Abstract
Enterococci are nosocomial pathogen with multiple-drug resistance by intrinsic and extrinsic mechanisms. Aminoglycosides along with cell wall inhibitors are given clinically for treating enterococcal infections. 178 enterococcal isolates were analyzed in this study. E. faecalis is identified to be the predominant Enterococcus species, along with E. faecium, E. avium, E. hirae, E. durans, E. dispar and E. gallinarum. High level aminoglycoside resistance (HLAR) by MIC for gentamicin (GM), streptomycin (SM) and both (GM + SM) antibiotics was found to be 42.7%, 29.8%, and 21.9%, respectively. Detection of aminoglycoside modifying enzyme encoding genes (AME) in enterococci was identified by multiplex PCR for aac(6′)-Ie-aph(2′′)-Ia; aph(2′′)-Ib; aph(2′′)-Ic; aph(2′′)-Id and aph(3′)-IIIa genes. 38.2% isolates carried aac(6′)-Ie-aph(2′′)-Ia gene and 40.4% isolates carried aph(3′)-IIIa gene. aph(2′′)-Ib; aph(2′′)-Ic; aph(2′′)-Id were not detected among our study isolates. aac(6′)-Ie-aph(2′′)-Ia and aph(3′)-IIIa genes were also observed in HLAR E. durans, E. avium, E. hirae, and E. gallinarum isolates. This indicates that high level aminoglycoside resistance genes are widely disseminated among isolates of enterococci from Chennai.
1. Introduction
Enterococci have emerged as an important multiple-drug resistant nosocomial pathogen reported worldwide. Its resistance to wider range of antimicrobial agents particularly, aminoglycosides, glycopeptides and beta-lactams had increasingly been documented [1]. Although enterococci are intrinsically resistant to low levels of aminoglycosides, high level resistance to aminoglycosides (MIC ≥ 2000 μg/mL) is mediated by acquisition of genes encoding AMEs. High level gentamicin resistance (MIC ≥ 500 μg/mL) in enterococci is predominantly mediated by aac(6′)-Ie-aph(2′′)-Ia, which encodes the bifunctional aminoglycoside modifying enzyme AAC(6′)-APH(2′′). The action of this enzyme in enterococci eliminates the synergistic activity of gentamicin when combined with a cell wall active agent, such as ampicillin or vancomycin. Recently, newer AME genes such as aph(2′′)-Ib, aph(2′′)-Ic, and aph(2′′)-Id have been detected as those conferring gentamicin resistance in enterococci. High level streptomycin and kanamycin resistance in enterococci are mediated by aph(3′)-IIIa gene encoding aminoglycoside phosphotransferase enzyme, APH(3′)-IIIa [2]. In India, high level aminoglycoside resistance has been reported from different centers; however, studies on prevalence of these resistance genes are limited. The goal of this study is to determine, the rate of high level aminoglycoside resistance and aminoglycoside resistance encoding genes in enterococcal isolates collected from different specimen sources in Chennai, India.
2. Materials and Methods
2.1. Bacterial Strains
A total of 178 nonidentical clinical isolates of enterococci were obtained from clinical specimens from various tertiary care centers from Chennai, during a period of 2010–2012. Appropriate inpatient details were collected and recorded to avoid identical isolates from the same patient. An Institutional ethical clearance was obtained for conducting this study (reference number: 1168). The strains were initially grown on MacConkey agar (MV082) and Enterococcus confirmatory agar (M392) (HiMedia, Mumbai, India). Species characterization was carried out by carbohydrate fermentation test using 1% sugars such as sucrose, sorbose, sorbitol, mannitol, glucose, pyruvate, inulin, ribose, melibiose, raffinose, arabinose, and arginine. All the isolates were confirmed for genus and species by standard protocols [3]. Enterococcus faecium and Enterococcus faecalis were further confirmed by PCR analysis using specific ddl E.faecium and ddl E.faecalis genes, respectively [4].
2.2. Minimum Inhibitory Concentration for Aminoglycosides
The isolates were confirmed as high level aminoglycoside resistant enterococci (HLARE) by considering growth ≥512 μg/mL for gentamicin and ≥2048 μg/mL for streptomycin. The overnight bacterial cultures were adjusted to 0.5 McFarland's turbidity and the inoculum was spot inoculated on the surface of brain heart infusion agar with increasing concentrations of gentamicin and streptomycin antibiotics (HiMedia, Mumbai, India). The plates were incubated at 37°C for 24 hrs and inspected for more than one colony forming units in the spotted area. Enterococcus faecalis ATCC 29212 was used as a negative control strain.
