Abstract
In humans, HLA-DR alleles sharing amino acids at the third hypervariable region with DRB1*0401(shared epitope) are associated with a predisposition to rheumatoid arthritis, whereas DRB1*0402 is not associated with such a predisposition. Both DRB1*0402 and DRB1*0401 occur in linkage with DQ8 (DQB1*0302). We have previously shown that transgenic (Tg) mice expressing HLA-DRB1*0401 develop collagen-induced arthritis. To delineate the role of “shared epitope” and gene complementation between DR and DQ in arthritis, we generated DRB1*0402, DRB1*0401.DQ8, and DRB1*0402.DQ8 Tg mice lacking endogenous class II molecules, AEo. DRB1*0402 mice are resistant to develop arthritis. In double-Tg mice, the DRB1*0401 gene contributes to the development of collagen-induced arthritis, whereas DRB1*0402 prevents the disease. Humoral response to type II collagen is not defective in resistant mice, although cellular response to type II collagen is lower in *0402 mice compared with *0401 mice. *0402 mice have lower numbers of T cells in thymus compared with *0401 mice, suggesting that the protective effect could be due to deletion of autoreactive T cells. Additionally, DRB1*0402 mice have a higher number of regulatory T cells and show increased activation-induced cell death, which might contribute toward protection. In DRB1*0401.DQ8 mice, activated CD4+ T cells express class II genes and can present DR4- and DQ8-restricted peptides in vitro, suggesting a role of class II+ CD4 T cells locally in the joints. The data suggest that polymorphism in DRB1 genes determines predisposition to develop arthritis by shaping the T cell repertoire in thymus and activating autoreactive or regulatory T cells.
Rheumatoid arthritis (RA)3 is a chronic autoimmune disease that affects ~1% of the population in the United States. Predisposition to RA has been associated with an expression of certain HLA class II haplotypes (1, 2). Association of HLA-DR4 with susceptibility to RA has been studied extensively ever since the initial observations made by Stastny in 1978 (3–5). Genetic susceptibility to RA in most populations has been explained by the presence of an amino acid motif known as “shared epitope” (6, 7). According to this hypothesis, amino acid positions 67–74 of DRB1 molecules is shared among all the alleles responsible for susceptibility either by selection of T cell repertoire or by being able to present similar Ags leading to development of arthritis. On the other hand, motif I/D/E/A (DRB1*0402) at positions 67, 70, 71, and 74 was found to be associated with nonsusceptibility to RA. Although the mechanisms by which these alleles predispose to RA are not known, positive selection of potentially autoreactive T cells by susceptible class II alleles or, conversely, negative selection of autoreactive T cells by nonsusceptible alleles may be playing a role (8, 9). Naturally processed peptides presented by class II molecules can be derived from endogenous class II molecules (10, 11). Thus, peptides from DRB1 alleles may be presented by HLA-DQ molecules in thymus, and polymorphism in DRB1 genes can lead to shaping of the T cell repertoire (12). However, it is difficult to experimentally assess the role of the shared epitope in humans.
We have previously shown that both the DQ and DR alleles might be involved in susceptibility or protection from disease (13). Binding studies have shown that RA-associated DR alleles bind limited numbers of human type II collagen (CII) peptides compared with HLA-DQ8 (DQB1*0302), which binds multiple peptides (14, 15). HLA-DQ occurs in linkage disequilibrium with DR genes and thus is inherited en bloc as a haplotype (16). HLA-DQ8 is found in linkage with DRB1*0401 and has been shown to occur with increased frequency in RA patients of some ethnic groups (17, 18).
Rheumatoid arthritis is a chronic inflammatory disease that is characterized by synovial inflammation and erosion of bone and cartilage that leads to destruction of joints. Autoreactivity to type II collagen has been shown to be involved in the pathogenesis of RA (19, 20). Patients with arthritis produce autoantibodies like rheumatoid factor (RF) and Abs to cyclic citrullinated peptides (21). Our initial studies with mice lacking endogenous class II molecules (Aβo) and expressing HLA-DQ8 (HLA-DQA1*0301/DQB1*0302) showed that they were highly susceptible to collagen-induced arthritis (CIA) (22). The DQ8-restricted development of arthritis could be modulated by the presence of various DR molecules similar to those observed in humans (23). We generated DR4.AEo and DQ8.AEo mice that lacked all four classical class II murine chains (Aα, Aβ, Eα, and Eβ) due to a deletion of the entire class II region. CIA studies showed that DRB1*0401.AEo mice develop milder arthritis with lower incidence than DQ8 mice (24).
In this study we investigated the role of DRB1*0402 in the susceptibility/resistance to develop arthritis. Additionally, we sought to determine the role of DR/DQ gene complementation in predisposition to arthritis by using double-transgenic (Tg) mice expressing DRB1*0401.DQ8 and DRB1*0402.DQ8. We hypothesized that DRB1*0401 should complement DQ8-restricted CIA while DRB1*0402 should protect DQ8 mice from developing arthritis. All Tg mice were studied in vivo using the CIA protocol. DRB1*0401.DQ8 mice develop severe arthritis while DRB1*0402 mice are resistant and DRB1*0402.DQ8 mice are protected from arthritis. Our data suggest that the DRB1 shared epitope predisposes to develop arthritis by activation of the autoreactive T cells and protects from arthritis by deletion of autoreactive T cells and generation of regulatory T cells.
Materials and Methods
Tg mice
The generation of DRB1*0401and DRB1*0402 Tg mice has been described previously (25). Aβo.DRB1*0401 and Aβo.DRB1*0402 mice were mated with MHC-IIΔ/Δ (AEo) mice (26) to generate AEo.DRB1*0401 (24) and AEo.DRB1*0402 mice. Similarly, AEo.DQ8 mice were generated. AEo-.DRB1*0401 and AEo.DRB1*0402 mice were mated with AEo.DQ8 mice to generate double-Tg AEo.DRB1*0401.DQ8 and AEo.DRB1*0402.DQ8 mice. Mice of both sexes (8–12 wk of age) used in this study were bred and maintained in the pathogen-free Immunogenetics Mouse Colony at the Mayo Clinic (Rochester, MN) in accordance with the Animal Use and Care Committee. All the experiments included littermate controls and were conducted with the approval of the Institutional Animal Care and Use Committee.
For convenience, DRB1*0402 mice are referred to as *0402, DRB1*0401 Tg mice as *0401, and double-Tg mice expressing both DR4 and DQ8 molecules as *0401.DQ8 and *0402.DQ8.
Antibodies
The expression of DR, DQ, and TCR Vβ-chains were analyzed by flow cytometry using mAbs: L227 (anti-DR), IVD12 (anti-DQ), 14-4-4s (anti-Eα), B20.6 (anti-Vβ2), KT4-10 (anti-Vβ4), MR9-4 (anti-Vβ5.1.2), MR9-8 (anti-Vβ5.1), 44-22-1 (anti-Vβ6), TR310 (anti-Vβ7), KJ-16 (anti-Vβ8.1.2), F23.2 (anti-Vβ8.2), MR10-2 (anti-Vβ9), KT11 (anti-Vβ11), 14.2 (anti-Vβ14), and KJ23a (anti-Vβ17) as previously described (24). Conjugated specific anti-CD3, CD4, CD8, CD25, B220, CD11c, CD11b, DR, and DQ (BD Pharmingen) were also utilized. Expression analysis experiments utilized anti-DR conjugated with PE and DQ with FITC. All cell-surface markers were analyzed with cells pooled from 2 mice/strain and repeated two to three times.
Intracellular staining for FoxP3 was performed using Abs obtained from eBioscience as per the manufacturer’s instructions. PE-conjugated rat IgG2a (BD Pharmingen) was used as the isotype control for FoxP3 staining.
Induction and evaluation of CIA
Pure native chick type II collagen was obtained by multiple-step purification as described elsewhere (27). Tg mice and negative littermates were immunized with chick CII as previously described for CIA protocol (24). Mice were monitored for the onset and progression of CIA from 3 to 12 wk postimmunization. The arthritic severity of mice was evaluated as described previously with a grading system for each paw from 0 to 3 (28). The mean arthritic score was determined using arthritic animals only.
