Abstract
A number of marine natural products are potent inhibitors of proteases, an important drug target class in human diseases. Hence, marine cyanobacterial extracts were assessed for inhibitory activity to human cathepsin L. Herein, we have shown that gallinamide A potently and selectively inhibits the human cysteine protease, cathepsin L. With 30 min of preincubation, gallinamide A displayed an IC50 of 5.0 nM, and kinetic analysis demonstrated an inhibition constant of ki = 9000 ± 260 M−1 s−1. Preincubation-dilution and activity-probe experiments revealed an irreversible mode of inhibition, and comparative IC50 values display a 28- to 320- fold greater selectivity toward cathepsin L than closely related human cysteine cathepsins V or B. Molecular docking and molecular dynamics simulations were used to determine the pose of gallinamide in the active site of cathepsin L. These data resulted in the identification of a pose characterized by high stability, a consistent hydrogen bond network, and the reactive Michael acceptor enamide of gallinamide A positioned near the active site cysteine of the protease, leading to a proposed mechanism of covalent inhibition. These data reveal and characterize the novel activity of gallinamide A as a potent inhibitor of human cathepsin L.
Marine cyanobacteria are exceptionally rich in biologically active natural products.1 From a biochemical perspective, their metabolites are highly diverse, often deriving from mixtures of nonribosomal peptide synthetase (NRPS), polyketide synthase (PKS), terpene, and sugar biosynthetic pathways, and are commonly further decorated with halogen atoms, methyl groups, and interesting oxidations. As a result, they have been actively investigated for their therapeutic potential for a number of years, especially for anticancer activity. One such recently approved agent for anaplastic large cell lymphoma and Hodgkin’s lymphoma, brentuximab vedotin, was inspired by the marine cyanobacterial metabolite dolastatin 10.2,3 In recent years, the biological evaluation of marine-derived natural products has broadened to include inflammation, infectious and parasitic diseases, and neurological diseases.4
In this latter regard, an emergent trend in the pharmacological mechanism of action of cyanobacterial natural products is that many are potent inhibitors of various classes of proteases.5,6 Proteases have been implicated in the pathogenesis of many human diseases, including cancer,7,8 neurological disorders such as Alzheimer’s Disease,9,10 and parasitic diseases;11 thus the therapeutic modulation of proteolytic activity offers an attractive potential treatment modality. However, with myriad proteases and many potential therapeutic applications, discovery of agents with selectivity for specific proteases is crucial to the development of truly useful pharmaceuticals in this class.
Whereas freshwater cyanobacteria have yielded a number of protease inhibitors,12 their marine relatives represent an under-explored resource for modulators of this enzyme class. Hence, we have initiated a program to survey marine cyanobacterial extracts, fractions and newly isolated pure compounds for interesting profiles of protease inhibition, with a special focus on enzymes in the cysteine cathepsin and proteasome classes. We have recently reported the structures of the carmaphycins, low nanomolar epoxyketone proteasome inhibitors from the Curaçao cyanobacterium Symploca sp., and previously had identified the depsipeptide symplocamide A as a potent serine protease inhibitor.5,13
Our recent efforts in this regard have focused on the human cysteine cathepsin L protease, an important lysosomal endopeptidase with exceptionally high proteinase activity. Aside from its traditional role in protein degradation, cathepsin L is responsible for many specialized roles that make it an interesting target for drug discovery. It is upregulated in multiple cancer cell types, and has been strongly implicated in bone resorption, bone pit formation, and invasion of bone tissue by osteoclasts due to its high level of secretion and efficient hydrolysis of bone matrix proteins.14 Multiple studies have shown significant reduction in tumor invasiveness and metastasis with treatment of pan cysteine protease or selective cathepsin L inhibitors.8 Furthermore, related cysteine proteases have been identified and targeted in various infectious diseases, including malaria, leishmaniasis, trypanosomiasis and others.15 Finally, recent evidence has mounted to elucidate the role of murine cathepsin L in proneuropeptide processing, with knockout (KO) and siRNA studies indicating a particularly important role in the production of the dynorphins and neuropeptide Y.16,17
Despite the multitude of disease implications associated with cathepsin L, few selective inhibitors have been described, and even fewer have appropriate pharmaceutical properties for potential clinical application. Herein, we report that the evaluation of cyanobacterial extracts led to the identification of gallinamide A (1)18 as a potent and selective inhibitor of human cathepsin L, and thus provides an active structure for developing agents with highly desired subtype selectivity within the cysteine proteases. Thus, this study describes the re-isolation and identification of gallinamide A, inhibitory potency to cathepsin L and related cysteine proteases, kinetic inhibition properties, and analyses of molecular docking to cathepsin L that indicates a Michael addition-based inhibition as supported by biochemical data. The molecular features of gallinamide A will assist future structure-based optimization efforts for effective inhibitors of human cathepsin L and members of the cysteine cathepsin protease family.
RESULTS AND DISCUSSION
Screening efforts of fractionated extracts from various marine cyanobacteria for modulation of human cathepsin L activity identified a number of active fractions. One such fraction from a collection of a re-tipped Schizothrix sp. showed 97% inhibition of cathepsin L at 3 µg/mL. This sample, eluting with 2:3 hexanes/EtOAc, is adjacent to the fraction that yielded gallinamide A (1) as described by Linington et al.19, and thus was fractionated by solid phase extraction (SPE) to produce eight subfractions. The subfractions eluting with 2:3 hexanes/EtOAc and 1:4 hexanes/EtOAc showed 99% and 99% inhibition, respectively, of cathepsin L at 3 µg/mL and were further fractionated by RP-HPLC to give five compounds, E45A-E45E. Compound E45A was inactive against cathepsin L, while compounds E45B, E45C, E45D and E45E displayed 97–99% inhibition of cathepsin L at 3 µg/mL. The major component, E45C, yielded a mass of 0.70 mg (0.14% of the crude extract), while the minor constituents E45B, E45D, and E45E yielded 0.33 mg, 0.40 mg, and 0.38 mg, respectively (0.059%, 0.072%, and 0.068% of crude extract).
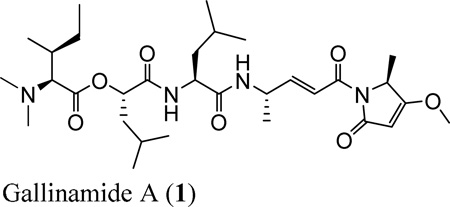
The HR-ESITOFMS spectrum of the major compound E45C gave an [M+H]+ ion at m/z 593.3908, which was consistent with a molecular formula of C31H53N4O7. This formula for a protonated molecule matched that of gallinamide A (1),19 and 1H NMR analysis was used to elucidate partial structural features which matched the previously reported subunits of gallinamide A. Furthermore, tandem LC-MS/MS analysis provided structural data consistent with the linear structure of gallinamide A (Figure 1A), and the observed specific rotation [α]25 d -12 (c 0.07, MeOH) was consistent with the literature value, confirming its absolute configuration to be the same as previously described.20
Figure 1.
