Abstract
Genetic and environmental components as well as their interaction contribute to the risk of schizophrenia, making it highly relevant to include environmental factors in genetic studies of schizophrenia. This study comprises genome-wide association (GWA) and follow-up analyses of all individuals born in Denmark since 1981 and diagnosed with schizophrenia as well as controls from the same birth cohort. Furthermore, we present the first genome-wide interaction survey of single nucleotide polymorphisms (SNPs) and maternal cytomegalovirus (CMV) infection. The GWA analysis included 888 cases and 882 controls, and the follow-up investigation of the top GWA results was performed in independent Danish (1396 cases and 1803 controls) and German-Dutch (1169 cases, 3714 controls) samples. The SNPs most strongly associated in the single-marker analysis of the combined Danish samples were rs4757144 in ARNTL (P=3.78 × 10−6) and rs8057927 in CDH13 (P=1.39 × 10−5). Both genes have previously been linked to schizophrenia or other psychiatric disorders. The strongest associated SNP in the combined analysis, including Danish and German-Dutch samples, was rs12922317 in RUNDC2A (P=9.04 × 10−7). A region-based analysis summarizing independent signals in segments of 100 kb identified a new region-based genome-wide significant locus overlapping the gene ZEB1 (P=7.0 × 10−7). This signal was replicated in the follow-up analysis (P=2.3 × 10−2). Significant interaction with maternal CMV infection was found for rs7902091 (PSNP × CMV=7.3 × 10−7) in CTNNA3, a gene not previously implicated in schizophrenia, stressing the importance of including environmental factors in genetic studies.
Keywords: CTNNA3, gene-environment interaction, GWAS, GWIS, region-wise analysis, ZEB1
Introduction
Schizophrenia is a severe life-long mental disorder, which affects approximately 1% of the population worldwide. Several studies have documented a strong genetic component in the etiology and the heritability is estimated to be around 80%.1 Besides the genetic component, environmental factors as well as gene–environment interactions are believed to contribute to the disease risk.2 Numerous linkage, candidate gene studies and genome-wide association (GWA) studies have been performed in order to elucidate the genetic architecture of the disease.3, 4, 5, 6, 7, 8 These studies have implicated several genes in disease risk but seldom unambiguously across different studies and populations. In the GWA studies, only a few loci have passed the generally accepted level of P<5 × 10−8 for genome-wide significance.4, 5, 9, 10, 11 From the GWA studies, it can be concluded that only moderate levels of association of common variants with schizophrenia can be expected, and recent results suggest that a high number of common susceptibility variants of small effect are involved, collectively capturing around 30% of the genetic risk.12, 13 The remaining genetic risk could involve de novo mutations, rare variants and gene–environment interactions.14, 15, 16, 17, 18 Due to the low-effect sizes of the common risk variants, the heritability that can be accounted for by those identified so far has been estimated to be between 1% and 2%.19, 20
It is well documented that the environment has an important role in the development of schizophrenia.21 Especially early in life the susceptibility to environmental risk factors may be increased, supported by several studies demonstrating an association of maternal infection with increased risk of the child developing schizophrenia later in life.22, 23, 24, 25 It has also been reported that interaction between genetic variation in the offspring and markers of maternal infection (maternal antibodies) may influence the risk of schizophrenia,26 stressing the importance of taking environmental factors into account in genetic studies.
The Danish population is, in general, considered ethnically homogenous with only recent immigration of non-Caucasian individuals, which makes it suitable for genetic studies. In Denmark, all newborn babies (around 65,000 each year) are screened for metabolic diseases and since 1981 the surplus of the analyzed blood spot samples have been stored in the Danish Newborn Screening Biobank (DNSB).27 Coupling with information from the Danish Psychiatric Central Research Register28 allows for the unique opportunity to obtain DNA from all individuals who have been diagnosed with schizophrenia since 1981. Furthermore, as the DNSB samples are obtained from babies before they are able to produce their own antibodies and hence the antibodies in the blood reflect the mother's antibodies, it is possible to investigate how maternal infections and their interactions with genetic variations in the offspring influence the risk of schizophrenia. Infection with cytomegalovirus (CMV), a neurotrophic virus of the herpesvirus family, has been associated with schizophrenia in several studies, and interactions between selected genes and CMV have been reported,29, 30
Here we report the results of a GWA study and follow-up investigation of all Danish individuals born since 1981 and diagnosed with schizophrenia, including single variant as well as regional analyses. In addition, as the first genome-wide gene–environment interaction study in schizophrenia, we examine interaction between single nucleotide polymorphisms (SNPs) and maternal CMV infection (maternal anti-CMV immunoglobulin G (IgG) antibody titer).
Materials and methods
Study design and power calculation
A two-stage design was applied in this study. In stage 1, a GWA analysis of 888 cases and 882 controls was performed and, in stage 2, a follow-up analysis of the strongest associated SNPs was performed on an independent sample consisting of 1396 cases and 1803 controls. Combined association analysis of both samples achieves a power of 80% to detect a disease allele with a frequency of 0.36 and odds ratio (OR) of 1.35 assuming prevalence of 0.01 at a significance level of 5 × 10−8.31 The SNPs strongest associated with schizophrenia were analyzed further by combining stage 1 and stage 2 individuals with a German-Dutch sample, a sample genetically closely related to the Danish.32
Study subjects and phenotype definition
Stage 1: It was possible to identify the samples of interest based on the unique personal identification number (CPR-number), which is assigned to all live-born babies in Denmark. This number is stored in the Danish Civil Registration System (DCRS)33 and is used in all the contacts with the public sector. In this study, information from the DCRS was linked with the information stored in the nationwide Danish Psychiatric Central Register28 in order to identify all individuals born in 1981 and onwards that in 2006 had been diagnosed with schizophrenia according to ICD-10-DCR (The ICD-10 Classification of Mental and Behavioural Disorders Diagnostic Criteria for Research; F20). For each schizophrenia case, one matched control individual was randomly selected with the same gender, date of birth and age and with no history of schizophrenia on the date of first diagnosis of schizophrenia of the case. Using this procedure, 915 cases and 915 controls were identified and subsequently dried blood spots from the individuals were obtained from the DNSB.
Stage 2: All individuals born since 1981 and onwards diagnosed with schizophrenia according to ICD-10-DCR, F20 between 2006 and 2010 and matched controls were identified as described above. In all, 1149 cases and 1303 controls were identified and subsequently blood spots were obtained from the DNSB. Furthermore, a sample of 247 schizophrenia cases fulfilling ICD-10 criteria and 500 controls were included as described previously.7, 26 The cases and controls were Danish Caucasians.
German-Dutch replication sample: A total of 1169 schizophrenia cases (464 German and 705 Dutch) and 3714 ethnically matched controls (1272 German and 2442 Dutch) were used in the replication analysis (details on criteria for inclusion can be found in Rietschel et al.5). Descriptive data for the three samples can be found in Supplementary Table S1. This study has been approved by the Danish Data Protection Agency and the local ethics committees in Denmark and abroad.
