I. OVERVIEW
Germ cell tumors (GCTs) are cancers of the testis, ovary, or extragonadal sites that occur in infants, children and adults. Although rare overall, they account for 15% of the cancers diagnosed in childhood and adolescence. Testicular GCT is the most common malignancy in young men aged 15-40, and the incidence of GCT around the world is rapidly increasing for unclear reasons (Frazier and Amatruda, 2009). Clinically, GCTs are treated with cisplatin-containing chemotherapy, surgery and in some cases radiotherapy. While the results are overall excellent, these patients undergoing these treatments often suffer long-term adverse effects, including cardiovascular disease, secondary malignancies, kidney dysfunction and hearing loss. In addition, current regimens still fail to cure about 15-20% of patients (Frazier and Amatruda, 2009). Together, these problems indicate a pressing need for improved, targeted therapies for GCTs. However, the poor understanding of the molecular basis of GCTs, and the lack of suitable animal models, represent an impediment to the development of new therapies. The many advantages of zebrafish for genetic analysis and disease modeling suggest that fish models of GCTs could have great translational impact. Similar to Wilms tumor, neuroblastoma, and medulloblastoma, GCTs are “embryonal” tumors, in which misregulation of developmental signaling pathways is likely to play a critical role. Therefore, better understanding of GCT biology can potentially also reveal mechanisms of normal germline development.
II. GERMLINE DEVELOPMENT
The earliest cells of the germ cell lineage are the Primordial Germ Cells (PGCs) (Kunwar et al., 2006; Molyneaux and Wylie, 2004; Molyneaux et al., 2001; Wylie, 2000). In most multicellular organisms, PGCs arise at distant sites and must migrate through the developing embryo to reach the site at which the gonad will develop. Throughout migration and development, PGCs are able to maintain their underlying pluripotency program while repressing somatic differentiation (van de Geijn et al., 2009; Western, 2009). This specialized function enables PGCs to ultimately fulfill their role when, upon fertilization, they reactivate their differentiation program to give rise to the next generation. Studies in Drosophila melanogaster, zebrafish, and mice have revealed conserved mechanisms of PGC specification, migration and development (Kunwar et al., 2006; Raz, 2003; Richardson and Lehmann, 2010; Williamson and Lehmann, 1996).
A. Specification of Primordial Germ Cells
Primordial germ cell specification in most organisms requires two conserved molecular mechanisms: repression of the somatic differentiation program and reactivation of pluripotency-associated gene expression. In zebrafish, primordial germ cell specification is predetermined by the inheritance of maternal RNA and proteins located in the germ plasm (Houston and King, 2000; Raz, 2003; Saffman and Lasko, 1999; Williamson and Lehmann, 1996). In contrast to other organisms, zebrafish PGCs arise in four distinct random locations with respect to the developmental axis of the embryo (Raz, 2003; Weidinger et al., 1999; Weidinger et al., 2002; Yoon et al., 1997). The identification of vasa as a germline cell marker in zebrafish was an important discovery that facilitated the study of PGC/germline development (Olsen et al., 1997; Yoon et al., 1997). In zebrafish, vasa expression is first detected in four strips of electron-dense germ plasm along the first two cleavage planes in the embryo. By the 4K cell stage, the vasa enriched germ plasm is distributed into the cytoplasm of four closely associated cells that then become PGCs. The four newly specified PGCs undergo multiple rounds of division to generate 25-50 PGCs that migrate to the genital ridges by the end of the first day (Braat et al., 1999; Knaut et al., 2000; Weidinger et al., 1999; Yoon et al., 1997).
Mice and other mammals lack germ plasm and require inductive signaling for PGC specification (Lawson et al., 1999; Tam and Zhou, 1996; Ying et al., 2001; Ying and Zhao, 2001). At E6.5, bone morphogenetic proteins 4, 8b, and 2 (BMP4/8b/2) and unidentified proteins signal from the extraembryonic ectoderm and visceral endoderm to pluripotent epiblast cells to induce fragilis/Ifitm3 expression (Saitou et al., 2002; Ying et al., 2001; Ying and Zhao, 2001; Zhao and Garbers, 2002). fragilis/Ifitm3 expression is required for the proximal epiblast cells to achieve competence to become PGC precursor cells (Lange et al., 2003; Saitou et al., 2002; Tanaka and Matsui, 2002; Tanaka et al., 2004; Tanaka et al., 2005). BMP4, BMP2, and BMP8b null mice lack or have severely reduced numbers of PGCs due to the failure to generate PGC precursor cells (de Sousa Lopes et al., 2004; Itman et al., 2006; Lawson et al., 1999; Ying et al., 2001; Ying and Zhao, 2001; Zhao and Garbers, 2002 2004).
An important molecular mechanism for PGC specification that is common to many organisms is the transcriptional silencing of somatic gene expression (Ohinata et al., 2005; Saitou et al., 2002; Yabuta et al., 2006). The fragilis/Ifitm-positive proximal epiblast cells also express somatic mesodermal genes including Hoxb1, Fgf8, and Snail (Ancelin et al., 2006; Hayashi et al., 2007; Yabuta et al., 2006). In these cells, B lymphocyte-induced maturation protein 1 (BLIMP1, also known as PRDM1), a transcriptional repressor, plays significant roles in the somatic gene repression as well as promoting upregulation of PGC-specific genes such as stella (Ohinata et al., 2005; Saitou et al., 2005; Vincent et al., 2005). The loss of Blimp1 in mutant mice results in reduced somatic gene silencing, loss of founder PGCs, and lack of PGC migration (Kurimoto et al., 2008; Yamaji et al., 2008). By E7.25, there are approximately 40 Blimp1 positive, specified PGCs. (Ohinata et al., 2005). These cells are characterized by their transcriptional silencing of somatic genes, the expression of PGC-specific genes, and maintenance or upregulation of pluripotency-associated genes such as Oct4, Sox2, and Nanog (Saitou et al., 2002; Scholer et al., 1990; Yabuta et al., 2006; Yamaguchi et al., 2005; Yeom et al., 1996).
B. Primordial Germ Cell Migration
In most organisms the PGCs arise in a location distal to the genital ridges where the PGCs will eventually reside. To arrive at the gonads the PGCs must gain motility and migrate through the embryo to their final location. Similar to PGC specification, there are conserved mechanisms for migration amongst different organisms, but there are also important distinctions and modes of migration.
Zebrafish have four clusters of PGCs originating in dispersed locations in the embryo that must migrate to the genital ridges (Weidinger et al., 2003; Weidinger et al., 1999; Weidinger et al., 2002; Yoon et al., 1997). The PGCs use a “run” and “tumble” system in which they migrate “run” short distances to intermediate stops where they remain stationary “tumble” for a short period of time to realign themselves to attractant chemokine signals that are guiding them through the embryo (Raz and Reichman-Fried, 2006; Reichman-Fried et al., 2004). The initiation of migration requires multiple steps in which the PGCs gain motility. Initially, zebrafish PGCs are morphologically indistinguishable from the surrounding somatic cells, exhibiting a smooth, round morphology. However, in the first 30 minutes after specification the PGCs start to exhibit a ruffled edge appearance with the extension of short cellular protrusions in all directions. At this time, the PGCs have not gained the ability to migrate and these protrusions are eventually lost as the PGCs divide. This is followed by a one hour transitional phase in which the PGCs become polarized and extend out broad pseudopodia for directional migration (Blaser et al., 2005). An RNA binding protein, Dead End (Dnd), is critical for PGC polarization and extension of the broad directional protrusions (Weidinger et al., 2003). The loss of Dnd in PGCs results in absence of PGC migration due to loss of polarization and protrusion extension, and ultimately in the death of the PGCs (Blaser et al., 2005). After becoming motile, zebrafish PGCs start to actively migrate towards attractant signals provided by somatic cells. Stromal-derived factor-1alpha (SDF-1a) has been identified as a critical component guiding the PGCs along the migratory pathway. SDF-1a binds to chemokine receptor, CXCR4b, which is expressed in PGCs. It was demonstrated that PGCs will migrate to ectopic locations in response to aberrant SDF-1a secreting cells (Doitsidou et al., 2002; Knaut et al., 2003).
Mouse PGCs are initially located in the primitive streak and must follow a migratory path through the posterior embryonic endoderm, extraembryonic endoderm, and finally through the allantois and hindgut to reach the genital ridges (Anderson et al., 2000). Several molecules/pathways have been identified as important mediators of proper PGC migration. Similar to zebrafish PGCs, guidance is provided by the SDF1 chemokine and CXCR4 interaction (Ara et al., 2003; Molyneaux et al., 2003). The c-kit receptor tyrosine kinase and its ligand steel were discovered to facilitate migration by regulating PGC motility (Gu et al., 2009). In addition, E-cadherin and β-1 integrin are required for proper exiting from the hindgut and migration into the gonads (Anderson et al., 1999a; Anderson et al., 1999b; Bendel-Stenzel et al., 2000).
