylation of histone H3 lysine 56 (H3K56Ac) [24-27], a modification on newly synthesized H3 [28]. Moreover, genetic studies indicate that Asf1’s role in nucleosome assembly and other processes in budding yeast are likely due to its ability to regulate H3K56Ac [26]. In human cells, there are two sequence homologs of Asf1, Asf1a and Asf1b. Both Asf1a and Asf1b are required for H3K56Ac in human cells, with Asf1a being the predominant form in regulating H3K56Ac [29, 30]. In addition, it is known that Asf1a and Asf1b associate with H3-H4 prior to the import of newly synthesized H3-H4 into the nucleus, suggesting that Asf1 in human cells may regulate nuclear import of new H3-H4 [31]. In addition to binding newly synthesized H3-H4 and promoting RC nucleosome assembly of newly synthesized H3-H4, Asf1 has other functions. First, yeast cells lacking Asf1 exhibit increased supercoiling of an endogenous 2μ plasmid, suggesting a role for Asf1 in nucleosome disassembly [32]; however, Asf1’s role in nucleosome disassembly is unclear. It is possible that Asf1 alone disassembles nucleosomes by removing H3-H4. Alternatively, Asf1 may collaborate with ATP dependent chromatin remodeling complexes to disassemble nucleosomes. Second, in S. pombe, Asf1 binds the histone deacetylase Clr6 and is required for deacetylation of histone H3 lysine 9 [33]. In S. cerevisiae, however, Asf1 is required for efficient acetylation of H3 lysine 9 [34]. These distinct functions of Asf1 in budding yeast and fission yeast are further highlighted in the fact that Asf1 is essential for cell growth of S. pombe, whereas S. cerevisiae cells lacking Asf1 are viable. Third, human Asf1 binds histones with marks representing parental histones, suggesting that Asf1 may have a role in parental H3-H4 transfer [35]. How Asf1 is involved in parental histone transfer remains to be determined.
In S. cerevisiae, Rtt106 is another H3-H4 histone chaperone found to be important for mediating de novo histone deposition. Rtt106’s function as a histone chaperone was originally identified in a yeast genetic screen for enhancers of silencing defects of mutant PCNA cells defective for CAF-1 binding [36]. Genetic and biochemical evidence suggest that Rtt106 functions in parallel with CAF-1 to promote RC nucleosome assembly [37, 38]. However, it remains to be resolved exactly how Rtt106 functions in parallel with CAF-1 in nucleosome assembly. It is possible that CAF-1 and Rtt106 deposit H3-H4 at distinct chromatin regions. Alternatively, CAF-1 and Rtt106 may function distinctly as histone chaperones for either the leading or lagging strand. Further supporting distinct roles for CAF-1 and Rtt106, Rtt106 binds dsDNA with no sequence specificity, and this, along with the histone binding ability, appears to be important for Rtt106’s function in transcriptional silencing [39]. Therefore, Rtt106 may have roles outside of RC nucleosome assembly.
As the assembly of newly-synthesized H3-H4 is tightly coupled with DNA replication, it is not surprising that several of the histone chaperones involved in RC nucleosome assembly exhibit interactions with DNA replication proteins, suggesting that the replication fork machinery recruit histone chaperones to the fork for histone deposition. CAF-1 is recruited to the replication fork through a direct interaction between the large subunit of CAF-1, p150, and PCNA [40, 41], the processivity factor for the DNA polymerases, an interaction conserved from yeast to humans [42]. In yeast, Asf1 directly binds RFC, a protein complex that loads PCNA onto DNA [43]. In mammalian cells, both Asf1a and Asf1b are found to complex with the replicative helicase MCM, and this association is required for fork progression [35]. It is still unclear as to how Rtt106 is recruited to the replication fork.
Furthermore, there appears to be significant coordination among the H3-H4 histone chaperones in order to effectively deposit newly-synthesized H3-H4 at the replication fork. Various studies support a model in which newly synthesized H3-H4 molecules bind Asf1. Asf1 binds H3 through the same H3 interface involved in formation of (H3-H4)2 tetramers, and in vitro, Asf1 disrupts (H3-H4)2 tetramers [44]. This raises the following questions, how are (H3-H4)2 tetramers, the first building block of a nucleosome, formed during RC nucleosome assembly? Pull-down experiments have revealed that in mammalian cells, the Asf1-H3-H4 heterotrimer resides in a complex with CAF-1 and HIRA [13]. We have shown that Asf1 is required for an efficient association of H3-H4 with the histone chaperones Rtt106 and CAF-1 in yeast [38]. Therefore, Asf1 may transfer histone H3-H4 dimers to other histone chaperones for deposition onto chromatin [38, 45]. Two possible models help explain on how newly synthesized (H3-H4)2 tetramers are formed (Figure 1B). First, two H3-H4 dimers are deposited sequentially by a histone chaperone onto replicating DNA to form the (H3-H4)2 tetramer for nucleosome formation. Second, a histone chaperone may bind a single (H3-H4)2 tetramer, which in turn, is deposited onto DNA to promote nucleosome formation. Several studies suggest that (H3-H4)2 tetramers can be deposited by histone chaperones via oligomerization of histone chaperone proteins. For instance, Xenopus p150, the large subunit of CAF-1, can form dimers in vitro, and the ability of p150 to dimerize is important for nucleosome assembly [46]. More recently, two members of the NAP (nucleosome assembly protein) family, Nap1 and Vps75, bind histone H3-H4 in a tetramer fashion [47]; however, Nap1 in cells is a H2A-H2B chaperone [48] and Vps75 is a component of Rtt109-Vps75 complex [49]. Therefore, the significance of Nap1 and Vps75 binding H3-H4 remains to be further investigated. Recent studies from our lab indicate that Rtt106 dimerizes and binds a (H3-H4)2 tetramer (Fazly et al., unpublished), further supporting the possibility that (H3-H4)2 tetramers are formed prior to the deposition of H3-H4 onto DNA. Furthermore, Rtt106 directly interacts with CAF-1 [37]. These studies highlight the complex interactions occurring among the histone chaperones for coordinated deposition of histones. Future studies are needed to address how each histone H3-H4 chaperone bind dimeric or tetrameric forms of H3-H4 and how the coordination among the histone chaperones and these different H3-H4 forms enables proper nucleosome assembly (Figure 1B).