Histopathology
Mice were sacrificed after 10–12 wk of immunization and paws were decalcified and fixed. Sections were stained with H&E and examined for infiltration and erosions.
Autoantibodies
Levels of anti-chick and anti-mouse CII IgG Abs were measured in sera obtained 35 days following CII immunization by a standard ELISA and are shown as OD. Briefly, microtiter plates were coated overnight with CII (6 µg/well in KPO4 (pH 7.6)) at 4°C, washed, and blocked with 1% BSA in PBS/0.05% Tween 20. Sera were added in 4-fold dilution (1/100 to 1/65,000) and incubated overnight at 4°C. The plates were washed, and peroxidase-conjugated goat anti-mouse IgG (Organon Teknika) was added for another overnight incubation at 4°C. After washing, O-phenylenediamine was added and the colorimetric change was measured at 410 nm.
Rheumatoid factor was measured by ELISA in Tg mice as previously described (24).
T cell proliferation assay
Mice were immunized with 200 µg of CII emulsified 1:1 in CFA (Difco) intradermally at the base of the tail and in one hind footpad. Ten days postimmunization, draining lymph nodes/spleen were removed and cultured in vitro. Lymph node cells (LNCs, 1 × 106) were cultured in HEPES-buffered RPMI 1640 containing 5% heat-inactivated horse serum and streptomycin and penicillin in 96-well flat-bottom tissue culture plates. Cells were challenged by adding 100 µl of RPMI 1640 medium (negative control), Con A (20 µg/ml, positive control), and native collagen (50 µg/ml). To determine CD4-mediated response, GK1.5 (anti-CD4) Ab was used for blocking. The cells were incubated for 48 h at 37°C. During the last 18 h the cells were pulsed with [3H]thymidine and the tritium incorporation was determined by liquid scintillation counting. Results are calculated as Δ cpm (i.e., mean cpm of triplicate cultures containing Ag − mean cpm of medium).
HLA class II Tg mice and negative littermates were tested for T cell response to self-DR peptide DW10 (65–79) and DW4 (65–79) and type II collagen-derived peptides spanning amino acids 254–273 and 284–303, which were synthesized and purified at the Mayo Clinic Peptide Facility. The mice were primed with 200 µg of peptide and challenged in vitro with 100 µg/ml of the peptide.
For presentation of CII-derived peptides 254–273 and 284–303 by CD4+ cells, cells were sorted by FACSorter from LNCs isolated from primed mice and cultured in vitro in the presence or absence of APCs. Up to 5 × 105 CD4+ sorted cells were used for culturing alone with CII (50 µg/ml). The other cultures utilized 5 × 105 of irradiated spleen cells as APCs. Cells were cultured in RPMI 1640 media as stated above and cell proliferation determined by thymidine incorporation. A stimulation index of 2 or more was taken as a positive response. The experiment was done twice with cells pooled from 2 to 3 mice/experiment.
Activation-induced cell death (AICD)
To study the sensitivity of the proliferating cells to CII-induced AICD, CD3+ cells were stained with 7-aminoactinomycin D conjugated with FITC after in vitro stimulation and analyzed by FACS.
BrdU Labeling
Mice were given BrdU (Sigma-Aldrich) in their drinking water at a concentration of 0.8 mg/ml. BrdU was dissolved in sterile water and was changed daily. BrdU incorporated into DNA was detected using a BrdU flow kit with an allophycocyanin-conjugated anti-BrdU Ab (BD Biosciences) following the manufacturer’s instructions.
Cytokines
Capture ELISA was done for measuring cytokines IL-10, IL-18, and TNF-α using kits according to the manufacturer’s instructions (BD Pharmingen). Cytokines were also measured using the Bio-Plex protein array system with the mouse cytokine 23-plex panel as per the manufacturer’s instructions and analyzed with Bio-Plex manager 2.0 software (Bio-Rad Laboratories).
Real-time PCR
Levels of TGF-β and IL-23 mRNA in vitro cultures were analyzed using real-time PCR. RNA was extracted from cells using RNAeasy columns (Qiagen), and cDNA was prepared using RNase H-reverse transcriptase (Invitrogen). cDNA was analyzed by real-time quantitative PCR in triplicates by using SYBR GreenER qPCR reagent system (Invitrogen). The expression level of each gene was quantified using the threshold cycle (Ct) method normalized for the housekeeping gene GAPDH. The primers for genes encoding IL-23 and GADPH were synthesized as described previously (29, 30).
Statistical analysis
The difference in the incidence of arthritis between groups was analyzed using the χ2 test. Ab and cytokine levels, onset of arthritis, and mean scores for arthritic mice were compared using Student’s t test.
Results
Tg mice express functional DRB1*04 and DQ8 molecules
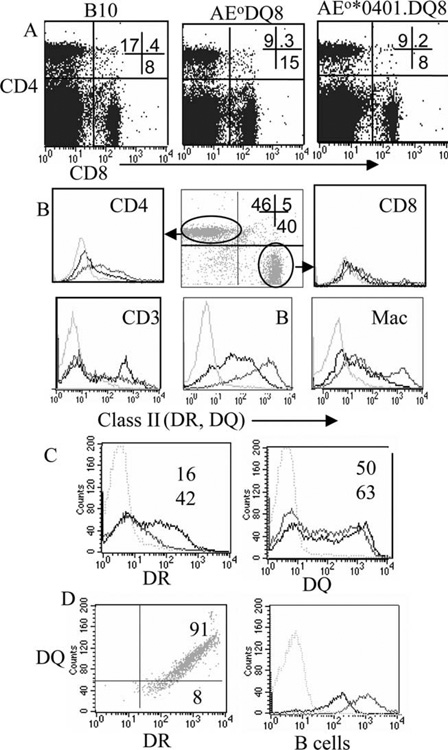
Single Tg mice *0402 and double-Tg mice expressing *0402.DQ8 and *0401.DQ8 were analyzed for expression of DR and DQ molecules in splenic APCs, macrophages and B cells, and CD3+ cells (Fig. 1). All mice showed normal expression of the transgene. APCs as well as CD3 cells expressed both DR and DQ molecules. Expression of DR and DQ in single- and double-Tg mice showed similar DQ expression, although DR was expressed much higher in double-Tg mice. Similar to humans who express both DQ and DR molecules on the same cell surface, B cells from peripheral blood of DR/DQ mice express both DR and DQ on the same cell (Fig. 1). T cell repertoire analysis suggested that transgene human class II molecules could select various Vβ T cells, as both the double-Tg mice showed partial deletion of Vβ5 and Vβ11 compared with DQ8 mice.
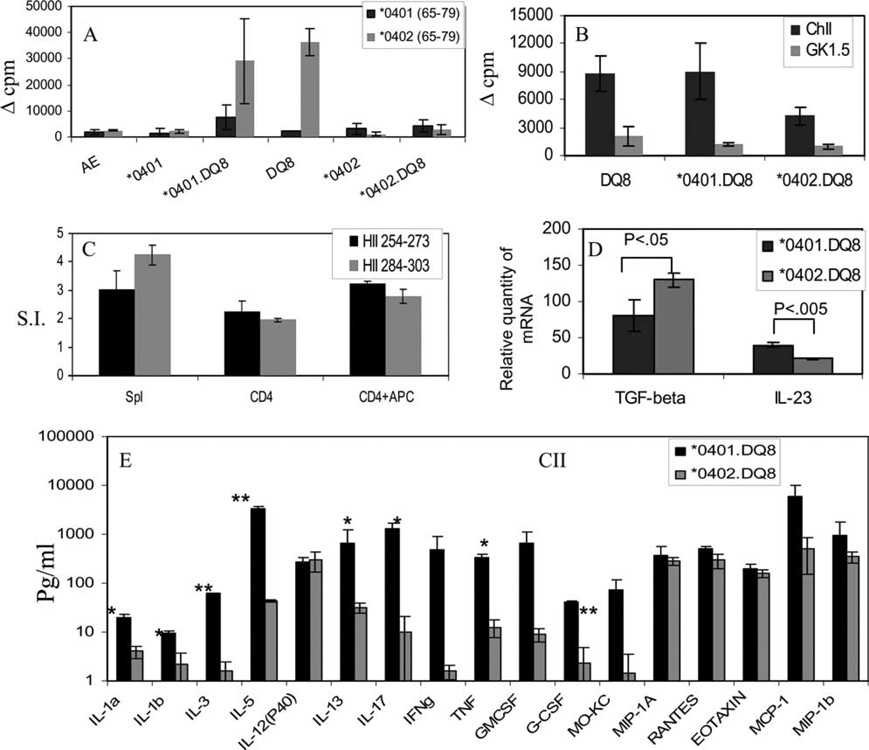
FIGURE 1.
