Abstract
Successful defence of plants against colonisation by fungal pathogens depends on the ability to prevent initial penetration of the plant cell wall. Here we report that the pathogen-induced (1,3)-β-glucan cell wall polymer callose, which is deposited at sites of attempted penetration, directly interacts with the most prominent cell wall polymer, the (1,4)-β-glucan cellulose, to form a three-dimensional network at sites of attempted fungal penetration. Localisation microscopy, a super-resolution microscopy technique based on the precise localisation of single fluorescent molecules, facilitated discrimination between single polymer fibrils in this network. Overexpression of the pathogen-induced callose synthase PMR4 in the model plant Arabidopsis thaliana not only enlarged focal callose deposition and polymer network formation but also resulted in the exposition of a callose layer on the surface of the pre-existing cellulosic cell wall facing the invading pathogen. The importance of this previously unknown polymeric defence network is to prevent cell wall hydrolysis and penetration by the fungus. We anticipate our study to promote nanoscale analysis of plant-microbe interactions with a special focus on polymer rearrangements in and at the cell wall. Moreover, the general applicability of localisation microscopy in visualising polymers beyond plant research will help elucidate their biological function in complex networks.
Cell wall thickenings at sites of attempted fungal penetration, so called papillae, were the first plant defence response that has been investigated on a cellular level starting 150 years ago1. Mangin reported in 18952 that the (1,3)-β-glucan polymer callose was the main constituent in the forming papillae. Within a few hours after initial landing of a fungal spore on the leaf surface, pre-existing, pathogen-induced callose synthases are recruited in vesicle-like bodies from the plasma membrane to sites of attempted fungal penetration3. There, callose synthases with their multi membrane-spanning domains4 are reintegrated into the plasma membrane to initiate callose biosynthesis and deposition of this (1,3)-β-glucan polymer between the plasma membrane and the cellulosic cell wall. As part of the plant's innate immunity5,6, the deposition of the (1,3)-β-glucan polymer callose strengthens the cell wall at sites of attempted penetration7. Fungal pathogens try to overcome this penetration barrier by secretion of cell wall degrading enzymes8.
However, the extent to which callose finally contributes to establish a physical barrier in order to prevent or slow pathogen penetration was subject of a controversial debate. Findings in A. thaliana mutants that lacked pathogen-induced callose formation but revealed increased resistance to invading powdery mildew species9 challenged an active role of callose in penetration resistance.
Nevertheless, we could recently directly confirm that localised callose deposition can prevent pathogen infection. We observed complete penetration resistance to the adapted powdery mildew Golovinomyces cichoracearum and the non-adapted powdery mildew Blumeria graminis f.sp. hordei in A. thaliana lines that overexpressed the pathogen-induced callose synthase PMR4 (POWDERY MILDEW RESISTANT4). Penetration resistance in these lines is based on an elevated early callose deposition at sites of attempted fungal penetration compared to wild-type plants3.
Results and discussion
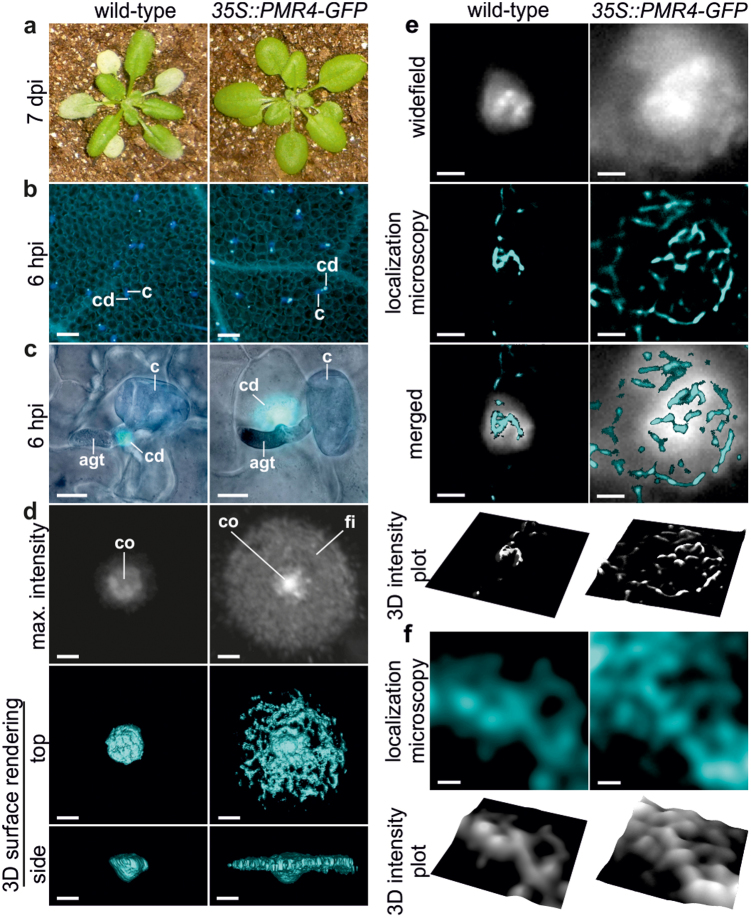