C. Epigenetic Reprogramming of Primordial Germ Cells
Until the time of PGC migration, all cells in a developing embryo have a bi-parental pattern of genomic imprinting. Genomic imprinting is an epigenetic phenomenon in which DNA methylation controls expression of a limited number of genes that are dependent on parental origin (McLaren, 2003; Surani, 2001). Approximately 100-200 genes in the human genome are imprinted such that only one allele, either maternal or paternal, is expressed (Lucifero et al., 2004; Paoloni-Giacobino and Chaillet, 2004). In PGCs, the bi-parental pattern of genomic imprinting must be erased, followed by the establishment of a uni-parental pattern in order to ensure that proper sex-specific imprinting is passed on to the next generation. This process, called genome-wide epigenetic reprogramming, occurs in migratory and post-migratory PGCs in mammals. Zebrafish and Xenopus, however, do not exhibit imprinting and do not undergo paternal genome reprogramming (Macleod et al., 1999; Reik et al., 2001). Once the PGCs (now referred to as gonocytes) arrive at the gonads, they begin differentiating into either pro-spermatogonia or oogonia based on the signals they receive from the gonadal microenvironment and surrounding somatic cells, and, in mammals, on their chromosomal constitution (XY or XX). Then at the appropriate time, depending on the sex of the organism, the process of gametogenesis (spermatogenesis if male and oogenesis if female) is initiated to generate fully mature gametes that are capable of producing the next generation upon fertilization. Oogenesis in zebrafish and medakafish has been recently reviewed (Clelland and Peng, 2009; Lessman, 2009; Saito and Tanaka, 2009). In the next section, we review the process of spermatogenesis.
D. Spermatogenesis
Spermatogenesis is a dynamic process occurring in a series of highly organized steps that culminates in the production of fully mature spermatozoa. Spermatogenesis is initiated during puberty and is maintained continuously throughout the life of most organisms. In most vertebrates, spermatogenesis occurs within seminiferous tubules that are composed of a basement membrane, basal compartment, and adluminal compartment. Blood vessels, Leydig cells, lymphatic epithelium, and macrophages surround each tubule in the interstitial space (as reviewed in Yoshida, 2008a). On the inside of the basement membrane reside the Sertoli cells. Representing the only somatic cell within the tubules, Sertoli cells support the germ cells by providing nourishment, growth factors, and function to organize the spermatogenic process (Griswold, 1998). One distinguishing characteristic of spermatogenesis in lower vertebrates (also known as anamniotes) including fish is that in the seminiferous tubules, spermatogenesis occurs within cysts that are formed by Sertoli cell cytoplasmic extensions (Schulz et al., 2010). In contrast to higher vertebrates (retiles, birds, and mammals) where one Sertoli cell is associated with several spermatogonial germ cells at different developmental stages, cystic spermatogenesis is represented by having one Sertoli cell associated with a single spermatogonial germ cell (Matta et al., 2002; Schulz et al., 2005).
The process of spermatogenesis is much conserved amongst different species including flies, fish, and mammals with relatively few minor distinctions. In the mouse, Type A undifferentiated spermatogonia (Asingle) are believed to be the stem cells of the testis and are closely associated with the basement membrane in the basal compartment (Chiarini-Garcia et al., 2001; Chiarini-Garcia and Russell, 2001; Hess et al., 2006; Oatley and Brinster, 2006; Ogawa et al., 2005; Ryu et al.). The Asingle spermatogonia are thought to undergo self-renewal as well as to give rise to specialized daughter cells that will divide and differentiate to ultimately become spermatozoa (Oatley and Brinster, 2006). Asingle spermatogonia give rise to the undifferentiated Type A spermatogonia (Aundiff), Apaired and Aaligned. These undifferentiated cells, including the Asingle cells are tightly clustered together and only account for 1% of testicular cells. The Aundiff cells will give rise to the next class of spermatogonia, the differentiated Type A1 spermatogonia. The differentiated Type A1 spermatogonia will undergo successive rounds of mitosis. The number of rounds is dependent on the organism. In rodents there are typically six rounds of mitotic division which results in Type A2-4 differentiated spermatogonia, intermediate spermatogonia, Type B spermatogonia, and finally the preleptotene primary spermatocytes (de Rooij and Russell, 2000). Zebrafish has a total of nine rounds of mitosis that is initiated by type A undifferentiated primary spermatogonia and results in three type A differentiated spermatogonia generations, five type B spermatogonia generations, and one generation of primary preleptotene spermatocytes (Leal et al., 2009).
Still closely associated with the basement membrane, the preleptotene spermatocytes initiate meiosis. During a long prophase, the leptotene and zygotene spermatocytes will move away from the basement membrane and will enter the adluminal compartment through the tight junctions that are formed by the Sertoli cells. The exchange of genetic information occurs during the recombination event in pachytene spermatocytes, after which the completion of meiosis I results in 2N secondary spermatocytes. These will then undergo meiosis II to produce four haploid (1N) round spermatids (Hess and Renato de Franca, 2008; Schulz et al., 2010; Yoshida, 2008b).
The round spermatids are transformed into elongated, highly condensed, mature spermatozoa through a process called spermiogenesis that includes four different stages; golgi, capping, acrosomal, and maturation (Hess and Renato de Franca, 2008). The mature spermatozoa are released into the lumen and upon ejaculation exit the body through the rete testes, epididymis, and vas deferens. In zebrafish, mature spermatozoa are present in the lumen and are released through the efferent duct (Leal et al., 2009).
III. GERM CELL TUMORS (GCTS)
A. Human Germ Cell Tumors
In multicellular organisms, proper development of the germ cells is essential for the transmission of genetic information to the next generation, and ultimately for the survival of the species. For this reason development of the germline is highly regulated to control germ cell proliferation, migration, differentiation, and survival (Chuva de Sousa Lopes and Roelen, 2010; Ewen and Koopman, 2009). Disruption of this regulation can lead to disorders such as infertility, chromosomal abnormalities, and germ cell tumors. Germ cell tumors (GCTs) are malignant tumors of the germline that occur in neonates, infants, children and adults (Frazier and Amatruda, 2009). GCTs arise primarily in the testes and ovaries, though they also occur in extragonadal sites along the midline of the body and the brain. Testicular GCT is the most common cancer in young men aged 15-40, and is increasing in incidence worldwide for unknown reasons. In humans, GCTs differ in clinical and histological presentation and are classified into five distinct groups based on differences in age of onset, histology, cell of origin, chromosomal constitution, and pattern of genomic imprinting (Oosterhuis and Looijenga, 2005). Type I GCTs consist of the teratomas and yolk sac tumors of neonates and young children. Type II GCTs encompass the seminomas and non-seminomas of adolescents and adults. Spermatocytic seminomas that affect men > 50 years old make up the Type III GCT. Finally, the ovarian dermoid cyst and uterine hydatidiform mole are classified as the type IV and type V GCTs respectively.
1. Type I Germ Cell Tumors
Type I germ cell tumors include teratomas and yolk sac tumors that affect neonates and children < 5 years of age (Oosterhuis and Looijenga, 2005). They most commonly occur in the testes, ovaries, sacrococcygeal area, retroperitoneum, head and neck, and in the hypophyseal region of the brain. The type I GCTs are relatively rare with an incidence of 0.12/100,000; sacral teratomas are the most commonly diagnosed type I GCT (Rescorla, 1999). Based on a partially erased biparental pattern of genomic imprinting, an early primordial germ cell is thought to be the cell of origin in type I germ cell tumors (Bussey et al., 2001; Schneider et al., 2001). Typically, the type I teratomas are benign; however, ovarian teratomas can be malignant and teratomas that are incompletely surgically removed can progress to yolk sac tumors, which have the potential to be metastatic (Gobel et al., 2000). Type I teratomas primarily have a normal chromosomal constitution while type I yolk sac tumors are characteristically aneuploid. Recurrent chromosomal abnormalities in yolk sac tumors include loss of 1p, 4, and 6q and gain of 1q, 12(p13), 20q, and 22 (Mostert et al., 2000; Perlman et al., 2000; Schneider et al., 2001)
Type I animal models include various mouse models of teratomas. Spontaneous testicular teratomas arise in the inbred 129-strain of mice that closely resemble human type I teratomas (Noguchi and Noguchi, 1985; Stevens, 1973). Mutations in Dead-end/Ter in the 129-strain mice cause significant PGC loss and increased type I testicular germ cell tumor susceptibility (Youngren et al., 2005). Targeted deletion of Pten in mouse PGCs leads to greater risk for testicular teratomas, increased germ cell proliferation, and greater capacity to generate embryonic germ cells in culture; thus indicating an important role for Pten in regulating germ cell proliferation and differentiation (Kimura et al., 2003).