Characterization of AEo.DRB1*0401.DQ8 Tg mice. A, Selection of CD4 and CD8 population in AEo.DRB1*0401.DQ8 and controls, B10 and AEo.DQ8 mice, shows that both CD4 and CD8 cells are positively selected. B, Expression of DR and DQ molecules by splenic CD4 and CD8 cells (gated on CD3+ cells), CD3 and APCs, and B cells and macrophages (Mac) in DRB1*0401.DQ8 mice; gray line indicates DQ; black line, DR; light gray, control. C, Percentage expression of DR4 and DQ8 in cells isolated from spleen of AEo.DRB1*0401.DQ8 mice shows more cells expressing the DR and DQ transgene compared with single-Tg mice; gray line indicates DRB1*0401.DQ8; black line, DRB1*0401 mice in DR (left) and DQ8 mice in DQ (right). Numbers are percentage positive; top number indicates single-Tg mice; bottom number, DRB1*0401.DQ8 mice. D, Expression of DQ and DR molecules on B cells isolated from peripheral blood shows that B cells express both DQ and DR on the same cell (left) although the level of expression of DR is much higher (right); gray line indicates DR; black line, DQ. All experiments were done three times with cells pooled from two to three mice/experiment.
*0402 protects mice from developing arthritis by deleting cells in thymus
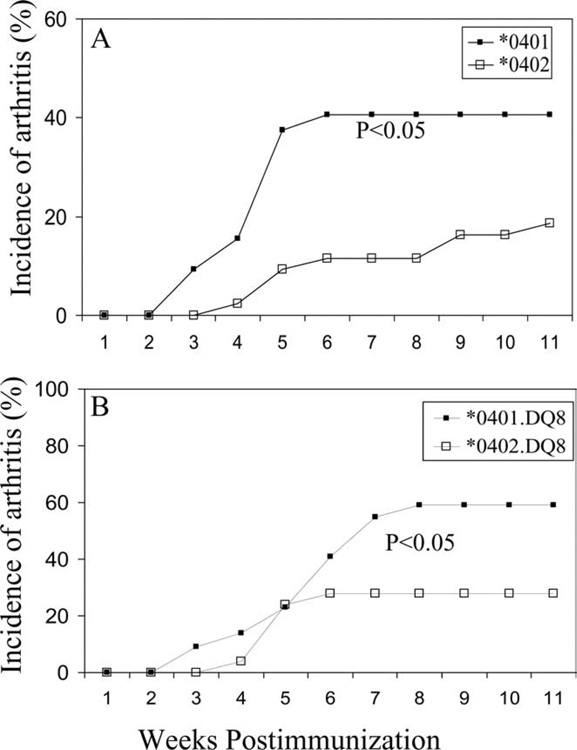
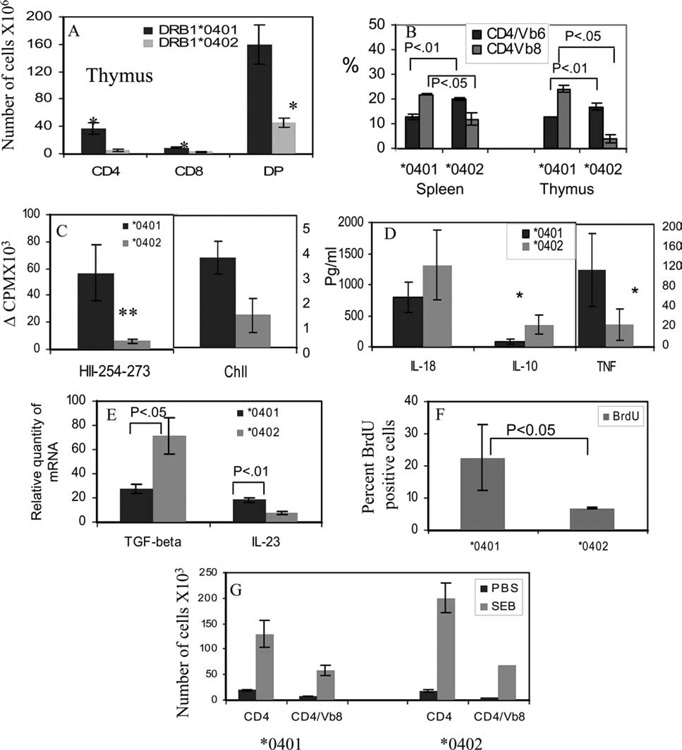
Only 19% (8 of 43) of *0402 mice developed CIA compared with 40% reported in *0401 mice (Fig. 2A), suggesting that *0402 provides significant protection from developing arthritis (p < 0.05). Onset of arthritis in *0402 mice was delayed when compared with *0401 mice (p < 0.008), although severity between the two strains did not differ significantly as studied by phenotype (Table I). MHC molecules are involved in positive and negative selection of T cells including selection of autoreactive T cells (31). One way of providing protection against disease could be by deleting autoreactive T cells in thymus. To determine whether selection of cells in thymus is different between the two strains, we compared the total number of thymic CD3 cells and its subsets of single-positive CD4 and CD8 cells and double-positive CD4+CD8+ cells in naive animals. *0402 mice have lower numbers of single-positive CD4 and CD8 cells as well as double-positive CD4+CD8+ cells compared with those found in *0401 mice, suggesting increased thymic deletion of T cells (Fig. 3A). This difference was also reflected in splenic CD4 and CD8 populations (data not shown). Since CD4+ T cells expressing Vβ6 and Vβ8 have been implicated in the pathogenesis of arthritis (32), we explored if CD4 T cells of *0401 mice carry these TCRs more often compared with resistant *0402 mice. In thymus and spleen, the difference was significant for Vβ6 and Vβ8 CD4+ T cells, with *0402 mice having a significantly lower percentage of Vβ8 (p < 0.04) but higher levels of Vβ6 CD4 T cells than do *0401 mice ( p < 0.01) (Fig. 3B). In *0402 mice, CD4 T regulatory cells had a higher percentage of Vβ6 cells compared with *0401 mice (p < 0.05) (data not shown).
FIGURE 2.
Incidence of collagen-induced arthritis in DRB1*0401 and DRB1*0402 mice (A), and double-Tg mice expressing DRB1*0401.DQ8 and DRB1*0402.DQ8 (B). DRB1*0402 mice are resistant to develop arthritis compared with *0401, *0402 vs *0401 (p < 0.05). *0401.DQ8 mice are susceptible while *0402.DQ8 mice are protected from arthritis, *0402.DQ8 vs *0401.DQ8 (*, p < 0.05).
Table I.
Incidence of collagen-induced arthritis in DRB1*0401 and *0402 single- and double-Tg mice expressing DQ8
| Mice | Incidence (%) | Onset | Severity |
|---|---|---|---|
| *0402 | 8/43 (19) | 6.7 ± 2.5 | 4.5 ± 2.9 |
| *0402/DQ8 | 7/25 (28) | 5 ± 0.5 | 6.8 ± 1.6 |
| *0401/DQ8 | 13/22 (59) | 5.5 ± 1.6 | 6.4 ± 1.2 |
| DQ8 | 14/20 (70) | 5.8 ± 2.3 | 6.7 ± 2.5 |
| *0401 | 14/36 (39) | 5.2 ± 1.4 | 3.5 ± 1.2a |
For *0402/DQ8 vs *0401/DQ8 and *0402 vs *0401, p < 0.05; *0402.DQ8 vs DQ8, p < 0.01; *0402 vs DQ8, p < 0.001.
From Ref. 24.
FIGURE 3.
