Abstract
Current molecular analysis of cells and tissues routinely relies on separation, enrichment, and subsequent measurements by various assays. Here we demonstrate a platform of hyperspectral stimulated Raman scattering microscopy for fast, quantitative and label-free imaging of biomolecules in intact tissues using spectroscopic fingerprints as the contrast mechanism.
Keywords: Vibrational spectroscopy, Stimulated Raman scattering imaging, Lipids
Cholesterol is an indispensable structural constituent of membranes and also plays prominent roles in biosynthesis of steroid hormones. Excessive cholesterol storage is a hallmark of atherosclerotic cardiovascular diseases. The cholesterol crystals have not only been found in advanced atherosclerotic lesions,[1] but also are recognized as inflammatory stimuli appearing in very early stage of atherosclerosis.[1c, 1d, 2] Abnormal storage of cholesterol also occurs in steroidogenic cells[3] and Niemann-Pick type C disease.[4] However, quantitative imaging of cholesterol and its esterified form in an intact biological tissue remains a challenge. The widely applied hematoxylin & eosin histology only allows indirect mapping of cholesterol crystal clefts.[1a] Lipid-soluble Sudan dyes such as Oil Red O and Bromine-Sudan black are widely used to stain neutral lipid droplets containing cholesteryl ester and triglyceride,[5] but these dyes neither stain cholesterol crystal nor tell the composition of lipid droplets. Filipin is a gold standard for staining free cholesterol in fixed cells but is unable to label either large cholesterol crystal or cholesteryl ester containing lipid droplets.[6] Confocal laser reflection microscopy[2, 7] and micro-optical coherence tomography[8] have been successfully implemented to visualize reflective substances such as cholesterol and silica, but neither has the capability for chemical identification.
Spontaneous Raman spectroscopy is an established method for imaging cholesterol and other bio-chemicals in isolated cells or tissue samples in a label free manner,[9] but suffers from slow imaging speed and strong background auto-fluorescence. Nonlinear coherent Raman microscopy[10] based on coherent anti-Stokes Raman scattering (CARS)[11] and stimulated Raman scattering (SRS)[12] significantly improved image acquisition speed. Furthermore, Multiplex / hyperspectral CARS[13] and hyperspectral SRS[14] have been developed to resolve spectrally overlapped Raman bands and employed to map cholesterol in atherosclerotic plaques.[13e, 13g, 14a] So far, CARS and SRS imaging of cholesterol have predominantly relied on the carbon-hydrogen (C-H) stretching region where lipids exhibit strong vibrational signals. Nevertheless, quantitative identification of free and esterified cholesterol from other lipids using C-H stretching bands is difficult because these bands are highly crowded and spectrally overlapped in the spectral window from 2800 to 3100 cm−1.[14a, 15] Additionally, these overlapped C-H stretching bands, even from the same molecule, were shown to exhibit varied dependence on excitation polarization,[14a] making quantitative mapping especially difficult.
Here, we demonstrate label-free quantitative imaging of cholesterol storage in intact atherosclerotic arterial tissues by detection of fingerprint Raman bands on a hyperspectral SRS microscope (Supplementary Figure S1). In particular, we employ the sterol C=C band at 1669 cm−1 as a characteristic band of cholesterol. Other significant Raman bands in the same spectral window, including the acyl C=C band, the ester C=O band and the amide I band, were recorded simultaneously. Quantitative chemical maps of each chemical group were generated by multivariate curve resolution (MCR)[16] analysis (Supplementary Figure S2). Hyperspectral SRS imaging and multivariate analysis in the fingerprint region, as reported here, open new research opportunities by providing chemical maps of key molecules in intact cells and tissues.
We first demonstrated the feasibility of our method via SRS imaging of a mixture of cholesterol crystal, glyceryl trioleate, and bovine serum albumin (BSA) powder, for which the molecular structures are presented in Figure 1a. As indicated, each glyceryl trioleate molecule has a total of three cis C=C bonds (termed as acyl C=C bond) in the three lipid acyl chains, and each cholesterol has one C=C bond (termed as sterol C=C bond) in the ring. Importantly, these two types of C=C bonds are spectrally separated in the Raman spectrum, peaked at 1655 cm−1 for acyl C=C and 1669 cm−1 for sterol C=C, respectively (Figure 1b). In the same spectral region, BSA, a protein representative, gives a broad amide I band. Because these Raman bands have either different peak positions or exhibit different profiles, selective mapping of triglyceride, cholesterol, and protein is possible through hyperspectral SRS imaging and MCR analysis. A hyperspectral stack of 60 images at wavenumbers ranging from 1620 to 1720 cm−1 were obtained in total acquisition time of less than 40 sec (Supplementary Movie S1). The X-Y-Ω image stack was analyzed by MCR algorithm, which retrieved both spectra and concentration maps corresponding to glyceryl trioleate, cholesterol and BSA.[14f] Figure 1c shows the MCR optimized spectra for each component, which match the spontaneous Raman spectra shown in Figure 1b. The reconstructed concentration maps of glyceryl trioleate, cholesterol and BSA are presented in Figure 1e–g and the overlay image is shown in Figure 1d. These data collectively demonstrate the applicability of SRS microscopy and MCR analysis for mapping biomolecules of overlapped Raman bands.