Genotyping and quality control (QC)
Stage 1: Sufficient biological material was available for 909 cases and 899 controls. DNA was extracted from the dried blood spots using Extract-N-Amp Blood PCR kit (Sigma Aldrich, Seelze, Germany) and subsequently whole genome-amplified in triplicates using the RepliG kit (Qiagen, Venlo, The Netherlands).34 The three separate reactions were pooled before genotyping, which was done using the Illumina Human 610-quad beadchip (San Diego, CA, USA). In all, 1774 individuals (892 cases, 882 controls) with gender in concordance with the register information were successfully genotyped with a call rate >0.97. Stringent QC was applied to data from samples with a call rate >0.97. The QC excluded SNPs with a call rate <0.99, SNPs with a deviation from Hardy–Weinberg equilibrium (P<0.0001 in controls) and a minor allele frequency (MAF) <0.0015. Furthermore, test for relatedness, estimation of individual heterozygosity and test for non-random missingness of SNPs between cases and controls were conducted (Supplementary Table S2). After QC, 1770 individuals (882 controls, 888 cases) and 541,148 SNPs were left for further analysis.
Stage 2: DNA from the 1149 cases and 1303 controls obtained from the DNSB was extracted and whole genome-amplified using the kits described above. DNA was isolated from blood samples from the additional 247 cases and 500 controls following standard procedures. In all, 193 follow-up SNPs (Supplementary Table S3) were genotyped as well as five SNPs on the sex-chromosomes, using the Sequenom MassARRAY genotyping platform (Sequenom, San Diego, CA, USA) following the protocol described in Nyegaard et al.7 It was checked that the estimated gender, based on the genotypic information, was in concordance with the gender given in the DCRS. In order to exclude the presence of identical samples, an identity by state analysis was performed using the software Graphical Relationship Representation.35 After QC, 3142 individuals (1370 cases, 1772 controls) with a call rate >0.8 were genotyped for 168 SNPs (Supplementary Table S3). The SNPs had a call rate >0.9, no significant deviation from Hardy–Weinberg equilibrium (P>0.0001 in controls) and a MAF>0.0015.
German-Dutch replication sample: The individuals were genotyped using the Illumina HumanHap550v3 BeadArray (Illumina). After QC genotypes for 475,427 SNPs were available in 1169 cases and 3714 controls (for details on genotyping and QC, see Rietschel et al.5).
Antibody measurements
Measurements of type-specific IgG antibodies to CMV were obtained by enzyme immunoassay36 for a subsample of stage 1 individuals (488 cases (216 females and 272 males) and equally many controls. The blood spots stored in the DNSB were taken when the neonates were 2–7 days old. At that age a child has not yet produced any significant amount of IgG antibodies, but while in utero maternal IgG antibodies are transferred across the placenta to the fetus. Hence, the antibodies measured can be assumed to be mainly maternal.37 The measurements were dichotomized at 0.2 optical density units, yielding a prevalence consistent with those measured in European populations.38
Statistical analysis
All analyses were performed using the software PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/)39 unless otherwise stated.
Population stratification
In order to minimize the effect of spurious association originating from population, stratification association analysis was performed using logistic regression with principal component one as covariate derived from principal component analysis40 (Supplementary Figure S2). This lowered the genomic inflation factor from 1.047 to 1.013. A deviation of λ from 1 can be expected under polygenic inheritance even when there is no population structure,41 so in order to avoid unnecessary correction only principal component one was used to correct for population stratification (more information is provided in the Supplementary Information).
Single-marker association analysis
GWA analysis was performed using logistic regression with principal component one as a covariate applying an additive genetic model. The SNPs demonstrating the strongest association with schizophrenia in the GWA analysis (2843 SNPs with a P-value <0.005), were further evaluated in a meta-analysis by including data from a German-Dutch replication sample.5 The meta-analysis implemented in PLINK was used, and a fixed effect model was considered.
Two types of analysis were applied in order to identify follow-up SNPs for genotyping in the stage 2 sample: (1) based on the GWA analysis, a set of 100 SNPs were identified using a top-down approach, and (2) based on the meta-analysis, a set of 100 SNPs were identified using a top-down approach. Association with schizophrenia was analyzed in the stage 2 sample by logistic regression using an additive genetic model. A binomial sign test was performed in order to test for evidence of directionally consistent replication.
A meta-analysis (as described above) was used to test for association of follow-up SNPs with schizophrenia in two combined data sets: (1) the combined stage 1 and the stage 2 samples, and (2) the combined stage 1, stage 2 and German-Dutch replication sample (referred to as extended meta-analysis from now), which in total included 3453 cases and 6399 controls.
Region-wise association analysis
All chromosomes were divided into overlapping regions of 100 kb, each overlapping its neighboring regions by 50 kb. For each region, a combined P-value was calculated by Fisher's method:42 X=−2∑ki=1loge(pi), where k is the number of SNPs in the region and pi is the P-value for each SNP calculated by a standard χ2 test, using an additive model. The P-value for each region was calculated by permutation test shuffling the case–control status. (see supplementary Material for details).
SNP × maternal CMV infection interaction analysis
The two-step method of Murcray et al.43 was applied in order to test whether genetic variation interacts with maternal CMV infection influencing the risk of schizophrenia in the offspring. First, the full set of SNPs was screened for association with maternal CMV infection in the combined sample of cases and controls at a significance level of 0.05. Second conditional logistic regression with inclusion of an interaction term in the regression on the m SNPs selected in step 1, was performed using Stata 10.0, College Station, TX, USA: StataCorp LP. Because the tests performed in steps 1 and 2 are (asymptotically) independent,43 Bonferroni correction for the m tests in step 2 preserves the family-wise error rate.