DRB1*0402 deletes more CD3+ thymic cells than does DRB1*0401. A, Number of CD4, CD8, and double-positive cells in thymus of Tg mice. B, Number of splenic and thymic Vβ6 and Vβ8 cells gated on CD4+ population. In vitro T cell response to (C) CII and human CII-derived peptide 254–273 (HII-254–273) in mice primed with respective Ags show that DRB1*0401 mice mount stronger response to both Ags when compared with DRB1*0402 mice. LNCs were used as described in Materials and Methods. D, Cytokines measured by ELISA in supernatants of cells cultured in the presence of CII-derived peptide 254–273. *0401 mice produce higher levels of TNF-α and lower levels of IL-10 compared with *0402 mice. E, Expression of TGF-β and IL-23 in Tg mice was quantified by real-time PCR. Expression of GADPH was measured as an internal control. The expression of different cytokines in LNCs stimulated with CII relative to that in medium control was calculated by the ΔΔCt method. Data are presented as means ± SD of at least four different mice. F, BrdU staining of splenic CD4+ cells shows lesser proliferation to CII in *0402 than in *0401 in vivo. G, Both DRB1*0401 and DRB1*0402 mice mount a strong in vitro response to superantigen and staphylococcal enterotoxin B (SEB), and they expand Vβ8+ T cells. Mice injected with saline did not show any proliferation in both strains. All experiments were repeated two to three times with cells pooled from two mice/strain. *0401 vs *0402: *, p < 0.05; **, p < 0.01.
CIA-resistant DRB1*0402 mice are defective in Ag-specific response
To understand if resistance to develop arthritis in *0402 mice is due to defective T cell response to CII, we studied in vivo and in vitro Ag-specific proliferation. In vitro proliferation of primed LNCs of *0402 mice mounted a milder response when challenged with CII compared with *0401 mice (Fig. 3C). Since CII-derived peptide 254–273 has been shown to bind both *0401 and *0402 molecules, we determined response to this peptide in both strains. The difference in response to CII-derived DR4-restricted immunodominant peptide 254–273 was much more marked, with *0401 cells mounting a robust immune response compared with cells isolated from *0402 mice (p < 0.001) (Fig. 3C). In response to peptide 254–273, *0402 mice produced higher levels of IL-10 and TGF-β and lower levels of TNF-α and IL-23 as compared with *0401 mice ( p < 0.05) (Fig. 3, D and E). We further confirmed this defect in Ag-specific proliferation in vivo by measuring BrdU incorporation. Mice were injected with CII and given BrDU. As depicted in Fig. 3, *0402 mice showed a lower proliferation to CII compared with *0401 mice (p < 0.05) (Fig. 3F). To ensure that the low T cell response to CII in DRB1*0402 mice was not due to a generalized defect in cellular response, we tested response to superantigen. Non-Ag-specific response in *0402 mice was not defective, as they responded strongly to superantigen, staphylococcal enterotoxin B, and showed expansion of the CD4+ Vβ8 population (Fig. 3G).
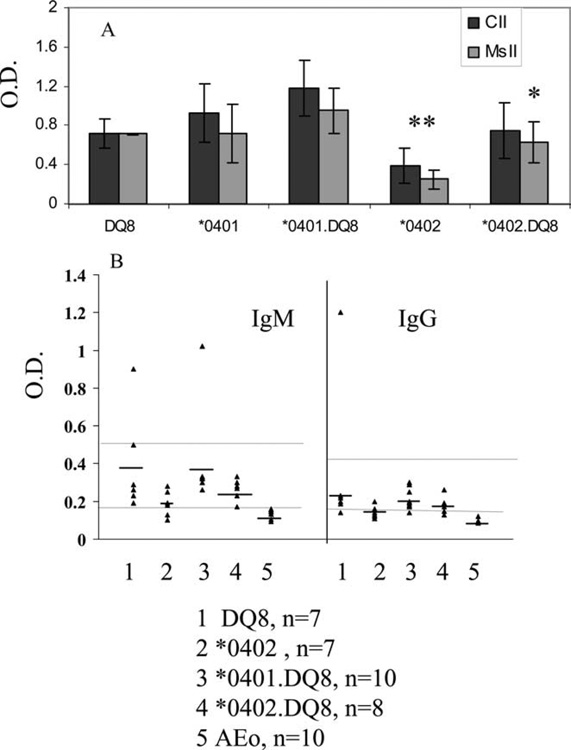
Since B cell activation requires T cells help, we next examined if a low cellular response to CII is accompanied by a defective humoral response in *0402 mice. For this purpose, we measured anti-CII Abs to immunizing CII as well as self-CII. All immunized mice made Abs to immunizing and self-CII (Fig. 4A). Additionally, all AEo Tg mice produced rheumatoid factor upon immunization with CII (Fig. 4B). However, the levels of autoantibodies produced by *0402 were lower than those of *0401 mice, although the difference was only significant for anti-CII Abs (p < 0.005).
FIGURE 4.
Humoral response in Tg mice primed with type II collagen. A, Abs to chick type II collagen (CII) and self-collagen (mouse type II collagen, MsII) in sera of mice primed with chick type II collagen. B, IgM and IgG rheumatoid factors produced by CII-primed mice. Upper line denotes positive control, Mrl-Lpr mice, and lower line denotes negative control, CII-primed B10 mice. Groups are: 1) DQ8 (n = 7), 2) DRB1*0402 (n = 7), 3) DRB1*0401.DQ8 (n = 10), 4) DRB1*0402.DQ8 (n = 8), and 5) AEo (n = 10). CII and MsII, *0401.DQ8 vs *0402.DQ8 (*, p < 0.01), *0402 vs *0401.DQ8 and *0402.DQ8 (**, p < 0.005). For IgG and IgM RF, all mice showed significantly higher levels only when compared with AEo mice.
*0402 protects while *0401 is permissive in vivo in DR/DQ Tg mice
To understand the role of RA-susceptible and -resistant haplotypes (DRB1*0401.DQ8 and DRB1*0402.DQ8, respectively), we studied the double-Tg mice for collagen-induced arthritis. *0401.DQ8 double-Tg mice developed CIA with increased incidence compared with *0401 mice (Table I). Introduction of DQ8 into *0401 mice led to much more severe disease than *0401 alone (p < 0.001). Since *0402 mice did not develop arthritis, we used *0402.DQ8 mice to investigate if *0402 would protect DQ8 mice from developing arthritis. The incidence of CIA in *0402.DQ8 mice was 28% compared with 70% in DQ8 single-Tg mice (p < 0.01), confirming its protective role (Fig. 2 and Table I).
It has been suggested that self-MHC-derived peptide can modulate predisposition to certain diseases, and CD4+ T cell responses to self-MHC Ags have been related to suppression of various autoimmune diseases (33). We have previously shown that in DQ8 mice, peptide (aa 65–74) derived from DRB1*0402 is presented much more robustly than that of DRB1*0401. In double-Tg mice we hypothesized that if DQ8 can present self-derived DR peptides, DRB1*0402.DQ8 mice should not respond to DRB1*0402-derived peptide since those cells should be deleted in thymus. All Tg mice were immunized with the DR-derived peptides, and primed cells were challenged in vitro with self or the non-self peptide. The splenic cells isolated from mice primed with DRB1*0402 peptide comprising aa 64–75 showed that *0401.DQ8 mice mounted T cell response to *0402 peptide while cells isolated from *0402.DQ8 mice did not respond to the peptide derived from *0402, suggesting that these cells were deleted in thymus (Fig. 5A).
FIGURE 5.
A, T cell proliferation to self-peptides spanning 65–79 of *0401 and *0402 in Tg mice. LNCs isolated from immunized mice were challenged in vitro with the priming peptide. B, *0402.DQ8 mice mount a low T cell response in vitro to CII compared with *0401.DQ8 and DQ8 mice (*, p < 0.05). Response to CII in all mice is CD4-restricted. C, CD4+ T cells were sorted from DRB1*0401.DQ8 mice primed with DR-restricted human type II collagen, HII-254 –273, and DQ-restricted, HII-284 –303, peptides. CD4 cells were challenged in vitro in the absence or presence of APCs. Splenic cells were used as controls. S.I. indicates stimulation index. D, Expression of TGF-β and IL-23 double-Tg mice were quantified by real-time PCR. Expression of GADPH was measured as an internal control. The expression of different cytokines in LNCs stimulated with CII relative to that in medium control was calculated by the ΔΔCt method. Data are presented as means ± SD of at least four different mice. E, Cytokine and chemokine production in response to in vitro challenge to CII shows higher levels of Th1, Th17, and Th2 cytokines produced by *0401.DQ8 mice compared with *0402.DQ8 mice. Most of the chemokines were not significantly different between the two strains. The y-axis is a logarithmic scale. *0401.DQ8 vs *0402.DQ8: *, p < 0.05; **, p < 0.005.