Figure 1.
Hyperspectral SRS imaging and MCR analysis of mixture of cholesterol, triglyceride and BSA. a) Chemical structure of glyceryl trioleate, Cholesterol and BSA. Acyl C=C bond, sterol C=C bond, and amide I group are indicated. b) Spontaneous Raman spectrum for each chemical group in the spectral window from 1620 to 1720 cm−1. c) Output spectra from hyperspectral SRS imaging and MCR analysis. d) Color overlay image produced by combining images e–g, which are MCR retrieved concentration maps of triglyceride (liquid), cholesterol (crystal) and BSA (powder). Scale bar: 50 µm.
We further developed a strategy for quantitation of cholesterol storage in lipid droplets. Under realistic biological circumstance, excess cholesterol exists either in the form of cholesterol crystal or in the esterified form in which an acyl chain is linked to the cholesterol via an ester bond. Cholesteryl ester is usually mixed with triglyceride and stored in lipid droplets. Quantifying the molar percentage of cholesteryl ester in lipid droplets is important to evaluate cholesterol metabolism. Though peaks of the acyl and sterol C=C bands are separated, the triglyceride molecule has various number of acyl C=C bonds in its three acyl chains depending on the degree of unsaturation. In addition, cholesteryl ester may contain zero (in cholesteryl palmitate), one (in cholesteryl oleate) or two (in cholesteryl linoleate) acyl C=C bonds in its acyl chain. Therefore, it is difficult to use the C=C bonds alone to quantify the molar percentage of cholesteryl ester in a lipid droplet. To address this difficulty, we developed a new strategy for cholesteryl ester quantification via counting the ester group C=O bond which gives a Raman band peaked at 1745 cm−1. It is known that triglyceride molecules have three ester C=O bonds which link glycerol with three acyl chains as shown in Figure 1a. Meanwhile, each cholesteryl ester molecule contains one sterol ring and one acyl chain linked by one ester group C=O bond. Given that y is molar fraction of cholesteryl ester in a triglyceride / cholesteryl ester mixture and x is measured concentration ratio of sterol C=C to C=O, we can derive the following equation,
| (2) |
Here, 3(1−y) is the relative concentration of C=O bonds in triglyceride. Based on Eq. (2), the molar fraction of cholesteryl ester is
| (3) |
Thus, if we perform hyperspectral SRS imaging and MCR analysis of acyl C=C, sterol C=C, and ester group C=O bonds, the above model will enable us to calculate the molar fraction of cholesteryl ester in a mixture. Moreover, the degree of unsaturation of the lipid droplet can be evaluated as the concentration ratio of acyl C=C to C=O.