Based on our recent results showing that enlarged, pathogen-induced callose deposits can effectively prevent fungal penetration3, we wanted to test whether additional factors might support callose-dependent penetration resistance. Therefore, we inoculated A. thaliana wild-type and PMR4-overexpressing lines with the adapted powdery mildew G. cichoracearum. As expected from our previous study, we did not observe macroscopic disease symptoms on PMR4-overexpressing plants 7 dpi (Fig. 1a); and microscopic analysis of callose deposits stained with (1,3)-β-glucan-specific aniline blue fluorochrome (ABF)10 revealed an elevated callose deposition at sites of attempted fungal penetration in the powdery mildew-resistant PMR4-overexpressing lines compared to wild-type (Fig. 1b, c). Additional confocal laser-scanning microscopy (CLSM) of ABF-stained callose deposits indicated that besides the time-point and quantity also the altered structure of callose deposits might influence penetration resistance. At 6 h post-inoculation (hpi) with G. cichoracearum, a cone-like, dense callosic matrix pointed towards the cytosol at sites of attempted fungal penetration, which was similar in wild-type and PMR4-overexpressing lines (Fig. 1d, see Supplementary Video S1 online). The cone-like shape resembled previous observations of callose-containing papillae of cross-sectioned, infected epidermal leaf cells in transmission electron microscopy9. Only callose deposits of the PMR4-overexpressing lines revealed an additional field of callose that surrounded the dense core region. After three-dimensional (3D) surface rendering of confocal Z-stack images, the top view onto these enlarged callose deposits revealed a network of callose fibrils whereas the side view resembled a UFO-like shape (Fig. 1d, see Supplementary Video S2). We then applied localisation microscopy11,12,13 on ABF-stained callose deposits to obtain structural information about callose fibrils. The prerequisites for an application of localisation microscopy were stochastic intensity fluctuations and photoblinking of the organic fluorophore ABF that we observed on stained callose (see Supplementary Fig. S1 online). Localisation microscopy of complete callose deposits showed that PMR4-overexpressing lines had distinct structural differences in the orientation of callose macrofibrils, which had a mean diameter of 250 nm (see Supplementary Fig. S2 online). In the central core of the callose deposit, callose macrofibrils formed an irregular structure whereas the surrounding field was characterised by a radial orientation of callose macrofibrils. The field of callose was enclosed by a ring of circular-oriented macrofibrils. In wild-type plant deposits, callose macrofibrils revealed the same irregular structure and diameter as in the core region of callose deposits of the resistant A. thaliana lines (Fig. 1e, see Supplementary Fig. S2 online). Because localisation microscopy facilitated a nanoscale resolution of callose structures, we were able to visualise the macrofibril-forming network of microfibrils. The diameter of single microfibrils with a mean value of 44 nm (see Supplementary Fig. S2 online) corresponded to the size of callose microfibrils synthesised in vitro by detergent extracts from A. thaliana14. We did not observe differences in the structure, orientation or diameter of callose microfibrils comparing wild-type and PMR4-overexpressing lines (Fig. 1f, see Supplementary Fig. S2 online). The results from ABF-stained callose micro- and macrofibril networks were confirmed in an additional localisation microscopy approach where we immunolabelled callose with a specific anti-callose antibody in combination with a secondary antibody coupled with the fluorophore CAGE552 (see Supplementary Fig. S3 online). This fluorophore belongs to class of caged rhodamines that has been successfully tested in localisation microscopy15. The advantage of direct labelling of a polymer versus immunolabelling was the higher resolution. Whereas direct ABF-staining of callose resulted in a maximum resolution of 32 nm (mean resolution 44 nm), a maximum resolution of 44 nm (mean resolution 58 nm) was obtained by immunolabelling of callose (see Supplementary Fig. S2 online). The difference of 12 nm in maximum resolution due to direct ABF-labelling of callose microfibrils was in the range of the linkage error of about 10 – 15 nm caused by the size of the primary and secondary antibody that was recently described16.