We further explored if Ag-specific cellular and humoral response in double-Tg mice support the in vivo observations. *0401.DQ8 mice produced higher amounts of anti-self-CII Abs compared with *0402.DQ8 mice (p < 0.01) (Fig. 4A). Although *0402.DQ8 mice produced lower amounts of IgM-RF and IgG-RF than did *0401.DQ8 mice, the difference was not statistically significant (Fig. 4B). Similar to *0402 mice, *0402.DQ8 mice mounted much weaker T cell response to CII compared with *0401.DQ8 mice (Figs. 3C and 5B). We have previously shown that activated and sorted CD4+ T cells can present DR-restricted CII-derived peptide 254–273 in *0401 Tg mice (24). In this study we determined if double-Tg mice present both DR- and DQ-restricted peptides (254–273 and 284–303, respectively). CD4+ T cells were sorted from *0401.DQ8 mice primed with 254–273 or 284–303 peptide and cultured alone or in the presence of APCs (Fig. 5C). Both the DR- and DQ-restricted peptides were presented by CD4+ T cells, suggesting both DR and DQ molecules function efficiently in double-Tg mice.
DRB1*0401.DQ8 mice produce high levels of Th17 cytokines
Since inflammatory cytokines and chemokines play an important role in development of RA, we analyzed levels of different Th1, Th2, and Th17 cytokines and chemokines from supernatants of cultures of LNCs of CII-primed animals stimulated with CII Ag in vitro. Disease-susceptible *0401.DQ8 mice produced significantly higher levels of proinflammatory cytokines IL-17, IFN-γ, IL-1, TNF-α, and IL-23 compared with resistant *0402.DQ8 mice (Fig. 5, D and E). Besides the Th1 and Th17 cytokines, *0401.DQ8 mice also produced higher levels of immunomodulatory cytokines IL-3, IL-5, and IL-13. The protected *0402.DQ8 mice produced very low levels of most of the cytokines except for TGF-β, which was higher than in *0401.DQ8 mice, and IL-12 (p40) was similar in both double-Tg mice. Both strains produced similar levels of chemokines except G-CSF, which was significantly higher in *0401.DQ8 mice compared with *0402.DQ8 mice, suggesting that immune response to CII is initiated in both strains but that the Ag-specific immune response in *0402.DQ8 mice is low, with low levels of proinflammatory cytokines making them resistant to develop arthritis.
CIA-resistant mice harbor high levels of regulatory T cells
Studies have suggested that CD4+CD25+FoxP3 regulatory T cells produce antiinflammatory cytokines TGF-β and suppress immune response and autoimmune disease. Since both *0402 and *0402.DQ8 mice are protected from arthritis and produce high levels of TGF-β with lower cellular response to CII, we tested if DRB1*0402 can positively select higher numbers of regulatory T cells in thymus compared with *0401, which would suggest that central tolerance by MHC molecules can modulate disease. Alternatively, the cytokine milieu in the periphery could lead to generation of regulatory T cells leading to protection from arthritis. Splenic and thymic cells from naive and CII-primed mice were isolated and total numbers of CD4+CD8−CD25+FoxP3+ cells were enumerated. Naive *0402 and *0402.DQ8 mice had significantly higher number of regulatory CD4+CD25+FoxP3+ T cells compared with *0401 and *0401/DQ8 mice in spleen (p < 0.0001) (Fig. 6A). *0402.DQ8 mice had higher numbers of regulatory cells compared with *0402 mice (p < 0.01). After immunization with CII, the difference between the two strains was significant for regulatory cells, with *0402 mice having higher numbers than *0401 mice (p < 0.02). In naive thymic cells, CD4+CD25+FoxP3 T cells were consistently much higher in *0402 and *0402/DQ8 mice compared with *0401 and *0401/DQ8 mice, although the difference was not significant (Fig. 6B).
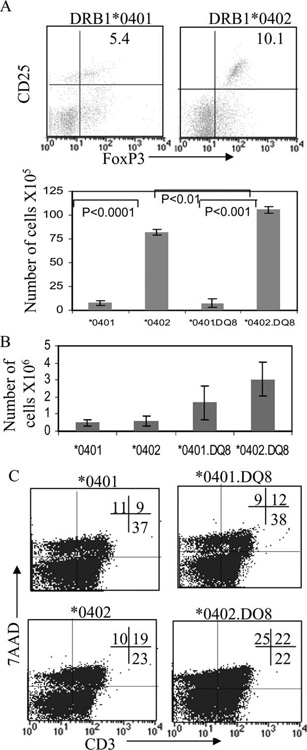
FIGURE 6.
CIA-resistant mice, DRB1*0402 and DRB1*0402.DQ8, have more regulatory cells and show increased AICD. A, FACS analysis showing regulatory cells, CD25+FoxP3+ cells gated on CD4+ cells in *0401 and *0402 mice. Histogram shows number of regulatory cells, CD4+CD25+FoxP3+ (mean ± SD), in single- and double-Tg mice. *0401 vs *0402, p < 0.0001; *0401.DQ8 vs *0402.DQ8, p < 0.001; and *0402 vs *0402.DQ8, p < 0.01. B, Histogram shows number of regulatory cells, CD4+CD25+FoxP3+ (mean ± SD) in Thymus of naive Tg mice. C, Splenic cells isolated from CII-primed mice were stained for 7-aminoactinomycin D (7AAD) and CD3 Abs and analyzed by FACS. *0402 and *0402.DQ8 mice showed increased AICD of CD3+ cells compared with *0401 and *0401.DQ8 mice: *0401 vs *0402 and *0401.DQ8 vs *0402.DQ8, p < 0.05.
Resistant mice show high AICD
Since DRB1*0402 mice had higher numbers of regulatory cells, we tested if after CII challenge, higher numbers of cells undergo AICD. Splenic cells were challenged in vitro with CII and tested for AICD. DRB1*0402 mice showed significantly increased AICD in CD3+ cells compared with *0401 mice (< 0.05) (Fig. 6C), suggesting increased AICD after Ag challenge and higher number of regulatory cells may be the reason for protection from arthritis. Similar to *0402, double-Tg *0402.DQ8 mice showed significantly higher AICD compared with *0401.DQ8 mice after CII immunization (p < 0.05).