Fragmentation patterns for (A) gallinamide A (1) by ESI-MS/MS.
Gallinamide A Potently and Selectively Inhibits Cathepsin L
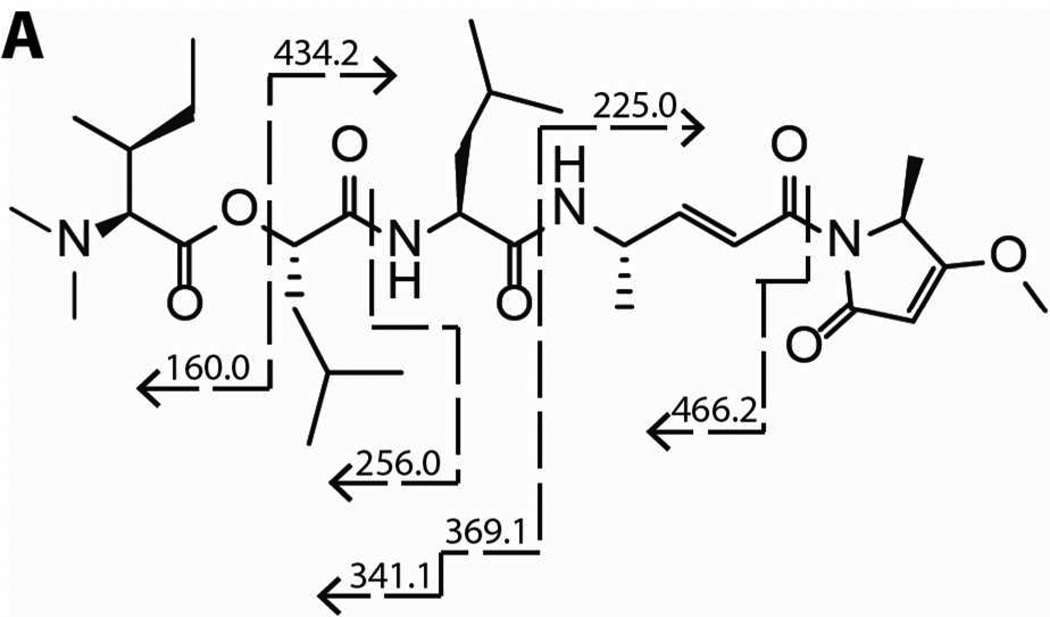
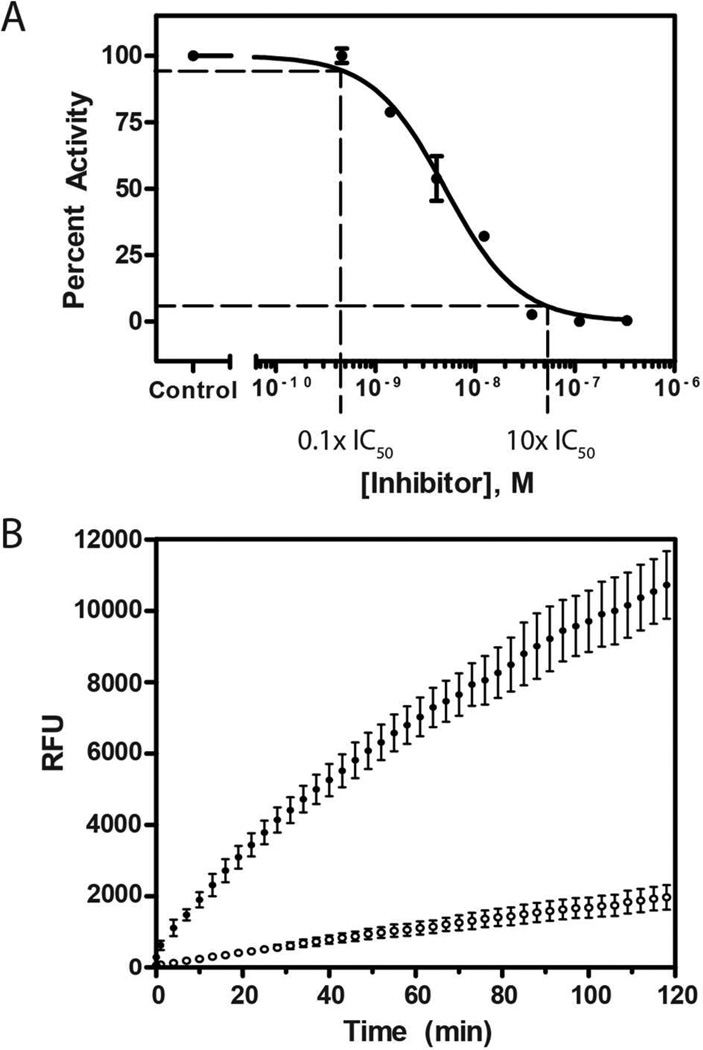
Next, we demonstrated that gallinamide A (1) blocks the binding of the activity based probe (ABP) DCG-04, a biotin labeled derivative of the potent cysteine cathepsin inhibitor E-64-c.21 Unlike experiments measuring enzyme inhibition, ABP studies investigate the ability of a compound to compete with and block binding of a potent active site-directed probe. Human recombinant cathepsin L was preincubated with compound 1 for 30 min, followed by visualization of residual active enzyme by the labeled probe. CLIK 148, a cathepsin L selective derivative of E-64-c, was included as a positive control. Samples were subsequently run on gel electrophoresis and transferred to a Hybond Nitrocell membrane for detection with avidin-HRP and a chemiluminescent substrate. Cathepsin L is a known target of DCG-04, and thus a reduction in band optical density revealed blockade of the active site. Compound 1 displayed a concentration-dependent inhibition of ABP labeling (Figure 2), with decreased labeling at 111 nM and complete loss of probe labeling at 333 nM. Interestingly, the concentration of enzyme in this assay was 100 nM; thus gallinamide A shows a high degree of efficiency in its inhibition, reducing binding at a 1:1 molar ratio of inhibitor and enzyme.
Figure 2.
Representative blot of competitive activity-based probe labeling of cathepsin L. Gallinamide A showed reduced labeling at 111 nM and complete inhibition at 333 nM.