2.2. Roles of modifications on newly synthesized H3 and H4 in RC nucleosome assembly
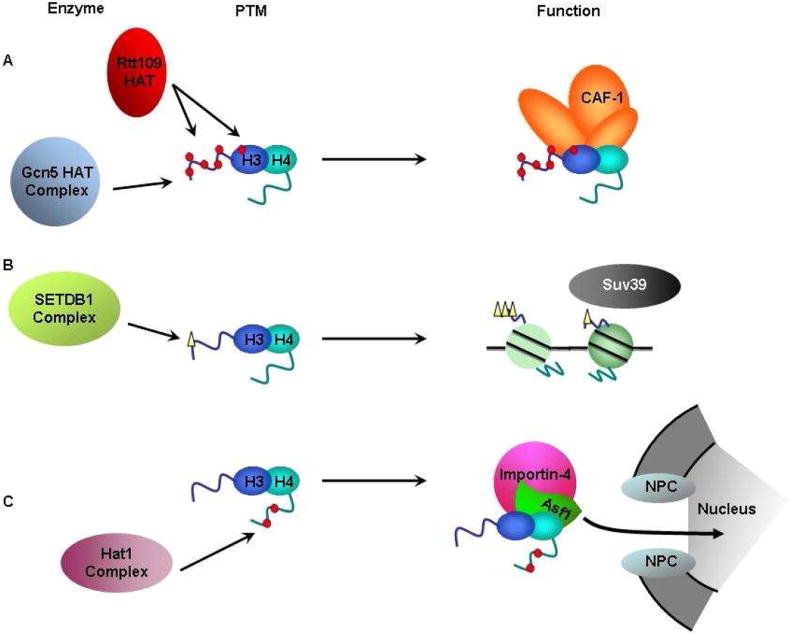
Histone proteins contain a number of post-translational modifications. These modifications aid in regulating a host of cellular processes [50]. It was observed long time ago that newly synthesized histones were acetylated and that these acetylation marks were removed shortly after deposition [51, 52]. Three acetylation marks on newly-synthesized histones are relatively conserved among species: H4 lysine residues 5 and 12 (H4K5,12Ac), H3 N-terminal tail acetylation and H3 lysine 56 (H3K56Ac) [28, 53-55]. In addition to acetylation, monomethylation of histone H3 K9 (H3K9me) is present on H3.1 prior to deposition in mammalian cells [56]. These modifications likely regulate RC nucleosome assembly via one of the following mechanisms. First, histone modifications may regulate the interactions between histones and histone chaperones. Second, histone modifications on newly synthesized histones may be important for regulating histone import as histones are shuttled from the cytoplasm into the nucleus for deposition. Finally, histone modifications on newly synthesized histones may facilitate the inheritance of marks on histones by aiding in the transfer of information from parental histones to the newly synthesized histones (Figure 2 and Table 1). Discussed below are the known and/or predicted functions of specific modifications found on newly synthesized histones.
Figure 2.
Roles for post-translational modifications on newly-synthesized histones in nucleosome assembly. Described are the enzymes, post-translational modification (PTM) and function associated with known modifications observed on newly-synthesized H3-H4. (A) Acetylation of the H3 N-terminal tail by Gcn5 and Rtt109 and H3K56 (within the H3 core domain) by Rtt109 regulates the interaction between histone chaperones (like CAF-1) and H3-H4. (B) H3K9me, catalyzed by SetDB1 (in complex with CAF-1 and HP1alpha), serves as a substrate for Suv39, and thereby contributing to the epigenetic inheritance of the H3K9me3 mark. (C) Hat1 catalyzes H4K5,12Ac, which may help regulate histone import from the cytoplasm into the nucleus.
The most highly conserved mark of newly synthesized histones is diacetylation of histone H4 at lysine residues 5 and 12 (H4K5,12Ac) [53]. This mark is catalyzed by the HAT1 acetyltransferase complex [57-59]. While this modification has been known for a long time, only recently has the function of this modification in RC nucleosome assembly come to light. A detailed study of histone complexes formed from histone synthesis to deposition indicate that H4K5, K12Ac occurs within the cytoplasm before histones associate with Asf1 and before newly-synthesized histones are imported into the nucleus [31]. Furthermore, an analysis of recombinant protein incorporation into the macropasmodia of Physarum revealed that the nonacetylable, H4K5,12R form of H3-H4 inhibits H3-H4 nuclear import while the import of an acetylation mimic, H4K5,12Q, is improved over wild type H3-H4 [60]. In addition, H4K5,12Ac is present on H4 co-purified with histone chaperone CAF-1 [18, 19]. These results suggest H4K5, K12Ac may have multiple roles in nucleosome assembly. First, it may facilitate the nuclear import of H3-H4. Second, it may regulate the function of CAF-1 in nucleosome assembly. In addition, Hat1 also co-purifies with H3.3, raising the possibility that this modification on H4 also impacts the nucleosome assembly of H3.3 [13, 61]. Future studies are needed to determine to what extent H4K5, K12Ac and Hat1 regulate RC nucleosome assembly via nuclear import of H3.1-H4 and/or the regulation of the association between histone chaperones and histones.