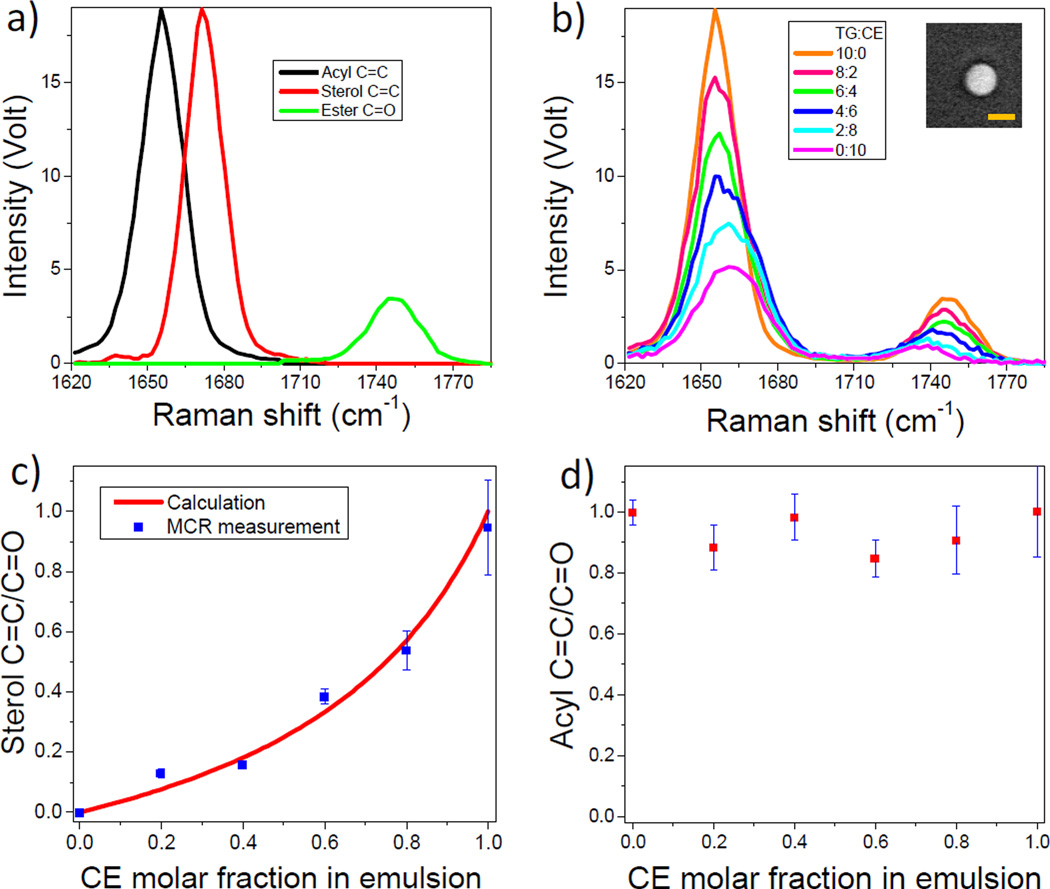
To experimentally validate the above strategy, we performed hyperspectral SRS imaging of emulsions composed of known molar ratios of glyceryl trioleate as a triglyceride representative and cholesteryl oleate as a cholesteryl ester representative. The spectral window covered Raman shift from 1620 to 1800 cm−1 for quantitative imaging of acyl C=C, ester group C=O, and sterol C=C bond. Figure 2a shows their SRS spectra recorded from pure glyceryl trioleate and pure cholesterol crystal. The ester group C=O band was found to be 5.4 times weaker in amplitude than the C=C band and has a separated single peak at 1745 cm−1. Hyperspectral SRS imaging of emulsions with various molar ratios of glyceryl trioleate to cholesteryl oleate produced spectra for all emulsions as shown in Figure 2b. With the increasing percentage of cholesteryl oleate in emulsion, SRS spectra exhibited a transition from a single narrow peak of acyl C=C bond (1655 cm−1, FWHM =17 cm−1) to an overlapped peak consisting of both acyl and sterol C=C bands. In the emulsion of pure cholesteryl oleate which contains one acyl and one sterol C=C bond, we observed a relatively broad Raman band contributed by acyl C=C band and sterol C=C band at equal intensity (colored magenta in Figure 2b). Such spectral profiles were confirmed by spontaneous Raman spectroscopy (Supplementary Figure S3). To quantify the cholesteryl ester percentage in mixed emulsions, hyperspectral SRS spectra of emulsions and the standard SRS spectra of acyl C=C, sterol C=C, and C=O bond were used as inputs to the MCR algorithm, serving as data matrix and initial spectra estimation, respectively. The concentration ratio of sterol C=C to C=O are plotted in Figure 2c as a function of actual cholesteryl ester percentage in each emulsion. The results are in good agreement with the calculation based on Eq. (2). By calculating the concentration ratio of acyl C=C to C=O, MCR quantification also provided the lipid unsaturation degree in all emulsions (Figure 2d). The degree of unsaturation is close to 1.0 for all emulsions. This result is consistent with the composition of the emulsions where both cholesteryl oleate and glyceryl trioleate are mono-unsaturated. These results collectively established an approach for quantifying the percentage of cholesteryl ester and degree of unsaturation in a lipid droplet based on SRS imaging of C=O and C=C bonds. Of note, Raman shift of the ester group C=O slightly varied with increasing percentage of cholesteryl ester, as shown in Figure 2b. However, MCR as a soft-modeling approach[17] allowed treatment of varying spectra in data sets and derivation of correct concentration profiles.
Figure 2.