2. Type II Germ Cell Tumors
Type II GCTs occur in adolescents and adults and testicular GCTs represent the most common malignancy found in men 20-40 years of age (McIntyre et al., 2008). Type II GCTs largely occur in the testes and ovaries but also occur in extragonadal sites such as the mediastinum and in the brain (Oosterhuis and Looijenga, 2005). Although type II GCTs are often diagnosed in females, they predominantly affect males and are referred to as testicular germ cell tumors (TGCTs). Type II GCTs are further divided into two subgroups based on histological and clinical variations; seminomas and nonseminomas (Figure 1). Seminomas (SE) are composed of primitive, undifferentiated germ cells that resemble PGCs/gonocytes and occur in the testes. They are also called dysgerminomas when present in the ovaries and germinomas when found extragonadally in the brain. Nonseminomas (NS) include GCTs that are further along the differentiation program than SE such as embryonal carcinomas (EC), choriocarcinomas (CC), yolk sac tumors (YST), and mature teratomas (TE) (Oosterhuis and Looijenga, 2005; van de Geijn et al., 2009).
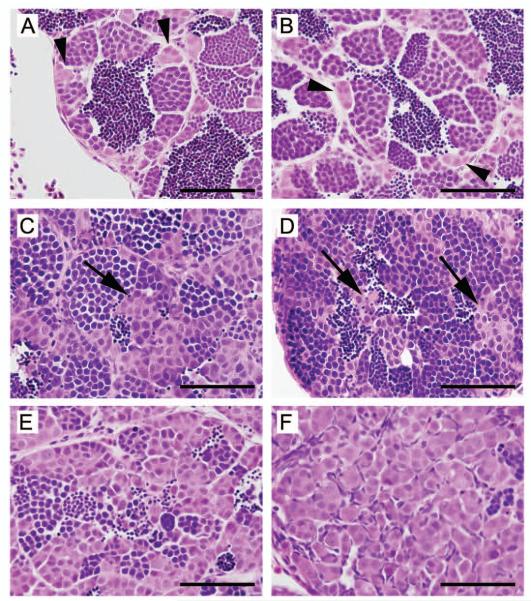
Figure 1. Histology of zebrafish germ cell tumors.
Hematoxylin & eosin stained sections from formalin-fixed, paraffin-embedded zebrafish testes. A, B: normal testis. The testis consists of cysts or lobules of spermatogenic cells surrounded by a basement membrane and somatic cells. Small clusters of spermatogonia are seen adjacent to the basement membrane (arrowheads). Successive stages of differentiation are also evident, including primary and secondary spermatocytes, spermatids and mature spermatozoa. C-F: testicular germ cell tumors. The images shown are from the tgct germ cell tumor mutant line; similar histology can be found in carcinogen-treated fish and in males of advanced age. C-F: loss of orderly architecture and impaired differentiation. Ectopic clusters of spermatogonia are seen in the center of the lobule, distant from the basement membrane (arrows). In E, a severe reduction in spermatogenesis is evident. F: complete loss of spermatocytic differentiation. The architecture of the lobule is overrun by a proliferation of primitive, spermatogonial-like cells. (Modified, with permission, from (Neumann et al., 2009)).
A fully erased pattern of biparental genomic imprinting suggests that type II GCTs arise from slightly later PGCs/gonocytes than the type I GCTs (Bussey et al., 2001; Oosterhuis and Looijenga, 2005; Schneider et al., 2001). Carcinoma in situ (CIS) is a noninvasive precursor lesion that gives rise to all type II TGCTs (Hoei-Hansen et al., 2005; Rajpert-De Meyts et al., 2003). CIS has an incidence rate similar to the type II GCTs and indicates that all CIS lesions will eventually progress to invasive TGCTs (Oosterhuis and Looijenga, 2005; van de Geijn et al., 2009). CIS cells show phenotypic characteristics similar to PGCs such as morphology, pluripotent gene expression, and genomic imprinting. A confirmed marker for CIS is Oct3/4, a gene that is known for a role in maintaining pluripotency. Oct3/4 is expressed in all CIS, seminomas, and the embryonal carcinoma component of nonseminomas which suggests that all SE and NS share similar pathogenesis pathways (Hoei-Hansen et al., 2005; Rajpert-De Meyts et al., 2003). Oct3/4 is expressed in normal PGCs during development but then decreases in the germ cells post-natally (Honecker et al., 2004; Rajpert-De Meyts et al., 2004; Stoop et al., 2005).
Seminomas exhibit an accumulation of undifferentiated PGC/gonocyte-like germ cells that share a similar morphology to CIS cells. Nonseminomas can be composed of different histological components that represent the differentiation of a truly totipotent cell. Embryonal carcinomas (EC) consist of undifferentiated stem cells and it is thought that ECs may arise from seminomas that have undergone reprogramming to activate underlying pluripotency to become ECs (Looijenga et al., 1999; Oosterhuis et al., 2003). ECs undergo differentiation to give rise to other nonseminoma components including choriocarcinomas and yolk sac tumors that are differentiated into extraembryonic components (trophoblast and yolk sac, respectively) and mature teratomas that have undergone somatic differentiation. Thus the EC represents the neoplastic counterpart to embryonic germ cells and the true cancer stem cell.
Type II TGCTs are typically aneuploid and show a consistent pattern of recurrent chromosomal abnormalities including the loss of chromosomes 4, 5, 11, 13, 18, and Y, and gain of chromosomes 7, 8, 12p, and X (Castedo et al., 1989; Looijenga et al., 2000; Oosterhuis and Looijenga, 2005; Ottesen et al., 1997; Summersgill et al., 1998). The gain of 12p is characteristic of all invasive type II TGCTs and interestingly is not consistently present in the preinvasive lesion, CIS (Looijenga et al., 2007; Looijenga et al., 2000; Oosterhuis et al., 1997; Summersgill et al., 2001; van Echten et al., 1995). This indicates that gain of 12p plays a significant role in the transformation of CIS into invasive TGCTs.
Very few mutations associated with TGCT development have been identified, primarily due to lack of large pedigrees for analysis and the lack of suitable animal models for type II TGCTs (Oosterhuis and Looijenga, 2005). However, activating mutations in c-Kit exon17, in particular at codon 816, are associated predominantly with bilateral TGCTs, which only account for up to 5% of TGCTs (Dieckmann et al., 2007a; Dieckmann et al., 2007b; Kemmer et al., 2004; Looijenga et al., 2003; Nakai et al., 2005; Oosterhuis and Looijenga, 2005; Sakuma et al., 2003; Tian et al., 1999). Two recent Genome-Wide Association Studies implicated KITLG (the kit ligand), DMRT1 and SPRY4 in familial testicular cancer (Kanetsky et al., 2009; Rapley et al., 2009; Turnbull et al., 2010).
B. Germ Cell Tumors in Zebrafish
1. Carcinogenesis and Reverse Genetic Models of GCT
Previous studies have described the spontaneous development of gonadal neoplasms in male zebrafish >2 years of age. Moore et al. described the tumor spectrum at 30-34 months of age in wild-type zebrafish and in carriers of the gin genomic instability phenotype (Moore et al., 2006). Testicular hyperplasias (enlarged testes containing all stages of spermatogenesis) were found in 48% of wildtypes and 25% of gin heterozygotes. “Benign seminomas”, which they defined as tumors of predominantly one cell type derived from an early stage in spermatogenesis, were seen in 17% of wildtype fish at 30-34 months of age; the incidence was 53% in gin carriers. In a survey of nearly 10,000 2 year-old zebrafish, Amsterdam et al. documented the tumor spectrum. Of 473 tumors found, approximately 40% were described as seminomas of the testis (Amsterdam et al., 2009). The testis has also been identified as a target of carcinogens in multiple fish species, including rainbow trout, medaka and zebrafish (Bailey et al., 1984; Hawkins et al., 1985; Neumann et al., 2009). Spitsbergen and colleagues reported testicular neoplasms of 5 of 68 juvenile fish treated with the carcinogens MNNG, and in 1 of 99 juveniles treated with DMBA (Spitsbergen et al., 2000a; Spitsbergen et al., 2000b). Other teleosts, such as rainbow trout and medaka are also susceptible to testicular carcinogenesis (Bailey 1984, Hawkins 1985). Recently, transgenic expression of large T antigen or the scl gene in the testis was found to result in germ cell tumors by 36 months of age (Gill et al., 2010).