In addition to acetylation of new H4, newly synthesized histone H3 is also acetylated. While acetylation of H3 lysine residues appears to be conserved among species, the acetylation patterns observed are different [53]. In S. cerevisiae, the predominant acetylation on newly synthesized histones is H3K56Ac, a residue located at the DNA entry/exit site of the nucleosome [28]. In the budding yeast, H3K56Ac is catalyzed by the Rtt109-Vps75 complex that utilizes Asf1-H3-H4 complex as the substrate [25-27, 49, 62-64]. While Vps75 is not required for H3K56Ac in vivo [49], Vps75 is required for the nuclear import of Rtt109 as well as the stability of Rtt109 [65, 66]. Following deposition, H3K56Ac is deacetylated by Hst3 and Hst4, members of the NAD dependent family of histone deacetylase (HDAC) enzymes, during late S or G2/M phase of the cell cycle [67, 68]. The primary role of H3K56Ac is to regulate RC nucleosome assembly. Supporting this idea, H3K56Ac peaks during S phase [28]. Using chromatinimmunoprecipitation (ChIP) assay, we have shown that H3K56Ac is deposited only onto replicating DNA, but not non-replicating DNA, during early S phase of the cell cycle [38]. This assay recapitulates early observations that nucleosome assembly is coupled to DNA replication [4]. Therefore, this assay is likely to be useful in determining to what extent a gene is involved in RC nucleosome assembly in budding yeast. Finally, H3K56Ac increases the binding affinity of H3-H4 with two histone chaperones, CAF-1 and Rtt106, suggesting that H3K56Ac regulates RC nucleosome assembly by increasing the binding affinity of H3-H4 with specific histone chaperones [38].
Cells lacking Rtt109 or those expressing a non acetylation state H3K56R mutant are much more sensitive towards DNA damaging agents than cells lacking both Rtt106 and CAF-1 [38]. This result suggests that H3K56Ac may also promote nucleosome assembly mediated by other histone chaperones in addition to CAF-1 and Rtt106. Alternatively, it implies that H3K56Ac has other functions in addition to its role in RC nucleosome assembly. Indeed, H3K56Ac is also important for nucleosome assembly following DNA repair, which is important for the recovery from checkpoint activation [69]. Furthermore, H3K56Ac has been shown to be involved in gene transcription [70, 71] as well as the regulation of RI nucleosome assembly (see discussion below)
In mammalian cells, H3K56Ac abundance is relatively low compared to yeast cells [30, 67, 72], leading one to inquire to what extent this modification plays a regulatory role in RC nucleosome assembly. Compared to H3K56Ac in budding yeast, the regulation of H3K56Ac in mammalian cells appears to be more complicated. For instance, while this modification is dependent on Asf1a and Asf1b in mammalian cells [29, 30], there are at least two acetyltransferases, p300/CBP and Gcn5, reported to acetylate mammalian H3K56 [30, 73], whereas Gcn5 in budding yeast is not required for H3K56Ac [74]. In addition, several H3K56Ac deacetylases including Sirt1, Sirt6, Hdac1 and Hdac2 have been reported [29, 30, 75, 76]. Finally, there are conflicting reports on whether this modification is regulated during S phase of the cell cycle and how H3K56Ac levels change in response to DNA damage agents [29, 30, 73]. These conflicting reports likely result from the following possibilities. First, since the levels of H3K56Ac are low in mammalian cells, much more specific antibodies are needed to detect this modification by Western blot in mammalian cells. Therefore, the antibodies used by different groups from different sources may contribute to such conflicting results. Second, compared to yeast cells that have only one form of H3, human cells contain H3.1 and H3.3, which cannot be differentiated by Western blot. Therefore, different enzymes catalyzing H3K56Ac may target H3.1 and H3.3 differently. Future studies are needed to resolve these issues.
In addition to H3K56Ac within the H3 core domain, acetylation of lysine residues at the N-terminus of H3 also regulates RC nucleosome assembly [77]. Acetylation patterns on the N terminal tails of new H3 vary among species. For instance, in yeast, H3K9 and K27 are acetylated, in Drosophila, H3K9 and K14, and in humans, H3K9 and H3K18 [53, 55, 72]. Both yeast Elp3 and Gcn5, two acetyltransferases with well-established roles in gene transcription, function with Rtt109 to acetylate the H3 N-terminal tail of newly-synthesized histones and promote nucleosome assembly in a manner parallel to H3K56Ac [74, 78]. Supporting this idea, Gcn5, although it does not directly regulate levels of H3K56Ac in yeast, is important for proper deposition of H3K56Ac at the replication fork. Furthermore, while H3K56Ac is important for both CAF-1 and Rtt106 to bind H3-H4, Gcn5 and H3 N-terminal tail acetylation appear to only regulate the interaction between CAF-1 and histones H3-H4 [74].
