Abstract
In healthy humans, 60–70% of the B lymphocytes produce kappa light chains, while the remaining cells produce lambda light chains. Malignant transformation and clonal expansion of B lymphocytes lead to an altered kappa : lambda expression ratio, which is an important diagnostic criteria of lymphomas. Here, we compared three methods for clonality determination of suspected B cell lymphomas. Tumor biopsies from 55 patients with B cell malignancies, 5 B-lymphoid tumor cell lines, and 20 biopsies from patients with lymphadenitis were analyzed by immunohistochemistry, flow cytometry, and reverse transcription quantitative real-time PCR. Clonality was determined by immunohistochemistry in 52/53 cases, flow cytometry in 30/39 cases, and reverse transcription quantitative real-time PCR in 33/55 cases. In conclusion, immunohistochemistry was superior to flow cytometry and reverse transcription quantitative real-time PCR for clonality identification. Flow cytometry and reverse transcription quantitative real-time PCR analysis has complementary values. In a considerable number of cases tumor cells produced both kappa and lambda light chain transcripts, but only one type of light chain peptide was produced.
1. Introduction
B lymphocytes produce immunoglobulins consisting of a heavy chain and either a kappa (IGKC) or a lambda (IGLC) light chain. Each B lymphocyte decides early by the rearrangement of its immunoglobulin genes which light chain to produce [1]. The excluded light chain gene is not properly rearranged or remains in germ line configuration [2]. In healthy humans 60–70% of the B cells produce kappa chains and the rest produce lambda chains [1, 3]. Normal lymphoid tissues therefore contain a mixture of B cells that express IGKC and IGLC at a ratio of about 60 : 40 = 1.5.
Tumors of B cell origin are monoclonal and arise from one transformed cell. The single cell origin of malignant clones results in exclusive expression of IGKC or IGLC light chains in the vast majority of all B cell malignancies although B cell tumors that produce both kappa and lambda chains have been reported [4]. The clonal expression of IGKC or IGLC is thus used as an important diagnostic marker for B cell malignancies and currently determined on protein level by immunohistochemistry (IHC), flow cytometry (FC), or enzyme-linked immunosorbent assay techniques. Previously, we used reverse transcription quantitative real-time PCR (RT-qPCR) to quantify IGKC and IGLC gene transcripts in a small set of lymphomas and found that also gene expression level clonality was frequently evident [5]. In the present study we have used the same RT-qPCR method together with IHC and FC to analyze a larger cohort of 39 non-Hodgkin lymphomas, 16 chronic lymphatic leukemias, and 5 B cell derived tumor cell lines. The non-Hodgkin lymphomas consisted of 20 diffuse large B cell lymphomas, 16 follicular lymphomas, and 3 mantle cell lymphomas.
2. Material and Methods
2.1. Biopsies, Flow Cytometry, and Immunohistochemistry
The samples were transported from the operation theatre in ice-water-chilled boxes, handled in the laboratory within 30 min, and stored at −140°C. Parts of the tissues were fixed in formalin and embedded in paraffin according to the routine protocols of the pathology laboratory. Diagnosis was reached by a combination of microscopic histological evaluation, IHC of several markers, including the κ and λ chains, and in some cases by FC. Series of 5 μm tissue sections were cut from each biopsy, deparaffinized, rehydrated, and stained with the following antibodies: rabbit anti-human kappa light chains and rabbit anti-human lambda chains (A0191 and A0194, DAKO). The antibodies were used at a dilution of 1 : 400 and bound antibodies were visualized using the second labeled antibody streptavidin biotin peroxidase system (DAKO). Stained sections were examined with a light microscope. For FC, a routine protocol for preparation of lymphoid cells from lymphoma tissue was used. Direct labeled antibodies specific for immunoglobulin kappa and lambda chains (A0191 and A0194, DAKO) were used for staining of recovered cells. First a gate for lymphocytes was set using forward and side scatter, followed by gates for kappa and lambda staining cells. We used a kappa : lambda ratio less than 0.4 or greater than 5 to prove FC monoclonality [6]. The samples were classified according to the Revised European-American Lymphoma and World Health Organization classification system.