Figure 1. Nanoscale resolution of callose polymer fibrils in pathogen-induced cell wall deposits.
A. thaliana wild-type and pathogen-resistant PMR4-overexpressing lines (35S::PMR4-GFP) were inoculated with G. cichoracearum. (a), Disease phenotype at 7 d post-inoculation (dpi). (b), Micrographs of pathogen-induced callose deposits in epidermal leaf cells 6 hpi. Callose stained with aniline blue fluorochrome (ABF). c: conidium, cd: callose deposit. Scale bars = 50 μm. (c), Merged brightfield and epi-fluorescence micrographs of callose at sites of attempted fungal penetration 6 hpi. agt: appressorial germ tube, c: conidium, cd: callose deposit. Scale bars = 10 μm. (d), High-resolution, confocal laser-scanning micrographs of ABF-stained callose. co: core of callose deposit, fi: field of callose. Scale bars = 2 μm. (e–f), Localisation microscopy of ABF-stained callose fibrils in pathogen-induced deposits. 3D intensity plots highlight network formation of callose fibrils. (e), Overview of callose macrofibril orientation. Scale bars = 2 μm. (f), Nanoscale network of callose microfibrils. Scale bars = 50 nm.
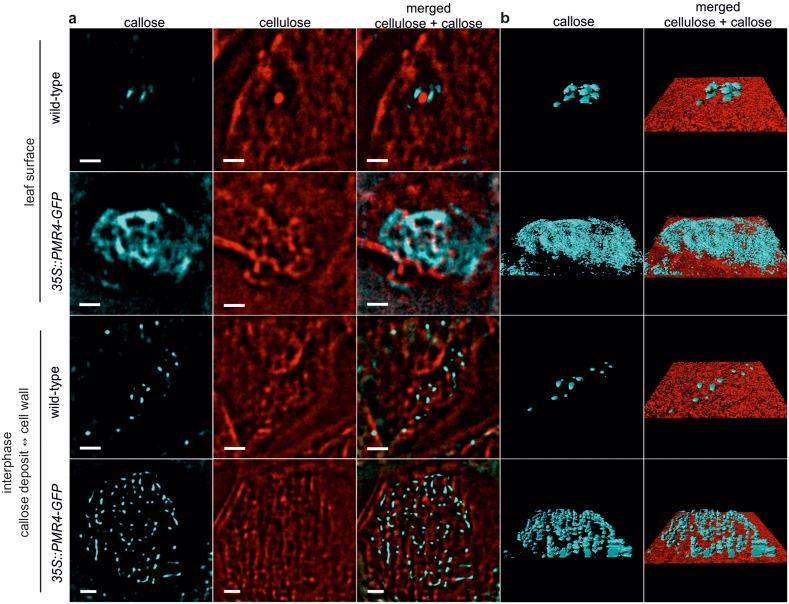
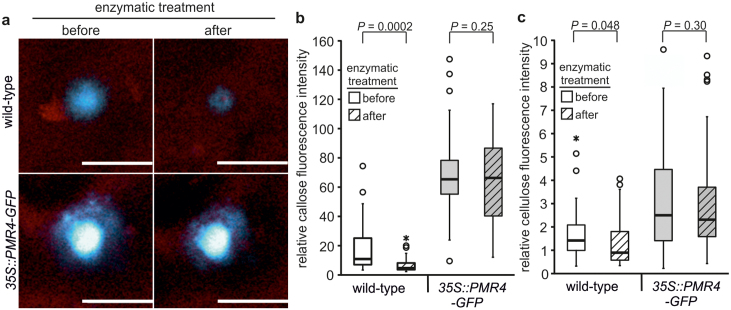
We then established localisation microscopy with the cellulose-specific fluorescent dye pontamine fast scarlet 4B (S4B)17 to enable discrimination of the two polymeric glucans cellulose and callose in complex networks. Similar to ABF, we observed stochastic intensity fluctuations and photoblinking for S4B that allowed an application of localisation microscopy on stained cellulose (see Supplementary Fig. S1 online). In both CLSM and localisation microscopy, the S4B-stained cellulose revealed a structural orientation (see Supplementary Fig. S4 and Supplementary Videos S3 and S4 online) that resembled the orientation in scanning electron microscopy of cellulosic cell walls from A. thaliana18. We determined a mean diameter of 474 nm for the cellulose macrofibrils (see Supplementary Fig. S5 online), which is in accordance to the analysis of atomic force microscopy19. CLSM examination of the interphase of the callose deposit and the cellulosic cell wall after ABF and S4B co-staining gave first indication of a callose/cellulose polymer interaction where callose fibrils might migrate into and permeate the pre-existing cellulosic cell wall (see Supplementary Fig. S6 and Supplementary Videos S5 and S6 online). Our assumption of a glucan polymer interaction was confirmed when we applied localisation microscopy at the callose/cellulosic cell wall interphase and detected callose fibrils that migrated into and permeated the cellulosic cell wall layer. We observed this direct callose/cellulose polymer interaction in wild-type and resistant PMR4-overexpression lines (Fig. 2a). However, 3D rendering of ABF- and S4B-stained callose/cellulose interphase indicated that the permeation of the complete cellulosic cell wall by callose fibrils was especially pronounced in PMR4-overexpression plants (Fig. 2b, see Supplementary Videos S7 and S8 online). Whereas we observed a mean of 10 callose