In addition to a role in nucleosome assembly, Gcn5 and acetylation of the H3 N-terminus are likely to have a role in the initiation of DNA replication [79, 80]. The levels of acetylation at multiple lysine residues on H3 and H4 in nucleosomes surrounding a replication origin increase during S phase. Interestingly, in the case of H3, multiple sites tend to be acetylated together. Mutational analysis suggests that acetylation at these sites functions redundantly in origin firing as the quantitative number of acetylation marks is more important in regulating origin firing than acetylation at a particular site [79]. Because nucleosome positioning at origins affects origin firing [81], it would be challenging to separate the roles for Gcn5 and acetylation of lysine residues at the H3 N-terminus in origin firing from their roles in nucleosome assembly.
Although histone lysine acetylation is predominantly a mark of newly synthesized histones, mammalian H3.1 is monomethylated at H3 lysine 9 (H3K9me) by SETDB1 on more than one third of the H3.1 pool [56]. SETDB1 forms a complex with both CAF-1 and HP1 during S-phase and preferentially methylates free histones over nucleosomal histones, suggesting it methylates histones prior to deposition. It is proposed that H3K9me may serve as a precursor to set up H3K9me3, catalyzed by Suv39, at pericentric heterochromatin sites and thus play a role in the propagation of this histone mark. Consistent with this idea, knockdown of SETDB1 results in lower levels of H3K9me3 [82]. Furthermore, kinetic studies on the levels of methylation of newly synthesized histones suggest that monomethylation of many residues most likely occurs concurrent with or shortly after deposition to enable further higher order methylation at particular sites [83]. A recent report suggests that H3K9me1 is one of the first modifications observed on H3.1 following synthesis, and the levels of H3K9me1 are sharply reduced in conjunction with the appearance of H4K5,12Ac, suggestive of a role of this modification in earlier histone processing [31]. Despite these advances in understanding how this mark occurs on newly synthesized histones, the exact role of this abundant modification on newly-synthesized H3 remains to be determined.
In summary, modifications on newly synthesized H3 and H4 likely play multiple roles in the regulation of RC nucleosome assembly, whether it be through the regulation of histone nuclear import, histone chaperone and histone interactions, and/or other functions.
3. Replication independent (RI) nucleosome assembly
While canonical histone H3 is strictly incorporated into chromatin during S phase of the cell cycle, the majority of histone H3 variant H3.3 is assembled into nucleosomes in a replication independent (RI) manner [15, 84]. This RI process likely involves the exchange of parental H3 including both canonical H3 and H3 variant H3.3, with new H3.3 to compensate for nucleosome loss and/or replacement of damaged histones in non-dividing cells such as neurons. In addition, RI nucleosome assembly is also important during gene transcription and other cellular processes.
Most of the early studies on RI histone exchange focused on histone H2A-H2B exchange. H2A-H2B exchange occurs namely at actively transcribed regions and the 5’ end of the genes, raising the possibility that histone exchange may be an important form of transcriptional regulation [85, 86]. While the frequency of H3-H4 incorporation is much lower than H2A-H2B, H3-H4 exchange does occur. The observation that H3-H4 incorporation independent of DNA replication occurs arose from an elegant study using a chromatin fractionation assay in immature erythrocytes [87]. In addition, histone H3-H4 exchange can be detected in the presence of the transcription inhibitor actinomycin D [88], suggesting that mechanisms other than gene transcription are likely involved in RI histone exchange. Supporting this idea, genome wide mapping of H3.3 shows that H3.3 is enriched at both active and inactive gene promoters [89, 90]. Increased histone exchange is also observed at boundary elements that block the spread of chromatin states in yeast and Drosophila [91, 92]. Together, these studies suggest that histone exchange may have an important regulatory role in chromatin dynamics.
Similar to the problem arising during DNA replication, nucleosomes serve as barriers for gene transcription, and therefore, changes in chromatin structure are often a prerequisite for the initiation of gene transcription and transcriptional elongation. Genome wide nucleosome positioning mapping has revealed that two well-positioned nucleosomes flank the transcription start site (TSS) with a nucleosome-depleted region (NFR) in between (reviewed in [93]). In mammalian cells, the NFR is likely to be marked by an unstable nucleosome containing H3.3 and H2AZ [90]. The dynamics of nucleosomes at the promoter likely provide a mechanism for regulating gene expression.
During transcriptional elongation, nucleosomes are temporally disassembled in front of the RNA Polymerase (Pol) II complex to facilitate passage of the transcriptional machinery. Following gene transcription, nucleosomes must be reassembled by recycling the parental histone (H3-H4)2 and/or incorporation of newly synthesized histones (replacement/exchange). Two different mechanisms are proposed to efficiently restore the chromatin structure following passage of RNA Pol II [94]. First, partial loss or exchange of core histones was observed at highly active transcribed gene regions [95-100]. Second, H2A-H2B exchange, but not H3-H4 exchange, was detected at moderately transcribed gene regions [101, 102]. As mentioned above, genome wide mapping of H3.3 occupancy reveals that H3.3 is enriched at gene bodies of both active and inactive genes in Drosophila and mammalian cells [84, 90], suggesting that nucleosome assembly and the exchange of H3.3 occurs in both a transcription dependent and independent manner. Consistent with this idea, despite having only one form of H3, histone H3-H4 replacement/exchange also occurs at both active and inactive gene promoter regions in budding yeast [103]. Collectively, these studies suggest that histone deposition and/replacement during gene transcription may regulate gene expression and/or maintain gene expression state.