Results
Single-marker association analysis
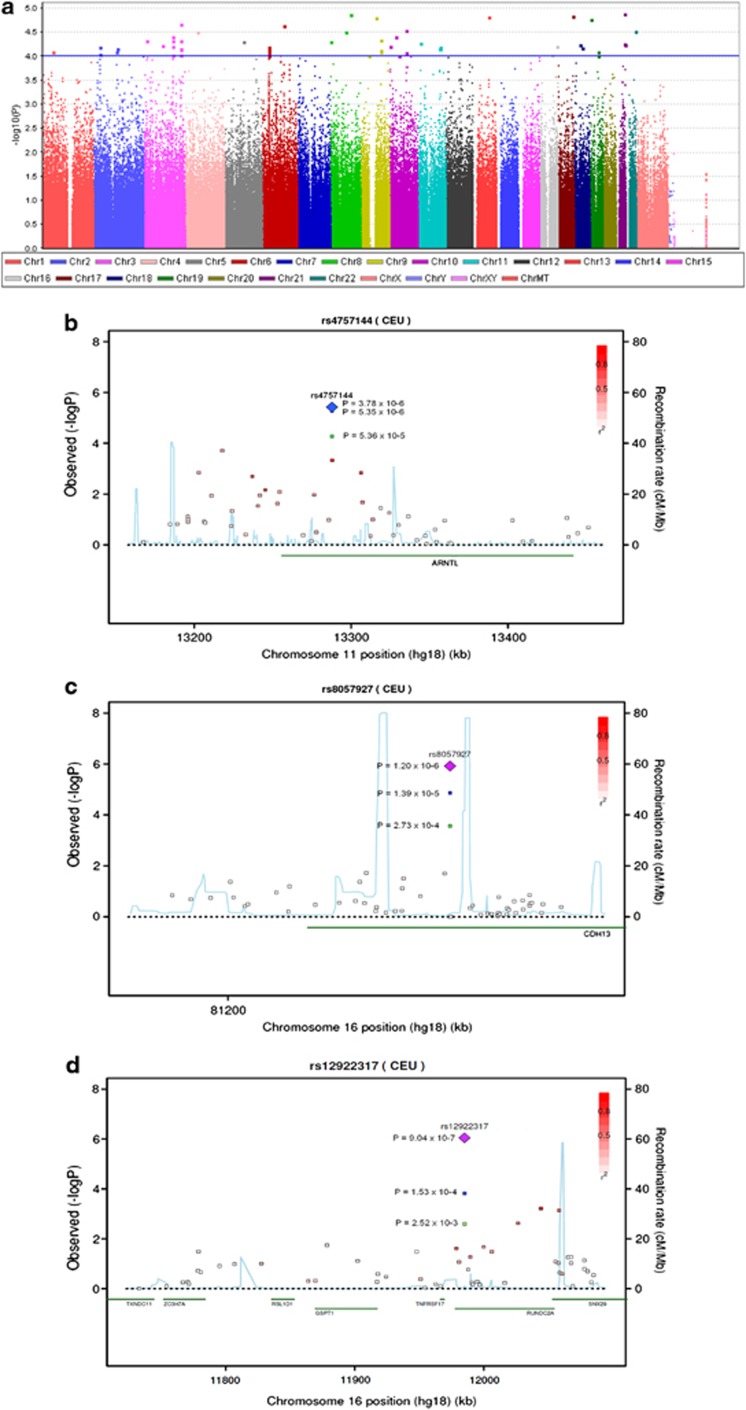
In the GWA analysis 26,863 SNPs demonstrated an association with schizophrenia with a P-value <0.05 (Figure 1a). In all, 54 SNPs showed P-values <1 × 10−4, and the SNP demonstrating the strongest association with schizophrenia was rs2836518 (P=1.32 × 10−5), located on chromosome 21q22 in the intron of ERG (list of all SNPs with P-values <1 × 10−4 can be found in Supplementary Table S5).
Figure 1.
(a) Manhattan plot of genome-wide association (GWA) analysis. The blue line indicates P=1 × 10−4. (b) Regional association plot of rs4757144 located in ARNTL. (c) Regional association plot of rs8057927 located in CDH13. (d) Regional association plot of rs12922317 located in RUNDC2A. The P-values in green are from the GWA analysis, P-values marked in blue are from the combined analysis of Danish individuals, P-values marked in purple are from the extended meta-analysis. The linkage disequilibrium (LD; r2) between the SNP in focus and its flanking markers genotyped in the GWA study are demonstrated in red (high LD) to white (low LD). The recombination rate is plotted in blue according to HapMap (CEU).
In the stage 2 sample, 165 SNPs were successfully genotyped, of which nine demonstrated a directionally consistent association at nominal significance (Table 1). At the experiment level, the associations of the SNPs genotyped in stage 2 demonstrated significant directional consistent evidence of replication (binomial sign test P<0.0096). The SNP showing the strongest association in the stage 2 sample was rs4757144 (P=0.0059) located on chromosome 11 in the intron of ARNTL on chromosome 11p15.
Table 1. P-values from association analysis of stage 1, stage 2 and German-Dutch samples and combined analysis of stage 1+stage 2 and stage 1+stage 2+German Dutch samples. The odds ratio (OR) is given and minor allele frequency (MAF) in cases and controls is also given.
|
Stage 1 (GWA sample) |
Stage 2 (follow-up sample) |
Meta-analysis stage 1+stage 2 |
German-Dutch replication sample |
Extended meta-analysis stage 1+stage 2+German-Dutch |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
MAF |
MAF |
MAF |
||||||||||||||||||
| SNP | Chr | Nearest gene | SNP type | Allelea | Cases | Controls | OR | P | Cases | Controls | OR | P | OR | P | Cases | Controls | OR | P | OR | P |
| rs4757144 | 11 | ARNTL | Intron | G/A | 0.435 | 0.365 | 1.324 | 5.36 × 10−5 | 0.427 | 0.392 | 1.155 | 5.90 × 10−3 | 1.214 | 3.78 × 10−6 | 0.438 | 0.418 | 1.081 | 1.03 × 10−1 | 1.154 | 5.35 × 10−6 |
| rs8057927 | 16 | CDH13 | Intron | C/T | 0.086 | 0.054 | 1.651 | 2.73 × 10−4 | 0.074 | 0.057 | 1.324 | 7.13 × 10−3 | 1.435 | 1.39 × 10−5 | 0.084 | 0.069 | 1.244 | 1.38 × 10−2 | 1.342 | 1.20 × 10−6 |
| rs2121783 | 3 | FOXP1 | Intron | C/T | 0.431 | 0.366 | 1.269 | 3.93 × 10−4 | 0.429 | 0.402 | 1.122 | 2.81 × 10−2 | 1.175 | 8.86 × 10−5 | 0.397 | 0.411 | 0.951 | 3.04 × 10−1 | 1.077 | 1.93 × 10−2 |
| rs3123688 | 10 | ZEB1 | Upstream | A/C | 0.018 | 0.004 | 3.915 | 1.71 × 10−3 | 0.009 | 0.004 | 2.334 | 1.16 × 10−2 | 2.831 | 9.05 × 10−5 | NA | NA | NA | NA | NA | NA |
| rs12741964 | 1 | ST3GAL3 | Intron | A/G | 0.155 | 0.199 | 0.737 | 7.22 × 10−4 | 0.168 | 0.191 | 0.858 | 2.09 × 10−2 | 0.813 | 1.12 × 10−4 | 0.163 | 0.176 | 0.911 | 1.44 × 10−1 | 0.852 | 9.56 × 10−5 |
| rs10828623 | 10 | CACNB2 | Intron | T/C | 0.156 | 0.199 | 0.749 | 1.23 × 10−3 | 0.163 | 0.186 | 0.851 | 1.90 × 10−2 | 0.812 | 1.29 × 10−4 | 0.153 | 0.163 | 0.928 | 2.54 × 10−1 | 0.857 | 2.36 × 10−4 |
| rs12922317 | 16 | RUNDC2A | Intron | G/A | 0.364 | 0.313 | 1.244 | 2.52 × 10−3 | 0.366 | 0.336 | 1.144 | 1.37 × 10−2 | 1.179 | 1.53 × 10−4 | 0.374 | 0.339 | 1.167 | 1.71 × 10−3 | 1.174 | 9.04 × 10−7 |
| rs6878606 | 5 | STC2 | Upstream | G/A | 0.354 | 0.303 | 1.253 | 1.54 × 10−3 | 0.335 | 0.309 | 1.125 | 3.34 × 10−2 | 1.172 | 2.87 × 10−4 | 0.316 | 0.292 | 1.122 | 2.55 × 10−2 | 1.151 | 2.58 × 10−5 |
| rs10741058 | 10 | CACNB2 | Intron | T/C | 0.121 | 0.156 | 0.745 | 3.02 × 10−3 | 0.118 | 0.138 | 0.842 | 2.64 × 10−2 | 0.804 | 3.50 × 10−4 | 0.117 | 0.126 | 0.923 | 2.82 × 10−1 | 0.850 | 5.76 × 10−4 |
Chr, chromosome; GWA, genome-wide association; NA, not available due to missing or failed genotyping; SNP, single nucleotide polymorphism.