2.2. RNA Extraction and RT-qPCR
RNA was extracted by use of the Fast Prep System (FastRNA Green; Qbiogene). We mixed 10 μg of total RNA with 2 μg of poly (dT) oligomers (Pharmacia) and incubated the mixture at 65°C for 5 min. First-strand cDNA synthesis was performed by adding 0.05 mol/L Tris-HCl (pH 8.3), 0.075 mol/L KCl, 3 mmol/L MgCl2, 0.01 mol/L dithiothreitol, 10 U/mL Moloney murine leukemia virus reverse transcriptase (Life Technologies), 0.05 units/mL RNA guard (Life Technologies), and 10 mmol/L of each deoxyribonucleotide (Life Technologies) to a final volume of 20 μL and incubating the samples at 37°C for 1 h. The reaction was terminated by incubation at 65°C for 5 min, and samples were stored at −80°C. Real-time PCR measurements were performed on a Rotorgene 3000 (Corbett Research). Each PCR reaction contained 10 mmol/L Tris-HCl pH 8.3, 50 mmol/L KCl, 4 mM MgCl2, 400 μmol/L each dNTP, 300 nmol/L each primer, 0.04 U/μL Jumpstart Taq (Sigma), and 0.25x SYBR Green (Molecular Probes) in a 20 μL reaction volume. The temperature profile was 95°C for 3 min followed by 40 cycles of amplification (95°C for 20 sec, 60°C for 20 sec, and 72°C for 20 sec). Primer sequences and detailed assay performance for IGKC and IGLC are reported elsewhere [5]. Formation of correctly sized PCR products was confirmed by agarose gel electrophoresis for all assays and melting curve analysis for all samples. RT-qPCR and statistical analysis of the data were performed as previously described [5]. A 95% confidence region for the IGKC : IGLC ratio using the negative lymphadenitis was used to prove RT-qPCR monoclonality.
2.3. RT-PCR Cloning and Sequencing
PCR products were obtained as for QPCR analysis and fractionated by agarose electrophoresis. Fragments were excised from the gels and used as templates for DNA sequencing as previously described [7].
3. Results and Discussion
The samples were analyzed by IHC, FC, and RT-qPCR for expression of IGKC and IGLC light chains (Tables 1 and 2). IHC indicated an exclusive or heavily dominant expression of IGKC or IGLC chains indicative of a monoclonal origin in all but one analyzed samples (98%). RT-qPCR analysis indicated a monoclonal dominance in 33 of 55 (60%) analyzed samples, while FC scored monoclonal dominance in 30 of 39 (77%) analyzed samples. Thus the IHC method was superior to FC and RT-qPCR in detecting monoclonality. The divergent results may in part be explained by the fact that biopsies, besides the tumor cells, often contain considerable numbers of normal lymphocytes that could contribute to the RNA samples analyzed by RT-qPCR and the cell population studied by flow cytometry. IHC analysis, on the other hand, allows for selective analysis of representative parts of the tumor tissue thus avoiding contribution of normal B lymphocytes. Furthermore, it is also known that 60–70% of non-Hodgkin lymphomas lack surface expression of IGLK and IGLC [8].
Table 1.
Detailed patient information.