Quantification of cholesteryl ester molar percentage in a lipid emulsion. a) SRS spectra of acyl C=C bond (black), sterol C=C bond (red) and ester group C=O bond (green). b) SRS spectra of emulsions composed of glyceryl trioleate and cholesteryl oleate at molar ratio of 10:0, 8:2, 6:4, 4:6, 2:8 and 0:10, respectively. Spectra were obtained from averaging of spectra in 100 pixels for each emulsion. Inset: SRS image of emulsion at TG and CE ratio of 8:2. CE and TG represent cholesteryl ester and triglyceride, respectively. Scalar bar: 5 µm. c) MCR quantification of cholesteryl ester molar fraction in emulsion. Red solid curve shows the calculation from Eq. (2). d) MCR measured unsaturation degree of emulsions.
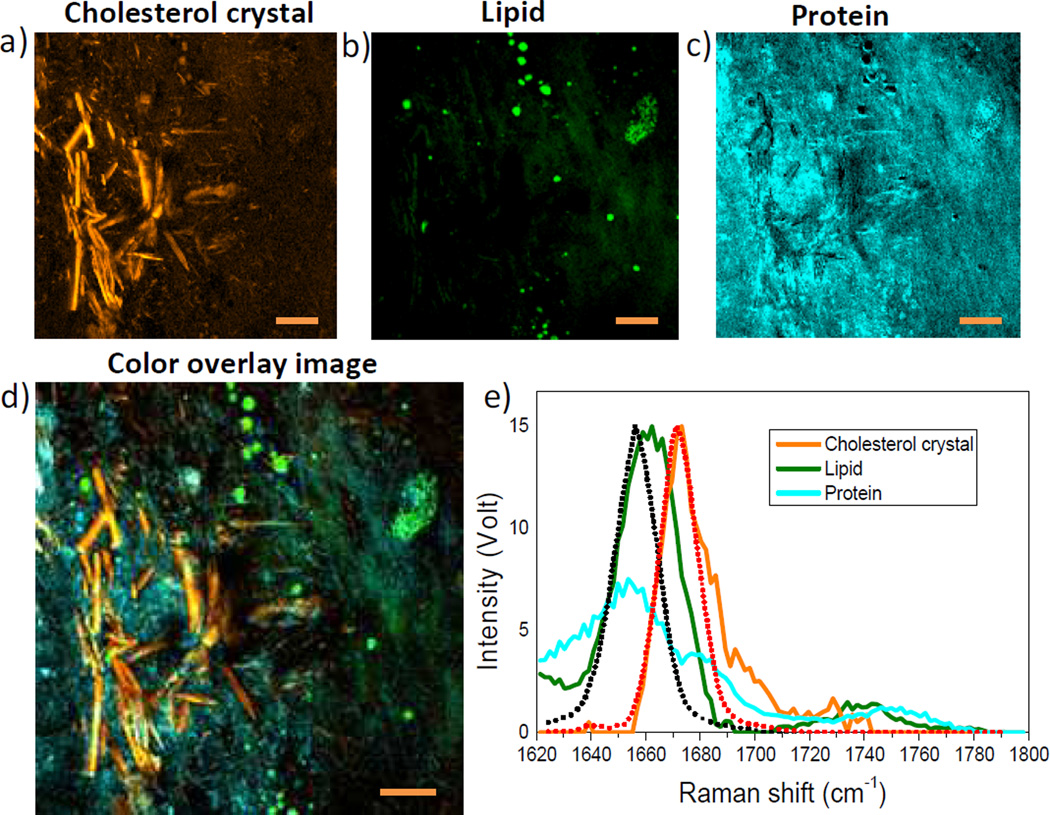
With the method established through the phantom studies, we further demonstrated quantitative concentration mapping of cholesterol crystal, lipid droplets, and proteins in intact arteries harvested from an atherosclerotic pig. An XY-Ω image stack was recorded (Supplementary Movie S2) in the fingerprint window between 1620 and 1800 cm−1. We then decomposed this stack into concentration maps of cholesterol crystal (Figure 3a), lipid (Figure 3b), and protein (Figure 3c), respectively. The overlay of three maps is shown in Figure 3d. Specifically, Figure 3a demonstrates the reconstructed concentration map of cholesterol crystal formed in the atherosclerotic plaque. The corresponding MCR optimized spectrum of sterol C=C band is peaked at 1669 cm−1 (Figure 3e, colored in orange), in accordance with the SRS spectrum recorded from pure cholesterol crystal (orange dashed line). Abundant needle-like cholesterol crystals are recognized in the reconstructed image. Such capability is important because cholesterol crystallization is well recognized to be a major determinant of plaque rupture in acute coronary syndrome[1b]. Figure 3b presents the map of lipid droplets, with corresponding MCR retrieved spectrum shown in Figure 3e (colored in green). Compared with the Raman spectrum of acyl C=C in pure glyceryl trioleate (black dashed line), the lipid spectrum exhibited a significantly broadened profile towards higher wavenumber, which implies that these lipid droplets contain a significant amount of cholesteryl ester. Figure 3c shows the concentration image of protein, which is ubiquitous but more concentrated in cholesterol crystal rich area. Taken together, the MCR analysis allowed distinctive mapping of cholesteryl ester rich lipid droplets, cholesterol crystal and protein in an intact atherosclerotic artery.