fibrils permeating the cellulosic cell wall layer at callose/cellulose interphases in wild-type, the number of permeation events increased to 50 in resistant PMR4-overexpression lines. However, the density of callose permeation events did not differ between wild-type and PMR4-overexpression lines (see Supplementary Fig. S7 online). These results indicated that the higher total number of permeation events in PMR4-overexpression lines was due to the lateral expansion of the callose deposit. To evaluate the extent to which callose fibres would permeate the cellulosic cell wall layers, we performed localisation microscopy on the surface of ABF- and S4B-stained epidermal leaf cells at 6 hpi with G. cichoracearum. Whereas only single callose fibres completely permeated the cellulosic cell wall of wild-type epidermal cells, a callose layer was formed on top of the pre-existing cellulosic cell wall in resistant PMR4-overexpression lines (Fig. 2b, see Supplementary Videos S9 and S10 online). The 3D model highlights the observed differences of the pathogen-induced callose deposit, of the callose/cellulose network within the plant cell wall, and in the formation of superficial callose layers, which directly face intruding fungal pathogens (Fig. 3). Digital cross-sections of the ABF- and S4B-stained callose/cellulose polymer network at sites of attempted powdery mildew infection suggest that callose fibrils migrated into and permeated the cellulosic layers through internal cell wall nanopores20,21 (see Supplementary Fig. S9 online). A permeation of the cell wall through nanopores suggest a gel-forming property of callose, which is known for (1,3)-β-glucans22,23. Because these nanopores have been predicted as putative entry sites for cell wall degrading enzymes (CWDE)24, we anticipated an increased resistance of the compact and extended callose/cellulosic polymer network to β-glucan hydrolases in powdery mildew resistant PMR4-overexpression lines. To test our hypothesis, we applied a harsh hydrolysing treatment to A. thaliana leaves at 6 hpi with the powdery mildew G. cichoracearum. After an Accellerase treatment, a cell wall degrading, hydrolysing enzyme complex used for biotechnological biomass conversion, at 50°C for 1 h, we observed a strong degradation of the pathogen-induced callose/cellulosic polymer network at sites of attempted fungal penetration in wild-type leaves whereas the enhanced glucan polymer networks remained intact in the resistant PMR4-overexpressing lines (Fig. 4). This suggests that the complete penetration resistance of PMR4-overexpressing A. thaliana lines is based on a physical strengthening of the cell wall at infection sites, which includes the establishment of a physical barrier against pathogen-secreted cell wall hydrolases25. In our model of the penetration resistance of the PMR4-overexpressing lines, we consider the defence-related glucan polymer callose to seal cell wall nanopores and to form a callose layer on the cellulosic cell wall surface (Fig. 3), which prevents ingress of CWDE secreted by the invading pathogens.