| Sample ID | Classification | Gender | Age | FC | IHC | RT-qPCR |
|---|---|---|---|---|---|---|
| 111 | CLL | F | 70 | ND | Lambda | Lambda |
| 112 | CLL | F | 82 | ND | Lambda | MNP |
| 113 | CLL | M | 58 | Lambda | Lambda | Lambda |
| 115 | CLL | F | 61 | Lambda | Lambda | Lambda |
| 116 | CLL | F | 57 | Kappa | Kappa | Kappa |
| 117 | CLL | F | 75 | Kappa | Kappa | Kappa |
| 118 | CLL | F | 85 | Kappa | Kappa | Kappa |
| 119 | CLL | F | 48 | ND | Lambda | Lambda |
| 131 | CLL | M | 72 | ND | Lambda | Lambda |
| 132 | CLL | F | 59 | ND | Lambda | Lambda |
| 145 | CLL | F | 76 | Kappa | Kappa | MNP |
| 146 | CLL | M | 46 | Lambda | Lambda | Lambda |
| 147 | CLL | F | 74 | Kappa | Kappa | Kappa |
| 149 | CLL | F | 76 | Kappa | Kappa | MNP |
| 189 | CLL | F | 65 | Kappa | Kappa | MNP |
| 191 | CLL | M | 60 | Lambda | Lambda | Lambda |
| 120 | DLBL | M | 80 | ND | Kappa | MNP |
| 121 | DLBL | M | 81 | ND | Kappa | Kappa |
| 123 | DLBL | M | 64 | Kappa | Kappa | Kappa |
| 124 | DLBL | F | 77 | Kappa | Kappa | Kappa |
| 125 | DLBL | M | 57 | ND | MNP | Lambda |
| 126 | DLBL | M | 70 | Lambda | Lambda | Lambda |
| 127 | DLBL | M | 70 | Kappa | Kappa | Kappa |
| 129 | DLBL | M | 58 | ND | Kappa | MNP |
| 133 | DLBL | M | 72 | MNP | Lambda | MNP |
| 135 | DLBL | F | 75 | MNP | Kappa | Kappa |
| 136 | DLBL | F | 78 | ND | Kappa | MNP |
| 137 | DLBL | M | 70 | Kappa | Kappa | Kappa |
| 138 | DLBL | M | 72 | ND | Kappa | MNP |
| 139 | DLBL | M | 33 | Kappa | Kappa | Kappa |
| 140 | DLBL | F | 78 | Kappa | Kappa | kappa |
| 153 | DLBL | M | 19 | ND | Kappa | Kappa |
| 156 | DLBL | M | 58 | Kappa | Kappa | Kappa |
| 182 | DLBL | F | 75 | MNP | Kappa | MNP |
| 184 | DLBL | M | 55 | Kappa | Kappa | Kappa |
| 186 | DLBL | F | 66 | Kappa | Kappa | Kappa |
| 157 | FL | F | 54 | Kappa | Kappa | MNP |
| 158 | FL | F | 51 | ND | Kappa | MNP |
| 159 | FL | M | 77 | ND | Kappa | MNP |
| 160 | FL | F | 60 | ND | Lambda | MNP |
| 161 | FL | M | 70 | Lambda | Lambda | MNP |
| 162 | FL | M | 69 | Kappa | Kappa | Kappa |
| 163 | FL | M | 52 | Kappa | Kappa | Kappa |
| 164 | FL | M | 53 | ND | Kappa | Kappa |
| 165 | FL | F | 59 | Kappa | Kappa | MNP |
| 166 | FL | M | 61 | MNP | Kappa | MNP |
| 167 | FL | F | 30 | MNP | Lambda | MNP |
| 168 | FL | M | 51 | MNP | Kappa | Kappa |
| 208 | FL | F | 83 | MNP | ND | MNP |
| 209 | FL | F | 55 | Lambda | Lambda | Lambda |
| 210 | FL | F | 46 | MNP | Kappa | MNP |
| 211 | FL | M | 64 | Lambda | ND | MNP |
| 204 | MCL | M | 61 | Lambda | Lambda | Lambda |
| 206 | MCL | F | 78 | Kappa | Kappa | Kappa |
| 207 | MCL | F | 70 | Kappa | Kappa | MNP |
| 101 | LA | F | 62 | ND | MNP | Control |
| 102 | LA | M | 25 | ND | MNP | Control |
| 103 | LA | M | 25 | ND | MNP | Control |
| 104 | LA | M | 16 | ND | MNP | Control |
| 105 | LA | F | 58 | ND | MNP | Control |
| 106 | LA | M | 59 | ND | MNP | Control |
| 107 | LA | M | 41 | ND | MNP | Control |
| 108 | LA | M | 7 | ND | MNP | Control |
| 109 | LA | F | 33 | ND | MNP | Control |
| 110 | LA | F | 28 | ND | MNP | Control |
| 192 | LA | F | 1 | MNP | MNP | Control |
| 193 | LA | F | 40 | MNP | MNP | Control |
| 194 | LA | M | 28 | MNP | MNP | Control |
| 195 | LA | M | 20 | MNP | MNP | Control |
| 196 | LA | M | 13 | MNP | MNP | Control |
| 197 | LA | M | 30 | MNP | MNP | Control |
| 198 | LA | F | 10 | MNP | MNP | Control |
| 199 | LA | F | 58 | MNP | MNP | Control |
| 200 | LA | M | 49 | MNP | MNP | Control |
| 201 | LA | F | 57 | MNP | MNP | Control |
CLL: chronic lymphocytic leukemia; DLBL: diffuse large B cell lymphoma; FL: follicular lymphoma; LA: lymphadenitis; F: female; M: male; MNP: monoclonality not proven; FC: flow cytometry; IHC: immunohistochemistry.