Minor/major.
In the combined analysis of stage 1 and stage 2, four SNPs showed P-values <1 × 10−4. The SNP demonstrating the strongest association in this analysis was also the ARNTL SNP rs4757144 (P=3.78 × 10−6; Figure 1b). The other three markers were rs8057927 located in the intron of CDH13 on chromosome 16q23 (P=1.39 × 10−5; Figure 1c), rs2121783 located in the intron of FOXP1 on 3p13 (P=8.86 × 10−5) and rs3123688 on 10p11 located between ZEB1 and ZNF438 upstream transcription start for both genes (P=9.05 × 10−5; Table 1).
In the extended meta-analysis, including both the Danish and German-Dutch individuals, four SNPs demonstrated P-values <1 × 10−5: rs12922317 located in the intron of RUNDC2 on 16p13 (P=9.04 × 10−7; Figure 1d), rs8057927 located in the intron of CDH13 on 16q23 (P=1.20 × 10−6), rs6485671 located upstream CREB3L1 on 11p11 (P=5.08 × 10−6) and rs4757144 in the intron of ARNTL (P=5.35 × 10−6; Table 1). Additional results of the combined analyses can be found in Supplementary Table S6.
Region-wise association analysis
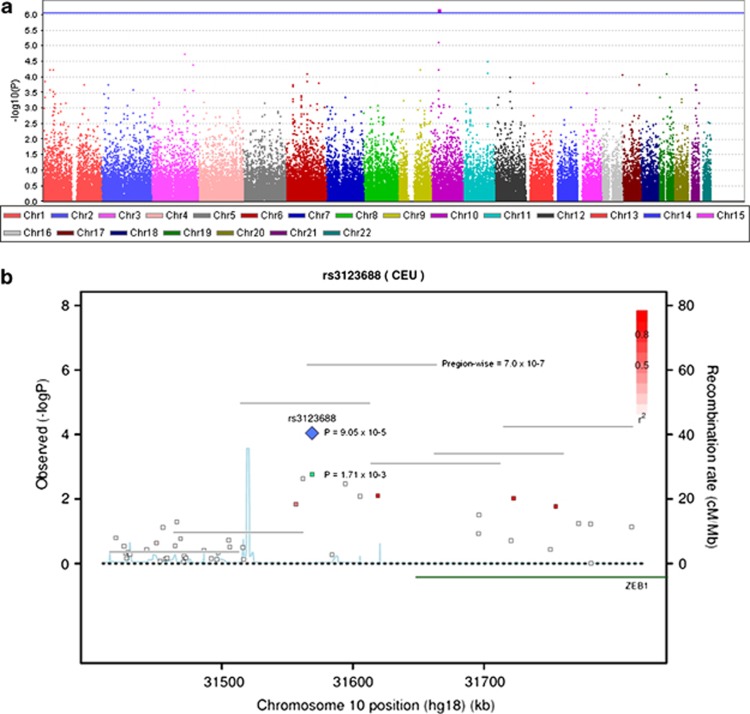
In total, 55,561 overlapping regions were tested for association with schizophrenia (Figure 2), requiring a region-based genome-wide significant level of P=9.6 × 10−7 if Bonferroni correction is applied. One region at chromosome 10p11:31,566,070–31,666,070 (hg18) overlapping ZEB1 was genome-wide significant (P=7.0 × 10−7; Figure 2, Supplementary Table S7). Five genotyped SNPs were located in this region (rs1314004, rs7083727, rs1314013, rs12242798 and rs3123688). Two of the SNPs are in high linkage disequilibrium (rs3123688 and rs12242798, r2=0.642374). The five SNPs were genotyped in the stage 2 sample. However, rs12242798 failed genotyping and was therefore imputed using the software MaCH 1.0 (http://www.sph.umich.edu/csg/abecasis/MACH/)44 with HapMap phase III, Release #2, CEU, as reference population. The resulting genotypes were imputed with good quality (quality=0.99, Rsq=0.34). Significant region-wise association was found in the stage 2 sample (P=0.023), thereby establishing formal replication of this locus in the Danish population.
Figure 2.
(a) Manhattan plot of region-wise association analysis. The blue line indicates genome-wide significance (P=9.0 × 10−7). (b) Regional association plot of rs3123688 located upstream of ZEB1. The P-values in green are from the genome-wide association (GWA) analysis and P-values marked in blue are from the combined analysis of Danish individuals. The vertical lines represent the region-wise P-values. The linkage disequilibrium (LD; r2) between the single nucleotide polymorphism in focus and its flanking markers genotyped in the GWA study are demonstrated in red (high LD) to white (low LD). The recombination rate is plotted in blue according to HapMap (CEU).
SNP × maternal CMV infection interaction analysis
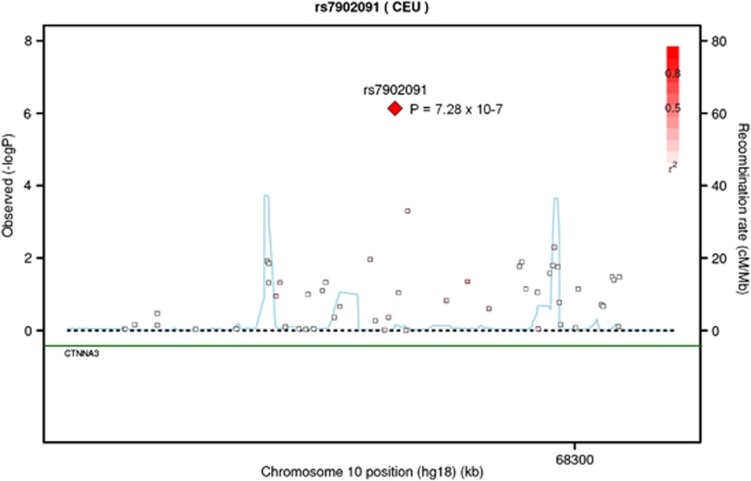
Of the case mothers, 73.2% were CMV positive while that was the case for 70.7% of the control mothers, corresponding to an OR of 1.13 (0.85−1.50), P=0.39, for CMV with respect to schizophrenia in the offspring. A total of 29,082 SNPs passed step 1 inducing a Bonferroni significance level of P=1.72 × 10−6 at step 2. A single SNP, rs7902091 (MAF 0.16 in cases and 0.15 in controls) located in an intron of CTNNA3 on chromosome 10q21, demonstrated experiment-wide significant interaction with maternal CMV infection, with an interaction P-value of 7.3 × 10−7 and interaction OR of 5.3 under an additive genetic model (Figure 3, Supplementary Table S8). On its own, rs7902091 showed no association with schizophrenia (OR=1.04, P=0.67). For non-carriers of the minor allele, the risk of schizophrenia from maternal CMV was not observed (OR=0.72, P=0.11) whereas for carriers the risk increased to OR=5.0 (P=3.8 × 10−2). Furthermore, a neighboring SNP, rs7919083 located 2206 bp from rs7902091, demonstrated a relatively low interaction P-value (P=5 × 10−4) practically independent of rs7902091 (r2=0.08).