Discussion
The humanized mouse model for arthritis described herein shows production of autoantibodies like RF and cyclic citrullinated peptides and carries HLA transgene expression on T cells, similar to that in humans, and develops arthritis predominantly in female mice (present data, and Ref. 24). Activated T cells can present peptides that can potentially play a role in pathogenesis as locally activated T cells in joints of patients with rheumatoid arthritis may present peptides generating more inflammation. Thus, these HLA class II Tg mice take us one step closer to the human disease. Our previous data with AEo DRB1*0401 Tg mice showed that DRB1*0401 renders mice susceptible to develop CIA. HLA-DQ8 occurs in linkage with DRB1*0401 and has been shown to occur with increased frequency in RA patients of some ethnic groups (17, 18). Similarly, DRB1*0402 occurs in linkage with DQ8 but does not occur in patients with significant frequency (34) suggesting DR can modulate DQ-restricted disease. Present data show *0402 Tg mice are resistant to develop arthritis. To determine the role of haplotype in predisposition to develop CIA, we studied double-Tg mice expressing RA-susceptible and RA-resistant DR4/DQ8 haplotypes. While *0401.DQ8 Tg mice are susceptible to CIA, *0402.DQ8 mice are resistant to CIA, suggesting that *0402 protects DQ8 mice from developing arthritis. Arthritis observed in *0401.DQ8 mice shares similarities with RA as they produce rheumatoid factor. Even though *0402.DQ8 mice were protected from arthritis, the few mice that developed arthritis showed a delayed onset compared with *0401.DQ8 mice, but there was no difference in severity of arthritis as studied by clinical scoring. TNF-α and IL-1b have been suggested to regulate disease severity in Ab-induced arthritis (35). We observed significantly higher levels of both cytokines in CIA-susceptible mice, suggesting that regulation of disease severity may be different in Ag-induced vs Ab-induced arthritis. Since these mice carry both DR and DQ it is possible that presentation of various peptides of CII by DQ8 may lead to epitope spreading, thus generating similar disease severity in both strains. Regulation of disease severity by regulatory T cells is controversial since some studies support it while others do not (36, 37). In the present data, *0402.DQ8 mice have higher numbers of regulatory cells, suggesting that they may influence incidence and onset of disease but not severity. Mice were scored for severity by phenotype and not based on histologic or radiographic parameters. Decreased incidence of arthritis in *0402.DQ8 mice is reminiscent of the model proposed for diabetes (38), according to which MHC molecules provide dominant resistance to a given autoimmune disease by deleting the most pathogenic autoreactive T cells rather than all autoreactive T cells. Thus, the reason that RA patients carrying *0402 are rare might be explained by the fact that most of the pathogenic autoreactive cells have been deleted in these individuals. Alternatively, increased numbers of positively selected T cells may be regulatory cells. Our observations in naive mice show that resistant mice have higher numbers of regulatory T cells in the periphery as well as in thymus compared with susceptible mice. This suggests that MHC molecules are associated with protection by positively selecting regulatory T cells in thymus and deleting autoreactive T cells while MHC susceptible alleles select lower numbers of regulatory cells. This is in line with previous observations that MHC molecules play a role in selecting autoreactive cells (31). In the periphery, TGF-β can induce generation of regulatory T cells. We did observe higher levels of TGF-β in CIA-protected *0402 mice compared with susceptible *0401 mice. Functionally, these regulatory cells can lead to a decrease in incidence or severity of disease in vivo. BrdU staining in vivo showed that *0402 mice produced a low T cell response to CII, which could be due to high number of regulatory cells. Experiments for the in vivo role of these regulatory cells are underway. Regulatory T cells have been shown to suppress T cell proliferation and also modulate Th1/Th2 cytokines by effector T cells in vitro, and in vivo they can inhibit organ-specific autoimmune pathology by depletion of, as well as transfer of, regulatory cells (36, 39, 40).
The cytokine data in this study agree with the above observations. All mice produced a strong Th1 response when challenged in vitro with CII. However, *0402 mice produced significantly lower amounts of TNF-α and higher Th2 cytokines. This could tip the balance toward protection in *0402 mice. Comparison between double-Tg mice showed that susceptible mice produced both pro- and antiinflammatory cytokines similar to those reported in rheumatoid arthritis (41). IL-17 is a newly described inflammatory cytokine and has been shown to play an important role in disease pathogenesis of arthritis since IL-17−/− mice are protected from developing collagen-induced arthritis (42–45). IL-23 drives the expansion and survival of IL-17 producing Th17 cells and promotes chronic inflammation via IL-17 and TNF. We observed significantly increased levels of IL-17, TNF-α, and IL-23 in *0401.DQ8 mice compared with *0402.DQ8 mice. Additionally, there was a small increase of IFN-γ in susceptible mice, suggesting that while IL-17/IL-23 are important for pathogenesis of arthritis, IFN-γ may also have a role in inflammation. Recent studies in RA patients positive for the DRB1*0401.DQ8 haplotype have shown an increased production of IFN-γ in response to in vitro challenge with type II collagen (46). Th2 cytokines, such as IL-5 and IL-13, were also significantly increased in susceptible mice. IL-13 has been shown to contribute to DC growth from progenitors and induction of T cell-attracting chemokine CCL18 in RA patients (47, 48). G-CSF and IL-3, significantly increased in *0401.DQ8 mice, have been shown to regulate proliferation and survival of neutrophils (49). Elevated levels of G-CSF are found in serum and synovial fluid of RA patients (50). Neutrophils stimulated by G-CSF release BAFF (B cell-activating factor belonging to the TNF family), which is important for proliferation and maturation of B cells. We did observe a higher level of autoantibodies in *0401.DQ8 mice compared with CIA-resistant *0402.DQ8 mice.
In vitro, both *0401 and *0401.DQ8 mice could present CII, even though double-Tg mice mounted stronger response than single-Tg mice, which could be due to the fact that DQ8 can present multiple epitopes from CII (15) while DRB1*0401 can present only one immunodominant epitope (51). Binding studies have demonstrated that RA-associated HLA-DR alleles bind fewer human CII peptides compared with HLA-DQ8 allele (14), suggesting that HLA-DQ molecules may be a major factor in conferring susceptibility to develop arthritis. In CIA-susceptible mice, pathogenic response has been mapped to the CB11 region of CII, with aa 260–270 being the immunodominant region (52). Interestingly, both Tg strains, *0402 and *0401, mount response to peptide 254–270 of human type II collagen although a significantly lower response was observed in *0402 mice. Both *0401 and *0402 have been shown to be able to bind this peptide, albeit with different anchors in P1 pocket (53). Double Tg mice can respond to both DR- and DQ-restricted peptides. Thus, presentation of this peptide in double-Tg mice might lead to an increase in proliferation. Moreover, activated CD4+ cells can present both DR- and DQ-restricted peptides. This might explain increased incidence of arthritis in *0401.DQ8 mice compared with single Tgs.
The AEo DR and DQ Tg mice develop normally and have no gross phenotypic abnormalities. MHC molecules have been shown to be important in positive and negative selection of various Vβ T cell repertoires in thymus (8, 9) that might affect the clinical outcome of the disease (25). Our data show that DR4 can positively and negatively select the TCR Vβ profile, thus suggesting that it can present peptides in thymus. In double-Tg mice, DRB1*0401 was able to negatively select TCR Vβ that are known to be positively selected by DQ8. DR-derived self-peptides could be presented by DQ molecules in thymus that could lead to positive and negative selection of various TCRs. This is supported by the present data showing that *0402 peptide is presented by DQ8 in *0401.DQ8 mice, but not by *0402.DQ8 mice. Since *0402 peptide is a self-peptide, it is presented in thymus, resulting in negative selection of self-reactive cells.
The resistant mice were not defective in humoral immune response as they produced anti-CII Abs, although *0402 mice produced much lower levels of Abs than did other strains. Ag-specific response to CII was significantly lower in resistant mice compared with susceptible strains; *0402 mice responded strongly to superantigens, suggesting that *0402 individuals may be able to respond to bacterial Ags much stronger than do *0401 mice. Indeed, HLA polymorphism is known to influence response to bacterial superantigens (54). *0402.DQ8 mice mount a higher proliferative and cytokine responses to a mixture of streptococcal superantigens compared with DQ6 mice. Recently, DRB1*04 has been associated with resistance to develop leprosy (55).
The Tg mice expressing *0401 and DQ8 molecules simulate human haplotype and are a good model to study the mechanism of pathogenesis of RA. HLA in Tg mice is expressed similar to humans. As shown in the present study, one APC can express both DR and DQ, and the expression of DR is higher than DQ molecules. The *0402.DQ8 haplotype is known to be resistant to arthritis in humans and mice. Studies in CIA-resistant *0402 mice can provide the answers to how individuals carrying this haplotype are protected from arthritis. This study shows that the protection from arthritis in *0402 mice could be due to 1) negative selection of autoreactive cells in thymus, 2) positive selection of regulatory T cells in thymus, 3) generation of higher numbers of regulatory cells in periphery that can lead to lower proliferation and low proinflammatory cytokines, and 4) increased AICD. All of the factors together may be important for resistance to the development of disease.
Acknowledgments
We thank Julie Hanson and her staff in the Mayo immunogenetic mouse colony for breeding and care of the mice. We thank Michele Smart for tissue typing of Tg mice. We are indebted to Drs. C. Benoist and D. Mathis for the class II-deficient (Aβo and MHCIIΔ/Δ) mice.
Footnotes
This study was supported by National Institutes of Health Grant AR 30752 and by the Mayo Foundation.