2.3. Molecular Analysis of Aminoglycoside Modifying Genes by PCR
The primers for AME genes such as aac(6′)-Ie-aph(2′′)-Ia; aph(2′′)-Ib; aph(2′′)-Ic; aph(2′′)-Id and aph(3′)-IIIa included in this study were previously described [5].
PCR was carried out with reaction tube containing 1 μL template DNA prepared from boiling lysis of bacterial suspension added to a 50 μL reaction mixture containing 25 mM Tris/HCl, 50 mM KCl, 1.5 mM MgCl2, 0.2 Mm of each dNTP (Bangalore Genei, India), and 1.5 U Taq polymerase (Bangalore Genei, India). First reaction with a pair of 25 pmol each of primers for aac(6′)-Ie-aph(2′′)-Ia and aph(3′)-IIIa (Sigma Aldrich, USA) and second reaction with aph(2′′)-Ib, aph(2′′)-Ic, aph(2′′)-Id primer sets separately.
Amplification was performed with PCR system (Eppendorf, Germany) and the cycling programs consisted of an initial denaturation (95°C, 5 min) followed by 32 cycles each of denaturation (95°C, 1 min), annealing (58°C, 1 min) and extension (72°C, 1 min), with a final extension of (72°C, 5 min). Each amplification product was resolved by electrophoresis with a 100-base pair molecular weight marker (Real Biotech Corporation, Taiwan) in a 1.2% agarose-Tris-borate-EDTA gel stained with ethidium bromide (0.5 μg/mL) and visualized under gel documentation system (BioRad, USA). Table 1 shows the product size of all the genes analyzed.
Table 1.
Primers and their sequences for aminoglycoside resistance encoding genes used in multiplex PCR.
| Genes | Primer sequences (5′-3′) | Size of PCR product (bp) | |
|---|---|---|---|
| aac(6′)-Ie-aph(2′′ )-Ia | F: CAGGAATTTATCGAAAATGGTAGAAAAG | R: CACAATCGACTAAAGAGTACCAATC | 369 |
| aph(2′′)-Ib | F: CTTGGACGCTGAGATATATGAGCAC | R: GTTTGTAGCAATTCAGAAACACCCTT | 867 |
| aph(2′′)-Ic | F: CCACAATGATAATGACTCAGTTCCC | R: CCACAGCTTCCGATAGCAAGAG | 444 |
| aph(2′′)-Id | F: GTGGTTTTTACAGGAATGCCATC | R: CCCTCTTCATACCAATCCATATAACC | 641 |
| aph(3′)-IIIa | F: GGCTAAAATGAGAATATCACCGG | R: CTTTAAAAAATCATACAGCTCGCG | 523 |
3. Results and Discussion
3.1. Identification of Enterococcus Species
Since early 1970s, Enterococci were considered as nosocomial pathogens. The incidences of high level GM/SM resistance have been disseminated in many Enterococcus species. Since then, the high level aminoglycoside resistance has become a serious problem in most of the health care settings; identification of clinical isolates of enterococci up to species level is essential for an appropriate management of the infection. The predominant species observed in our study was E. faecalis 86/178 (48.3%) as observed in previous studies [6] in our region. Other than E. faecium which was 80/178 (44.9%), we have also obtained E. avium (2%), E. hirae (1.6%), E. durans (0.6%), E. gallinarum, and E. dispar (1%). The species distribution and specimen source of isolates were listed in Table 2.
Table 2.
Distribution of Enterococcus species from various clinical specimens.
| Source of the isolates | Distribution of Enterococcus spp. (n = 178) | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| E. faecalis | E. faecium | E. durans | E. avium | E. hirae | E. dispar | E. gallinarum | ||
| Urine | 62 | 38 | — | 2 | 3 | 2 | — | 107 |
| DFU† | 9 | 2 | 1 | 1 | — | — | — | 13 |
| Blood | 7 | 35 | — | — | — | — | 2 | 44 |
| Pus | 5 | 3 | — | 1 | — | — | — | 9 |
| CSF | — | 1 | — | — | — | — | — | 1 |
| Vaginal/semen swab | 3 | 1 | — | — | — | — | — | 4 |
|
| ||||||||
| Total | 86 | 80 | 1 | 4 | 3 | 2 | 2 | 178 |
†DFU: diabetic foot ulcer isolates.