Figure 3.
Hyperspectral SRS imaging and MCR analysis of atherosclerotic artery. a–c) Reconstructed concentration images of crystalized free cholesterol (yellow), lipid droplets (green) and protein background (cyan). d) Overlay image of a–c. Scalar bars: 20 µm. e, MCR output spectra of cholesterol crystal, lipid droplets, and protein. Orange and dark dashed lines represent SRS spectra of acyl C=C and sterol C=C bonds, respectively.
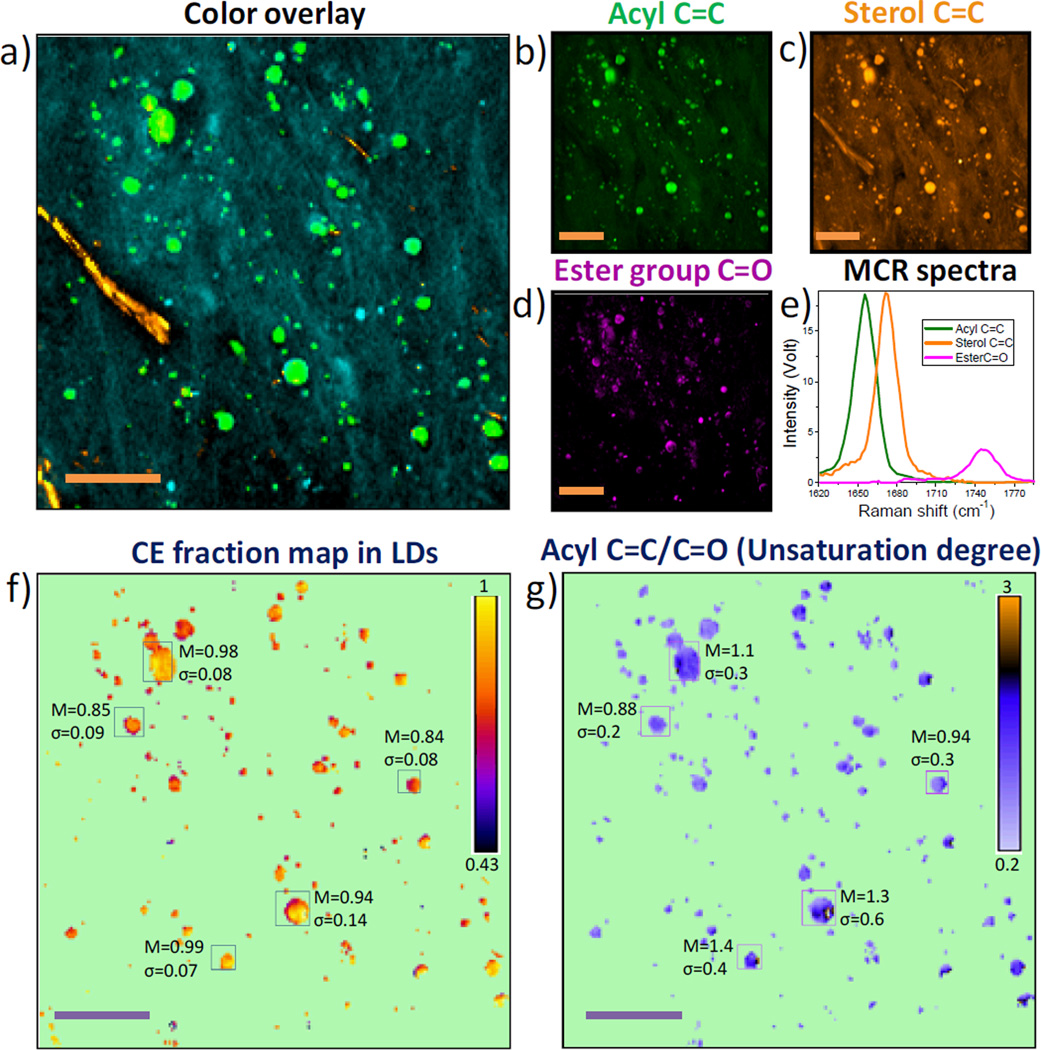
Using the strategy shown in Figure 2, we further quantified the molar fraction of cholesteryl ester and the degree of unsaturation for the lipid droplets in the atherosclerotic artery. Figure 4a shows an area with abundant lipid droplets (colored in green), distributed protein (colored in cyan), and a few cholesterol crystals (colored in yellow). To quantify the fraction of cholesteryl ester in these lipid droplets, concentration maps of acyl C=C, sterol C=C and ester group C=O bonds (Figure. 4b–d) were reconstructed from the hyperspectral SRS stack (Supplementary Movie S3) and the SRS spectra of pure component shown in Figure 2a. The corresponding MCR optimized spectra are shown in Figure 4e. Compared to the input spectrum, we note that the C=O band is slightly shifted to lower wavenumber due to presence of cholesteryl ester in the lipid droplets.