Table 2.
Monoclonal populations detected in lymphoma and lymphadenitis biopsiesa.
| RT-qPCR | FC | IHC | |
|---|---|---|---|
| CLL | 12/16 | 11/11 | 16/16 |
| DLBL | 14/20 | 10/13 | 19/20 |
| FL | 5/16 | 6/12 | 14/14 |
| MCL | 2/3 | 3/3 | 3/3 |
| CLL/lymphoma total | 33/55 | 30/39 | 52/53 |
| LA | 0/20 | 0/10 | 0/20 |
aNumber of samples with clonality proven/total number of samples is shown. CLL: chronic lymphocytic leukemia; DLBL: diffuse large B cell lymphoma; FL: follicular lymphoma; MCL: mantle cell lymphoma; LA: lymphadenitis (polyclonal control).
FC was more efficient than RT-qPCR in identifying clonal populations. However, there was an overall good correlation in expression levels of IGKC and IGLC between FC and RT-qPCR data despite the fact that FC data reflects cell number and RT-qPCR data reflects transcript numbers (Figure 1). In two cases (samples 135 and 168, Table 1), RT-qPCR detected clonal populations where FC failed. RT-qPCR may therefore serve as a valuable complement to IHC and FC in detection of monoclonal B cell populations. Moreover, RT-qPCR analysis can also be employed on minute fine needle aspirate samples that are not sufficient for flow cytometry or IHC and the same sample may also be analyzed simultaneously for expression of more marker genes.
Figure 1.
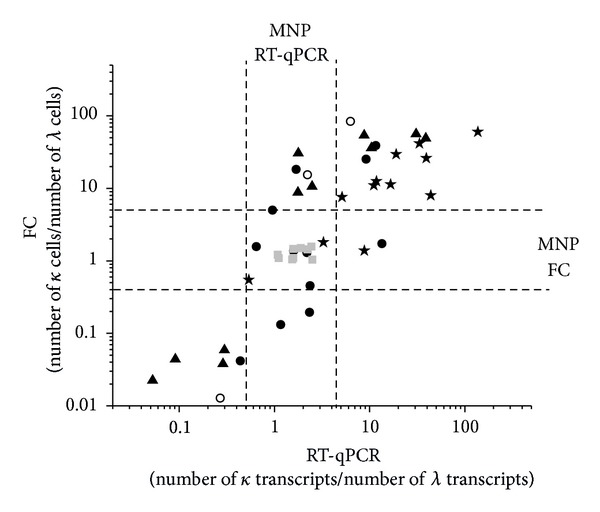
Comparison of IGKC : IGLC ratio between FC and RT-qPCR. The Pearson correlation coefficient is 0.65 (P < 0.01). The FC ratio refers to cell number, while the RT-qPCR ratio refers to transcript number. The areas where no monoclonality (MNP) could be proven are shown as dashed lines. Grey square, lymphadenitis; stars, diffuse large B cell lymphoma; triangles, chronic lymphocytic leukemia; dot, follicular lymphoma; circles, mantle cell lymphoma; κ, IGKC; λ, IGLC.
Several of the IGKC and IGLC cotranscribing tumors appeared by IHC and FC to consist of homogenous tumor cell populations with no or few normal lymphocytes suggesting that some malignant B cell clones were dual producers of light chain mRNA (Figure 2). Dual production of IGLC chain mRNAs and proteins has been reported for various types of malignant B cell clones and normal B lymphocytes [4, 9]. These reports and our data prompted us to analyze five monoclonal B cell derived tumor cell lines (Table 3). These cell lines produce only one light chain species as analyzed by immunofluorescence, but four of them contained both IGKC and IGLC transcripts. To further confirm this result we isolated PCR products from two of the cell lines and sequence analysis showed that IGKC and IGLC transcripts were indeed produced simultaneously in these cell lines. Thus we conclude that human monoclonal B-lymphoid populations frequently simultaneously produce IGKC and IGLC transcripts.