Abbreviations used in this paper: RA, rheumatoid arthritis; AICD, activation-induced cell death; CIA, collagen-induced arthritis; CII, type II collagen; LNC, lymph node cell; RF, rheumatoid factor; Tg, transgenic.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Newton JL, Harney SMJ, Wordsworth BP, Brown MA. A review of the MHC genetics of rheumatoid arthritis. Genes Immun. 2004;5:151–157. doi: 10.1038/sj.gene.6364045. [DOI] [PubMed] [Google Scholar]
- 2.Taneja V, Giphart MJ, Verduijn W, Naipal A, Malaviya AN, Mehra NK. Polymorphism of HLA-DRB, -DQA1, and -DQB1 in rheumatoid arthritis in Asian Indians: association with DRB1*0405 and DRB1*1001. Hum. Immunol. 1996;46:35–40. doi: 10.1016/0198-8859(95)00165-4. [DOI] [PubMed] [Google Scholar]
- 3.Stastny P. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N. Engl. J. Med. 1978;298:869–871. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 4.Nepom GT, Nepom BS. Prediction of susceptibility to rheumatoid arthritis by human leukocyte antigen genotyping. Rheum. Dis. Clin. N. Am. 1992;18:785–792. [PubMed] [Google Scholar]
- 5.Wordsworth BP, Lanchbury JS, Sakkas LI, Welsh KI, Panayi GS, Bell JI. HLA-DR4 subtype frequencies in rheumatoid arthritis indicate that DRB1 is the major susceptibility locus within the HLA class II region. Proc. Natl. Acad. Sci. USA. 1989;86:10049–10053. doi: 10.1073/pnas.86.24.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis: an approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 7.Winchester R, Dawyer E, Rose S. The genetic basis of rheumatoid arthritis: the shared epitope hypothesis. Rheum. Dis. Clin. N. Am. 1992;18:761–783. [PubMed] [Google Scholar]
- 8.Mitchison NA. Specialization, tolerance, memory, competition, latency, and strife among T cells. Annu. Rev. Immunol. 1992;10:1–12. doi: 10.1146/annurev.iy.10.040192.000245. [DOI] [PubMed] [Google Scholar]
- 9.Moller E, Bohme J, Valugerdi MA, Ridderstad A, Olerup O. Speculations on the mechanism of HLA association with autoimmune diseases and the specificity of autoreactive T lymphocytes. Immunol. Rev. 1990;118:5–19. doi: 10.1111/j.1600-065x.1990.tb00811.x. [DOI] [PubMed] [Google Scholar]
- 10.Kirschmann DA, Duffin KL, Smith CE, Welply JK, Howard SC, Schwartz BD, Woulfe SL. Naturally processed peptides from rheumatoid arthritis associated and non-associated HLA-DR alleles. J. Immunol. 1995;155:5655–5662. [PubMed] [Google Scholar]
- 11.Snijders A, Elferink DG, Geluk A, van Der Zanden AL, Vos K, Schreuder GM, Breedveld FC, de Vries RR, Zanelli EH. An HLA-DRB1-derived peptide associated with protection against rheumatoid arthritis is naturally processed by human APCs. J. Immunol. 2001;166:4987–4993. doi: 10.4049/jimmunol.166.8.4987. [DOI] [PubMed] [Google Scholar]
- 12.Zanelli E, Gonzalez-Gay MA, David CS. Could DRB1 be the protective locus in rheumatoid arthritis? Immunol. Today. 1995;16:274–278. doi: 10.1016/0167-5699(95)80181-2. [DOI] [PubMed] [Google Scholar]
- 13.Taneja V, David CS. HLA class II transgenic mice as humanized mouse models disease and immunity. J. Clin. Invest. 1998;101:921–926. doi: 10.1172/JCI2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsushita S, Nishi T, Oiso M, Yamaoka K, Yone K, Kanai T, Nishimura Y. HLA-DQ-binding motifs: I. Comparative binding analysis of type II collagen-derived peptides to DR and DQ molecules of rheumatoid arthritis-susceptible and non-susceptible haplotypes. Int. Immunol. 1996;8:757–764. doi: 10.1093/intimm/8.5.757. [DOI] [PubMed] [Google Scholar]
- 15.Krco CJ, Pawelski J, Harders J, McCormick I, Griffiths MM, Luthra HS, David CS. Characterization of the antigenic structure of human type II collagen. J. Immunol. 1996;156:2761–2768. [PubMed] [Google Scholar]
- 16.Begovich AB, McClure GR, Suraj VC, Helmuth RC, Fildes N, Bugawan TL, Erlich HA, Klitz W. Polymorphism recombination and linkage disequilibrium within the HLA class II region. J. Immunol. 1992;148:249–258. [PubMed] [Google Scholar]
- 17.Taneja V, Mehra NK, Chadreshekaran AN, Ahuja RK, Singh YN, Malaviya AN. HLA-DR4-DQw8 but not DR4-DQw7 haplotypes occur in Indian patients with rheumatoid arthritis. Rheumatol. Int. 1992;11:251–255. doi: 10.1007/BF00301502. [DOI] [PubMed] [Google Scholar]
- 18.Laivoranta-Nyman S, Mottonen T, Hermann R, Tuokko J, Hakala M, Luukainen R, Hannonen P, Korpela M, Yli-Kerttula U, Toivanen A. HLA-DR-DQ haplotypes and genotypes in Finnish patients with rheumatoid arthritis. Ann. Rheum. Dis. 2004;63:1406–1412. doi: 10.1136/ard.2003.009969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cook AD, Gray R, Ramshaw J, Mackay IR, Rowley MJ. Antibodies against the CB10 fragment of type II collagen in rheumatoid arthritis. Arthritis Res. Ther. 2004;6:R477–R483. doi: 10.1186/ar1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He X, Kang AH, Stuart JM. Accumulation of T cells reactive to type II collagen in synovial fluid of patients with rheumatoid arthritis. J. Rheumatol. 2000;27:589–593. [PubMed] [Google Scholar]
- 21.Nishimura K, Sugiyama D, Kogata Y, Tsuji G, Nakazawa T, Kawano S, Saigo K, Morinobu A, Koshiba M, Kuntz KM, et al. Meta-analysis: diagnostic accuracy of anti-cyclic citrullinated peptide antibody and rheumatoid factor for rheumatoid arthritis. Ann. Intern. Med. 2007;146:797–808. doi: 10.7326/0003-4819-146-11-200706050-00008. [DOI] [PubMed] [Google Scholar]
- 22.Taneja V, Taneja N, Paisansinsup T, Behrens M, Griffiths MM, Luthra HS, David CS. CD4 and CD8 T cells in susceptibility/protection to collagen-induced arthritis in HLA-DQ8 transgenic mice: implications for rheumatoid arthritis. J. Immunol. 2002;168:5867–5875. doi: 10.4049/jimmunol.168.11.5867. [DOI] [PubMed] [Google Scholar]
- 23.Taneja V, Griffiths MM, Luthra HS, David CS. Modulation of HLA-DQ-restricted collagen-induced arthritis by HLA-DRB1 polymorphism. Int. Immunol. 1998;10:1449–1457. doi: 10.1093/intimm/10.10.1449. [DOI] [PubMed] [Google Scholar]
- 24.Taneja V, Behrens M, Mangalam A, Griffiths MM, Luthra HS, David CS. New humanized HLA-DR4-transgenic mice that mimic the sex bias of rheumatoid arthritis. Arthritis Rheum. 2007;56:69–78. doi: 10.1002/art.22213. [DOI] [PubMed] [Google Scholar]
- 25.Taneja V, Taneja N, Behrens M, Pan S, Trejo T, Griffiths MM, Luthra HS, David CS. HLA-DRB1*0402 (*0402) transgene protects collagen-induced arthritis-susceptible H2Aq and DRB1*0401 (*0401) transgenic mice from arthritis. J. Immunol. 2003;171:4431–4438. doi: 10.4049/jimmunol.171.8.4431. [DOI] [PubMed] [Google Scholar]
- 26.Kouskoff V, Fehling HJ, Lemeur M, Benoist C, Mathis D. A vector driving the expression of foreign cDNAs in the MHC class II-positive cells of transgenic mice. J. Immunol. Methods. 1993;166:287–291. doi: 10.1016/0022-1759(93)90370-m. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths MM, Eichwald EJ, Martin JH, Smith CB, DeWitt CW. Immunogenetic control of experimental type II collagen induced arthritis. Arthritis Rheum. 1981;24:781–789. doi: 10.1002/art.1780240605. [DOI] [PubMed] [Google Scholar]