3.1 Histone chaperones involved in replication-independent nucleosome assembly
Like RC nucleosome assembly, RI nucleosome assembly/exchange is mediated by histone chaperones. Multiple histone chaperones including Asf1, Hir1, Rtt106 and Spt6 have been shown to be involved in RI nucleosome assembly in budding yeast. The Hir1 complex (consisting of Hir1, Hir2, Hir3, and Hpc2 in budding yeast) is the major histone chaperone involved in RI nucleosome replacement [104, 105]. In higher eukaryotes, HIRA, the sequence homolog of Hir1, is well known to promote the assembly of histone H3.3 into chromatin independently of DNA replication [13, 106, 107]. Similar to RC nucleosome assembly mediated by CAF-1, Asf1 is proposed to deliver H3-H4 to HIRA for subsequent nucleosome assembly.
In human cells, Asf1a and Asf1b appear to have distinct functions in regard to the regulation of RI nucleosome assembly. For instance, HIRA preferentially binds Asf1a over Asf1b [108]. Moreover, depletion of Asf1b, but not Asf1a, results in cell proliferation defects [109]. Interestingly, both Asf1a and HIRA promote the formation of senescence associated heterochromatin foci [110]. These results raise the possibility that Asf1a is the primary histone chaperone that transfers histones to HIRA for nucleosome assembly.
While it is still not clear why cells need so many histone chaperones for RI nucleosome assembly, based on studies of HIRA and Daxx (Death domain containing protein) in mammalian cells, it is possible that these histone chaperones promote RI nucleosome assembly at distinct chromatin regions to regulate gene expression at different chromatin domains. Daxx has been identified as another H3.3 histone chaperone in mammalian cells [61, 89, 111]. In contrast to HIRA which is required for deposition of H3.3 at genic regions, Daxx is required for the localization of H3.3 at telomeric repeats [89]. Interestingly, Daxx contains a Rtt106-like domain at the C terminus, and this domain contributes to the H3.3 binding affinity [61]. Therefore, it is possible that while Daxx does not share significant homology with Rtt106, Daxx may be the functional homolog of Rtt106 in human cells. In addition to Daxx and HIRA, other H3.3 chaperones will likely be identified because it has been shown that the H3.3 occupancy at regulatory elements is independent of either Daxx or HIRA [89].
Unlike RC nucleosome assembly where the coordination between DNA replication and nucleosome assembly is carried out by the recruitment of histone chaperone complexes via direct interactions with the replication machinery, it is still unclear as to how the histone chaperones involved in RI nucleosome assembly are recruited to chromatin. Accumulating evidence suggests that histone chaperones likely interact with transcriptional machinery and chromatin remodeling complexes to promote RI nucleosome assembly and regulate gene expression. For instance, Spt6 interacts with RNA Pol II physically and promotes nucleosome disassembly in order to facilitate the passage of RNA Pol II followed by nucleosome reassembly [112]. Similarly, the histone chaperone complex FACT, which has multiple roles, was first indentified through co-purification with RNA Pol II and is important for both transcription initiation and elongation (Reviewed in [113]). Second, Drosophila Asf1 interacts with the Brahma chromatin remodeling complex [114]. Third, HIRA interacts with the chromatin remodeling complex Chd1 [115]. Lastly, Daxx interacts with the SWI/SNF type chromatin-remodeling factor ATRX (α-thalassemia/mental retardation syndrome protein), and the deposition of H3.3 onto telomere and pericentric DNA repeat regions depends on ATRX [61, 89]. Supporting this idea, it has been shown that ATRX is localized at repetitive DNA sequences [116]. It is worth noting that some histone chaperones, such as DEK, can serve as transcription coactivators [117]. These studies suggest that multiple factors may aid in recruiting histone chaperone complexes for RI nucleosome assembly and such events may serve as another form of regulation. Further studies should be carried out to determine how these different modes of RI nucleosome assembly pathway impact gene transcription.
3.2 Role of histone modifications in RI nucleosome assembly
Unlike modifications on H3 and H4 that function in RC nucleosome assembly, the roles of modifications on new H3 and H4 in RI nucleosome assembly has not been well explored. However, several studies suggest that posttranslational modifications on H3 and H4 also likely regulate RI nucleosome assembly as well. For instance, genome wide measurements of H3K56Ac turnover in yeast cells reveal that H3K56Ac marks the most dynamic nucleosomes [103]. This result suggests that in addition to its role in RC nucleosome assembly, H3K56Ac likely also impacts RI nucleosome assembly. Supporting this idea, Rtt109 and Asf1, two regulators of H3K56Ac, promote histone replacement/exchange during gene transcription. Interestingly, Vps75, a histone chaperone facilitating the enzymatic activity of Rtt109, inhibits histone replacement, suggesting a possible feedback mechanism for nucleosome replacement [103]. In mammalian cells, quantitative mass spectrometry analysis of histone modifications uncovered that histone turnover rates depend upon both site-specific histone modifications and the amino acid sequence of histone variants [118]. In general, histones containing active marks have a faster turnover rate than histones containing repressive marks, consistent with the idea that gene transcription influences histone turnover and/or assembly. Interestingly, the turnover rate of the H3K56Ac peptide is faster than many other histone modifications [118]. Despite these interesting observations, detailed studies on the role of modifications on H3 and H4 in RI nucleosome assembly are needed to understand to what extent modifications on H3 and H4 function in RI nucleosome assembly/exchange. Furthermore, since specific chromatin domains have distinct nucleosomal histone modifications [119, 120], it is also possible that these histone modifications also regulate histone turnover rates and drive the histone deposition pathways. Recently, our laboratory has found that phosphorylation of histone H4 S47 (H4S47ph), catalyzed by the Pak2 kinase, promotes HIRA mediated nucleosome assembly of H3.3-H4 and inhibits CAF-1 mediated nucleosome assembly of H3.1-H4. This suggests that modifications on histone H4 can help specify nucleosome assembly of H3.1 and H3.3 [121].