Figure 2.
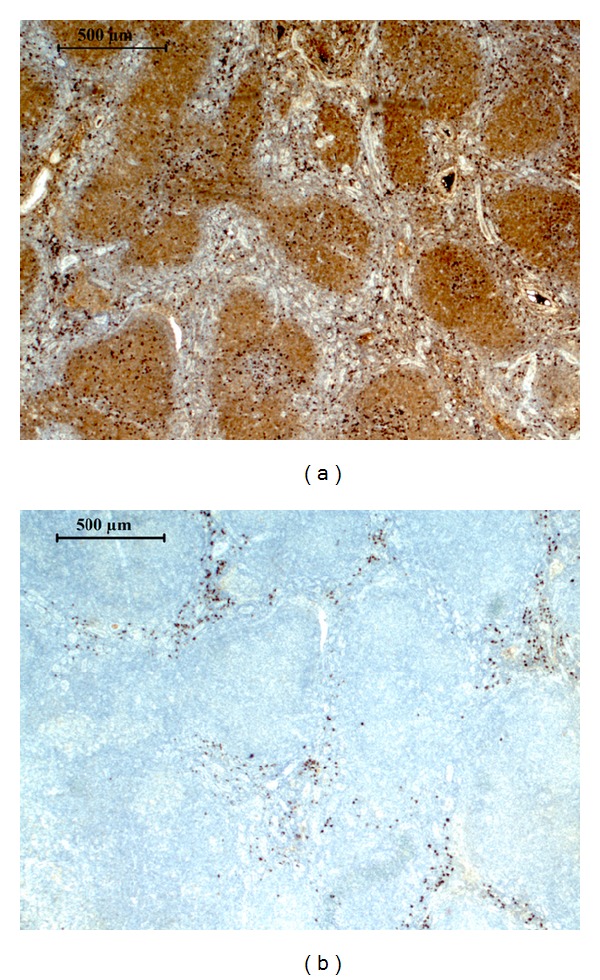
Immunohistochemistry of sample 165. The follicular lymphoma sample 165 was stained for (a) IGKC and (b) IGLC. Using RT-qPCR no clonality could be proven for this IGKC IHC positive sample.
Table 3.
RT-qPCR data for 5 B-lymphoid tumor cell lines.
| Cellinje | Kappa Cq | Lambda Cq | Kappa noRT Cq | Lambda noRT Cq | mRNA expression | IHC |
|---|---|---|---|---|---|---|
| U698 | 10.3 | 13.8 | 27 | 23.5 | Kappa > Lambda | Kappa |
| DG75 | 10.4 | 15.9 | 25.2 | 24.2 | Kappa > Lambda | Kappa |
| MC116 | 34.8 | 10.4 | Not detected | 23.5 | Lambda only | Lambda |
| RPMI8226 | 12.2 | 14.4 | 25.1 | 22.2 | Kappa > Lambda | Lambda |
| CCRF-SB | 12.3 | 11.2 | 26.7 | 24.4 | Lambda </≈ Kappa | Kappa |
| Neg. control | >33 | ~30 |
Cq: cycle of quantification (log transcript numbers); noRT: samples without reverse transcriptase; IHC: immunohistochemistry.
We searched for possible correlations between dual light chain producers and tumor type. Dual producers were most common in the follicular lymphoma group in which 11/16 tumors failed to score for monoclonality based on RT-qPCR analysis. This could possibly be explained by larger abundance of normal cells in follicular lymphomas since FC showed highest failure rate for these samples, and may also depend on a higher frequency of dual producers.
Eleven of twelve cases where monoclonality was not detected by RT-qPCR produced IGKC protein as judged by IHC. This is more than expected from the normal ratio of IGKC and IGLC producers among normal B lymphocytes and lymphoma populations. In the present cohort of IHC investigated lymphomas, 30% were lambda producers. Thus, it is possible that dual mRNA producers arise more often among kappa protein producing cells. This is surprising since kappa chains are considered to be rearranged first and IGLC rearrangement is pursued only if IGKC rearrangement fails to produce a functional IGKC protein [10]. The activation of IGLC transcription is most likely a result of rearrangement that occurs after IGKC protein is produced and may therefore indicate a failure to inactivate the rearrangement mechanism. Failure to regulate a potentially risky DNA rearrangement process may also contribute to the frequent multiple genetic rearrangements in lymphomas. Age and gender were not correlated to the determined clonality (data not shown).
