Autism is a highly heterogeneous neurodevelopmental disorder with impaired language, social communication, and restricted and repetitive interests and behavior. Monogenic developmental brain disorders with autism features such as Rett syndrome, Angelman syndrome, Fragile X syndrome, and others provide important tractable models of relevance to severe autism. In addition to observing autism symptoms in the monogenic condition, if the gene responsible is also significantly misregulated in the brains of people with idiopathic autism, then these data provide substantial independent support for the importance of the gene in autism pathophysiology.
Given this logic, we set out to test if there were gene expression changes in postmortem brain tissue from patients with idiopathic autism in the genes of interest that encode the Na+/H+ exchanger family of proteins, with particular interest in the forms of exchangers localized to endosomes, namely, NHE6 and NHE9. Mutations in the X-linked endosomal Na+/H+ Exchanger 6 (NHE6, also known as SLC9A6) represent a novel neurogenetic syndrome with variable expressivity.1 In a systematic, large-scale resequencing screen of X-chromosome coding exons in >200 pedigrees consistent with X-linked intellectual disability, NHE6 was among the top six most recurrently mutated genes.2 The ‘Christianson syndrome', based on the initial clinical description, reported an association with autistic symptoms as has been reported subsequently.3, 4 In parallel to the description of autistic symptoms associated with mutations in NHE6, Morrow et al.5 published mutations in the highly related endosomal protein NHE9 in severe autism with epilepsy. Endosomal processes, such as would be suggested by mutations in NHE6 and NHE9, represent an important cellular mechanism for investigations regarding disorders of cognitive development. Interestingly, of the six top genes implicated in the Tarpey et al.2 study, two of the genes, NHE6 and AP1S2, are known to be involved in endosomal mechanisms.
We therefore investigated whether the expression of NHE genes, NHE6 and NHE9, were altered in autism cerebral cortex. We did this by renormalizing and analyzing publically available microarray data. Specifically, we first analyzed data from Voineagu et al.,6 which were previously used to compare autism and control cortex (n=29 of each). We renormalized the data as described in Voineagu et al.,6 except for when we wished to analyze a subset of NHE genes that were excluded from the analysis because of low expression. Among their results, Voineagu et al.6 reported that synapse-associated genes were downregulated in autism cortex,6 and we confirmed this result by performing differential gene expression analysis7 between autism and control cortex (for detailed Methods, please see Supplementary Information). We then performed DAVID functional annotation clustering analysis8, 9 on the 197 genes with at least 1.3-fold reduction in autism cortex and P<0.05 (after Benjamini-Hochberg adjustment10), which were the differential expression cutoffs described by Voineagu et al.6 Indeed, the most significant gene set in the top scoring cluster of the DAVID results was the Gene Ontology term ‘synapse,' for which 21 genes overlapped with those downregulated in autism cortex (BH-adjusted overlap P-value=2.2 × 10−7). This confirmed that synapse-related genes, such as GABRA1 and CHRM1, were downregulated in autism cortex in the Voineagu et al.6 data set (Supplementary Tables 1 and 2 for description of the downregulated synapse genes). We next tested whether NHE genes (NHE1–11) were differentially expressed between autism and control cortex using a t-test (Supplementary Table 3). Notably, NHE1 and NHE6 were significantly downregulated in autism cortex (P=0.0030 and 0.0042, respectively) and NHE9 was significantly upregulated (P=0.00075), yet other NHE genes were not significantly differentially expressed between autism and control cortex (P>0.15).