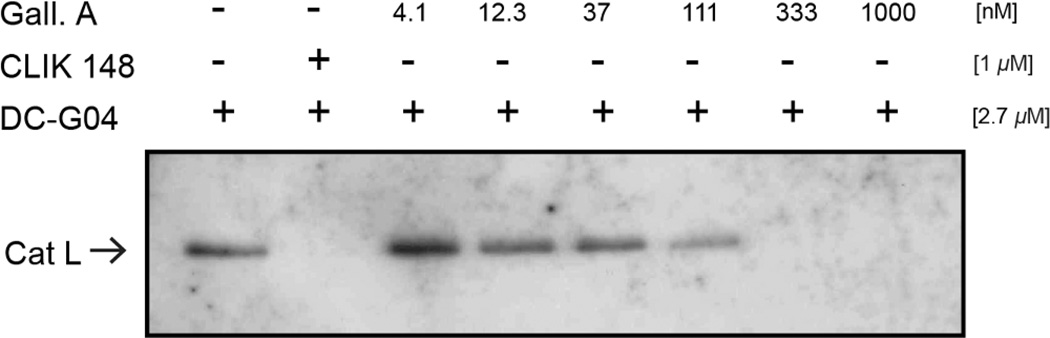
The potency of enzyme activity inhibition, commonly reported as an IC50 (inhibitory concentration for 50% activity), was found to be time-dependent in preincubation dose-response experiments. With immediate mixing of enzyme, substrate, and inhibitor, compound 1 inhibited human cathepsin L with an IC50 of 47 nM. Following a preincubation of enzyme and inhibitor for 30 min prior to addition of substrate, compound 1 displayed increased potency, with an IC50 of 5.0 nM (Figure 3). Time-dependent inhibition is a hallmark of slow-binding inhibitors,22 a finding that directed the subsequent characterization of the mode of inhibition and binding affinity of gallinamide A.
Figure 3.
Dose response curves following gallinamide A preincubation with cathepsin L for 0 min (▲) and 30 min (●). The measured IC50 following immediate mixing is 47 nM, while 30 min preincubation results in an IC50 of 5.0 nM. IC50 data are significantly different (p<0.0001).
To assess enzyme selectivity, compound 1 was tested for inhibitory activity against the highly homologous cysteine proteases cathepsin V (human) and cathepsin B (human), as well as the cysteinyl exopeptidase cathepsin H. IC50 values were obtained with and without preincubation of inhibitor and enzyme, as described for cathepsin L. A selectivity index was calculated as a ratio of the IC50 of the assayed protease to that of cathepsin L for each incubation condition, summarized in Table 1. Interestingly, gallinamide A displays a 10-fold increase in potency for cathepsin L relative to cathepsin V without preincubation, and this metric increases to 28-fold after allowing the inhibitor to associate with the protease for 30 min. Cathepsin V is a highly homologous cysteine protease, and inhibitors capable of distinguishing between these two enzymes are currently limited. Selectivity is exaggerated for the more distantly related cathepsin B, which shows a 320-fold higher IC50 following 30 min of preincubation. Gallinamide A was inactive against the exopeptidase cathepsin H at the highest assay concentrations, further exemplifying its selectivity within the cathepsin protease family. While previous studies have demonstrated moderate inhibition of murine cathepsin L by compound 1 as well as potent inhibition of the Plasmodium falciparum falcipain proteases,23 a full panel of related cysteine proteases was not able to be evaluated due to insufficient material in the current study. To fully appreciate the selectivity of this inhibitor, additional enzymes in this family should be evaluated.
Table 1.
IC50 Values and Selectivity Indices of Gallinamide A (1) for Cathepsins L, V, B, and H.
| IC50a | Selectivity Index | |||
|---|---|---|---|---|
| Enzyme | 0 Min | 30 Min | 0 Min | 30 Min |
| µM | ||||
| Human Cathepsin L | 0.047 | 0.0050 | 1 | 1 |
| Human Cathepsin V | 0.46 | 0.14 | 10 | 28 |
| Human Cathepsin B | 4.2 | 1.7 | 91 | 320 |
| Human Cathepsin H | >30 | >30 | N/A | N/A |
95% confidence intervals: L(0)=0.0042–0.0060; L(30)=0.040–0.053; V(0)=0.42–0.50; V(30)=0.12–0.17; B(0)=3.7–4.7; B(30)=1.3–2.0
Taken together, these experiments reveal human cathepsin L as a major target of gallinamide A based on potency and selectivity. With increased focus on the discovery of these inhibitors, gallinamide A represents an interesting active compound containing easily modifiable structural features. With two total syntheses now published and a limited initial SAR study completed,20,23 the gallinamide structural class has great potential to yield analogs with improved potency, selectivity and pharmaceutical properties.
Gallinamide A is an Irreversible Inhibitor of Cathepsin L
As many slow-binding inhibitors work through covalent or tight-binding interactions,22 the reversibility of the enzyme-inhibitor complex was subsequently assessed. Irreversibility of gallinamide A (1) inhibition was determined using a preincubation-dilution experiment adapted from Copeland et al.,22 in which a concentrated sample of cathepsin L was incubated with concentrated compound 1 before a rapid dilution of the combined solution (Figure 4A). A preincubation of 30 min was used, and approximate levels of expected enzyme activity were obtained from the 30-min preincubation dose-response experiment (Figure 4B). Following rapid dilution, a shift in inhibitor concentration from 10-fold IC50 to 0.1-fold IC50 value should cause a fully reversible inhibitor to immediately dissociate from the enzyme, resulting in activity levels equivalent to 90% of control. Alternatively, a slowly-reversible inhibitor will display a reduced reaction rate initially but with a gradual increase over time, whereas an irreversible inhibitor will show an initial rate of approximately 10% of the control condition, which persists over time. Immediately following the dilution, cathepsin L activity was measured at 12% of the vehicle control. The reaction rate remained linear and was unchanged for 2 h following the dilution (Figure 4C). These data indicate that gallinamide is an irreversible inhibitor of the cathepsin L protease. This is congruent with previous studies showing that gallinamide A was a covalent, irreversible inhibitor of the related P. falciparum falcipain proteases based on kinetic model fitting.23
Figure 4.
A concentrated solution of enzyme and gallinamide A was incubated for 30 min and then then diluted. (A) The resulting shift in the enzymatic activity is based on the dose response curve. (B) The subsequent rate of the reaction was monitored for 2 h, and comparison of initial reaction rates showed 12% of the activity with preincubation of gallinamide A (○) as compared to the control (●). The reaction rate was constant over the course of the 2 h monitoring period, demonstrating an irreversible mode of inhibition.