- 28.Wooley PH, Luthra HS, Stuart JM, David CS. Type II collagen-induced arthritis in mice: I. Major histocompatibility complex (I region) linkage and antibody correlates. J. Exp. Med. 1981;154:688–700. doi: 10.1084/jem.154.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-β induces development of the TH17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 30.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 31.Stratmann T, Martin-Orozco N, Mallet-Designe V, Poirot L, McGavern D, Losyev G, Dobbs CM, Oldstone MBA, Yoshida K, Kikutani H, et al. Susceptible MHC alleles, not background genes, select an autoimmune T cell reactivity. J. Clin. Invest. 2003;112:902–914. doi: 10.1172/JCI18337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haqqi TM, Anderson GD, Banerjee S, David CS. Restricted heterogeneity in T-cell antigen receptor Vβ gene usage in lymph nodes and arthritic joints of mice. Proc. Natl. Acad. Sci. USA. 1992;89:1253–1255. doi: 10.1073/pnas.89.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaturvedi P, Agarwal B, Zechel M, Lee-Chan E, Singh B. A self MHC class II β-chain peptide prevents diabetes in nonobese diabetic mice. J. Immunol. 2000;164:6610–6620. doi: 10.4049/jimmunol.164.12.6610. [DOI] [PubMed] [Google Scholar]
- 34.Khani-Hanjani A, Lacaille D, Horne C, Chalmers A, Hoar DI, Balshaw R, Keown PA. Expression of QK/QR/RRRAA or DERAA motifs at the third hypervariable region of HLA-DRB1 and disease severity in rheumatoid arthritis. J. Rheumatol. 2002;29:1358–1365. [PubMed] [Google Scholar]
- 35.Ji H, Pettit A, Ohmura K, Ortiz-Lopez A, Duchatelle V, Degott C, Gravallese E, Mathis D, Benoist C. Critical roles for interleukin 1 and tumor necrosis factor α in antibody-induced arthritis. J. Exp. Med. 2002;196:77–85. doi: 10.1084/jem.20020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen LT, Jacobs J, Mathis D, Benoist C. Where FoxP3-dependent regulatory T cells impinge on the development of inflammatory arthritis. Arthritis Rheum. 2007;56:509–520. doi: 10.1002/art.22272. [DOI] [PubMed] [Google Scholar]
- 37.Bardos T, Czipri M, Vermes C, Finnegan A, Mikecz K, Zhang J. CD4+CD25+ immunoregulatory T cells may not be involved in controlling autoimmune arthritis. Arthritis Res. Ther. 2003;5:R106–R113. doi: 10.1186/ar624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt D, Verdaguer J, Averill N, Santamaria P. A mechanism for the major histocompatibility complex-linked resistance to autoimmunity. J. Exp. Med. 1997;186:1059–1075. doi: 10.1084/jem.186.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Majoly KJ, Powrie F. Regulatory T cells in the control of immune pathology. Nat. Immunol. 2001;2:816–822. doi: 10.1038/ni0901-816. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar S, Fox DA. Regulatory T cell defects in rheumatoid arthritis. Arthritis Rheum. 2007;56:710–713. doi: 10.1002/art.22415. [DOI] [PubMed] [Google Scholar]
- 41.Raza K, Falcani F, Curnow SJ, Ross EJ, Lee CY, Akbar AN, Lord JM, Gordon C, Buckley CD, Salmon M. Early rheumatoid arthritis is characterized by a distinct and transient synovial fluid cytokine profile of T cell and stromal cell origin. Arthritis Res. Ther. 2005;7:R784–R795. doi: 10.1186/ar1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McInnes IB, Liew FY. Cytokine networks: towards new therapies for rheumatoid arthritis. Nat. Clin. Pract. Rheumatol. 2005;1:31–39. doi: 10.1038/ncprheum0020. [DOI] [PubMed] [Google Scholar]
- 43.Nakae S, Nambu A, Sudo K, Iwakura Y. Suppression of immune induction of collagen-induced arthritis in IL-17-deficient mice. J. Immunol. 2003;17:6173–6177. doi: 10.4049/jimmunol.171.11.6173. [DOI] [PubMed] [Google Scholar]
- 44.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar S, Tesmer LA, Hindnavis V, Endres JL, Fox DA. Interleukin-17 as a molecular target in immune-mediated arthritis. Arthritis Rheum. 2007;56:89–100. doi: 10.1002/art.22311. [DOI] [PubMed] [Google Scholar]
- 46.Berg L, Ronnelid J, Sanjeevi CB, Lampa J, Klareskog L. Interferon γ production in response to in vitro stimulation with collagen type II in rheumatoid arthritis is associated with HLA-DRB1*0401 and HLA-DQ8. Arthritis Res. 2000;2:75–84. doi: 10.1186/ar71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokayer A, Carsons SE, Chokshi B, Santiago-Schwarz F. High levels of interleukin 13 in rheumatoid arthritis sera are modulated by tumor necrosis factor antagonist therapy: association with dendritic cell growth activity. J. Rheumatol. 2002;29:454–461. [PubMed] [Google Scholar]
- 48.van Lieshout A, van der Voort R, le Blanc LM, Roelofs MF, Schreurs BW, van Riel PL, Adema GJ, Radstake T. Novel insights in the regulation of CCL18 secretion by monocytes and dendritic cells via cytokines, Toll-like receptors and rheumatoid synovial fluid. BMC Immunol. 2006;7:23–35. doi: 10.1186/1471-2172-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geijsen N, Koenderman L, Coffer PJ. Specificity in cytokine signal transduction: lessons learned from the IL-3/IL-5/GM-CSF receptor family. Cytokine Growth Factor Rev. 2001;12:19–25. doi: 10.1016/s1359-6101(00)00019-8. [DOI] [PubMed] [Google Scholar]
- 50.Eyles JL, Roberts AW, Metcalf D, Wicks IP. Granulocyte colony-stimulating factor and neutrophils: forgotten mediators of inflammatory disease. Nat. Clin. Pract. Rheumatol. 2006;2:500–510. doi: 10.1038/ncprheum0291. [DOI] [PubMed] [Google Scholar]
- 51.Rosloniec EF, Whittington KB, Zaller DM, Kang AH. HLA-DR1 (DRB1*0101) and DR4 (DRB1*0401) use the same anchor residues for binding an immunodominant peptide derived from human type II collagen. J. Immunol. 2002;168:253–259. doi: 10.4049/jimmunol.168.1.253. [DOI] [PubMed] [Google Scholar]
- 52.Terato K, Hasty KA, Cremer MA, Stuart JM, Towens AS, Kang AH. Collagen induced arthritis in mice: localization of an arthritogenic determinant to a fragment of the type II collagen molecule. J. Exp. Med. 1985;162:637–646. doi: 10.1084/jem.162.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diab BY, Lambert NC, L’Faqihi FE, Loubet-Lescoulie P, de Preval C, Coppin H. Human collagen II peptide 256–271 preferentially binds to HLA-DR molecules associated with susceptibility to rheumatoid arthritis. Immunogenetics. 1999;49:36–44. doi: 10.1007/s002510050461. [DOI] [PubMed] [Google Scholar]
- 54.Nooh MM, El-Gengehi N, Kansal R, David CS, Kotb M. HLA transgenic mice provide a direct and dominant role of HLA class II variation in modulating the severity of streptococcal sepsis. J. Immunol. 2007;178:3076–3083. doi: 10.4049/jimmunol.178.5.3076. [DOI] [PubMed] [Google Scholar]
- 55.Vanderborght PR, Pacheco AG, Moraes ME, Antoni G, Romero M, Verville A, Thai VH, Huong NT, Ba NN, Schurr E, et al. HLA-DRB1*04 and DRB1*10 are associated with resistance and susceptibility, respectively, in Brazilian and Vietnamese leprosy patients. Genes Immun. 2007;8:320–324. doi: 10.1038/sj.gene.6364390. [DOI] [PubMed] [Google Scholar]