Figure 3.
Regional association plot of rs7902091. P-values of interaction between single nucleotide polymorphisms (SNPs) and maternal cytomegalovirus infection. The linkage disequilibrium (LD; r2) between the SNP in focus and its flanking markers genotyped in the genome-wide association study are demonstrated in red (high LD) to white (low LD). The recombination rate is plotted in blue according to HapMap (CEU).
Discussion
Here we report the results of a GWA study of schizophrenia using cases from a complete Danish birth cohort and follow-up investigations in additional samples, applying single variant and regional analyses, the latter identifying a novel locus at ZEB1. Moreover, conducting the first genome-wide gene-environment interaction survey in psychiatric disorders, we report significant interaction between CTNNA3 and maternal CMV infection.
In the single variant analysis, none of the analyzed SNPs passed the widely accepted genome-wide significance threshold of P=5 × 10−8. However, the results highlighted a number of loci with the strongest signals located on 10p11, 11p15, 16q23 and 16p13 (Table 1). The SNP rs4757144, located in an intron of the circadian rhythm-associated gene ARNTL on 11p15, demonstrated the strongest association in the combined analysis of Danish stage 1 and stage 2 individuals and was the fourth most associated SNP in the extended meta-analysis. ARNTL is expressed in several regions of the human brain.45 Circadian-rhythm abnormalities in schizophrenia patients have been reported.46, 47, 48, 49 Several candidate gene studies have investigated the involvement of circadian genes in schizophrenia and other psychiatric disorders, with some suggesting the involvement of ARNTL in disease risk50, 51, 52 and others not.53 The SNP rs4757144 reported in this study is in high linkage disequilibrium (r2>0.8, HapMap release 22), with two SNPs previously reported to be associated with schizophrenia (rs198235051) and bipolar disorder (rs4757142,51 rs198235052). ARNTL was also one of the top four candidate genes associated with bipolar disorder identified by convergent functional genomics data mining of existing GWAS data sets.54
The second most associated SNP in both the combined analysis of the Danish samples and in the extended meta-analysis was rs8057927 in the intron of CDH13 on 16q23. CDH13 encodes cadherin-13, a member of the cadherin super family of molecules that mediates Ca2+-dependent cell–cell adhesion in solid tissue.55, 56, 57 CDH13 is expressed in several parts of the adult human brain58 and appears to have a negative role in neural cell proliferation in the developing nervous system.58, 59, 60 The implication of CDH13 in other psychiatric disorders has been suggested. GWA studies of attention deficit/hyperactivity disorder identified CDH13 as one of the most associated genes,61, 62, 63, 64 and a meta-analysis of attention deficit/hyperactivity disorder linkage scans identified the region with CDH13 as the only genome-wide significant.65 GWA studies have also indicated the involvement of CDH13 in depression66 and autism,67 and a recent study implicated CNVs encompassing CDH13 in autism susceptibility.68 The important role of cadherin-13 during brain development and in maintaining neural circuitry together with the reports of involvement of CDH13 in other psychiatric disorders therefore support our result, which, for the first time, suggests the involvement of CDH13 in schizophrenia (discussion of other top hits from the extended meta-analysis can be found in the Supplementary Material).
One SNP (rs10828623) out of the 10 most associated SNPs in the combined analysis of Danish stage 1 and stage 2 individuals was nominal significantly associated with schizophrenia in the data from the Psychiatric Genomics Consortium (PGC; P=0.002),11 resulting in a P-value=4.54 × 10−6 in the combined analysis of Danish stage 1 and 2 individuals and the PGC samples. The German-Dutch sample was included in the PGC data. Thus, there was no overlap in the discovery and follow-up samples. The limited replication could be due to genetic heterogeneity between the Danish and PGC samples, reducing the power to detect variants with small effects. The single marker loci demonstrating the strongest association in this study are therefore only valid for Danish, German and Dutch populations.
Applying a regional analysis, summarizing independent signals in relatively small segments of overlapping regions of 100 kb, we found region-based genome-wide significant association at a region on 10p11 containing ZEB1. The applied significance level was based on Bonferroni correction of the total number of analyzed regions, which is analogous to how the conventional GWAS threshold for single-SNP association of 5 × 10−8 is deduced but which in this case is conservative due to the regions being 50% overlapping and therefore far from independent. This approach was able to identify a novel risk locus even though it was performed using a small sample compared with recent GWA studies, indicating that aggregating P-values in this fashion can be a powerful approach. This is supported by the accumulating observations of independent association signals from closely positioned SNPs (see for example, Steinberg et al.4 and Ripke et al.11). Moreover, the region showed significant association in the Danish stage 2 sample, providing independent replication of this locus in the Danish population. No replication was attempted in the German-Dutch GWA data set or the PGC data as not all SNPs (or proxies) in this region were present in these data sets. ZEB1 encodes an E-box binding zinc finger transcription factor, which is widely expressed in the central nervous system and has an important role in development of the brain69 and neuronal differentiation.70 The associated region includes the promoter of ZEB1 and could therefore be involved in or linked to variants involved in regulation of expression. This is intriguing as it has been demonstrated that the expression of ZEB1 is regulated by another transcription factor protein, TCF4,71 which is one of the best validated schizophrenia susceptibility genes. Two independent SNPs in this gene have passed the threshold for genome-wide significant association,4, 5, 9 and several studies have found TCF4 SNPs demonstrating close to genome-wide significance.5, 11, 12 Notably, studies have also found strong evidence for the involvement of both TCF4 and ZEB1 genetic variants in another disorder, namely Fuch's corneal dystrophy.72, 73 Interestingly, ZEB1 is involved in regulation of cadherin-13 expression by physically binding to the E2-box in the promotor region of CDH13 decreasing the expression of the gene;74 however, this finding still needs to be confirmed in nerve cells. Our results together with previous findings could therefore indicate that ZEB1, TCF4 and cadherin-13 are elements of a common pathway involved in schizophrenia.
The interaction analysis of SNPs with maternal CMV infection found a significant interaction at CTNNA3, using an efficient two-step method where only SNPs passing step 1 were tested for interaction.43 Thus, only SNPs showing nominal association with CMV infection in the pooled sample of cases and controls were tested for interaction. This amounted to around 29,000 SNPs distributed across the genome. The interacting SNP was rs7902091 located in an intron of CTNNA3, just upstream the gene LRRTM3, which is nested within CTNNA3. CTNNA3 encodes catenin alpha-3, which is predominantly expressed in heart and testis but expression of the gene in the brain has also been demonstrated.75 Catenin alpha-3 mediates cell–cell adhesion by functioning as a link between cadherin-based cell–cell adhesion complexes and the cytoskeleton.76, 77 Biologically the interaction of rs7902091 in CTNNA3 with maternal CMV makes sense, because CMV during infection may disrupt cell-to-cell connections by disconnecting the cadherin–catenin–actin complex within endothelial cells,78 and in a study of human CMV in transgenic Drosophila, expression of the regulatory virus genes caused abnormal embryonic development by interfering with cell-to-cell adherens junctions through an effect on catenins.79 The interaction observed suggests that the region around rs7902091 in concert with maternal CMV infection may have a role in the etiology of schizophrenia. However, this should be replicated in additional studies. It is noteworthy, though, that neighboring SNPs (in particular rs7919083) showed a low interaction P-value independently of rs7902091, supporting the involvement of this locus. CTNNA3 and its nested gene LRRTM3 (encoding the Leucine-rich repeat transmembrane neuronal protein 3) have both previously been found associated to Alzheimer's disease80, 81, 82, 83, 84 and with autism spectrum disorder.67, 85 In relation to Alzheimer's disease, CTNNA3 have been observed to have stronger effect in females than in males.82 We therefore performed a secondary analysis of gender differences in the interaction of CTNNA3 and CMV. The results are shown in the Supplementary Material (Supplementary Table S3).