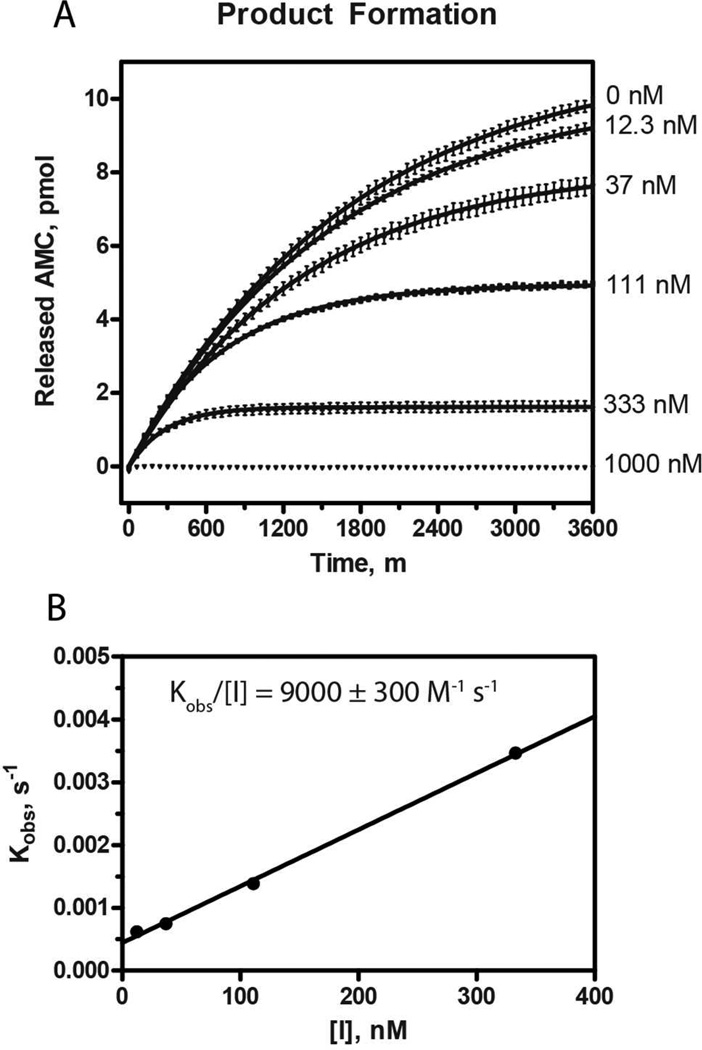
To more accurately characterize the inhibitory potency of gallinamide A, the substrate turnover kinetics of recombinant human cathepsin L were determined in the presence of various concentrations of inhibitor (Figure 5). Progress curves were fitted to a model of irreversible enzyme inactivation, and the calculated first order rate constants kobs were plotted against [I].24,25 The resulting relationship was linear, and the second order rate constant represented by kobs/[I] was determined to be 9000 ± 260 M−1 s−1. A kobs value was not determined for the [I] = 1000 nM condition, as the level of inhibition did not produce a progress curve capable of regression by the model. These data are consistent with two key aspects of this study: 1) gallinamide A is a potent and efficient inhibitor of human cathepsin L, and 2) the inactivation is irreversible.
Figure 5.
(A) Product formation from the turnover of substrate by cathepsin L in the presence of various concentrations of gallinamide A was monitored over time. The resulting plots were fitted to a model of irreversible inhibition, and the obtained kobs values were plotted against [I]. (B) This produced a linear relationship, the slope of which represents the second order inhibition constant, ki = 9000 ± 260 M−1 s−1.
Gallinamide A Inhibits Cathepsin L via Michael Addition
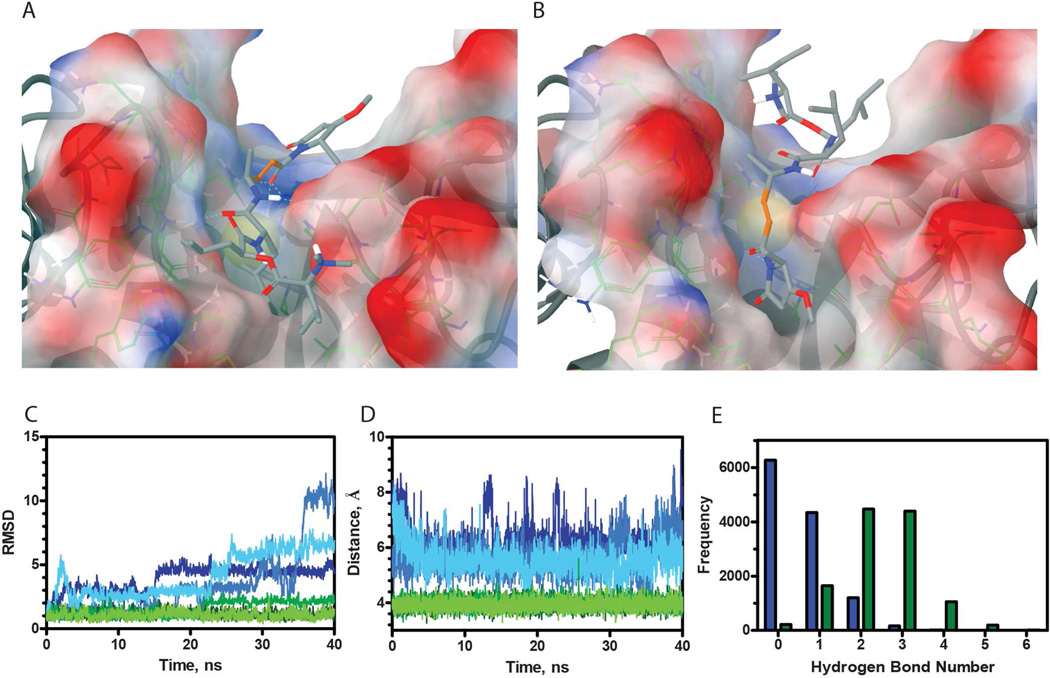
Leveraging the knowledge that gallinamide A binds irreversibly to cysteine cathepsins, a hypothesis that the reaction occurs via a Michael addition to the reactive gallinamide A enamide was generated. In previous studies, it has been shown that the top scoring pose in silico is not necessarily the correct pose in vitro.26 With this in mind, gallinamide A was docked with the crystal structure of the catalytic domain of human cathepsin L, and poses in which the enamide was positioned acceptably close (<4 Å) to the catalytic cysteine were selected. The induced fit docking protocol produced two different poses with similar predicted binding affinities, referred henceforth as ‘top’ (XPGScore −8.539) and ‘bottom’ (XPGScore −7.359) orientations (Figure 6A–B, respectively). In no circumstance did Glide position other potential reactive groups near the catalytic cysteine.
Figure 6.