3.2. High Level Aminoglycoside Resistance in Enterococcal Isolates
Aminoglycoside antibiotics are considered efficient in treating serious infections caused by both gram positive and gram negative organisms. Due to acquisition of extrinsic resistance to high level aminoglycoside antibiotics in enterococci, these strains gain importance in clinical settings. A total of 178 enterococcal isolates were screened by MIC method, 76/178 (42.7%) were HLGR (MIC ≥ 512 μg/mL) for gentamicin and 53/178 (29.8%) were HLSR (MIC ≥ 2048 μg/mL) for streptomycin (Table 3). Although the clinical use of streptomycin for enterococci has long been restricted due to intrinsic low level resistance (LLR), the present study revealed HLSR strains.
Table 3.
Results of minimum inhibitory concentration of enterococcal isolates to gentamicin and streptomycin (n = 178).
| MIC (μg/mL) | Gentamicin (n = 178) |
Streptomycin (n = 178) |
|---|---|---|
| 0.50 | 0 | 0 |
| 1 | 0 | 0 |
| 2 | 1 | 1 |
| 4 | 6 | 0 |
| 8 | 8 | 0 |
| 16 | 44 | 0 |
| 32 | 15 | 0 |
| 64 | 5 | 12 |
| 128 | 19 | 85 |
| 256 | 4 | 11 |
| >512 | 76 | 2 |
| 1024 | ND† | 14 |
| >2048 | ND† | 53 |
†ND: not done; HLGR > 500 μg/mL; HLSR > 2000 μg/mL [7].
A total of 129/178 (72.47%) high level aminoglycoside resistant enterococci (HLGR and HLSR) were observed among our study isolates.
Recent studies also indicated HLGR to be more common in all species of enterococci than HLSR. Similarly, we had observed HLGR to be more predominant than HLSR in our study isolates. One E. avium, E. hirae, E. durans, and E. gallinarum isolates were exhibiting MIC of ≥512 μg/mL for gentamicin and ≥2048 μg/mL for streptomycin antibiotics. The reports on resistance carried by species other than E. faecalis and E. faecium were observed from late 1990s [8]. A surveillance study that analyzed 20 European countries had reported 32% and 22% HLGR and 41% and 49% HLSR among gentamicin resistant E. faecalis and E. faecium, respectively [9]. A very recent study conducted in Iran [10] had reported around 60.45% HLGR strains in their region. This is higher than our present report. They were suggesting cotransfer of these resistance genes along with VRE for the higher percentage of HLGR in their study. However, studies on AME gene profile were not done frequently in our region.
3.3. PCR Identification of HLAR Genes in Enterococci
All 178 enterococcal isolates were analyzed for the presence of aminoglycoside modifying enzyme coding genes (Figure 1).
Figure 1.
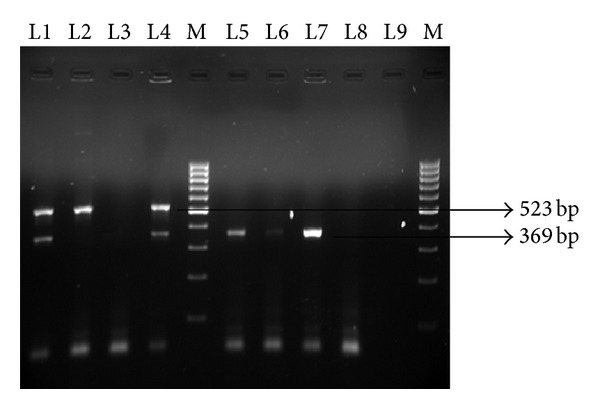
Amplified products of AME genes generated by multiplex PCR. L1, L2, L4-aph(3′)-IIIa positive (523 bp); L1, L4, L5, L6, L7 aac(6′)-Ie-aph(2′′)-Ia positive (369 bp); M-marker (100 bp DNA ladder).