Figure 2. Pathogen-induced nanoscale glucan polymer networks at sites of attempted fungal penetration.
A. thaliana wild-type and pathogen-resistant PMR4-overexpressing lines (35S::PMR4-GFP) were inoculated with G. cichoracearum. Localisation microscopy at the interphase of pathogen-induced callose deposits and the cellulosic cell wall and at the surface of epidermal cells after co-staining. Blue channel: fluorescence of aniline blue fluorochrome-stained callose, red channel: fluorescence of pontamine fast scarlet 4B-stained cellulose. (a), Permeation of callose fibrils through cellulosic cell wall layers and formation of callose layers on the cell wall surface at sites of attempted fungal penetration. Scale bars = 2 μm. (b), 3D surface rendering of callose/cellulose polymer networks.
Figure 3. 3D model of callose/cellulose interaction at the site of attempted fungal penetration.
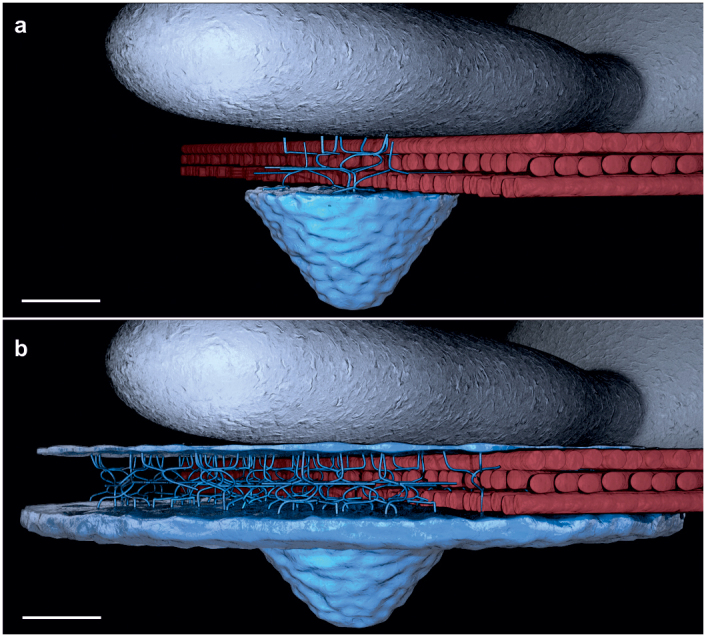
3D model represents the formation of callose deposits, callose/cellulose polymer networks, and superficial callose layers at sites of attempted fungal penetration in (a), A. thaliana wild-type and (b), penetration-resistant PMR4-overexpressing epidermal leaf cells at 6 h post-inoculation with powdery mildew. Blue colours: callose ((1,3)-β-glucan), red colours: cellulose ((1,4)-β-glucan), grey colours: fungal structures (hyphae and conidium). Scale bars = 2 μm.
Figure 4. Resistance of the pathogen-induced callose/cellulose polymer network to enzymatic degradation in penetration-resistant A. thaliana mutant.
A. thaliana wild-type and pathogen-resistant PMR4-overexpressing lines (35S::PMR4-GFP) were inoculated with G. cichoracearum. (a), Confocal laser-scanning micrographs of same callose deposits before and after enzymatic treatment. Blue channel: fluorescence of aniline blue fluorochrome (ABF)-stained callose, red channel: pontamine fast scarlet 4B (S4B)-stained cellulose. Scale bars = 20 μm. (b–c), Relative fluorescence intensity of ABF-stained callose (b) and of S4B-stained cellulose (c) in callose/cellulose polymer networks before and after enzymatic treatment. Box plots from n ≥ 35 measurements of four individual plants, whiskers: minimum and maximum value within the 1.5 x interquatrile range (IQR), circle: outlier, star: extreme outlier outside the 1.5 x IQR. P values of t-test.