(A–B) Induced fit docked poses of representative structures for two gallinamide A (1) conformations docked into cathepsin L resulted in ‘top’ and ‘bottom’ poses, respectively. In C–E, the scores for the ‘top’ pose are represented by green and the ‘bottom’ pose blue. (C) RMSD values were obtained for each pose, corresponding to differences between the structure at a given time and the original pose, and thus are inversely correlated with stability. (D) The distance between Cys25-SH and C* for gallinamide A bound to cathepsin L. (E) A histogram of hydrogen bond count for both orientations of gallinamide A docked into the cathepsin L active site.
Both of these poses possessed structurally desirable qualities for a reaction to occur. The ‘top’ orientation, in which the cyclic head group interacted with the S1’ pocket, was positioned such that the enamide carbonyl, which is negatively charged in the predicted reaction intermediate, is stabilized in an oxyanion hole by the Gln19 and positively charged His163 side chains, as well as the backbone NH of Cys25. Additionally, intermolecular hydrogen bonds are evident throughout the ligand-enzyme interface, which may help stabilize the reactive complex. In the ‘bottom’ orientation, the enamide carbonyl is stabilized by a hydrogen bond to Gly164. Additionally, the cyclic head group appears to be sterically accommodated very nicely into the S2 pocket of cathepsin L. Although the predicted hydrogen bond network was found to be less extensive in this orientation, the pose remained a viable option and was not discarded.
Due to the importance of identifying the correct pose for structure-guided lead optimization, molecular dynamic simulations were employed to elucidate the most probable ligand orientation. We hypothesized that a noncovalent complex must persist for a chemical reaction to occur between the enzyme and inhibitor. Specifically, a complex must exist where the ligand position does not significantly fluctuate and the Cys25 thiol involved in catalysis is continually well positioned to react with the gallinamide A enamide. The use of multiple simulations enabled a more robust assessment of stability than using a single trajectory, which might become trapped in a local minimum. As binding between gallinamide A and cysteine cathepsins does not occur 100% of the time, it is reasonable to expect that not every simulation would result in a stable binding pose. Consequently, it became our goal to assess whether one pose consistently provided a more stable, reaction-ready conformation. The first 10 ns of each trajectory were discarded for equilibration, which left 120 ns of usable simulation time for pose analysis. All trajectories were aligned by Cα RMSD to their average structure, and these aligned structures were used in the analysis. The stabilities of complexes were assessed using three metrics: RMSD of gallinamide A in the binding pocket (Figure 6C), distance between the Cys25-SH and the reactive Michael addition carbon, henceforward referred to as C* (Figure 6D), and hydrogen bond persistence (Figure 6E). In all three metrics, the orientation where the head group was oriented near the S1’ pocket appeared to be better suited for covalent reaction to occur between the enzyme and inhibitor.
To first assess stability of the complexes, the movement of gallinamide A within the binding pocket was analyzed. Theoretically, ligands that have more stable interactions with their corresponding receptors move less within a pocket. The RMSD, a metric computing similarity of two structures, of gallinamide A complexed to cathepsin L was used to compare the stability of each pose (Figure 6C). All three simulations of gallinamide A in the ‘top’ orientation proved to be more stable than in the ‘bottom’ orientation based on lower RMSD values.
The most important reaction criterion is that the Cys25 thiol must be positioned adjacent to a reactive center on gallinamide A. Gallinamide A has three potentially reactive centers: an enamide, a vinylogous carbamate, and an ester. In all of the computationally docked poses, only the β carbon of the enamide, C*, was positioned adjacent to Cys25-SH, likely due to the geometric requirements of the cathepsin binding pocket. To assess stability of the bound complexes, we first looked at the percentage of time Cys25-SH was within 4 Å of C* (Figure 6D). While this distance cutoff is not enough to ensure reactivity, it is close enough that slight changes in the conformation of the bound complex could lead to a productive reaction. Distances were measured for each frame in the trajectories (12000 per complex). For the ‘bottom’ pose, C* was positioned within 4 Å of Cys25-SH only 0.28% of the time, as opposed to 65.80% for the ‘top’ pose. It should be noted that these results were not the result of the initial bias toward the first frame of the analysis, as the latter half of all trajectories were not statistically different from the first half.
Additionally, hydrogen bond interactions contribute significantly to the binding free energy of a complex, and can heavily dictate selectivity. In this regard, the two distinct poses of a protein-ligand complex could result in similar contributions from other contributing forces (e.g. van der Waals), yet a protein should have a more persistent hydrogen bond network with the correctly oriented ligand than one incorrectly oriented. For all trajectories, hydrogen bonds between cathepsin L and gallinamide A were counted and a histogram was generated of hydrogen bond count (Figure 6E). Hydrogen bonds were more persistent in the ‘top’ orientation compared to the ‘bottom’ orientation (p <<< 0.001, T-test), further supporting the ‘top’ orientation.
Favorable attributes of a correct docking pose are also consistent with the ‘top’ orientation. Most importantly, the utilization of the oxyanion hole to stabilize the reaction is consistent with what is observed with the natural proneuropeptide substrates. In our simulations, the enamide carbonyl of gallinamide A was observed to hydrogen bond with the oxyanion hole 55.4% of the time in the ‘top’ orientation, as compared to 17.06% in the ‘bottom’ orientation. The positively charged His163 is an especially good stabilizing residue for the transition state compared to Gly24 in the ‘bottom’ orientation, as individual electrostatic interactions are stronger than individual hydrogen bonds. Additionally, the positively charged tertiary amine in gallinamide A is positioned relatively close to the S2 pocket, which accommodates a basic residue in many of its natural proneuropeptide substrates. This positive charge may also help confer selectivity toward cathepsin L over cathepsin V, as the S2 pocket in cathepsin V is not as negatively charged.
Other atomistic interactions between gallinamide A and cathepsin L lend support to the head group ‘top’ pose being the correct orientation. Stolze et al.23 conducted a limited SAR of gallinamide Ainhibition of a P. falciparum cysteine protease and found that the cyclic head group was essential for gallinamide A-based inhibition. In the ‘top’ orientation, the carbonyl in the cyclic head group was found to hydrogen bond to either Gln19 or His189 for 83.55% of the simulation. The high persistence of this hydrogen bond suggests that removal would significantly reduce the inhibitory potential of gallinamide A with cathepsin L. In contrast, hydrogen bonds with the cyclic head group were absent in the ‘bottom’ orientation, which would not support the aforementioned SAR. In other structural studies of cathepsin L inhibitors, Gly68 has been shown to hydrogen bond with inhibitors.27 In our simulations, a hydrogen bond between Gly68 and the carbonyl of the leucine residue appears 82.75% of the time in the ‘top’ pose, consistent with these findings.