Figure 4.
MCR Quantitation of cholesteryl ester level and unsaturation degree in lipid droplets of intact atherosclerotic artery. a) Overlay map of cholesterol crystal (yellow), lipid droplets (green) and protein (cyan). b–d) Concentration maps of acyl C=C, sterol C=C, and ester C=O bonds reconstructed by MCR. e) MCR output spectra corresponding to maps b–d. f) Image of lipid droplets based on molar fraction of cholesteryl ester in each lipid droplet. Calibrated cholesteryl ester level for some Lipid droplets is shown. g) Image of lipid droplets based on ratio between concentration of acyl C=C and that of ester group C=O. Unsaturation degree for some lipid droplets is shown. Scalar bars: 20 µm. M: mean value of cholesteryl ester percentage. σ: standard deviation.
We then used the map of ester bond C=O as an internal standard to normalize maps of both acyl C=C bond and sterol C=C bond. The ratio between maps of sterol C=C bond and ester bond C=O (sterol C=C/C=O) represents the number of sterol C=C bond per fatty acid chain. According to Eq. (3), we obtained the map of cholesteryl ester molar fraction (Figure 4f). The mean cholesteryl ester level (presented by M) and its standard deviation (σ) are given in the indicated areas. Our data show that the lipid droplets in the imaged area are highly abundant in cholesteryl ester. Figure 4g maps the ratio between maps of acyl C=C and ester group C=O, which reflects the unsaturation degree of the lipid droplets. The measured unsaturation degrees of indicated lipid droplets are close to 1.0. This result is consistent with literature showing that cholesteryl oleate is the major component inside the lipid droplets of atherosclerotic arteries.[18] These results together show that hyperspectral SRS and MCR are capable of compositional analysis of lipid droplets in intact biological specimens.
Methods
Details about experimental setup, specimen preparation, and MCR algorithm can be found in Supporting Information.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health grant R21EB015901 and R21GM104681.
Supporting information for this article is available on the WWW under http://www.angewandte.org.
Contributor Information
Dr. Ping Wang, Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47906 (USA)
Junjie Li, Department of Biological Sciences, Purdue University, West Lafayette, IN 47906 (USA).
Pu Wang, Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47906 (USA).
Chun-Rui Hu, Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47906 (USA).
Delong Zhang, Department of Chemistry, Purdue University, West Lafayette, IN 47906 (USA).
Prof. Michael Sturek, Department of Cellular and Integrative Physiology, Indiana University Medical School, Indianapolis, IN 46202 (USA).
Prof. Ji-Xin Cheng, Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN 47906 (USA).