Conclusion
In summary, we not only report about the first successful application of localisation microscopy on carbohydrate polymers to receive nanoscale, 3D structural information, which helped to explain the observed pathogen-resistant phenotype, but also the first successful application of localisation microscopy in intact plant tissue in general.
The advantages of localisation over atomic force microscopy, electron microscopy or electron tomography, which would represent alternative methods with a resolution high enough to visualise polymer microfibrils, are that i) the examination of callose deposited in papillae does not require the preparation of sections from embedded plant tissue with the risk of artefact production and ii) a discrimination of different types of polymer fibres is allowed due to staining with highly specific organic fluorophores.
Methods
Growth conditions, inoculations, and cytology
Cultivation of A. thaliana wild-type (Columbia) and PMR4-overexpression line 35S::PMR4-GFP from our previous study8 as well as inoculation of three-week-old plants with the powdery mildew G. cichoracearum (strain UCSC1) followed the description in Stein et al.26. Rosette leaves were harvested 6 h post-inoculation (hpi) and destained in ethanol prior glucan staining. Aniline blue fluorochrome (ABF) (Biosupplies, Bundoora, Australia) was used for specific callose staining according to manufacturer's instructions; and pontamine fast scarlet 4B (S4B) (Sigma-Aldrich, Steinheim, Germany) for specific cellulose staining according to Anderson et al.17.
Localisation microscopy of ABF- and S4B stained leaf samples
Datasets for localisation microscopy were acquired on a custom modified Nikon stochastic optical reconstruction microscope (N-STORM, Nikon GmbH, Düsseldorf, Germany). The microscope was equipped with an Apo TIRF 100x oil immersion objective with a numerical aperture of 1.49 (Nikon GmbH), an electron multiplying charge-coupled device (EMCCD) camera (iXon+ DU-897, Andor Technology Plc, Belfast, UK) and a quadband filter composed of a quad line beamsplitter (zt405/488/561/640rpc TIRF, Chroma Technology Corporation, Bellows Falls, VT, USA) and a quad line emission filter (brightline HC 446, 523, 600, 677, Semrock, Inc., Rochester, NY, USA). For excitation of ABF, a 100 mW 405 nm diode laser (CUBE 405-100C, Coherent Inc., Santa Clara, CA, USA) and for excitation of S4B a 150 mW 561 nm optically pumped semiconductor laser (Sapphire 561 LP, Coherent, Inc.) were used. Single colour datasets were acquired with continuous illumination. For two colour imaging, the lasers were switched on and off alternately controlled by an acoustooptic tunable filter (AOTF). The integration time of the EMCCD camera was set to 16 ms per frame with an EM gain of 300. The focus was kept stable during acquisition using Nikon's perfect focus system.
Super-resolution images were reconstructed from a series of 10,000 – 48,000 widefield images per channel using the N-STORM analysis module v. 2.0.0.76 of NIS Elements AR v. 4.00.07 (Laboratory imaging s.r.o., Prague, Czech Republic). 3D super-resolution microscopy was performed using an astigmatism based approach according to Huang et al.27. For each colour channel a separate 3D calibration curve was used. Intensity fluctuations and photoblinking of ABF and S4B were analysed in NIS Elements AR v. 4.00.07.
Bitplane Imaris 7.6.1. (Bitplane AG, Zurich, Switzerland) was used for visualization of 3D super-resolution images.
Localisation microscopy of immunolabelled leaf samples
Immunolabelling of deposited callose in destained A. thaliana leaves 6 h post-inoculation with a callose-specific monoclonal anti-(1,3)-β-glucan antibody (Biosupplies) as primary antibody and the fluorophore-labelled secondary antibody CAGE 552 (Abberior GmbH, Göttingen, Germany) was done as described in Ellinger et al.8.