The identification of the correct pose and proposed reaction mechanism between gallinamide A (1) and cathepsin L will serve as a platform on which to base the development of therapeutically relevant agents. The potential applications of gallinamide A as an antimalarial agent have been investigated since its discovery, though the potency and selectivity of this compound make it a valuable structural class in other fields. Inhibitors with selectivity between various human cysteine cathepsin proteases are lacking, and compounds with such a property may prove valuable in the elucidation of the relative roles of these enzymes in proneuropeptide processing.28 Furthermore, inhibitors of cathepsin L are under investigation for the reduction of tumor metastasis.14 In this capacity, compounds do not need to enter cells to work as effective agents, as cathepsin L promotes metastasis through its degradation of matrix proteins in the extracellular space.14 Due to the easily modifiable carbon skeleton of 1, this structure could serve as a starting scaffold for the development of compounds with such favorable pharmaceutical properties. Lastly, this study identifies potential off-target binding sites for this marine natural product, an important consideration in its development as a potential antimalarial agent, and thus will help guide the development of more selective and potent therapeutic agents.
EXPERIMENTAL SECTION
General Experimental Procedures
UV spectra were measured on a Beckman Coulter DU 800 Spectrophotometer. NMR spectra were collected on a Varian Unity 500 MHz (500 MHz and 125 MHz for 1H and 13C NMR respectively) using CDCl3 from Cambridge Isotope Laboratories, Inc. 99.8% D containing 0.03% v/v trimethylsilane (δH 0.0 and δC 77.16 as internal standards using trimethylsilane and CDCl3, respectively). LCMS data were obtained with a Phenomenex Kinetex 5 µm C18(2) 100Å column (4.6 × 250 mm) with a Thermo Finnigan Surveyor Autosampler-Plus/LC-Pump-Plus/PDA-Plus system and a Thermo Finnigan LCQ Advantage Max mass spectrometer. HPLC purification was carried out with a Waters 515 HPLC Pump with a Waters 996 Photoiode Array Detector using Empower Pro software. All solvents were HPLC grade except for 99.8% acetone from Fisher which was distilled before use, and H2O which was purified by a Millipore Milli-Q system before use.
Collection, Extraction and Isolation
The collection, extraction and VLC fractionation information can be found in the original isolation report.19 The fraction eluting with 4:6 hexanes/EtOAc (12.3 mg) showed the strongest inhibition (3% activity remaining) of cathepsin L at 3 µg/mL. LC-MS and 1H- NMR spectra were obtained, and then the sample was further fractionated using 200 mg Si NP solid phase extraction (SPE), producing eight subfractions. These eluted with 3 mL washes of: 9:1 hexanes/EtOAc; 8:2 hexanes/EtOAc; 6:4 hexanes/EtOAc; 4:6 hexanes/EtOAc; 2:8 hexanes/EtOAc; 100% EtOAc; 1:1 EtOAc/MeOH; 100% MeOH. LC-MS and 1H NMR traces were again obtained for each sub-fraction. Sub-fractions 6 through 8 were combined based on similarities in their LC-MS and NMR data, and all sub-fractions were subjected to the cathepsin L inhibition assay. The fourth and fifth fractions, eluting with 4:6 hexanes/EtOAc (1.64 mg) and 2:8 hexanes/EtOAc (0.52 mg), respectively, demonstrated the greatest inhibition of cathepsin L (99% and 99%, respectively), and were thus selected for additional purification. These two fractions were separately subjected to C18 RP-HPLC (Phenomenex Luna C18 4.6 × 250 mm RP-HPLC column, 5 µm ACN/H2O 40:60, 1 mL/min) under both neutral and acidic (0.01% TFA) conditions. Separation was more robust under acidic conditions, and collections were tested in the cathepsin L inhibition assay to ensure protonation did not affect bioactivity. Collections from both neutral and acidic conditions demonstrated inhibition of cathepsin L (data not shown). Analytical RP-HPLC was used to profile each subfraction, and preparative HPLC was performed to isolate and collect the major components (Phenomenex Luna C18 4.6 × 250 mm, 5 µm, 35% ACN/65% H2O/0.01%TFA, 1 mL/min); 0.70 mg of pure gallinamide A was obtained, 0.14% of the crude extract mass.
Gallinamide A (1)
colorless amorphous solid; [α]25 d −12 (c 0.07, MeOH); UV, 1H NMR, and high resolution ESITOFMS matched reported values within experimental error and are available in the Supporting Information.
Cathepsin L Assay
Z-Phe-Arg-AMC substrate and E-64-c were purchased from Bachem Americas. Human recombinant cathepsin L was purchased from R&D Systems. Assays were carried out using 20 µM Z-Phe-Arg-AMC and 3.0 ng/mL human recombinant cathepsin L. Assay buffer consisted of 50 mM sodium acetate, 100 mM NaCl, 1.0 mM EDTA and 4 mM dithiotreitol, pH 5.5. The enzymatic reaction (25°C) was monitored on a SpecraMax Gemini or SpectraMax microplate reader (PerkinElmer Life Sciences) and the fluorescent signal was measured at the excitation and emission wavelengths of 365 and 450 nm, respectively.
Inhibitor Potency Determination
An 8-point, 3-fold serial dilution dose-response assay was performed in triplicate. Each well of a 96-well assay plate contained 50 µL of 2× concentrated assay buffer, 20 µL of H2O, and 10 µL of inhibitor in 10% DMSO. Positive and negative controls of 10 µL of 10 µM E-64-c or 10% DMSO, respectively, replaced inhibitor as internal controls. A 10 µL aliquot of a 30 ng/mL cathepsin L solution and 10 µL of 200 µM Z-Phe-Arg-AMC in assay buffer were added sequentially to initiate the protease reaction. Assay buffer (10 µL) was added in place of cathepsin L as a substrate blank for baseline correction. The resultant dose response concentration range was 333 nM to 0.46 nM inhibitor in a 100 µL final reaction volume. Data were scaled to internal controls and a four-parameter logistic model (GraphPad vs. 5.0, Prism) was used to fit the measured data and determine IC50 values.
Selectivity
Gallinamide was tested for inhibition of human cathepsin V (100 ng/mL; R&D Systems),human cathepsin B (100 ng/mL; R&D Systems), and human cathepsin H (50 ng/mL; Athens Research and Technology). For cathepsins B and V: reactions were performed in 50 mM sodium acetate buffer containing 100 mM NaCl, 4 mM DTT, and 1 mM EDTA, pH 5.5, using 10 µM Z-Phe-Arg-AMC substrate. For cathepsin H reactions were performed in 50 mM potassium phosphate buffer containing 3 mM cysteine, 1 mM EDTA, pH 6.4, using 200 µM Arg-AMC substrate. IC50 values were determined for both 0 min and 30 min preincubations, and selectivity index values were calculated relative to cathepsin L. All values were obtained in technical triplicate, as compound supply was insufficient to perform replicate plates.