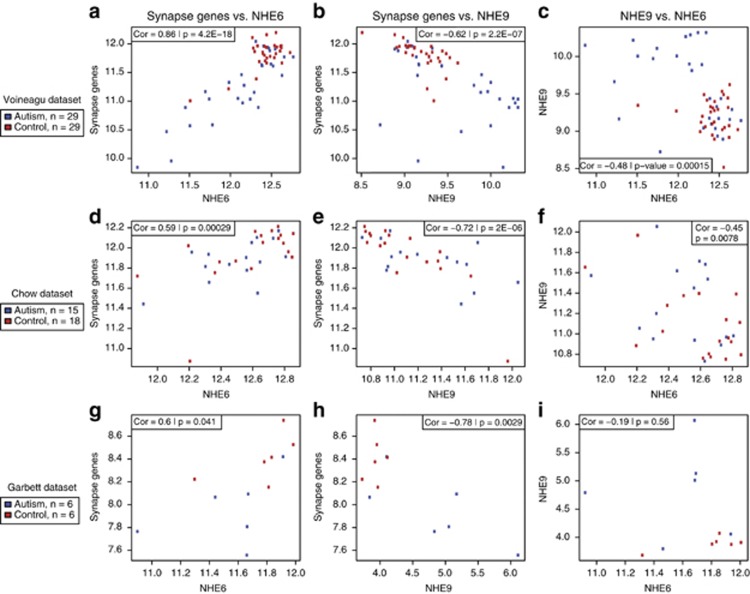
We further hypothesized that changes in these genes were reflective of broader changes in gene expression, such as downregulation of synapse genes. To investigate the functional changes associated with NHE1, NHE6 and NHE9 gene expression, we found Pearson correlation coefficients between each of these genes and the average expression of the 21 synapse-related genes. NHE1 was not significantly correlated with the synapse genes (r=0.15, P=0.28), but NHE6 and NHE9 both were strongly correlated with the synapse genes (r=0.86, P=4.2 × 10−18 for NHE6; r=−0.62, P=2.2 × 10−7 for NHE9—see Figure 1). Furthermore, the sub-population of samples with the lowest NHE6 expression was almost entirely autism cases, as was the sub-population of high NHE9 expression. Additionally, NHE6 and NHE9 were negatively correlated (P=0.00015), and when NHE6 expression was high, NHE9 was tightly regulated. A similar decrease in NHE9 variability could be seen with high synapse gene expression (Figure 1). Thus, a sub-population within autism is not only associated with lower synapse gene expression, but also with lower NHE6 expression and increased and potentially misregulated NHE9 expression. We also studied the correlation of NHE6 and NHE9 with the downregulated synapse gene set during normal brain development. We capitalized on the large mRNA-seq data set from human brain made available by the Allen Brain Institute (http://www.developinghumanbrain.org). We find that NHE6 clusters strongly during development with this synapse gene group (see Supplementary Information). NHE9 expression was far lower embryonically, increased postnatally but did not appreciably cluster with this synapse group in typical brain development (Supplementary Figures 1–3).
Figure 1.
Expression plots of NHE6 versus synapse genes, NHE9 versus synapse genes and NHE6 versus NHE9 in three independent data sets. We compared the average log base 2 expression of 21 synapse genes with that of NHE6 and NHE9 expression in microarray data sets published by Voineagu et al.6 (a–c, n=58), Chow et al.11 (d–f, n=33) and Garbett et al.12 (g–i, n=12). Each data set was designed to compare gene expression in autism and control cerebral cortex. The synapse genes used were the 21 synapse genes downregulated in autism cortex compared with control in the Voineagu et al.6 data set (Supplementary Tables 1 and 2).