2. Forward Genetic Screens for Gonadal Phenotypes and GCT
Forward genetic approaches have also been used to generate GCTs in zebrafish. Bauer and Goetz identified 11 mutations that caused gonadal phenotypes in either males or females during a mutational screen using N-ethyl N-nitrosourea (Bauer and Goetz, 2001). The males had altered spermatogenesis in which the testes contained predominantly spermatogonia and/or spermatocytes, similar to the benign seminomas that were described by Moore et al (Bauer and Goetz, 2001; Moore et al., 2006)). During a forward genetic screen to identify cancer susceptibility mutations, we identified a highly heritable testicular germ cell tumor (tgct) mutant (Neumann et al., 2009). Homozygous and heterozygous adult male fish from this line develop bilateral TGCTs that can grow to occupy the entire abdominal cavity. The tumors are notable for impaired spermatocytic differentiation that leads to the accumulation of undifferentiated spermatogonial-like cells that disrupt normal testicular architecture (Figure 1). Homozygous mutant females display an oocyte maturation defect that affects fertility and causes an increase of stage I and II immature oocytes and lack of fully mature stage V ova. In the Medaka fish mutant, hotei, a nonsense mutation in anti-Mullerian hormone receptor II (AMHRII) impairs gonadal development and results in a phenotype somewhat overlapping that of the zebrafish tgct mutant. Hotei (hot) mutant fish develop enlarged gonads; males exhibit hypertrophic testes with disorganized spermatogenesis, whereas females display arrested follicular development.
IV. METHODS FOR STUDYING ZEBRAFISH GERM CELLS
Methods that can be used to study germline development and disease in zebrafish provide certain advantages over other model organisms. Techniques such as fusing GFP with nanos 3′ UTR, in vitro germ cell differentiation assays, and the use of germ cell specific promoters (i.e. ziwi), allows studies of germ cell specification and migration, interactions with the microenvironment, and germ cell specific gene expression. In this section, we provide protocols for several different methods to study germ cells in zebrafish.
A. Visualization of Primordial Germ Cells
Studies on germ cells in zebrafish are significantly enhanced by the ability to visualize germ cells both in vitro and in vivo. The identification of germ cell specific genes such as vasa and nanos provided a means to visualize PGCs to examine germ cell specification, migration, and development (Koprunner et al., 2001; Yoon et al., 1997). In situ hybridization probes against vasa and nanos have frequently been used to look for germ cell specific phenotypes. It was found that the 3′UTR of vasa and nanos is essential to germ cell specific gene expression. Constructs that fuse GFP to the vasa or nanos 3′UTR allows visualization of PGCs in living embryos (Wolke et al., 2002) (Figure 2). These constructs not only can be used to conduct screens to identify genes required for germ cell development but they can also be used to perform germ cell transplantation assays to study germ cell interactions with the microenvironment . Recently, Leu and Draper achieved robust and specific expression of transgenes in the early cells of the testis and ovary using the ziwi promoter (Leu and Draper). Visualizing germ cells will undoubtedly remain critical to the study of germ cells and the identification of genes and pathways that are essential for proper germline development.
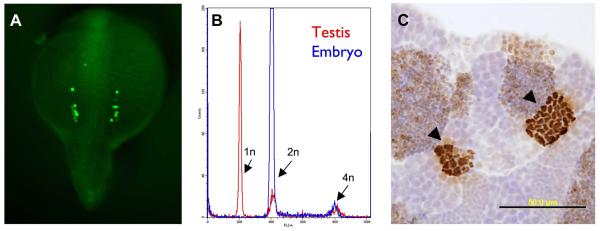
Figure 2. Methods to visualize germ cells.
A, embryo injected at the 1-cell stage with GFP-nanos 3′UTR mRNA (gift of Gilbert Weidinger). Bilateral clusters of PGCs are visible in this dorsal view of a 15-somite stage embryo. B, FACS DNA content analysis of the testis. Red line indicates testis, with haploid (1n), diploid (2n) and mitotic (4n) cells. Blue line is a normal, diploid embryo DNA content profile for comparison. C, anti-phosphohistone H3 immunohistochemistry of adult, wild-type testis. Clusters of synchronously-dividing spermatocytes are seen (arrowheads).
B. In vitro culture of the testis
Dr. Noriyoshi Sakai and colleagues published pioneering studies describing the in vitro culture of zebrafish testis (Sakai, 2002; Sakai, 2006). Unlike mammalian testis, zebrafish germ cells are capable of undergoing meiosis in vitro to produce functional sperm. Below, we present a protocol for culture of testicular germ cells (modified from (Sakai, 2006)). Note that variations of this protocol are possible, including the use of feeder cell layers (Sakai, 2006).
Sacrificing Animals
Anesthetize fish in 0.2% Tricaine solution for 2 minutes
Dip fish in 100% EtOH to disinfect the carcass.
Testis Dissection
3) Decapitate the fish and use surgical scissors to open the ventral surface. Expose the testis by carefully removing the gut and swim bladder.
-
4) Using fine-tipped tweezers, carefully remove the bilateral testis from the body cavity.
Note: It is recommended to use the testes from 6-8 fish to obtain enough cells to set up primary cultures.
Primary Culture from Testis
Note: All of these steps should be done in a sterile cell culture hood to avoid contamination of the primary cultures.
5) Add dissected testis to a 6-well plate with 2mL 1x Phosphate-buffered saline (PBS).
6) Wash 3 times with sterile 1x PBS.
7) Mince large pieces of the tissue with surgical scissors and pass through a pipette to break up the large pieces.
8) Transfer the sample into a 15mL conical tube and bring the volume up to 5mL with sterile 1x PBS.
9) Gently centrifuge the sample 5 minutes, 1000 rpm.
10) Carefully remove the supernatant, being sure not the dislodge the cell pellet.
11) Add 3mL Dispase to the cell pellet for enzymatic disaggregation of the sample.
12) Incubate 37°C for 30 minutes with gentle agitation to break up the tissue further.
13) Once enzymatic digestion is completed, quench Dispase with 12mL DMEM/F12 Complete and shake well.
14) Spin the sample 5 minutes, 1000 rpm.
15) Remove the supernatant, leaving about 500μL over the cell pellet.
16) Resuspend the cell pellet gently with a pipette.
17) Bring the volume up to 6mL with DMEM/F12 Complete.
18) Filter the sample through a nylon mesh cell strainer (40μm) and into a 50mL conical tube.
19) Add 15mL of DMEM/F12 Complete.
20) Spin down the samples 5 minutes, 1000 rpm.
21) Remove the supernatant, being careful not to disturb the small, loose cell pellet.
22) Resuspend the pellet one last time in 4mL DMEM/F12 Complete HI-TS, FBS.
-
23) Add 1mL sample to each of the 4 wells of a gelatin coated 12-well plate.
Note: Adjust volume to plate about 2 × 105 cells per well
24) Incubate cells 28 °C. / 5% CO2 over night
25) Remove media and plate floating, non-adherent cells into a new gelatin coated well to grow.
26) Feed the cells new DMEM/F12 Complete HI-TS, FBS every ~3 days, split cells 1:2 when 80-90% confluence is reached.
C. Profiling testis DNA content by FACS
DNA content profiling by Fluorescence-Activated Cell Sorting (FACS) is a rapid and convenient method to assess the capacity of zebrafish germ cells to differentiate to haploid spermatocytes and sperm (Figure 2).
Sacrificing Animals
Anesthetize fish in 0.2% Tricaine solution for 2 minutes.
Dip fish in 100% EtOH to disinfect the carcass.
Testis Dissection
3) Decapitate the fish and use surgical scissors to open the ventral surface. Expose the testis by carefully removing the gut and swim bladder.
4) Using fine-tipped tweezers, carefully remove the bilateral testis from the body cavity.
Preparation of Samples for FACS sorting
5) Add dissected testis to 250μL DMEM in a 1.5mL tube.
6) Using a pestle, grind the testis to disaggregate the tissue.
7) Add 750μL DMEM to bring the sample volume up to 1mL.
8) Disaggregate the sample further but pipetting up and down 5-10 times.
9) Pass the sample through a piece of 40μm mesh and into a new tube to remove any clumps of tissue left over.
10) Gently spin down the sample for 4 minutes, 1200 rpm.