4. Conclusions
We conclude that IHC, FC, and RT-qPCR analysis of human B-lymphoid tumors gives partially divergent results. This may in part be explained by contribution of normal B lymphocytes into the tumor tissue. Our results also suggest that dual transcription of IGKC and IGLC loci is common in these tumors. This has to be taken in consideration when RT-qPCR analysis of IGKC and IGLC is used for diagnostic purpose. RT-qPCR remains an interesting complement for diagnostic purpose when only small samples, such as from fine needle cytology, are available.
Acknowledgments
This work was supported by Swedish Society for Medical Research, Swedish Cancer Foundation, Swedish Children Cancer Foundation, Swedish Research Council, LUA-ALF grants, BioCARE National Strategic Research Program at University of Gothenburg, Ministry of Health Grant-IGA-NS (NS 9976-3), and Grant Agency of the Czech Republic (IAA500970904).
Conflict of Interests
The IGKC-IGLC RT-qPCR test is patent granted (WO02099135).
References
- 1.Levy R, Warnke R, Dorfman RF, Haimovich J. The monoclonality of human B cell lymphomas. Journal of Experimental Medicine. 1977;145(4):1014–1028. doi: 10.1084/jem.145.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang H-E, Hsu L-Y, Cado D, Schlissel MS. Variegated transcriptional activation of the immunoglobulin κ locus in pre-B cells contributes to the allelic exclusion of light-chain expression. Cell. 2004;118(1):19–29. doi: 10.1016/j.cell.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 3.Barandun S, Morell A, Skvaril F, Oberdorfer A. Deficiency of kappa or λ type immunoglobulins. Blood. 1976;47(1):79–89. [PubMed] [Google Scholar]
- 4.Xu D. Dual surface immunoglobulin light-chain expression in B-cell lymphoproliferative disorders. Archives of Pathology and Laboratory Medicine. 2006;130(6):853–856. doi: 10.5858/2006-130-853-DSILEI. [DOI] [PubMed] [Google Scholar]
- 5.Ståhlberg A, Åman P, Ridell B, Mostad P, Kubista M. Quantitative real-time PCR method for detection of B-lymphocyte monoclonality by comparison of κ and λ immunoglobulin light chain expression. Clinical Chemistry. 2003;49(1):51–59. doi: 10.1373/49.1.51. [DOI] [PubMed] [Google Scholar]
- 6.Chizuka A, Kanda Y, Nannya Y, et al. The diagnostic value of kappa/lambda ratios determined by flow cytometric analysis of biopsy specimens in B-cell lymphoma. Clinical and Laboratory Haematology. 2002;24(1):33–36. doi: 10.1046/j.1365-2257.2002.00175.x. [DOI] [PubMed] [Google Scholar]
- 7.Göransson M, Elias E, Ståhlberg A, Olofsson A, Andersson C, Åman P. Myxoid liposarcoma FUS-DDIT3 fusion oncogene induces C/EBP β-mediated interleukin 6 expression. International Journal of Cancer. 2005;115(4):556–560. doi: 10.1002/ijc.20893. [DOI] [PubMed] [Google Scholar]
- 8.Picker LJ, Weiss LM, Medeiros LJ. Immunophenotypic criteria for the diagnosis of non-Hodgkin’s lymphoma. The American Journal of Pathology. 1987;128(1):181–201. [PMC free article] [PubMed] [Google Scholar]
- 9.Giachino C, Padovan E, Lanzavecchia A. κ+ λ+ Dual receptor B cells are present in the human peripheral repertoire. Journal of Experimental Medicine. 1995;181(3):1245–1250. doi: 10.1084/jem.181.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alt FW, Enea V, Bothwell ALM, Baltimore D. Activity of multiple light chain genes in murine myeloma cells producing a single, functional light chain. Cell. 1980;21(1):1–12. doi: 10.1016/0092-8674(80)90109-9. [DOI] [PubMed] [Google Scholar]