High level gentamicin resistance is primarily due to the presence of bifunctional enzyme aac(6′)-Ie-aph(2′′)-Ia which also confers high level resistance to amikacin, tobramycin, kanamycin, netilmicin, and dibekacin except streptomycin [11]. aph(2′′)-Ib was first detected from E. faecium and E. coli and confers high level resistance to gentamicin, tobramycin, amikacin, kanamycin, netilmicin, and dibekacin but not to amikacin. aph(2′′)-Ic confers HLR to gentamicin, tobramycin, and kanamycin while the strains carrying them can be treated with amikacin, netilmicin, and streptomycin in combination with cell wall inhibitors. Earlier this gene was shown to be present in E. gallinarum; but it had also been reported in isolates obtained from farm animals and in E. faecalis and E. faecium [12]. aph(2′′)-Id was reported in E. casseliflavus and has similar mechanism to that of aph(2′′)-Ib [13].
aac(6′)-Ie-aph(2′′)-Ia gene was found in 38.2% of enterococcal isolates in our study. But, out of 76 strains of HLGR identified by MIC method, only 52 strains (68.4%) carried aac(6′)-Ie-aph(2′′)-Ia gene. However, 24/76 (31.57%) isolates that were high level gentamicin resistant and 12/53 (22.64%) isolates that were high level streptomycin resistant did not carry any of the genes tested. In a previous study [14], all the high level gentamicin resistant E. faecalis and E. faecium isolates were found to carry aac(6′)-Ie-aph(2′′)-Ia gene.
Newer aminoglycoside resistance genes such as aph(2′′)-Ib, aph(2′′)-Ic, and aph(2′′)-Id also found to encode high level resistance to gentamicin (>500 μg/mL) were not detected among our study isolates.
Another most important gene tested in our study, aph(3′)-IIIa (GenBank Accession number: KF550184) was detected among 40.4% (72/178) isolates of enterococci. Out of 53 high level streptomycin resistant isolates, 41 (77.4%) carried aph(3′)-IIIa gene.
20.2% (39/178) of strains carried both aac(6′)-Ie-aph(2′′)-Ia and aph(3′)-IIIa genes in our study. Out of 39 (21.9%) HLAR isolates which were resistant to both gentamicin (MIC > 512 μg/mL) and streptomycin (MIC > 2048 μg/mL) by MIC, 19 (48.7%) isolates carried both the genes, and the remaining 38.5% isolates had one of the genes either aac(6′)-Ie-aph(2′)-Ia or aph(3′)-IIIa gene, while 12.8% isolates did not carry neither of the genes tested.
E. faecalis, the predominant isolate of our study was found to carry aac(6′)-Ie-aph(2′′)-Ia gene in 34/86 (39.5%) isolates and aph(3′)-IIIa gene in 34/86 (39.5%) isolates. Another predominant pathogenic species obtained among our study isolates was Enterococcus faecium. 30/80 (37.5%) E. faecium isolates with an MIC range between 128 μg/mL and 512 μg/mL carried the bifunctional enzyme coding gene aac(6′)-Ie-aph(2′′)-Ia.
Each of the HLAR E. avium, E. hirae, and E. durans had both the genes while another E. avium strain carried aph(3′)-IIIa with streptomycin MIC of 1024 μg/mL. One of the two E. gallinarum strains isolated was HLAR and it carried both aminoglycoside resistance genes aac(6′)-Ie-aph(2′′)-Ia and aph(3′)-IIIa, while the other strain carried aph(3′)-IIIa gene alone but with an MIC of ≥16 μg/mL for gentamicin and >64 μg/mL for streptomycin. One of the two E. dispar isolates in our study was found to be HLGR + HLSR and carried aph(3′)-IIIa gene (see Table 4).
Table 4.
Results of high level aminoglycoside resistance and distribution of aminoglycoside modifying enzyme encoding genes among Enterococcus spp.
| HLARE (MIC and detection of genes by PCR) |
Distribution of high level aminoglycoside resistance in Enterococcus spp. (n = 178) | Total | ||||||
|---|---|---|---|---|---|---|---|---|
|
E. faecalis
(86) |
E. faecium
(80) |
E. durans
(1) |
E. avium
(4) |
E. hirae
(3) |
E. dispar
(2) |
E. gallinarum
(2) |
||
| HLGR* | 32 | 39 | 1 | 1 | 1 | 1 | 1 | 76 |
| HLSR* | 21 | 27 | 1 | 1 | 1 | 1 | 1 | 53 |
| aac(6′)-Ie-aph(2′′ )-Ia | 34 | 30 | 1 | 1 | 1 | — | 1 | 68 |
| aph(2′′)-Ib | — | — | — | — | — | — | — | — |
| aph(2′′)-Ic | — | — | — | — | — | — | — | — |
| aph(2′′)-Id | — | — | — | — | — | — | — | — |
| aph(3′)-IIIa | 34 | 30 | 1 | 2 | 1 | 2 | 2 | 72 |
*HLGR; HLSR; MIC screening.