4. Summary and future directions
Over the last several years, we have witnessed explosive interest in addressing the regulation of nucleosome assembly pathways by histone chaperones and histone modifications and the functional implications of RC and RI nucleosome assembly in the inheritance of chromatin structure and regulation of gene expression. However, there is still a plethora of unanswered questions waiting to be addressed.
Emerging evidence indicates that nucleosomes are much more dynamic than previously recognized. In addition, deposition of new H3-H4 and transfer of parental H3-H4 during DNA replication and DNA repair proceed with rapid speed. Therefore, technical limitations have made it extremely difficult to spatially and temporally monitor the dynamics of histone deposition in live cells. Hopefully, advances in single cell imaging will enable us to design approaches to analyze how parental histones H3-H4 are transferred onto replicated DNA during S phase of the cell cycle and how multiple histone deposition pathways function coordinately to promote nucleosome assembly [122].
Second, in addition to regulating gene transcription, H3.3 likely has roles in heterochromatin formation [123]. Studies are needed to address how H3.3 performs these functions and to what extent these different functions are linked to histone chaperones that deposit H3.3 to distinct chromatin domains.
Third, yeast models have aided immensely in understanding regulatory mechanisms for nucleosome assembly, such as H3K56Ac and the regulation of histone chaperone interactions with H3-H4. It remains not well explored on how RC and RI nucleosome assembly pathways are regulated in higher eukaryotes. Given that nucleosome assembly in mammalian cells is much more complex, it is likely that histone modifications play a significant role in regulating RC and RI nucleosome assembly in mammalian cells. Indeed, we have recently discovered that phosphorylation of histone H4 promotes HIRA mediated nucleosome assembly and inhibits CAF-1 mediated nucleosome assembly [121]. Moreover, covalent modifications on histone chaperones are also detected and these modifications likely impact the function of histone chaperones [124].
Lastly, several histone chaperones have been shown to interact with ATP dependent nucleosome remodeling complexes for nucleosome assembly and/or gene regulation. We suspect that future studies will reveal additional interactions among histone chaperones and chromatin remodeling complexes. The challenge will be to determine how histone chaperones coordinate with chromatin remodeling complexes during different cellular processes.
Research highlights.
We review literatures on replication-coupled and replication-independent nucleosome assembly.
We highlight the functions of histone chaperones in nucleosome assembly.
We discuss the roles of histone modifications in nucleosome assembly.
We speculate the future direction of the nucleosome assembly field.
Acknowledgments
This work is supported by NIH grants to ZZ. ZZ is a scholar of the Leukemia and Lymphoma Society. RB is supported by a grant from American Heart Association. We apologize to those whose publications have not been cited in this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 2.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- 4.McKnight SL, Miller OL. Electron Microscopic Analysis of Chromatin Replication in the Cellular Blastoderm Drosophilia melanogaster Embryo. Cell. 1977;12:795–804. doi: 10.1016/0092-8674(77)90278-1. [DOI] [PubMed] [Google Scholar]
- 5.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 6.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enomoto S, McCune-Zierath PD, Gerami-Nejad M, Sanders MA, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 8.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 9.Myung K, Pennaneach V, Kats ES, Kolodner RD. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci U S A. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonne-Andrea C, Wong AMLM. In vitro replication through nucleosomes without histone displacement. Nature. 1990;343:719–726. doi: 10.1038/343719a0. [DOI] [PubMed] [Google Scholar]
- 11.Leffak IM, Grainger R, Weintraub H. Conservative assembly and segregation of nucleosomal histories. Cell. 1977;12:837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- 12.Jackson V, Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981;23:121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- 13.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 Complexes Mediate Nucleosome Assembly Pathways Dependent or Independent of DNA Synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 14.Russev G, Hancock R. Formation of hybrid nucleosomes containing new and old histones. Nucleic Acids Research. 1981;9:4129–4137. doi: 10.1093/nar/9.16.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of Histone H3-H4 Tetramers During DNA Replication,ÄìDependent Chromatin Assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 16.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 18.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 19.Zhou H, Madden BJ, Muddiman DC, Zhang Z. Chromatin assembly factor 1 interacts with histone H3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair. Biochemistry. 2006;45:2852–2861. doi: 10.1021/bi0521083. [DOI] [PubMed] [Google Scholar]
- 20.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 21.Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 23.Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, Kaufman PD, Allis CD. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci U S A. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 26.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 27.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, Freitas MA, Denu JM, Kaufman PD. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 29.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WH, Seepany H, Gao Z, Day LA, Greenblatt JF, Reinberg D. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adkins MW, Tyler JK. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J Biol Chem. 2004;279:52069–52074. doi: 10.1074/jbc.M406113200. [DOI] [PubMed] [Google Scholar]