We used two independent microarray data sets for autism and control cerebral cortex to validate both the differential gene expression of NHE6 and NHE9 in autism cases and these genes' associations with synapse genes. The validation data sets were from Chow et al.11 (n=15 autism, n=18 control) and Garbett et al.12 (n=6 of each; data set details in Supplementary Table 4). The Chow et al.11 data were downloaded from GEO (GSE28475), and the Garbett et al.12 data were provided by the authors. In the Garbett et al.12 data, NHE9 was significantly higher in autism than control cortex (P=0.039), and while NHE6 was lower on average in autism cortex, this was not statistically significant (P=0.22), although this may have been due to low sample size (Figure 1). In the Chow et al.11 data, although NHE6 and NHE9 had lower and higher expression in autism cortex compared with control, respectively, these trends were not significant (P=0.38 and 0.17). However, the mean expression of synapse genes was also not significantly different in the Chow et al.11 data set (P=0.46), suggesting that the Voineagu et al.6 and Chow et al.11 data sets represent different populations. Additionally, NHE6 and NHE9 were negatively correlated across data sets, except for in Garbett et al.,12 in which the correlation was not significant (P=0.56) but suggesting as yet unknown mechanisms of interaction. Despite this lack of significance in the independent data sets, these genes' associations with synapse genes remained strong in both additional data sets: NHE6 is positively associated with synapse genes (r=0.59, P=0.00029 for NHE6 in Chow et al.11 study; and r=0.6, P=0.041 for NHE6 in Garbett et al.12 study—see Figure 1) and NHE9 negatively associated (r=−0.72, P=2.0 × 10−6 for NHE9 in Chow et al.11 study; and r=−0.78, P=0.0029 for NHE9 in Garbett et al.12 study—see Figure 1). Finally, we tested for an association of gene expression for NHE6 and NHE9 and well-established autism-related genes such as SHANK2/3, NLGN4X, NRXN1 and PTEN. Notably, NHE6 and NHE9 each showed a strong association with NRXN1 (P=2 × 10−10 for NHE6, and P=2.9 × 10−8 for NHE9; Supplementary Figures 4 and 5).
In summary, we find interesting gene expression changes in endosomal NHE6 and NHE9 in postmortem autism brains. These gene expression changes are largely replicated across data sets or the trends of these changes are maintained given limitations in sample size. We also report a strong correlation of endosomal NHE6 and NHE9 gene expression with the synapse genes across all data sets. The strong correlation of endosomal NHEs with synapse genes suggests that changes in synapse genes in autism involves a cellular mechanism that involves endosomal NHE6 and NHE9 in at least some autism brains. In conclusion, these gene expression studies in postmortem brains from patients with idiopathic autism provide additional support, in addition to the association of autism symptoms with the single-gene mutations, that endosomal NHEs are mechanistically involved in the pathophysiology of autism.
Acknowledgments
EMM has received a Career Award in Medical Science from the Burroughs Wellcome Fund and support from NIMH 1K23MH080954-05. This work was supported by a grant from the Simons Foundation (SFARI #239834 to EMM), and also generous support to EMM from the Nancy Lurie Marks Foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Gilfillan GD, Selmer KK, Roxrud I, Smith R, Kyllerman M, Eiklid K et al. Am J Hum Genet. 2008; 82: 1003–1010. [DOI] [PMC free article] [PubMed]
- Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C et al. Nat Genet. 2009; 41: 535–543. [DOI] [PMC free article] [PubMed]
- Garbern JY, Neumann M, Trojanowski JQ, Lee VM, Feldman G, Norris JW et al. Brain 2010; 133(Pt 5): 1391–1402. [DOI] [PMC free article] [PubMed]
- Christianson AL, Stevenson RE, van der Meyden CH, Pelser J, Theron FW, van Rensburg PL. J Med Genet. 1999; 36: 759–766. [DOI] [PMC free article] [PubMed]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS et al. Science 2008; 321: 218–223. [DOI] [PMC free article] [PubMed]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S et al. Nature. 2011; 474: 380–384. [DOI] [PMC free article] [PubMed]
- Smyth GK. Stat Appl Genet Mol Biol. 2004; 3: Article 3.
- Huang da W, Sherman BT, Lempicki RA. Nucleic Acids Res. 2009; 37: 1–13. [DOI] [PMC free article] [PubMed]
- Huang da W, Sherman BT, Lempicki RA. Nat Protoc. 2009; 4: 44–57. [DOI] [PubMed]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Behav Brain Res. 2001; 125: 279–284. [DOI] [PubMed]
- Chow ML, Pramparo T, Winn ME, Barnes CC, Li HR, Weiss L et al. PLoS Genet. 2012; 8: e1002592. [DOI] [PMC free article] [PubMed]
- Garbett K, Ebert PJ, Mitchell A, Lintas C, Manzi B, Mirnics K et al. Neurobiol Dis. 2008; 30: 303–311. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.