References
- 1.a) Abela GS, Aziz K, Vedre A, Pathak DR, Talbott JD, DeJong J. Am. J. Cardiol. 2009;103:959. doi: 10.1016/j.amjcard.2008.12.019. [DOI] [PubMed] [Google Scholar]; b) Chen Z, Ichetovkin M, Kurtz M, Zycband E, Kawka D, Woods J, He X, Plump AS, Hailman E. Lipids Health Dis. 2010;9:61. doi: 10.1186/1476-511X-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rajamaki K, Lappalainen J, Oorni K, Valimaki E, Matikainen S, Kovanen PT, Eklund KK. Plos One. 2010:5. doi: 10.1371/journal.pone.0011765. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Abela GS. J. Clin. Lipidol. 2010;4:156. doi: 10.1016/j.jacl.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Duewell P, et al. Nature. 2010;464:1357. doi: 10.1038/nature08938. see Supporting Information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Hu J, Zhang Z, Shen WJ, Azhar S. Nutr. Metab. 2010;7:47. doi: 10.1186/1743-7075-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kraemer FB, Khor VK, Shen WJ, Azhar S. Mol. Cell. Endocrinol. 2013;371:15. doi: 10.1016/j.mce.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanier MT. Orphanet J. Rare Dis. 2010;5:16. doi: 10.1186/1750-1172-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Gent CM, Emeis JJ. Prog. Biochem. Pharmacol. 1977;13:262. [PubMed] [Google Scholar]
- 6.Hirsch D, Azoury R, Sarig S, Kruth HS. Calcif. Tissue Int. 1993;52:94. doi: 10.1007/BF00308315. [DOI] [PubMed] [Google Scholar]
- 7.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Nat. Immunol. 2008;9:847. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, Gardecki JA, Nadkarni SK, Toussaint JD, Yagi Y, Bouma BE, Tearney GJ. Nat. Med. 2011;17:1010. doi: 10.1038/nm.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lattermann A, Matthaus C, Bergner N, Beleites C, Romeike BF, Krafft C, Brehm BR, Popp J. J. Biophotonics. 2013;6:110. doi: 10.1002/jbio.201200146. [DOI] [PubMed] [Google Scholar]
- 10.Cheng JX, Xie XS. Coherent Raman Scattering Microscopy. New York: Taylor & Francis Group; 2012. [Google Scholar]
- 11.a) Duncan MD, Reintjes J, Manuccia TJ. Opt. Lett. 1982;7:350. doi: 10.1364/ol.7.000350. [DOI] [PubMed] [Google Scholar]; b) Zumbusch A, Holtom GR, Xie XS. Phys. Rev. Lett. 1999;82:4142. [Google Scholar]; c) Cheng J-X, Xie XS. J. Phys. Chem. B. 2004;108:827. [Google Scholar]; d) Andreas V. J. Phys. D: Appl. Phys. 2005;38:R59. [Google Scholar]; e) Rodriguez LG, Lockett SJ, Holtom GR. Cytometry A. 2006;69:779. doi: 10.1002/cyto.a.20299. [DOI] [PubMed] [Google Scholar]; f) Müller M, Zumbusch A. Chemphyschem. 2007;8:2156. doi: 10.1002/cphc.200700202. [DOI] [PubMed] [Google Scholar]; g) Hellerer T, Axang C, Brackmann C, Hillertz P, Pilon M, Enejder A. Proc. Natl. Acad. Sci. USA. 2007;104:14658. doi: 10.1073/pnas.0703594104. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Evans CL, Xie XS. Annu. Rev. Anal. Chem. 2008;1:883. doi: 10.1146/annurev.anchem.1.031207.112754. [DOI] [PubMed] [Google Scholar]; i) Pezacki JP, Blake JA, Danielson DC, Kennedy DC, Lyn RK, Singaravelu R. Nat. Chem. Biol. 2011;7:137. doi: 10.1038/nchembio.525. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Bélanger E, Henry FP, Vallée R, Randolph MA, Kochevar IE, Winograd JM, Lin CP, Côté D. Biomed. Opt. Express. 2011;2:2698. doi: 10.1364/BOE.2.002698. [DOI] [PMC free article] [PubMed] [Google Scholar]; k) Paar M, et al. J. Biol. Chem. 2012;287:11164. doi: 10.1074/jbc.M111.316794. see Supporting Information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Ploetz E, Laimgruber S, Berner S, Zinth W, Gilch P. Appl. Phys. B: Lasers Opt. 2007;87:389. [Google Scholar]; b) Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He CW, Tsai JC, Kang JX, Xie XS. Science. 2008;322:1857. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Nandakumar P, Kovalev A, Volkmer A. New J. Phys. 2009;11:033026. [Google Scholar]; d) Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS. Science. 