Reversibility
A preincubation-dilution experiment was adapted from Copeland et al.22 Cathepsin L at 100-fold its final assay concentration was incubated with gallinamide A at 10-fold its IC50 value for 30 min in a volume of 2 µL in a 96-well plate. This mixture was diluted 100-fold with assay buffer containing 10 µM Z-FR-AMC substrate to a final volume of 200 µL, resulting in a standard concentration of enzyme and 0.1-times the IC50 value of gallinamide A. A rapidly reversible inhibitor will dissociate from the enzyme to restore approximately 90% of enzymatic activity following the dilution event, while an irreversible inhibitor will maintain approximately 10% of enzymatic activity. Fluorescence intensities of the 200-µL wells were monitored continuously for AMC hydrolysis on a Specramax plate reader in kinetic mode for 2 hr.
Active-site Directed Probe Competition Binding Assay
Gallinamide A was prepared in a 6-point, 3-fold serial dilution in 10% DMSO. Assay buffer consisted of 100 mM sodium acetate, 1.0 mM EDTA, and 4 mM DTT, pH 5.5. One µL of 40 ng/µL cathepsin L solution was added to 8.5 µL of assay buffer and preincubated with 1.5 µL of inhibitor for 30 min at room temperature. Positive and negative controls of 1.5 µL of 100 µM CLIK-148 or 10% DMSO were included. After preincubation, 4 µL of 10 µM Biotin-Lys-C5 alkyl linker-Tyr-Leu-epoxide (DCG-04) was added and incubated for 30 min at room temperature. The incubated sample was mixed with 4× NuPAGE sample buffer and 50 mM DTT and denatured at 70 °C for 5 min. SDS-polyacrylamide gel separation was performed with Xcell SureLock system (Invitrogen) on 12% Bis-Tris gel (Invitrogen) at 200 V for 50 min. After transfer to a Hybond Nitrocell membrane (Amersham) at 30 V (1 h), the membrane was blocked in 5% milk in TBS + 0.05% Tween for 1 h and then incubated with avidin and biotinylated horse-radish peroxidase, followed by washing (4×) and detection with ECL+ chemiluminescent substrate (Amersham).
Kinetic Analysis
Kinetic characterization of the interaction between gallinamide A and cathepsin L was performed to obtain an accurate second order inhibition constant corresponding to kobs/[I]. Continuous monitoring of substrate hydrolysis in the presence of the inhibitor was used to measure the decrease in enzyme activity over time. Gallinamide A was tested in concentrations ranging from 12.3–1000 nM with simultaneous mixing of enzyme, substrate and inhibitor. Progression curves of product formation were fitted to a simple model of irreversible inhibition:
| Equation 1 |
in which [P] is concentration of product, νi is the initial reaction rate, kobs is the observed first order rate constant, and t is time in seconds. The observed first order inhibition constant Kobs was plotted against [I] to obtain a linear relationship, the slope of which represents the second order rate constant kinact/KI, a measure of affinity for the slow binding inhibitor. Kinetic analysis was performed in technical quadruplicate.
Receptor and Ligand Preparation
A cathepsin L structure (PDB ID: 2XU3)27 was downloaded from the Protein Data Bank.29 All ligands and H2O molecules were removed and hydrogens were subsequently added to the protein structure using PROPKA30–33 and PDB2PQR34 at pH 5.5. The proteins were then processed using Schrödinger Maestro’s Protein Preparation Wizard (www.schrodinger.com). Grids were generated using the catalytic cysteine as the center. The inner box, which imposes restrictions on the location of the center of the ligand, was set to a 10 Å cube and the outer box, which restricts the possible locations of all ligand atoms, was set to a 30 Å cube. Gallinamide A was built using Schrödinger Maestro’s 2D builder and processed using LigPrep at pH 5.5 +/− 2.0 to mimic assay conditions.
Docking Protocol
Gallinamide A was positioned into cathepsin L active site using the Induced Fit Docking module of Schrödinger’s glide.35 The center of the catalytic residues Cys25 and His163 was used as the box center. No hydrogen bond constraints were applied. Side chains within 5.0 Å of docked ligands were refined with Prime. XP Precision was used to score poses in the final re-docking step.
Molecular ynamics simulations
Selected complexes were extracted from the docking runs. In all poses, Na+ counter-ions were added to neutralize the solution, and the complex was solvated in a TIP4PEW water box extending 10 Å in all x,y,z directions beyond the complex.36 Minimization and equilibration were done similarly to previous MD studies.37 In brief, the system was first minimized to remove any steric issues. Harmonic restraints of 2.0 kcal/mol/Å2 were applied to the protein as the system was heated to 300K. A 1 ns simulation to equilibrate the system was then run with restraints removed. The SHAKE algorithm was used to constrain bonds involving hydrogens,38 and Particle Mesh Ewald was used to treat long-range electrostatics.39 Simulations of 50 ns duration were run in triplicate from the minimized structure. For each simulation, the first 10 ns were discarded for equilibration of the docked pose, resulting in 120 ns of simulation per system that was analyzed.
Simulation snapshots were taken every 10 ps. All analysis was performed using either the ptraj module of AMBER1224 or analysis tools in VMD.41 Trajectories were aligned by Cα RMSD to their average structure. Hydrogen bonds were determined for polar hydrogen acceptor atoms (O, N, S) with 3.0 Å and 120° cutoffs.
Supplementary Material
ACKNOWLEDGMENTS
Support for this work was provided by NIH T32DA007315 and NIH T32GM007752 (BM), USDA 2008-35621-04749 and NIH FIC U01 TW006634 (WHG), NIH R01MH077305 (VH), NIH 5T32GM007752-32 (AJF) and NIH GM31749, NSF MCB-1020765, and MCA93S013 (JAM). Support from the Howard Hughes Medical Institute, the NSF Supercomputer Centers, the San Diego Supercomputer Center, the W.M. Keck Foundation, the National Biomedical Computational Resource, and the Center for Theoretical Biological Physics is gratefully acknowledged. The DCG-04 probe was a gift from Mathhew Bogyo (Stanford University).