11) Remove and discard the supernatant.
12) Gently resuspend the cell pellet in 500μL 1x PBS.
13) Pass the sample through a new piece of 40μm mesh and into a FACS culture tube to remove any cell clumps.
14) Add 1.5mL PI/Triton X-100 Solution to the sample for a total volume of 2mL.
15) Add 4μL DNase-free RNase to the sample and place on ice in the dark for 15-20 minutes.
-
16) Samples are now ready to be analyzed.
Note: If performing FACS on cells from culture collect cells, gently pellet as in step 6 and continue protocol.
D. Detection of cell proliferation by anti-phosphohistone H3 immunohistochemistry
Phosphorylation of Histone H3 on serine 10 is correlated with the onset of chromatin condensation, and therefore marks mitotic cells. Immunohistochemistry of the testis with this marker marks clusters of synchronously-dividing spermatocytes (Figure 2). The protocol can easily be adapted to detect other antigens.
Immunohistochemical staining of zebrafish testis
Deparaffinize slides in Xylene, 2 × 10 minutes
- Rehydrate slides by placing in decreasing amounts of EtOH.
- 100% EtOH, 2 × 3 min
- 95% EtOH, 2 × 3 min
- dH2O, 2 × 3 min
Antigen Retrieval: Incubate slides in Trilogy reagent (Cell Marque) in a pressure cooker for 15 minutes. For other antigens, conditions for antigen retrieval may need to be individually optimized.
Cool slides in Trilogy reagent for 20 minutes.
Peroxidase Block: Place slides in 0.3% H2O2/H2O for 30 minutes. Rinse in dH2O for 1 minute.
Block in 2.5% horse serum (Immpress; Vector labs) for 30 minutes at room temperature. Do not let sections dry out at anytime during blocking or incubation with antibodies.
Apply antiphosphohistone H3 (Santa Cruz biotech) at 1:750 in horse serum for 2 hours at room temperature or overnight at 4 degrees.
Rinse quickly in 1X PBST (phosphate-buffered saline plus 0.1% Tween-20). Wash 4 × 5 minutes in 1X PBST.
Apply Immpress α-rabbit secondary antibody, enough to cover section. Incubate 30 minutes at room temperature.
Rinse quickly in 1X PBST. Wash 4 × 5 minutes in 1X PBST.
Apply 200 μL 1X DAB solution (1:10 DAB in DAB buffer; BD Pharmingen) until staining of sections is visible. Rinse quickly in 1X PBST before counterstaining.
Counterstain in hematoxylin for 2 minutes. Run tap water over slides until hematoxylin stain is evident by acquisition of a blue tint; typically 7-11 minutes.
- Dehydrate slides by placing in increasing amounts of EtOH.
- 95% EtOH, 2 × 1 minute
- 100% EtOH, 2 × 1 minute
- Xylene, 2 × 1 minute
Let slides air dry and then mount with DEPEX mounting solution.
E. Materials
PI/Triton X-100 Solution:
Dilute Triton X-100 1:10 (1μL Triton X-100 into 9μL PBS)
Add 1μL of 1:10 Triton X-100 to 5mL Propidium Iodide (50μg/mL in 0.1% Sodium Citrate)
2% Gelatin Coated Plates:
- Add 500μL Gelatin to necessary wells
- ○ Incubate 37°C for 15 minutes
- ○ Remove excess liquid from top of wells before use
DMEM/F12 Complete:
50% DMEM Media
50% F12 Media
1x Antibiotic-Antimyotic
1x MEM Vitamins
1x MEM Non-essential Amino Acids
2mM L-Glutamine
Note: Filter sterilize media before use
DMEM/F12 Complete HI-TS, FBS:
DMEM/F12 Complete
5% Fetal Bovine Serum
5% Heat Inactivated Trout Serum
Note: Filter sterilize media before use
Heat Inactivated Trout Serum:
10mL Trout Serum
- 10mL DMEM/F12 Complete
- ○ Heat 55°C for 30 minutes
- ○ Spin down
- ○ Collect supernatant
- ○ Filter sterilize before using
Dispase: BD Biosciences #354235
Trout Serum (“SeaGrow”): East Coast Bio #JJ80
2% Gelatin: Sigma Aldrich #G1393
DMEM/F12: Invitrogen #11039-047
Antibiotic-Antimyotic: Invitrogen #15240-062
MEM Vitamin Solution: Invitrogen #11120-052
MEM Non-Essential Amino Acids: Invitrogen #11140-050
200mM L-Glutamine: Invitrogen #25030-081
REFERENCES
- Amsterdam A, Lai K, Komisarczuk AZ, Becker TS, Bronson RT, Hopkins N, Lees JA. Zebrafish Hagoromo mutants up-regulate fgf8 postembryonically and develop neuroblastoma. Mol Cancer Res. 2009;7:841–50. doi: 10.1158/1541-7786.MCR-08-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–30. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Anderson R, Copeland TK, Scholer H, Heasman J, Wylie C. The onset of germ cell migration in the mouse embryo. Mech Dev. 2000;91:61–8. doi: 10.1016/s0925-4773(99)00271-3. [DOI] [PubMed] [Google Scholar]
- Anderson R, Fassler R, Georges-Labouesse E, Hynes RO, Bader BL, Kreidberg JA, Schaible K, Heasman J, Wylie C. Mouse primordial germ cells lacking beta1 integrins enter the germline but fail to migrate normally to the gonads. Development. 1999a;126:1655–64. doi: 10.1242/dev.126.8.1655. [DOI] [PubMed] [Google Scholar]
- Anderson R, Schaible K, Heasman J, Wylie C. Expression of the homophilic adhesion molecule, Ep-CAM, in the mammalian germ line. J Reprod Fertil. 1999b;116:379–84. doi: 10.1530/jrf.0.1160379. [DOI] [PubMed] [Google Scholar]
- Ara T, Nakamura Y, Egawa T, Sugiyama T, Abe K, Kishimoto T, Matsui Y, Nagasawa T. Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1) Proc Natl Acad Sci U S A. 2003;100:5319–23. doi: 10.1073/pnas.0730719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey GS, Hendricks JD, Nixon JE, Pawlowski NE. The sensitivity of rainbow trout and other fish to carcinogens. Drug Metab Rev. 1984;15:725–50. doi: 10.3109/03602538409041078. [DOI] [PubMed] [Google Scholar]
- Bauer MP, Goetz FW. Isolation of gonadal mutations in adult zebrafish from a chemical mutagenesis screen. Biol Reprod. 2001;64:548–54. doi: 10.1095/biolreprod64.2.548. [DOI] [PubMed] [Google Scholar]
- Bendel-Stenzel MR, Gomperts M, Anderson R, Heasman J, Wylie C. The role of cadherins during primordial germ cell migration and early gonad formation in the mouse. Mech Dev. 2000;91:143–52. doi: 10.1016/s0925-4773(99)00287-7. [DOI] [PubMed] [Google Scholar]
- Blaser H, Eisenbeiss S, Neumann M, Reichman-Fried M, Thisse B, Thisse C, Raz E. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J Cell Sci. 2005;118:4027–38. doi: 10.1242/jcs.02522. [DOI] [PubMed] [Google Scholar]
- Braat AK, Zandbergen T, van de Water S, Goos HJ, Zivkovic D. Characterization of zebrafish primordial germ cells: morphology and early distribution of vasa RNA. Dev Dyn. 1999;216:153–67. doi: 10.1002/(SICI)1097-0177(199910)216:2<153::AID-DVDY6>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Bussey KJ, Lawce HJ, Himoe E, Shu XO, Heerema NA, Perlman EJ, Olson SB, Magenis RE. SNRPN methylation patterns in germ cell tumors as a reflection of primordial germ cell development. Genes Chromosomes Cancer. 2001;32:342–52. doi: 10.1002/gcc.1199. [DOI] [PubMed] [Google Scholar]
- Castedo SM, de Jong B, Oosterhuis JW, Seruca R, te Meerman GJ, Dam A, Schraffordt Koops H. Cytogenetic analysis of ten human seminomas. Cancer Res. 1989;49:439–43. [PubMed] [Google Scholar]
- Chiarini-Garcia H, Hornick JR, Griswold MD, Russell LD. Distribution of type A spermatogonia in the mouse is not random. Biol Reprod. 2001;65:1179–85. doi: 10.1095/biolreprod65.4.1179. [DOI] [PubMed] [Google Scholar]
- Chiarini-Garcia H, Russell LD. High-resolution light microscopic characterization of mouse spermatogonia. Biol Reprod. 2001;65:1170–8. doi: 10.1095/biolreprod65.4.1170. [DOI] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes SM, Roelen BA. On the formation of germ cells: The good, the bad and the ugly. Differentiation. 2010;79:131–40. doi: 10.1016/j.diff.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Clelland E, Peng C. Endocrine/paracrine control of zebrafish ovarian development. Mol Cell Endocrinol. 2009;312:42–52. doi: 10.1016/j.mce.2009.04.009. [DOI] [PubMed] [Google Scholar]
- de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl. 2000;21:776–98. [PubMed] [Google Scholar]