Localisation microscopy of immunolabelled callose was performed with a Zeiss PS.1 super-resolution system (Carl Zeiss MicroImaging GmbH, Jena, Germany). The setup included the objective alpha Plan-Apochromat 100x/1.46 Oil DIC (Zeiss GmbH), an iXon 897 back-thinned EMCCD camera (Andor Technology Plc) and a multi-purpose filter set (Elyra filter set n. 3; Zeiss GmbH) for excitation and detection with direct STORM (dSTORM). In order to efficiently excite and detect CAGE 552 (Abberior GmbH), a 100 mW 561 nm optically pumped semiconductor laser (Coherent Inc.) and a BP 570/650 nm emission filter (Optics Balzers AG, Balzers, Liechtenstein) were used. The detection of the blinking fluorophore was achieved with an exposure time of 100 ms per frame and an EMCCD gain at level 30.
dSTORM super-resolution pictures were composed after a time series of 10,000 widefield images with Zeiss ZEN 2012 software and Zeiss software module Photoactivated Localisation Microscopy (PAL-M) (Zeiss, GmbH).
Confocal laser-scanning microscopy of ABF- and S4B stained leaf samples
Confocal laser-scanning microscopy of ABF-stained callose followed the description in Ellinger et al.8. The Zeiss LSM 780 microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany) was used. S4B was excited at 561 nm by using an optically pumped semiconductor laser (Coherent Inc.). Emission filtering was achieved using a 568 – 595-nm bandpass filter. Emission signals were gathered by a gallium-arsenite-phosphid nondescanned photodetector (Zeiss GmbH). Image processing, including maximum intensity 3D reconstruction, surface rendering, and video generation, was performed with integral functions of the ZEN 2010 (Zeiss GmbH) operating software.
Enzymatic treatment
A. thaliana leaves were harvested 6 hpi, destained in ethanol, and co-stained with ABF and S4B to measure the relative emitted fluorescence intensity of callose and cellulose at site of callose deposition and callose/cellulose polymer network formation before enzymatic treatment. For measuring the relative emitted fluorescence intensity, sites of callose deposition were manually selected on confocal laser-scanning micrographs by using the image processing software Adobe Photoshop CS6 Extended Edition (Adobe Systems Inc., San José, CA, USA) and the integral function of histogram and the average of luminescence as the channel for relative ABF-emission quantification and the average of channel red for relative S4B-emission quantification. Values are based on the RGB colour space ranging from 1 (black = no signal) to 255 (highest signal). Normalization of data was done against the ABF or S4B background fluorescence outside the callose deposit and callose/cellulose polymer network at sites of attempted fungal penetration.
Subsequently, leaves were incubated in 1 mL of 0.1 M sodium acetate buffer (pH 5.0) supplemented with 10 μL of Accellerase 1500 cellulase enzyme complex (Danisco, Copenhagen, Denmark) for lignocellulosic cell wall hydrolysis for 1 h at 50°C. Leaves were removed from Accellerase solution, washed twice with water, and co-stained again with ABF and S4B to measure the relative emitted fluorescence intensity at sites callose deposition and callose/cellulose polymer network formation after enzymatic treatment.
Statistical analysis
Descriptive statistics including the mean and whisker of boxplots along with the Tukey range test for multiple comparison procedures in conjunction with ANOVA and with the Student's t-test for pairwise comparison were used to determine significant differences. P < 0.05 was considered significant.
Author Contributions
D.E. performed localisation microscopy and collected, processed and refined localisation microscopy data; M.N. cultivated and inoculated plants, collected plant samples, performed CLSM and collected, processed and refined CLSM data; D.E., M.N. and R.R. were involved in study design and analysed data; C.A.V. designed the study, analysed data and wrote the paper. D.E. and M.N. contributed equally to the study.
Supplementary Material
Supplementary Information
Video S1
Video S2
Video S3
Video S4
Video S5
Video S6
Video S7
Video S8
Video S9
Video S10
Acknowledgments
We thank C. Falter from the University of Hamburg for preparing immunolabelled leave material; D. Furlani from Carl Zeiss Microscopy GmbH for his support in localization microscopy of immunolabelled samples with the Zeiss Elyra system; M. Holthaus from Slug Graphix for generating 3D model figures of interaction sites; and the Nikon Application Center Northern Germany (Nikon GmbH) for providing the Nikon N-STORM microscope. M.N. is funded by the German Federal ministry of Education and Research (grant no. FKZ 0315521A).