2010;330:1368. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Zhang D, Sipchenko MN, Cheng JX. J. Phys. Chem. Lett. 2011;2:1248. doi: 10.1021/jz200516n. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Min W, Freudiger CW, Lu S, Xie XS. Annu. Rev. Phys. Chem. 2011;62:507. doi: 10.1146/annurev.physchem.012809.103512. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Moger J, Garrett NL, Begley D, Mihoreanu L, Lalatsa A, Lozano MV, Mazza M, Schatzlein A, Uchegbu I. J. Raman Spectrosc. 2012;43:668. [Google Scholar]; h) Lee T, Mundoor H, Gann DG, Callahan TJ, Smalyukh II. Opt. Express. 2013;21:12129. doi: 10.1364/OE.21.012129. [DOI] [PubMed] [Google Scholar]; i) Mansfield JC, Littlejohn GR, Seymour MP, Lind RJ, Perfect S, Moger J. Anal. Chem. 2013;85:5055. doi: 10.1021/ac400266a. [DOI] [PubMed] [Google Scholar]; j) Wei L, Yu Y, Shen Y, Wang MC, Min W. Proc. Natl. Acad. Sci. USA. 2013 [Google Scholar]
- 13.a) Cheng J-X, Volkmer A, Book LD, Xie XS. J. Phys. Chem. B. 2002;106:8493. [Google Scholar]; b) Müller M, Schins JM. J. Phys. Chem. B. 2002;106:3715. [Google Scholar]; c) Rinia HA, Burger KNJ, Bonn M, Muller M. Biophys. J. 2008;95:4908. doi: 10.1529/biophysj.108.137737. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Bonn M, Müller M, Rinia HA, Burger KNJ. J. Raman Spectrosc. 2009;40:763. [Google Scholar]; e) Kim SH, Lee ES, Lee JY, Lee BS, Park JE, Moon DW. Circul. Res. 2010;106:1332. doi: 10.1161/CIRCRESAHA.109.208678. [DOI] [PubMed] [Google Scholar]; f) Chowdary PD, Benalcazar WA, Jiang Z, Marks DM, Boppart SA, Gruebele M. Anal. Chem. 2010;82:3812. doi: 10.1021/ac100222c. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Lim RS, Suhalim JL, Miyazaki-Anzai S, Miyazaki M, Levi M, Potma EO, Tromberg BJ. J. Lipid Res. 2011;52:2177. doi: 10.1194/jlr.M018077. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Lin CY, Suhalim JL, Nien CL, Miljkovic MD, Diem M, Jester JV, Potma EO. J. Biomed. Opt. 2011;16:021104. doi: 10.1117/1.3533716. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Bégin S, Burgoyne B, Mercier V, Villeneuve A, Vallée R, Côté D. Biomed. Opt. Express. 2011;2:1296. doi: 10.1364/BOE.2.001296. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Lee YJ, Moon D, Migler KB, Cicerone MT. Anal. Chem. 2011;83:2733. doi: 10.1021/ac103351q. [DOI] [PubMed] [Google Scholar]; k) Pegoraro AF, Slepkov AD, Ridsdale A, Moffatt DJ, Stolow A. J. Biophotonics. 2012 doi: 10.1002/jbio.201200171. [DOI] [PubMed] [Google Scholar]
- 14.a) Suhalim JL, Chung C-Y, Lilledahl Magnus B, Lim Ryan S, Levi M, Tromberg Bruce J, Potma Eric O. Biophys. J. 2012;102:1988. doi: 10.1016/j.bpj.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ozeki Y, Umemura W, Otsuka Y, Satoh S, Hashimoto H, Sumimura K, Nishizawa N, Fukui K, Itoh K. Nat. Photonics. 2012;6:844. [Google Scholar]; c) Fu D, Lu F-K, Zhang X, Freudiger C, Pernik DR, Holtom G, Xie XS. J. Am. Chem. Soc. 2012;134:3623. doi: 10.1021/ja210081h. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Kong LJ, Ji MB, Holtom GR, Fu D, Freudiger CW, Xie XS. Opt. Lett. 2013;38:145. doi: 10.1364/OL.38.000145. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Mansfield J, Moger J, Green E, Moger C, Winlove CP. J. Biophotonics. 2013 doi: 10.1002/jbio.201200213. [DOI] [PubMed] [Google Scholar]; f) Zhang D, Wang P, Slipchenko MN, Ben-Amotz D, Weiner AM, Cheng JX. Anal. Chem. 2013;85:98. doi: 10.1021/ac3019119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Movasaghi Z, Rehman S, Rehman IU. Appl. Spectrosc. Rev. 2007;42:493. [Google Scholar]
- 16.Jaumot J, Gargallo R, de Juan A, Tauler R. Chemometr. Intell. Lab. 2005;76:101. [Google Scholar]
- 17.de Juan A, Maeder M, Martinez M, Tauler R. Chemometr. Intell. Lab. 2000;54:123. [Google Scholar]
- 18.Degirolamo C, Shelness GS, Rudel LL. J. Lipid Res. 2009;50:S434. doi: 10.1194/jlr.R800076-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.