- 33.Yamane K, Mizuguchi T, Cui B, Zofall M, Noma K, Grewal SI. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J Biol Chem. 2007;282:1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- 35.Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G. Regulation of replication fork progression through histone supply and demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 36.Huang S, Zhou H, Katzmann D, Hochstrasser M, Atanasova E, Zhang Z. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc Natl Acad Sci U S A. 2005;102:13410–13415. doi: 10.1073/pnas.0506176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. Embo J. 2007;26:2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Huang H, Zhou BO, Wang SS, Hu Y, Li X, Liu J, Zang J, Niu L, Wu J, Zhou JQ, Teng M, Shi Y. Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing. J Biol Chem. 285:4251–4262. doi: 10.1074/jbc.M109.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 41.Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]
- 43.Franco AA, Lam WM, Burgers PM, Kaufman PD. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 2005;19:1365–1375. doi: 10.1101/gad.1305005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.English CM, Adkins MW, Carson JJ, Churchill MEA, Tyler JK. Structural Basis for the Histone Chaperone Activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das C, Tyler JK, Churchill ME. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci. 2010;35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quivy J-P, Grandi P, Almouzni G. Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J. 2001;20:2015–2027. doi: 10.1093/emboj/20.8.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowman A, Ward R, Wiechens N, Singh V, El-Mkami H, Norman DG, Owen-Hughes T. The Histone Chaperones Nap1 and Vps75 Bind Histones H3 and H4 in a Tetrameric Conformation, Molecular. Cell. 2011;41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito T, Bulger M, Kobayashi R, Kadonaga JT. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han J, Zhou H, Li Z, Xu RM, Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J Biol Chem. 2007;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- 50.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 51.Jackson V, Shires A, Tanphaichitr N, Chalkley R. Modifications to histones immediately after synthesis. J Mol Biol. 1976;104:471–483. doi: 10.1016/0022-2836(76)90282-5. [DOI] [PubMed] [Google Scholar]
- 52.Annunziato AT, Hansen JC. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 2000;9:37–61. doi: 10.3727/000000001783992687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, Strack CL, Cook RG, Mizzen CA, Annunziato AT. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- 55.Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 56.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 57.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 58.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 59.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 60.Ejlassi-Lassallette A, Mocquard E, Arnaud MC, Thiriet C. H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Mol Biol Cell. 2011;22:245–255. doi: 10.1091/mbc.E10-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes & Development. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J Biol Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- 64.Xhemalce B, Miller KM, Driscoll R, Masumoto H, Jackson SP, Kouzarides T, Verreault A, Arcangioli B. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J Biol Chem. 2007;282:15040–15047. doi: 10.1074/jbc.M701197200. [DOI] [PubMed] [Google Scholar]
- 65.Keck KM, Pemberton LF. Interaction with the Histone Chaperone Vps75 Promotes Nuclear Localization and HAT Activity of Rtt109 In Vivo. Traffic. 2011;12:826–839. doi: 10.1111/j.1600-0854.2011.01202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh MC, Greenblatt JF. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol. 2008;28:4342–4353. doi: 10.1128/MCB.00182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 68.Maas NL, Miller KM, DeFazio LG, Toczyski DP. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 69.Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, Grunstein M. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 72.Jasencakova Z, Scharf AN, Ask K, Corpet A, Imhof A, Almouzni G, Groth A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 73.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. Embo J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burgess RJ, Zhou H, Han J, Zhang Z. A role for Gcn5 in replication-coupled nucleosome assembly. Mol Cell. 37:469–480. doi: 10.1016/j.molcel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 78.Li Q, Fazly AM, Zhou H, Huang S, Zhang Z, Stillman B. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 2009;5:e1000684. doi: 10.1371/journal.pgen.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 2010;17:430–437. doi: 10.1038/nsmb.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Espinosa MC, Rehman MA, Chisamore-Robert P, Jeffery D, Yankulov K. GCN5 is a positive regulator of origins of DNA replication in Saccharomyces cerevisiae. PLoS One. 2010;5:e8964. doi: 10.1371/journal.pone.0008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 82.Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A, Nakatani Y, Dent SY, Almouzni G. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scharf AN, Barth TK, Imhof A. Establishment of histone modifications after chromatin assembly. Nucleic Acids Res. 2009;37:5032–5040. doi: 10.1093/nar/gkp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ahmad K, Henikoff S. The Histone Variant H3.3 Marks Active Chromatin by Replication-Independent Nucleosome Assembly. Molecular Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 85.Jackson V MS, Chalkley R. The sites of deposition of newly synthesized histone. Nucleic Acids Research. 1981;9:4563–4581. doi: 10.1093/nar/9.18.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perry CA, Dadd CA, Allis CD, Annunziato AT. Analysis of nucleosome assembly and histone exchange using antibodies specific for acetylated H4. Biochemistry. 1993;32:13605–13614. doi: 10.1021/bi00212a028. [DOI] [PubMed] [Google Scholar]