References
- deBary A. Recherches sur le développement de quelques champignons parasites. Annales des Sciences Naturelles. Botanique et Biologie végétale 20, 5–148 (1863). [Google Scholar]
- Mangin L. Recherches sur les Péronosporées. Bulletin de la Société d'Histoire Naturelle d'Autun 8, 55–108 (1895). [Google Scholar]
- Ellinger D. et al. Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol. 161, 1433–1444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. K. et al. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15, 2503–2513 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D. & Dangl J. L. The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- Schwessinger B. & Ronald P. C. Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Bio.l 63, 451–482 (2012). [DOI] [PubMed] [Google Scholar]
- Stone B. A. & Clarke A. E. Chemistry and Biology of (1→3)-β-glucans (La Trobe University Press, Melbourne, 1992). [Google Scholar]
- Juge N. Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 11, 359–367 (2006). [DOI] [PubMed] [Google Scholar]
- Nishimura M. T. et al. Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972 (2003). [DOI] [PubMed] [Google Scholar]
- Evans N. A., Hoyne P. A. & Stone B. A. Characteristics and specificity of the interaction of a fluorochrome from aniline blue (sirofluor) with polysaccharides. Carbohyd. Polym. 4, 215–230 (1984). [Google Scholar]
- Betzig E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006). [DOI] [PubMed] [Google Scholar]
- Rust M. J., Bates M. & Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176 (2008). [DOI] [PubMed] [Google Scholar]
- Him J. L., Pelosi L., Chanzy H., Putaux J. L. & Bulone V. Biosynthesis of (1–>3)-beta-D-glucan (callose) by detergent extracts of a microsomal fraction from Arabidopsis thaliana. Euro. J. Biochem. 268, 4628–4638 (2001). [DOI] [PubMed] [Google Scholar]
- Belov V. N., Wurm C. A., Boyarskiy V. P., Jakobs S. & Hell S. W. Rhodamines NN: a novel class of caged fluorescent dyes. Angew. Chem. Int. Ed. Engl. 49, 3520–3523 (2010). [DOI] [PubMed] [Google Scholar]
- Ries J., Kaplan C., Platonova E., Eghlidi H. & Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584 (2012). [DOI] [PubMed] [Google Scholar]
- Anderson C. T., Carroll A., Akhmetova L. & Somerville C. Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S. et al. Role of the putative membrane-bound endo-1,4-beta-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol. 42, 251–263 (2001). [DOI] [PubMed] [Google Scholar]
- Paiva A. T., Sequeira S. M., Evtuguin D. V., Kholkin A. L. & Portugal I. Modern Research and Educational Topics in Microscopy Méndez-Vilas A. & J. Díaz, J. (eds.) 726–733 (Formatex Research Center, Badajoz, 2007). [Google Scholar]
- Hubbe M. A., Rojas O. J., Lucia L. A. & Jung T. M. Consequences of the nanoporosity of cellulosic fibers on their streaming potential and their interactions with cationic polyelectrolytes. Cellulose 14, 655–671 (2007). [Google Scholar]
- Carles J. E. & Scallan A. M. The determination of the amount of bound water within cellulosic gels by NMR spectroscopy. J. Appl. Polym. Sci. 17, 1855–1865 (1973). [Google Scholar]
- Harada T., Misaki A. & Saito H. Curdlan: A bacterial gel-forming β-1, 3-glucan. Arch. Biochem. Biophys. 124, 292–298 (1968). [DOI] [PubMed] [Google Scholar]
- Saito H., Ohki T. & Sasaki T. A 13C-nuclear magnetic resonance study of polysaccharide gels. Molecular architecture in the gels consisting of fungal, branched (1→3)-β-D-glucans (lentinan and schizophyllan) as manifested by conformational changes induced by sodium hydroxide. Carbohyd. Res. 74, 227–240 (1979). [Google Scholar]
- Chundawat S. P. S. et al. Multi-scale visualization and characterization of lignocellulosic plant cell wall deconstruction during thermochemical pretreatment. Energy Environ. Sci. 4, 973–984 (2011). [Google Scholar]
- Zhao Z., Liu H., Wang C. & Xu J. R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 14, 274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M. et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18, 731–746 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Wang W., Bates M. & Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Video S1
Video S2
Video S3
Video S4
Video S5
Video S6
Video S7
Video S8
Video S9
Video S10