REFERENCES
- 1.Nunnery JK, Mevers E, Gerwick WH. Curr. Opin. Biotechnol. 2010;21:787–793. doi: 10.1016/j.copbio.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pettit GR, Kamano Y, Herald CL, Tuinman AA, Boettner FE, Kizu H, Schmidt JM, Baczynskyj L, Tomer KB, Bontems RJ. J. Am. Chem. Soc. 1987;109:6883–6885. [Google Scholar]
- 3.Luesch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. J. Nat. Prod. 2001;64:907–910. doi: 10.1021/np010049y. [DOI] [PubMed] [Google Scholar]
- 4.Villa FA, Gerwick L. Immunopharmacol. Immunotoxicol. 2010;32:228–237. doi: 10.3109/08923970903296136. [DOI] [PubMed] [Google Scholar]
- 5.Linington RG, Edwards DJ, Shuman CF, McPhail KL, Matainaho T, Gerwick WH. J. Nat. Prod. 2008;71:22–27. doi: 10.1021/np070280x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunasekera SP, Miller MW, Kwan JC, Luesch H, Paul VJ. J. Nat. Prod. 2010;73:459–462. doi: 10.1021/np900603f. [DOI] [PubMed] [Google Scholar]
- 7.Gondi CS, Rao JS. Expert Opin. Ther. Targets. 2013;17:281–291. doi: 10.1517/14728222.2013.740461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lankelma JM, Voorend DM, Barwari T, Koetsveld J, Van der Spek AH, De Porto APNA, Van Rooijen G, Van Noorden CJF. Life Sci. 2010;86:225–233. doi: 10.1016/j.lfs.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Hook VYH, Kindy M, Hook G. J. Biol. Chem. 2008;283:7745–7753. doi: 10.1074/jbc.M708362200. [DOI] [PubMed] [Google Scholar]
- 10.Sathya M, Premkumar P, Karthick C, Moorthi P, Jayachandran KS, Anusuyadevi M. Clin. Chim. Acta. 2012;414:171–178. doi: 10.1016/j.cca.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Rosenthal PJ. Emerg. Infect. Dis. 1998;4:49–57. doi: 10.3201/eid0401.980107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwan JC, Eksioglu EA, Liu C, Paul VJ, Luesch H. J. Med. Chem. 2009;52:5732–5747. doi: 10.1021/jm9009394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira AR, Kale AJ, Fenley AT, Byrum T, Debonsi HM, Gilson MK, Valeriote FA, Moore BS, Gerwick WH. ChemBioChem. 2012;13:810–817. doi: 10.1002/cbic.201200007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leto G, Sepporta MV, Crescimanno M, Flandina C, Tumminello FM. Biol. Chem. 2010;391:655–664. doi: 10.1515/BC.2010.069. [DOI] [PubMed] [Google Scholar]
- 15.Rosenthal PJ. In: Cysteine Proteases of Pathogenic Organisms. Robinson MW, Dalton JP, editors. Springer US: Advances in Experimental Medicine and Biology; 2011. pp. 30–48. [Google Scholar]
- 16.Funkelstein L, Toneff T, Hwang S-R, Reinkeckel T, Peters C, Hook V. J. Neurochem. 2008;106:384–391. doi: 10.1111/j.1471-4159.2008.05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minokadeh A, Funkelstein L, Toneff T, Hwang S-R, Beinfeld M, Reinkeckel T, Peters C, Zadina J, Hook V. Mol. Cell. Neurosci. 2010;43:98–107. doi: 10.1016/j.mcn.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Gallinamide A is also known as symplostatin 4 in the literature; see Taori M, Liu Y, Paul V, Luesch H. ChemBioChem. 2009;10:1634–1639. doi: 10.1002/cbic.200900192.
- 19.Linington RG, Clark BR, Trimble EE, Almanza A, Ureña L-D, Kyle DE, Gerwick WH. J. Nat. Prod. 2009;72:14–17. doi: 10.1021/np8003529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conroy T, Guo JT, Linington RG, Hunt NH, Payne RJ. Chem. Weinh. Bergstr. Ger. 2011;17:13544–13552. doi: 10.1002/chem.201102538. [DOI] [PubMed] [Google Scholar]
- 21.Yasothornsrikul S, Greenbaum D, Medzihradszky KF, Toneff T, Bundey R, Miller R, Schilling B, Petermann I, Dehnert J, Logvinova A, Goldsmith P, Neveu JM, Lane WS, Gibson B, Reinheckel T, Peters C, Bogyo M, Hook V. Proc. Natl. Acad. Sci. 2003;100:9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Copeland RA. Methods Biochem. Anal. 2005;46:1–265. [PubMed] [Google Scholar]
- 23.Stolze SC, Deu E, Kaschani F, Li N, Florea BI, Richau KH, Colby T, van der Hoorn RAL, Overkleeft HS, Bogyo M, Kaiser M. Chem. Biol. 2012;19:1546–1555. doi: 10.1016/j.chembiol.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian WX, Tsou CL. Biochemistry (Mosc.) 1982;21:1028–1032. doi: 10.1021/bi00534a031. [DOI] [PubMed] [Google Scholar]
- 25.Leytus SP, Toledo DL, Mangel WF. Biochim. Biophys. Acta BBA - Protein Struct. Mol. Enzym. 1984;788:74–86. doi: 10.1016/0167-4838(84)90299-1. [DOI] [PubMed] [Google Scholar]
- 26.Warren GL, Andrews CW, Capelli A-M, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. J. Med. Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 27.Hardegger LA, Kuhn B, Spinnler B, Anselm L, Ecabert R, Stihle M, Gsell B, Thoma R, Diez J, Benz J, Plancher J-M, Hartmann G, Banner DW, Haap W, Diederich F. Angew. Chem. Int. Ed Engl. 2011;50:314–318. doi: 10.1002/anie.201006781. [DOI] [PubMed] [Google Scholar]
- 28.Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang S-R. Annu Rev Pharmacol Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bas DC, Rogers DM, Jensen JH. Proteins. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Robertson AD, Jensen JH. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 32.Søndergaard CR, Olsson MHM, Rostkowski M, Jensen JH. J. Chem. Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 33.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 34.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 36.Horn HW, Swope WC, Pitera JW, Madura JD, Dick TJ, Hura GL, Head-Gordon T. J. Chem. Phys. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 37.Sinko W, de Oliveira C, Williams S, Van Wynsberghe A, Durrant JD, Cao R, Oldfield E, McCammon JA. Chem. Biol. Drug Des. 2011;77:412–420. doi: 10.1111/j.1747-0285.2011.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lippert RA, Bowers KJ, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Shaw DE. J. Chem. Phys. 2007;126:046101. doi: 10.1063/1.2431176. [DOI] [PubMed] [Google Scholar]
- 39.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 40.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 12. San Francisco: University of California; 2012. [Google Scholar]
- 41.Humphrey W, Dalke A, Schulten K. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.