As in any other observational study involving environmental factors, confounding cannot be ruled out. With the apparent risk from CMV being turned on and off by the presence or absence of the variant, any confounder of CMV–schizophrenia association would confound the interaction result. For instance, the prevalence of CMV infection has been reported to correlate with the prevalence of other infections, including other members of the Herpes family,86 and social-economic status87 that could be potential confounders. However, regardless of whether the CMV–schizophrenia association can be explained by confounding in part or completely, there is still interaction at the CTNNA3 locus identifying sub-populations of different risk profiles for schizophrenia.
We have reported the first GWA study and follow-up analysis of all the Danish individuals born since 1981 and diagnosed with schizophrenia up to 2010 and controls from the same birth cohort. Furthermore, we have followed up in an additional sample from a genetically related population. The results support the findings from other GWA studies, suggesting the involvement of many common variants each contributing only slightly to disease risk. Applying a region-wise analysis, a new risk locus (at ZEB1) was identified and replicated. Several other plausible susceptibility loci were also suggested. This is also the first genome-wide study analyzing how maternal CMV infection interacts with the genotype of the progeny affecting the risk of schizophrenia, identifying a significant interaction at CTNNA3, a gene not previously implicated with schizophrenia. The result stresses the importance of including environmental factors in the evaluation of disease risk. Moreover, this is, to our knowledge, the first significant gene–environment interaction identified in a genome-wide survey of a psychiatric disorder. Future studies should confirm the associations of the genomic regions with schizophrenia, demonstrating the strongest signals in this study, as well as enlarge, the inclusion of environmental factors when identifying genetic risk variants. The unique samples from the DNSB together with information from Danish register systems makes it possible to perform genetic studies with inclusion of a wealth of potential environmental risk factors. Future studies of the Danish population could therefore provide valuable insight into how gene–environment interactions influence the risk of schizophrenia.
Acknowledgments
The study was supported by grants from The Danish Strategic Research Council, H. Lundbeck A/S, The Faculty of Health Sciences at Aarhus University, Lundbeck Foundation, The Stanley Research Foundation and The Villum Kann Rasmussen Foundation. The German SCZ GWAS sample was supported by the German Federal Ministry of Education and Research (BMBF), within the context of the National Genome Research Network 2 (NGFN-2), the National Genome Research Network plus (NGFNplus) and the Integrated Genome Research Network (IG) MooDS (Grant 01GS08144 to SC and MMN, Grant 01GS08147 to MR). The Dutch GWAS sample from Utrecht was sponsored by NIMH funding, R01 MH078075.
Author contributions
Direction of study: ADB. Overall idea and strategy: ADB, OM and PBM. Collection and ascertainment of Danish study subjects: OM, MN, PBM, CBP and ADB. Sample preparation, laboratory experiments and genotyping: DD, MH, DMH, MN, AH, TØ, CW, MD and RHY. Statistical analyses: DD, JG, JP and ADB. German-Dutch GWA data: SC, MM, MMN, MR, GROUP, AU and RAO. Writing of manuscript: DD and ADB. Commenting and editing manuscript: All authors.
Appendix
Genetic Risk and Outcome in Psychosis (GROUP Investigators): René S Kahn1, Don H Linszen2, Jim van Os3, DurkbWiersma4, Richard Bruggeman4, Wiepke Cahn1, Lieuwe de Haan2, Lydia Krabbendam3, Inez Myin-Germeys3.
1Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, University Medical Center Utrecht, Utrecht, The Netherlands; 2Department of Psychiatry, Academic Medical Center University of Amsterdam, Amsterdam, The Netherlands; 3Maastricht University Medical Center, South Limburg Mental Health Research and Teaching Network, Maastricht, The Netherlands; 4Department of Psychiatry, University Medical Center Groningen, University of Groningen, Groningen, The Netherlands.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- van Os J, Rutten BP, Poulton R. Gene-environment interactions in schizophrenia: review of epidemiological findings and future directions. Schizophr Bull. 2008;34:1066–1082. doi: 10.1093/schbul/sbn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiu L, Mattingsdal M, Kahler AK, Brown A, Gustafsson O, Agartz I, et al. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res. 2010;44:748–753. doi: 10.1016/j.jpsychires.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S, de Jong S, Andreassen OA, Werge T, Borglum AD, Mors O, et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet. 2011;20:4076–4081. doi: 10.1093/hmg/ddr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel M, Mattheisen M, Degenhardt F, Kahn RS, Linszen DH, Os JV, et al. Association between genetic variation in a region on chromosome 11 and schizophrenia in large samples from Europe. Mol Psychiatry. 2012;17:906–917. doi: 10.1038/mp.2011.80. [DOI] [PubMed] [Google Scholar]
- Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyegaard M, Severinsen JE, Als TD, Hedemand A, Straarup S, Nordentoft M, et al. Support of association between BRD1 and both schizophrenia and bipolar affective disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:582–591. doi: 10.1002/ajmg.b.31023. [DOI] [PubMed] [Google Scholar]
- Holmans PA, Riley B, Pulver AE, Owen MJ, Wildenauer DB, Gejman PV, et al. Genomewide linkage scan of schizophrenia in a large multicenter pedigree sample using single nucleotide polymorphisms. Mol Psychiatry. 2009;14:786–795. doi: 10.1038/mp.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, DeCandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buizer-Voskamp JE, Muntjewerff JW, Strengman E, Sabatti C, Stefansson H, Vorstman JA, et al. Genome-wide analysis shows increased frequency of copy number variation deletions in dutch schizophrenia patients. Biol Psychiatry. 2011;70:655–662. doi: 10.1016/j.biopsych.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Mulle JG, Kaminsky EB, Sanders SJ, Myers SM, Adam MP, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens LN, De Rijk P, Reumers J, Van Den Bossche MJ, Glassee W, De Zutter S, et al. Sequencing of DISC1 pathway genes reveals increased burden of rare missense variants in schizophrenia patients from a Northern Swedish population. PLoS One. 2011;6:e23450. doi: 10.1371/journal.pone.0023450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- Crow TJ. ‘The missing genes: what happened to the heritability of psychiatric disorders?'. Mol Psychiatry. 2011;16:362–364. doi: 10.1038/mp.2010.92. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Buka SL, Cannon TD, Torrey EF, Yolken RH. Maternal exposure to herpes simplex virus and risk of psychosis among adult offspring. Biol Psychiatry. 2008;63:809–815. doi: 10.1016/j.biopsych.2007.09.022. [DOI] [PubMed] [Google Scholar]