- de Sousa Lopes SM, Roelen BA, Monteiro RM, Emmens R, Lin HY, Li E, Lawson KA, Mummery CL. BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Genes Dev. 2004;18:1838–49. doi: 10.1101/gad.294004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann KP, Kulejewski M, Pichlmeier U, Loy V. Diagnosis of contralateral testicular intraepithelial neoplasia (TIN) in patients with testicular germ cell cancer: systematic two-site biopsies are more sensitive than a single random biopsy. Eur Urol. 2007a;51:175–83. doi: 10.1016/j.eururo.2006.05.051. discussion 183-5. [DOI] [PubMed] [Google Scholar]
- Dieckmann KP, Linke J, Pichlmeier U, Kulejewski M, Loy V. Spermatogenesis in the contralateral testis of patients with testicular germ cell cancer: histological evaluation of testicular biopsies and a comparison with healthy males. BJU Int. 2007b;99:1079–85. doi: 10.1111/j.1464-410X.2006.06686.x. [DOI] [PubMed] [Google Scholar]
- Doitsidou M, Reichman-Fried M, Stebler J, Koprunner M, Dorries J, Meyer D, Esguerra CV, Leung T, Raz E. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–59. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- Ewen KA, Koopman P. Mouse germ cell development: from specification to sex determination. Mol Cell Endocrinol. 2009;323:76–93. doi: 10.1016/j.mce.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Frazier AL, Amatruda JF. Germ Cell Tumors. In: Fisher DE, Nathan D, Look AT, editors. Nathan and Oski’s Textbook of Pediatric Hematology-Oncology. Elsevier; London: 2009. [Google Scholar]
- Gill JA, Lowe L, Nguyen J, Liu PP, Blake T, Venkatesh B, Aplan PD. Enforced expression of Simian virus 40 large T-antigen leads to testicular germ cell tumors in zebrafish. Zebrafish. 2010;7:333–41. doi: 10.1089/zeb.2010.0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobel U, Schneider DT, Calaminus G, Haas RJ, Schmidt P, Harms D. Germ-cell tumors in childhood and adolescence. GPOH MAKEI and the MAHO study groups. Ann Oncol. 2000;11:263–71. doi: 10.1023/a:1008360523160. [DOI] [PubMed] [Google Scholar]
- Griswold MD. The central role of Sertoli cells in spermatogenesis. Semin Cell Dev Biol. 1998;9:411–6. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- Gu Y, Runyan C, Shoemaker A, Surani A, Wylie C. Steel factor controls primordial germ cell survival and motility from the time of their specification in the allantois, and provides a continuous niche throughout their migration. Development. 2009;136:1295–303. doi: 10.1242/dev.030619. [DOI] [PubMed] [Google Scholar]
- Hawkins WE, Overstreet RM, Fournie JW, Walker WW. Development of aquarium fish models for environmental carcinogenesis: tumor induction in seven species. J Appl Toxicol. 1985;5:261–4. doi: 10.1002/jat.2550050408. [DOI] [PubMed] [Google Scholar]
- Hayashi K, de Sousa Lopes SM, Surani MA. Germ cell specification in mice. Science. 2007;316:394–6. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- Hess RA, Cooke PS, Hofmann MC, Murphy KM. Mechanistic insights into the regulation of the spermatogonial stem cell niche. Cell Cycle. 2006;5:1164–70. doi: 10.4161/cc.5.11.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess RA, Renato de Franca L. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol. 2008;636:1–15. doi: 10.1007/978-0-387-09597-4_1. [DOI] [PubMed] [Google Scholar]
- Hoei-Hansen CE, Rajpert-De Meyts E, Daugaard G, Skakkebaek NE. Carcinoma in situ testis, the progenitor of testicular germ cell tumours: a clinical review. Ann Oncol. 2005;16:863–8. doi: 10.1093/annonc/mdi175. [DOI] [PubMed] [Google Scholar]
- Honecker F, Stoop H, de Krijger RR, Chris Lau YF, Bokemeyer C, Looijenga LH. Pathobiological implications of the expression of markers of testicular carcinoma in situ by fetal germ cells. J Pathol. 2004;203:849–57. doi: 10.1002/path.1587. [DOI] [PubMed] [Google Scholar]
- Houston DW, King ML. Germ plasm and molecular determinants of germ cell fate. Curr Top Dev Biol. 2000;50:155–81. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- Itman C, Mendis S, Barakat B, Loveland KL. All in the family: TGF-beta family action in testis development. Reproduction. 2006;132:233–46. doi: 10.1530/rep.1.01075. [DOI] [PubMed] [Google Scholar]
- Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, Letrero R, Ciosek SL, Doody DR, Smith LM, Weaver J, Albano A, Chen C, Starr JR, Rader DJ, Godwin AK, Reilly MP, Hakonarson H, Schwartz SM, Nathanson KL. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–5. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmer K, Corless CL, Fletcher JA, McGreevey L, Haley A, Griffith D, Cummings OW, Wait C, Town A, Heinrich MC. KIT mutations are common in testicular seminomas. Am J Pathol. 2004;164:305–13. doi: 10.1016/S0002-9440(10)63120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Suzuki A, Fujita Y, Yomogida K, Lomeli H, Asada N, Ikeuchi M, Nagy A, Mak TW, Nakano T. Conditional loss of PTEN leads to testicular teratoma and enhances embryonic germ cell production. Development. 2003;130:1691–700. doi: 10.1242/dev.00392. [DOI] [PubMed] [Google Scholar]
- Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149:875–88. doi: 10.1083/jcb.149.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut H, Werz C, Geisler R, Nusslein-Volhard C. A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 2003;421:279–82. doi: 10.1038/nature01338. [DOI] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–85. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar PS, Siekhaus DE, Lehmann R. In vivo migration: a germ cell perspective. Annu Rev Cell Dev Biol. 2006;22:237–65. doi: 10.1146/annurev.cellbio.22.010305.103337. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yamaji M, Seki Y, Saitou M. Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle. 2008;7:3514–8. doi: 10.4161/cc.7.22.6979. [DOI] [PubMed] [Google Scholar]
- Lange UC, Saitou M, Western PS, Barton SC, Surani MA. The fragilis interferoninducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol. 2003;3:1. doi: 10.1186/1471-213X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–36. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal MC, Cardoso ER, Nobrega RH, Batlouni SR, Bogerd J, Franca LR, Schulz RW. Histological and stereological evaluation of zebrafish (Danio rerio) spermatogenesis with an emphasis on spermatogonial generations. Biol Reprod. 2009;81:177–87. doi: 10.1095/biolreprod.109.076299. [DOI] [PubMed] [Google Scholar]
- Lessman CA. Oocyte maturation: converting the zebrafish oocyte to the fertilizable egg. Gen Comp Endocrinol. 2009;161:53–7. doi: 10.1016/j.ygcen.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Leu DH, Draper BW. The ziwi promoter drives germline-specific gene expression in zebrafish. Dev Dyn. 2010 doi: 10.1002/dvdy.22404. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, de Leeuw H, van Oorschot M, van Gurp RJ, Stoop H, Gillis AJ, de Gouveia Brazao CA, Weber RF, Kirkels WJ, van Dijk T, von Lindern M, Valk P, Lajos G, Olah E, Nesland JM, Fossa SD, Oosterhuis JW. Stem cell factor receptor (c-KIT) codon 816 mutations predict development of bilateral testicular germcell tumors. Cancer Res. 2003;63:7674–8. [PubMed] [Google Scholar]
- Looijenga LH, de Munnik H, Oosterhuis JW. A molecular model for the development of germ cell cancer. Int J Cancer. 1999;83:809–14. doi: 10.1002/(sici)1097-0215(19991210)83:6<809::aid-ijc20>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, Stoop HJ, Hersmus R, Oosterhuis JW. Chromosomes and expression in human testicular germ-cell tumors: insight into their cell of origin and pathogenesis. Ann N Y Acad Sci. 2007;1120:187–214. doi: 10.1196/annals.1411.000. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Rosenberg C, van Gurp RJ, Geelen E, van Echten-Arends J, de Jong B, Mostert M, Wolter Oosterhuis J. Comparative genomic hybridization of microdissected samples from different stages in the development of a seminoma and a non-seminoma. J Pathol. 2000;191:187–92. doi: 10.1002/(SICI)1096-9896(200006)191:2<187::AID-PATH584>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Chaillet JR, Trasler JM. Potential significance of genomic imprinting defects for reproduction and assisted reproductive technology. Hum Reprod Update. 2004;10:3–18. doi: 10.1093/humupd/dmh002. [DOI] [PubMed] [Google Scholar]