4. Conclusion
In our study, we had observed enterococcal isolates with phenotypic resistance towards high level gentamicin and streptomycin antibiotics without presence of respective AME gene. This may be due to the expression of genes other than genes analyzed in this study. The coexistence of aac(6′)-Ie-aph(2′′)-Ia and aph(3′)-IIIa was observed in 20.2% of the isolates.
Though an array of AMEs are responsible for HLAR status among Enterococcus species, we have demonstrated aac(6′)-Ie-aph(2′′)-Ia and aph(3′)-IIIa genes more frequently occurring than other genes. This observation emphasizes the restricted gene distribution and transfer of resistant genes within a geographical region. Hence, surveillance studies should be conducted among Enterococcus isolates from different sources in any given geographical area to document the AME gene profile. Our study is the first to report resistance gene analysis among the Enterococcus species in India.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Shindae S, Koppikar GV, Oommen S. Characterization and antimicrobial susceptibility pattern of clinical isolates of enterococci at a tertiary care hospital in Mumbai, India. Annals of Tropical Medicine and Public Health. 2012;5(2):85–88. [Google Scholar]
- 2.Kobayashi N, Mahbub Alam M, Nishimoto Y, Urasawa S, Uehara N, Watanabe N. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium . Epidemiology and Infection. 2001;126(2):197–204. doi: 10.1017/s0950268801005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Washington W, Allen S, Janda W, et al. Koneman's Color Atlas and Textbook of Diagnostic Microbiology. 6th edition 2006. [Google Scholar]
- 4.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. Journal of Clinical Microbiology. 1995;33(1):24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vakulenko SB, Donabedian SM, Voskresenskiy AM, Zervos MJ, Lerner SA, Chow JW. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrobial Agents and Chemotherapy. 2003;47(4):1423–1426. doi: 10.1128/AAC.47.4.1423-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sreeja S, Sreenivasa Babu PR, Prathab AG. The prevalence and the characterization of the Enterococcus species from various clinical samples in a tertiary care hospital. Journal of Clinical and Diagnostic Research. 2012;6(9):1486–1488. doi: 10.7860/JCDR/2012/4560.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical Laboratory Standards Institute. CLSI Document. M100-S21. Wayne, Pa, USa: Clinical and Laboratory Standards Institute; 2011. Performance standards for antimicrobial susceptibility testing, twenty-first informational supplement. M02-A10 and M07-A8. [Google Scholar]
- 8.Sahm DF, Boonlayangoor S, Schulz JE. Detection of high-level aminoglycoside resistance in enterococci other than Enterococcus faecalis . Journal of Clinical Microbiology. 1991;29(11):2595–2598. doi: 10.1128/jcm.29.11.2595-2598.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitz F-J, Verhoef J, Fluit AC. Prevalence of aminoglycoside resistance in 20 European university hospitals participating in the European SENTRY Antimicrobial Surveillance Programme. European Journal of Clinical Microbiology and Infectious Diseases. 1999;18(6):414–421. doi: 10.1007/s100960050310. [DOI] [PubMed] [Google Scholar]
- 10.Hasani A, Sharifi Y, Ghotaslon R, et al. Molecular screening of virulence genes in high level gentamicin resistant Enterococcus faecalis and Enterococcus faecium isolated from clinical specimens in northwest Iran. Indian Journal of Medical Microbiology. 2012;30(2):175–181. doi: 10.4103/0255-0857.96687. [DOI] [PubMed] [Google Scholar]
- 11.Chow JW. Aminoglycoside resistance in enterococci. Clinical Infectious Diseases. 2000;31(2):586–589. doi: 10.1086/313949. [DOI] [PubMed] [Google Scholar]
- 12.Donabedian SM, Thal LA, Hershberger E, et al. Molecular characterization of gentamicin-resistant Enterococci in the United States: evidence of spread from animals to humans through food. Journal of Clinical Microbiology. 2003;41(3):1109–1113. doi: 10.1128/JCM.41.3.1109-1113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vakulenko SB, Mobashery S. Versatility of aminoglycosides and prospects for their future. Clinical Microbiology Reviews. 2003;16(3):430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarrilli R, Tripodi M-F, Di Popolo A, et al. Molecular epidemiology of high-level aminoglycoside-resistant enterococci isolated from patients in a university hospital in southern Italy. Journal of Antimicrobial Chemotherapy. 2005;56(5):827–835. doi: 10.1093/jac/dki347. [DOI] [PubMed] [Google Scholar]