- Mortensen PB, Pedersen CB, Hougaard DM, Norgaard-Petersen B, Mors O, Borglum AD, et al. A Danish National Birth Cohort study of maternal HSV-2 antibodies as a risk factor for schizophrenia in their offspring. Schizophr Res. 2010;122:257–263. doi: 10.1016/j.schres.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Brown AS, Begg MD, Gravenstein S, Schaefer CA, Wyatt RJ, Bresnahan M, et al. Serologic evidence of prenatal influenza in the etiology of schizophrenia. Arch Gen Psychiatry. 2004;61:774–780. doi: 10.1001/archpsyc.61.8.774. [DOI] [PubMed] [Google Scholar]
- Brown AS, Schaefer CA, Quesenberry CP, Liu L, Babulas VP, Susser ES. Maternal exposure to toxoplasmosis and risk of schizophrenia in adult offspring. Am J Psychiatry. 2005;162:767–773. doi: 10.1176/appi.ajp.162.4.767. [DOI] [PubMed] [Google Scholar]
- Demontis D, Nyegaard M, Buttenschøn HN, Hedemand A, Pedersen CP, Grove J, et al. Association of GRIN1 and GRIN2A-D with schizophrenia and genetic interaction with maternal herpes simplex virus-2 infection affecting disease risk. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:913–922. doi: 10.1002/ajmg.b.31234. [DOI] [PubMed] [Google Scholar]
- Nørgaard-Pedersen B, Hougaard DM. Storage policies and use of the Danish Newborn Screening Biobank. J Inherit Metab Dis. 2007;30:530–536. doi: 10.1007/s10545-007-0631-x. [DOI] [PubMed] [Google Scholar]
- Mors O, Perto GP, Mortensen PB. The Danish Psychiatric Central Research Register. Scand J Public Health. 2011;39:54–57. doi: 10.1177/1403494810395825. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Shirts BH, Dayal M, Bacanu SA, Wood J, Xie W, et al. Are exposure to cytomegalovirus and genetic variation on chromosome 6p joint risk factors for schizophrenia. Ann Med. 2007;39:145–153. doi: 10.1080/07853890601083808. [DOI] [PubMed] [Google Scholar]
- Shirts BH, Kim JJ, Reich S, Dickerson FB, Yolken RH, Devlin B, et al. Polymorphisms in MICB are associated with human herpes virus seropositivity and schizophrenia risk. Schizophr Res. 2007;94:342–353. doi: 10.1016/j.schres.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen CB, Gotzsche H, Moller JO, Mortensen PB. The Danish Civil Registration System. A cohort of eight million persons. Dan Med Bull. 2006;53:441–449. [PubMed] [Google Scholar]
- Hollegaard MV, Grauholm J, Borglum A, Nyegaard M, Norgaard-Pedersen B, Orntoft T, et al. Genome-wide scans using archived neonatal dried blood spot samples. BMC Genomics. 2009;10:297. doi: 10.1186/1471-2164-10-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- Buka SL, Cannon TD, Torrey EF, Yolken RH, group TCS Maternal exposure to herpes simplex virus and risk of psychosis among adult offspring. Biol Psychiatry. 2008;63:809–815. doi: 10.1016/j.biopsych.2007.09.022. [DOI] [PubMed] [Google Scholar]
- Schelonka RL, Infante AJ. Neonatal immunology. Semin Perinatol. 1998;22:2–14. doi: 10.1016/s0146-0005(98)80003-7. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Hengel H. Epidemiological impact and disease burden of congenital cytomegalovirus infection in Europe. Euro Surveill. 2009;14:26–32. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AR.Statistical Methods for Research Workers13th edn,Oliver and Boyd: London; 1925 [Google Scholar]
- Murcray CE, Lewinger JP, Gauderman WJ. Gene-environment interaction in genome-wide association studies. Am J Epidemiol. 2009;169:219–226. doi: 10.1093/aje/kwn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Abecasis GR. Mach 1.0: rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- Cermakian N, Lamont EW, Boudreau P, Boivin DB. Circadian clock gene expression in brain regions of Alzheimer 's disease patients and control subjects. J Biol Rhythms. 2011;26:160–170. doi: 10.1177/0748730410395732. [DOI] [PubMed] [Google Scholar]
- Wulff K, Joyce E, Middleton B, Dijk DJ, Foster RG. The suitability of actigraphy, diary data, and urinary melatonin profiles for quantitative assessment of sleep disturbances in schizophrenia: a case report. Chronobiol Int. 2006;23:485–495. doi: 10.1080/07420520500545987. [DOI] [PubMed] [Google Scholar]
- Keshavan MS, Reynolds CF, Kupfer DJ. Electroencephalographic sleep in schizophrenia: a critical review. Compr Psychiatry. 1990;31:34–47. doi: 10.1016/0010-440x(90)90052-t. [DOI] [PubMed] [Google Scholar]
- Boivin DB. Influence of sleep-wake and circadian rhythm disturbances in psychiatric disorders. J Psychiatry Neurosci. 2000;25:446–458. [PMC free article] [PubMed] [Google Scholar]
- Rao ML, Gross G, Strebel B, Halaris A, Huber G, Braunig P, et al. Circadian rhythm of tryptophan, serotonin, melatonin, and pituitary hormones in schizophrenia. Biol Psychiatry. 1994;35:151–163. doi: 10.1016/0006-3223(94)91147-9. [DOI] [PubMed] [Google Scholar]
- Nievergelt CM, Kripke DF, Barrett TB, Burg E, Remick RA, Sadovnick AD, et al. Suggestive evidence for association of the circadian genes PERIOD3 and ARNTL with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:234–241. doi: 10.1002/ajmg.b.30252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour HA, Wood J, Logue T, Chowdari KV, Dayal M, Kupfer DJ, et al. Association study of eight circadian genes with bipolar I disorder, schizoaffective disorder and schizophrenia. Genes Brain Behav. 2006;5:150–157. doi: 10.1111/j.1601-183X.2005.00147.x. [DOI] [PubMed] [Google Scholar]
- Soria V, Martinez-Amoros E, Escaramis G, Valero J, Perez-Egea R, Garcia C, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology. 2010;35:1279–1289. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour HA, Talkowski ME, Wood J, Chowdari KV, McClain L, Prasad K, et al. Association study of 21 circadian genes with bipolar I disorder, schizoaffective disorder, and schizophrenia. Bipolar Disord. 2009;11:701–710. doi: 10.1111/j.1399-5618.2009.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Misaki A, Liang SB, Tachibana A, Hayashi N, Sonobe H, et al. Expression of T-cadherin (CDH13, H-Cadherin) in human brain and its characteristics as a negative growth regulator of epidermal growth factor in neuroblastoma cells. J Neurochem. 2000;74:1489–1497. doi: 10.1046/j.1471-4159.2000.0741489.x. [DOI] [PubMed] [Google Scholar]