- 87.Davie MJHaJR. Nucleosomal histones of transcriptionally active/competent chromatin preferentially exchange with newly synthesized histones in quiescent chicken erythrocytes. Biochem J. 1990;271:67–73. doi: 10.1042/bj2710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kimura H, Cook PR. Kinetics of Core Histones in Living Human Cells. J Cell Biol. 2001;153:1341–1354. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goldberg AD, Banaszynski LA, Noh K-M, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee Y-L, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of Replication-Independent Histone Turnover in Budding Yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 92.Mito Y, Henikoff JG, Henikoff S. Histone Replacement Marks the Boundaries of cis-Regulatory Domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 93.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kulaeva OI, Studitsky VM. Mechanism of histone survival during transcription by RNA polymerase II. Transcription. 2010;1:85–88. doi: 10.4161/trns.1.2.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kristjuhan A, Svejstrup JQ. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. Embo J. 2010;29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schwabish MA, Struhl K. Evidence for Eviction and Rapid Deposition of Histones upon Transcriptional Elongation by RNA Polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petesch SJ, Lis JT. Rapid Transcription-Independent Loss of Nucleosomes over a Large Chromatin Domain at Hsp70 Loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao J, Herrera-Diaz J, Gross DS. Domain-Wide Displacement of Histones by Activated Heat Shock Factor Occurs Independently of Swi/Snf and Is Not Correlated with RNA Polymerase II Density. Mol Cell Biol. 2005;25:8985–8999. doi: 10.1128/MCB.25.20.8985-8999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thiriet C, Hayes JJ. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes & Development. 2005;19:677–682. doi: 10.1101/gad.1265205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jamai A, Imoberdorf RM, Strubin M. Continuous Histone H2B and Transcription-Dependent Histone H3 Exchange in Yeast Cells outside of Replication. Molecular Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 102.Rufiange A, Jacques P-t, Bhat W, Robert F, Nourani A. Genome-Wide Replication-Independent Histone H3 Exchange Occurs Predominantly at Promoters and Implicates H3 K56 Acetylation and Asf1. Molecular Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 103.Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaufman PD, Cohen JL, Osley MA. Hir Proteins Are Required for Position-Dependent Gene Silencing in Saccharomyces cerevisiae in the Absence of Chromatin Assembly Factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sherwood PW, Tsang SV, Osley MA. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:28–38. doi: 10.1128/mcb.13.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 107.Ray-Gallet D, Quivy J-P, Scamps C, Martini EMD, Lipinski M, Almouzni G. HIRA Is Critical for a Nucleosome Assembly Pathway Independent of DNA Synthesis. Molecular Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 108.Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, Canutescu A, Dunbrack R, Adams PD, Marmorstein R. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol. 2006;13:921–929. doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Corpet A, De Koning L, Toedling J, Savignoni A, Berger F, Lemaitre C, O’Sullivan RJ, Karlseder J, Barillot E, Asselain B, Sastre-Garau X, Almouzni G. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 2011;30:480–493. doi: 10.1038/emboj.2010.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, Pehrson JR, Berger JM, Kaufman PD, Adams PD. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 111.Lewis PW, Elsaesser SJ, Noh K-M, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proceedings of the National Academy of Sciences. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. RNA Polymerase II Elongation Factors of Saccharomyces cerevisiae: a Targeted Proteomics Approach. Mol Cell Biol. 2002;22:6979–6992. doi: 10.1128/MCB.22.20.6979-6992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Formosa T. FACT and the reorganized nucleosome. Molecular BioSystems. 2008;4:1085–1093. doi: 10.1039/b812136b. [DOI] [PubMed] [Google Scholar]
- 114.Moshkin YM, Armstrong JA, Maeda RK, Tamkun JW, Verrijzer P, Kennison JA, Karch F. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 2002;16:2621–2626. doi: 10.1101/gad.231202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, Fyodorov DV. CHD1 Motor Protein Is Required for Deposition of Histone Variant H3.3 into Chromatin in Vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, Abbott A, Wilder SP, Taylor S, Santos GM, Cross J, Ayyub H, Jones S, Ragoussis J, Rhodes D, Dunham I, Higgs DR, Gibbons RJ. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 117.Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, Ueda T, Umetsu D, Ito T, Takeyama K-i, Kato S. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes & Development. 2010;24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zee Barry M, Levin Rebecca S, DiMaggio Peter A, Garcia BA. Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin. 2010;3:22. doi: 10.1186/1756-8935-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Iii, Gingeras TR, Schreiber SL, Lander ES. Genomic Maps and Comparative Analysis of Histone Modifications in Human and Mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 120.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 121.Kang B, Pu M, Hu G, Wen W, Dong Z, Zhao K, Stillman B, Zhang Z. Phosphorylation of H4 serine 47 promotes HIRA-mediated nucleosome assembly. Genes and Development. 2011:1359–1364. doi: 10.1101/gad.2055511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ito T, Umehara T, Sasaki K, Nakamura Y, Nishino N, Terada T, Shirouzu M, Padmanabhan B, Yokoyama S, Ito A, Yoshida M. Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors. Chemistry & Biology. 2011;18:495–507. doi: 10.1016/j.chembiol.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 123.Szenker E, Ray-Gallet D, Almouzni G. The double face of the histone variant H3.3. Cell Res. 2011;21:421–434. doi: 10.1038/cr.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Han J, Li Q, McCullough L, Kettelkamp C, Formosa T, Zhang Z. Ubiquitylation of FACT by the Cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes & Development. 2010;24:1485–1490. doi: 10.1101/gad.1887310. [DOI] [PMC free article] [PubMed] [Google Scholar]