- Macleod D, Clark VH, Bird A. Absence of genome-wide changes in DNA methylation during development of the zebrafish. Nat Genet. 1999;23:139–40. doi: 10.1038/13767. [DOI] [PubMed] [Google Scholar]
- Matta SL, Vilela DA, Godinho HP, Franca LR. The goitrogen 6-n-propyl-2-thiouracil (PTU) given during testis development increases Sertoli and germ cell numbers per cyst in fish: the tilapia (Oreochromis niloticus) model. Endocrinology. 2002;143:970–8. doi: 10.1210/endo.143.3.8666. [DOI] [PubMed] [Google Scholar]
- McIntyre A, Gilbert D, Goddard N, Looijenga L, Shipley J. Genes, chromosomes and the development of testicular germ cell tumors of adolescents and adults. Genes Chromosomes Cancer. 2008;47:547–57. doi: 10.1002/gcc.20562. [DOI] [PubMed] [Google Scholar]
- McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- Molyneaux K, Wylie C. Primordial germ cell migration. Int J Dev Biol. 2004;48:537–44. doi: 10.1387/ijdb.041833km. [DOI] [PubMed] [Google Scholar]
- Molyneaux KA, Stallock J, Schaible K, Wylie C. Time-lapse analysis of living mouse germ cell migration. Dev Biol. 2001;240:488–98. doi: 10.1006/dbio.2001.0436. [DOI] [PubMed] [Google Scholar]
- Molyneaux KA, Zinszner H, Kunwar PS, Schaible K, Stebler J, Sunshine MJ, O’Brien W, Raz E, Littman D, Wylie C, Lehmann R. The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development. 2003;130:4279–86. doi: 10.1242/dev.00640. [DOI] [PubMed] [Google Scholar]
- Moore JL, Rush LM, Breneman C, Mohideen MA, Cheng KC. Zebrafish genomic instability mutants and cancer susceptibility. Genetics. 2006;174:585–600. doi: 10.1534/genetics.106.059386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostert M, Rosenberg C, Stoop H, Schuyer M, Timmer A, Oosterhuis W, Looijenga L. Comparative genomic and in situ hybridization of germ cell tumors of the infantile testis. Lab Invest. 2000;80:1055–64. doi: 10.1038/labinvest.3780110. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Nonomura N, Oka D, Shiba M, Arai Y, Nakayama M, Inoue H, Nishimura K, Aozasa K, Mizutani Y, Miki T, Okuyama A. KIT (c-kit oncogene product) pathway is constitutively activated in human testicular germ cell tumors. Biochem Biophys Res Commun. 2005;337:289–96. doi: 10.1016/j.bbrc.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Neumann JC, Dovey JS, Chandler GL, Carbajal L, Amatruda JF. Identification of a heritable model of testicular germ cell tumor in the zebrafish. Zebrafish. 2009;6:319–27. doi: 10.1089/zeb.2009.0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Noguchi M. A recessive mutation (ter) causing germ cell deficiency and a high incidence of congenital testicular teratomas in 129/Sv-ter mice. J Natl Cancer Inst. 1985;75:385–92. [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. Spermatogonial stem cells. Methods Enzymol. 2006;419:259–82. doi: 10.1016/S0076-6879(06)19011-4. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Ohmura M, Ohbo K. The niche for spermatogonial stem cells in the mammalian testis. Int J Hematol. 2005;82:381–8. doi: 10.1532/IJH97.05088. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–13. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Olsen LC, Aasland R, Fjose A. A vasa-like gene in zebrafish identifies putative primordial germ cells. Mech Dev. 1997;66:95–105. doi: 10.1016/s0925-4773(97)00099-3. [DOI] [PubMed] [Google Scholar]
- Oosterhuis JW, Kersemaekers AM, Jacobsen GK, Timmer A, Steyerberg EW, Molier M, Van Weeren PC, Stoop H, Looijenga LH. Morphology of testicular parenchyma adjacent to germ cell tumours. An interim report. APMIS. 2003;111:32–40. doi: 10.1034/j.1600-0463.2003.11101061.x. discussion 41-2. [DOI] [PubMed] [Google Scholar]
- Oosterhuis JW, Looijenga LH. Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer. 2005;5:210–22. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- Oosterhuis JW, Looijenga LH, van Echten J, de Jong B. Chromosomal constitution and developmental potential of human germ cell tumors and teratomas. Cancer Genet Cytogenet. 1997;95:96–102. doi: 10.1016/s0165-4608(96)00275-0. [DOI] [PubMed] [Google Scholar]
- Ottesen AM, Kirchhoff M, De-Meyts ER, Maahr J, Gerdes T, Rose H, Lundsteen C, Petersen PM, Philip J, Skakkebaek NE. Detection of chromosomal aberrations in seminomatous germ cell tumours using comparative genomic hybridization. Genes Chromosomes Cancer. 1997;20:412–8. [PubMed] [Google Scholar]
- Paoloni-Giacobino A, Chaillet JR. Genomic imprinting and assisted reproduction. Reprod Health. 2004;1:6. doi: 10.1186/1742-4755-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman EJ, Hu J, Ho D, Cushing B, Lauer S, Castleberry RP. Genetic analysis of childhood endodermal sinus tumors by comparative genomic hybridization. J Pediatr Hematol Oncol. 2000;22:100–5. doi: 10.1097/00043426-200003000-00003. [DOI] [PubMed] [Google Scholar]
- Rajpert-De Meyts E, Bartkova J, Samson M, Hoei-Hansen CE, Frydelund-Larsen L, Bartek J, Skakkebaek NE. The emerging phenotype of the testicular carcinoma in situ germ cell. APMIS. 2003;111:267–78. doi: 10.1034/j.1600-0463.2003.11101301.x. discussion 278-9. [DOI] [PubMed] [Google Scholar]
- Rajpert-De Meyts E, Hanstein R, Jorgensen N, Graem N, Vogt PH, Skakkebaek NE. Developmental expression of POU5F1 (OCT-3/4) in normal and dysgenetic human gonads. Hum Reprod. 2004;19:1338–44. doi: 10.1093/humrep/deh265. [DOI] [PubMed] [Google Scholar]
- Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, Huddart RA, Renwick A, Hughes D, Hines S, Seal S, Morrison J, Nsengimana J, Deloukas P, Rahman N, Bishop DT, Easton DF, Stratton MR. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–10. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz E. Primordial germ-cell development: the zebrafish perspective. Nat Rev Genet. 2003;4:690–700. doi: 10.1038/nrg1154. [DOI] [PubMed] [Google Scholar]
- Raz E, Reichman-Fried M. Attraction rules: germ cell migration in zebrafish. Curr Opin Genet Dev. 2006;16:355–9. doi: 10.1016/j.gde.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Reichman-Fried M, Minina S, Raz E. Autonomous modes of behavior in primordial germ cell migration. Dev Cell. 2004;6:589–96. doi: 10.1016/s1534-5807(04)00074-7. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Rescorla FJ. Pediatric germ cell tumors. Semin Surg Oncol. 1999;16:144–58. doi: 10.1002/(sici)1098-2388(199903)16:2<144::aid-ssu6>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat Rev Mol Cell Biol. 2010;11:37–49. doi: 10.1038/nrm2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu BY, Orwig KE, Oatley JM, Avarbock MR, Brinster RL. Effects of aging and niche microenvironment on spermatogonial stem cell self-renewal. Stem Cells. 2006;24:1505–11. doi: 10.1634/stemcells.2005-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman EE, Lasko P. Germline development in vertebrates and invertebrates. Cell Mol Life Sci. 1999;55:1141–63. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito D, Tanaka M. Comparative aspects of gonadal sex differentiation in medaka: a conserved role of developing oocytes in sexual canalization. Sex Dev. 2009;3:99–107. doi: 10.1159/000223075. [DOI] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Saitou M, Payer B, O’Carroll D, Ohinata Y, Surani MA. Blimp1 and the emergence of the germ line during development in the mouse. Cell Cycle. 2005;4:1736–40. doi: 10.4161/cc.4.12.2209. [DOI] [PubMed] [Google Scholar]
- Sakai N. Transmeiotic differentiation of zebrafish germ cells into functional sperm in culture. Development. 2002;129:3359–65. doi: 10.1242/dev.129.14.3359. [DOI] [PubMed] [Google Scholar]