- Fredette BJ, Miller J, Ranscht B. Inhibition of motor axon growth by T-cadherin substrata. Development. 1996;122:3163–3171. doi: 10.1242/dev.122.10.3163. [DOI] [PubMed] [Google Scholar]
- Ciatto C, Bahna F, Zampieri N, VanSteenhouse HC, Katsamba PS, Ahlsen G, et al. T-cadherin structures reveal a novel adhesive binding mechanism. Nat Struct Mol Biol. 2010;17:339–347. doi: 10.1038/nsmb.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky-Su J, Neale BM, Franke B, Anney RJ, Zhou K, Maller JB, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- Neale BM, Lasky-Su J, Anney R, Franke B, Zhou K, Maller JB, et al. Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1337–1344. doi: 10.1002/ajmg.b.30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Medland S, Ripke S, Anney RJ, Asherson P, Buitelaar J, et al. Case-control genome-wide association study of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:906–920. doi: 10.1016/j.jaac.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Dempfle A, Arcos-Burgos M, Bakker SC, Banaschewski T, Biederman J, et al. Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1392–1398. doi: 10.1002/ajmg.b.30878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, et al. Genome-wide association scan of trait depression. Biol Psychiatry. 2010;68:811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Maruhashi M, Van De Putte T, Kondoh H, Huylebroeck D, Higashi Y. Complementary expression pattern of Zfhx1 genes Sip1 and deltaEF1 in the mouse embryo and their genetic interaction revealed by compound mutants. Dev Dyn. 2006;235:1941–1952. doi: 10.1002/dvdy.20799. [DOI] [PubMed] [Google Scholar]
- Ravanpay AC, Hansen SJ, Olson JM. Transcriptional inhibition of REST by NeuroD2 during neuronal differentiation. Mol Cell Neurosci. 2010;44:178–189. doi: 10.1016/j.mcn.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrado VR, Moreno-Bueno G, Cubillo E, Holt LJ, Nieto MA, Portillo F, et al. The class I bHLH factors E2-2A and E2-2B regulate EMT. J Cell Sci. 2009;122:1014–1024. doi: 10.1242/jcs.028241. [DOI] [PubMed] [Google Scholar]
- Baratz KH, Tosakulwong N, Ryu E, Brown WL, Branham K, Chen W, et al. E2-2 protein and Fuchs's corneal dystrophy. N Engl J Med. 2010;363:1016–1024. doi: 10.1056/NEJMoa1007064. [DOI] [PubMed] [Google Scholar]
- Riazuddin SA, Zaghloul NA, Al-Saif A, Davey L, Diplas BH, Meadows DN, et al. Missense mutations in TCF8 cause late-onset Fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am J Hum Genet. 2010;86:45–53. doi: 10.1016/j.ajhg.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y, Takeuchi T, Nagayama T, Ohtsuki Y, Furihata M. Zeb1-mediated T-cadherin repression increases the invasive potential of gallbladder cancer. FEBS Lett. 2009;583:430–436. doi: 10.1016/j.febslet.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Busby V, Goossens S, Nowotny P, Hamilton G, Smemo S, Harold D, et al. Alpha-T-catenin is expressed in human brain and interacts with the Wnt signaling pathway but is not responsible for linkage to chromosome 10 in Alzheimer's disease. Neuromolecular Med. 2004;5:133–146. doi: 10.1385/NMM:5:2:133. [DOI] [PubMed] [Google Scholar]
- Yap AS, Brieher WM, Gumbiner BM. Molecular and functional analysis of cadherin-based adherens junctions. Annu Rev Cell Dev Biol. 1997;13:119–146. doi: 10.1146/annurev.cellbio.13.1.119. [DOI] [PubMed] [Google Scholar]
- Janssens B, Goossens S, Staes K, Gilbert B, van Hengel J, Colpaert C, et al. alphaT-catenin: a novel tissue-specific beta-catenin-binding protein mediating strong cell-cell adhesion. J Cell Sci. 2001;114:3177–3188. doi: 10.1242/jcs.114.17.3177. [DOI] [PubMed] [Google Scholar]
- Scholz M, Blaheta RA, Vogel J, Doerr HW, Cinatl J. Cytomegalovirus-induced transendothelial cell migration. a closer look at intercellular communication mechanisms. Intervirology. 1999;42:350–356. doi: 10.1159/000053971. [DOI] [PubMed] [Google Scholar]
- Steinberg R, Shemer-Avni Y, Adler N, Neuman-Silberberg S. Human cytomegalovirus immediate-early-gene expression disrupts embryogenesis in transgenic Drosophila. Transgenic Res. 2008;17:105–119. doi: 10.1007/s11248-007-9136-5. [DOI] [PubMed] [Google Scholar]
- Majercak J, Ray WJ, Espeseth A, Simon A, Shi XP, Wolffe C, et al. LRRTM3 promotes processing of amyloid-precursor protein by BACE1 and is a positional candidate gene for late-onset Alzheimer's disease. Proc Natl Acad Sci USA. 2006;103:17967–17972. doi: 10.1073/pnas.0605461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Martin ER, Schnetz-Boutaud N, Bartlett J, Anderson B, Zuchner S, et al. Effect of heterogeneity on the chromosome 10 risk in late-onset Alzheimer disease. Hum Mutat. 2007;28:1065–1073. doi: 10.1002/humu.20567. [DOI] [PubMed] [Google Scholar]
- Miyashita A, Arai H, Asada T, Imagawa M, Matsubara E, Shoji M, et al. Genetic association of CTNNA3 with late-onset Alzheimer's disease in females. Hum Mol Genet. 2007;16:2854–2869. doi: 10.1093/hmg/ddm244. [DOI] [PubMed] [Google Scholar]
- Morgan AR, Hamilton G, Turic D, Jehu L, Harold D, Abraham R, et al. Association analysis of 528 intra-genic SNPs in a region of chromosome 10 linked to late onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:727–731. doi: 10.1002/ajmg.b.30670. [DOI] [PubMed] [Google Scholar]
- Edwards TL, Pericak-Vance M, Gilbert JR, Haines JL, Martin ER, Ritchie MD. An association analysis of Alzheimer disease candidate genes detects an ancestral risk haplotype clade in ACE and putative multilocus association between ACE, A2M, and LRRTM3. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:721–735. doi: 10.1002/ajmg.b.30899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa I, Clark TG, Holt R, Pagnamenta AT, Mulder EJ, Minderaa RB, et al. Polymorphisms in leucine-rich repeat genes are associated with autism spectrum disorder susceptibility in populations of European ancestry. Mol Autism. 2010;1:7. doi: 10.1186/2040-2392-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embil JA, Garner JB, Pereira LH, White FM, Manuel FR. Association of cytomegalovirus and herpes simplex virus infections of the cervix in four clinic populations. Sex Transm Dis. 1985;12:224–228. doi: 10.1097/00007435-198510000-00011. [DOI] [PubMed] [Google Scholar]
- Staras SA, Dollard SC, Radford KW, Flanders WD, Pass RF, Cannon MJ. Seroprevalence of cytomegalovirus infection in the United States, 1988–1994. Clin Infect Dis. 2006;43:1143–1151. doi: 10.1086/508173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.