- Sakai N. In vitro male germ cell cultures of zebrafish. Methods. 2006;39:239–45. doi: 10.1016/j.ymeth.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Sakurai S, Oguni S, Hironaka M, Saito K. Alterations of the c-kit gene in testicular germ cell tumors. Cancer Sci. 2003;94:486–91. doi: 10.1111/j.1349-7006.2003.tb01470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DT, Schuster AE, Fritsch MK, Hu J, Olson T, Lauer S, Gobel U, Perlman EJ. Multipoint imprinting analysis indicates a common precursor cell for gonadal and nongonadal pediatric germ cell tumors. Cancer Res. 2001;61:7268–76. [PubMed] [Google Scholar]
- Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germlinespecific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–95. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz RW, de Franca LR, Lareyre JJ, Le Gac F, Chiarini-Garcia H, Nobrega RH, Miura T. Spermatogenesis in fish. Gen Comp Endocrinol. 2010;165:390–411. doi: 10.1016/j.ygcen.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Schulz RW, Menting S, Bogerd J, Franca LR, Vilela DA, Godinho HP. Sertoli cell proliferation in the adult testis--evidence from two fish species belonging to different orders. Biol Reprod. 2005;73:891–8. doi: 10.1095/biolreprod.105.039891. [DOI] [PubMed] [Google Scholar]
- Spitsbergen JM, Tsai HW, Reddy A, Miller T, Arbogast D, Hendricks JD, Bailey GS. Neoplasia in zebrafish (Danio rerio) treated with 7,12-dimethylbenz[a]anthracene by two exposure routes at different developmental stages. Toxicol Pathol. 2000a;28:705–15. doi: 10.1177/019262330002800511. [DOI] [PubMed] [Google Scholar]
- Spitsbergen JM, Tsai HW, Reddy A, Miller T, Arbogast D, Hendricks JD, Bailey GS. Neoplasia in zebrafish (Danio rerio) treated with N-methyl-N’-nitro-Nnitrosoguanidine by three exposure routes at different developmental stages. Toxicol Pathol. 2000b;28:716–25. doi: 10.1177/019262330002800512. [DOI] [PubMed] [Google Scholar]
- Stevens LC. A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J Natl Cancer Inst. 1973;50:235–42. doi: 10.1093/jnci/50.1.235. [DOI] [PubMed] [Google Scholar]
- Stoop H, Honecker F, Cools M, de Krijger R, Bokemeyer C, Looijenga LH. Differentiation and development of human female germ cells during prenatal gonadogenesis: an immunohistochemical study. Hum Reprod. 2005;20:1466–76. doi: 10.1093/humrep/deh800. [DOI] [PubMed] [Google Scholar]
- Summersgill B, Goker H, Weber-Hall S, Huddart R, Horwich A, Shipley J. Molecular cytogenetic analysis of adult testicular germ cell tumours and identification of regions of consensus copy number change. Br J Cancer. 1998;77:305–13. doi: 10.1038/bjc.1998.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summersgill B, Osin P, Lu YJ, Huddart R, Shipley J. Chromosomal imbalances associated with carcinoma in situ and associated testicular germ cell tumours of adolescents and adults. Br J Cancer. 2001;85:213–20. doi: 10.1054/bjoc.2001.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA. Reprogramming of genome function through epigenetic inheritance. Nature. 2001;414:122–8. doi: 10.1038/35102186. [DOI] [PubMed] [Google Scholar]
- Tam PP, Zhou SX. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev Biol. 1996;178:124–32. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Matsui Y. Developmentally regulated expression of mil-1 and mil-2, mouse interferon-induced transmembrane protein like genes, during formation and differentiation of primordial germ cells. Mech Dev. 2002;119(Suppl 1):S261–7. doi: 10.1016/s0925-4773(03)00126-6. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Nagamatsu G, Tokitake Y, Kasa M, Tam PP, Matsui Y. Regulation of expression of mouse interferon-induced transmembrane protein like gene-3, Ifitm3 (mil-1, fragilis), in germ cells. Dev Dyn. 2004;230:651–9. doi: 10.1002/dvdy.20085. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Yamaguchi YL, Tsoi B, Lickert H, Tam PP. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev Cell. 2005;9:745–56. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Tian Q, Frierson HF, Jr., Krystal GW, Moskaluk CA. Activating c-kit gene mutations in human germ cell tumors. Am J Pathol. 1999;154:1643–7. doi: 10.1016/S0002-9440(10)65419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, Huddart RA, Bishop DT, Easton DF, Stratton MR, Rahman N. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–7. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Geijn GJ, Hersmus R, Looijenga LH. Recent developments in testicular germ cell tumor research. Birth Defects Res C Embryo Today. 2009;87:96–113. doi: 10.1002/bdrc.20140. [DOI] [PubMed] [Google Scholar]
- van Echten J, van Gurp RJ, Stoepker M, Looijenga LH, de Jong J, Oosterhuis W. Cytogenetic evidence that carcinoma in situ is the precursor lesion for invasive testicular germ cell tumors. Cancer Genet Cytogenet. 1995;85:133–7. doi: 10.1016/0165-4608(95)00151-4. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–25. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Weidinger G, Stebler J, Slanchev K, Dumstrei K, Wise C, Lovell-Badge R, Thisse C, Thisse B, Raz E. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol. 2003;13:1429–34. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- Weidinger G, Wolke U, Koprunner M, Klinger M, Raz E. Identification of tissues and patterning events required for distinct steps in early migration of zebrafish primordial germ cells. Development. 1999;126:5295–307. doi: 10.1242/dev.126.23.5295. [DOI] [PubMed] [Google Scholar]
- Weidinger G, Wolke U, Koprunner M, Thisse C, Thisse B, Raz E. Regulation of zebrafish primordial germ cell migration by attraction towards an intermediate target. Development. 2002;129:25–36. doi: 10.1242/dev.129.1.25. [DOI] [PubMed] [Google Scholar]
- Western P. Foetal germ cells: striking the balance between pluripotency and differentiation. Int J Dev Biol. 2009;53:393–409. doi: 10.1387/ijdb.082671pw. [DOI] [PubMed] [Google Scholar]
- Williamson A, Lehmann R. Germ cell development in Drosophila. Annu Rev Cell Dev Biol. 1996;12:365–91. doi: 10.1146/annurev.cellbio.12.1.365. [DOI] [PubMed] [Google Scholar]
- Wolke U, Weidinger G, Koprunner M, Raz E. Multiple levels of posttranscriptional control lead to germ line-specific gene expression in the zebrafish. Curr Biol. 2002;12:289–94. doi: 10.1016/s0960-9822(02)00679-6. [DOI] [PubMed] [Google Scholar]
- Wylie C. Germ cells. Curr Opin Genet Dev. 2000;10:410–3. doi: 10.1016/s0959-437x(00)00105-2. [DOI] [PubMed] [Google Scholar]
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M. Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod. 2006;75:705–16. doi: 10.1095/biolreprod.106.053686. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T. Nanog expression in mouse germ cell development. Gene Expr Patterns. 2005;5:639–46. doi: 10.1016/j.modgep.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–22. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–94. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Ying Y, Qi X, Zhao GQ. Induction of primordial germ cells from murine epiblasts by synergistic action of BMP4 and BMP8B signaling pathways. Proc Natl Acad Sci U S A. 2001;98:7858–62. doi: 10.1073/pnas.151242798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y, Zhao GQ. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev Biol. 2001;232:484–92. doi: 10.1006/dbio.2001.0173. [DOI] [PubMed] [Google Scholar]
- Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–65. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- Yoshida S. [“Flexible” stem cell-niche system in mouse spermatogenesis] Tanpakushitsu Kakusan Koso. 2008a;53:1125–32. [PubMed] [Google Scholar]
- Yoshida S. Spermatogenic stem cell system in the mouse testis. Cold Spring Harb Symp Quant Biol. 2008b;73:25–32. doi: 10.1101/sqb.2008.73.046. [DOI] [PubMed] [Google Scholar]
- Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, Rubin EM, Nadeau JH, Matin A. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–4. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GQ, Garbers DL. Male germ cell specification and differentiation. Dev Cell. 2002;2:537–47. doi: 10.1016/s1534-5807(02)00173-9. [DOI] [PubMed] [Google Scholar]