Abstract
Objective
To create a sense inventory of abbreviations and acronyms from clinical texts.
Methods
The most frequently occurring abbreviations and acronyms from 352 267 dictated clinical notes were used to create a clinical sense inventory. Senses of each abbreviation and acronym were manually annotated from 500 random instances and lexically matched with long forms within the Unified Medical Language System (UMLS V.2011AB), Another Database of Abbreviations in Medline (ADAM), and Stedman's Dictionary, Medical Abbreviations, Acronyms & Symbols, 4th edition (Stedman's). Redundant long forms were merged after they were lexically normalized using Lexical Variant Generation (LVG).
Results
The clinical sense inventory was found to have skewed sense distributions, practice-specific senses, and incorrect uses. Of 440 abbreviations and acronyms analyzed in this study, 949 long forms were identified in clinical notes. This set was mapped to 17 359, 5233, and 4879 long forms in UMLS, ADAM, and Stedman's, respectively. After merging long forms, only 2.3% matched across all medical resources. The UMLS, ADAM, and Stedman's covered 5.7%, 8.4%, and 11% of the merged clinical long forms, respectively. The sense inventory of clinical abbreviations and acronyms and anonymized datasets generated from this study are available for public use at http://www.bmhi.umn.edu/ihi/research/nlpie/resources/index.htm (‘Sense Inventories’, website).
Conclusions
Clinical sense inventories of abbreviations and acronyms created using clinical notes and medical dictionary resources demonstrate challenges with term coverage and resource integration. Further work is needed to help with standardizing abbreviations and acronyms in clinical care and biomedicine to facilitate automated processes such as text-mining and information extraction.
Keywords: Abbreviations as Topic*, Medical Records*, Natural Language Processing*, Word sense disambiguation, Clinical sense inventory
Introduction
Abbreviations and acronyms in biomedical and clinical documents are pervasive, and their use is expanding rapidly.1–5 With the accelerated adoption of electronic health record (EHR) systems and proliferation of clinical texts, there is an increasing need to deal with abbreviations and acronyms and to utilize electronic clinical documents for automated processes. In addition to electronic clinical notes that are traditionally created by dictation and transcription, many clinical notes are now created at the point of care where clinicians type, dictate using voice recognition software, enter notes using a semi-structured or templated document entry system, or use a hybrid of several of these approaches. This often results in the use of shortened word forms that often have multiple meanings and may present a challenge for subsequent automated information extraction from notes, potentially resulting in patient safety issues.6–8 Computational approaches to automated Word Sense Disambiguation (WSD) that rely on Natural Language Processing (NLP) can help resolve abbreviation ambiguity and improve information extraction from clinical texts.
Sense inventories of abbreviations and acronyms are important and considered an essential component for automated NLP systems. Abbreviation and acronym sense resolution, a special case of WSD,9–11 is most effectively achieved based on the presence of a consistent and complete sense inventory. Compiling sense inventories is a challenge, however, since this process is very labor intensive. As a consequence, the work on abbreviation and acronym sense inventories to date in the clinical domain is somewhat limited, resulting in limited availability of these inventories.
Although abbreviation and acronym sense inventories extracted from biomedical literature have been studied extensively, relatively little research has been devoted to the creation of a sense inventory of abbreviations and acronyms within clinical notes.12–13 In this paper, ‘biomedical literature’ is defined as the collective literature from various fields of biomedicine and healthcare in the form of abstracts and full-text articles. Here, ‘clinical documents’ are clinical notes created in the process of patient care. With biomedical literature,14–19 typically the first instance of a short form for the abbreviation or acronym co-occurs with the long form as a parenthetical expression or vice versa (eg, ‘mucosal ulcerative colitis (MUC)’).20 In contrast, clinical notes are informal in nature and the co-location of the long form and the short form in clinical text is rarely observed.4 12 Moreover, the development of any abbreviation and acronym sense inventory from clinical texts is hindered by issues of patient confidentiality and privacy that make sharing and using clinical notes for research purposes difficult.1 21 Not surprisingly, there are currently only small clinical sense inventory datasets (up to 7738) of abbreviations and acronyms (up to 16) available (eg, datasets by Xu et al12 and Joshi et al13).
The goal of this work was to create and release for public use a clinical sense inventory of clinical abbreviations and acronyms, harmonized with a medical dictionary Stedman's Medical Abbreviations, Acronyms & Symbols, 4th edition (Stedman's)22; the Unified Medical Language System (UMLS)23; and an abbreviation and acronym sense inventory from biomedical literature, Another Database of Abbreviations in Medline (ADAM).14 From this work, we sought to understand different usages of clinical abbreviations and acronyms and the relative coverage and degree of overlap across these resources.
Background
Unified Medical Language System
The UMLS is distributed through the National Library of Medicine as a set of medical terminology resources organized by concepts. In addition to providing a resource for identification of medical terms, the UMLS provides ontological relationship information consisting largely of concepts connected via an ‘is-a’ hierarchy.9 21 24 While the UMLS was chosen for use in this study, the National Center for Biomedical Ontology (NCBO) BioPortal25 is a complementary ontological resource with similar functionality including both ‘is-a’ and other relationships between concepts. There are a number of relational files and tools available to access and utilize the UMLS. For example, the National Library of Medicine provides the SPECIALIST Lexicon26 (including the LRABR file) and a part of the SPECIALIST Lexicon tool, Lexical Variant Generation (LVG),27 which allows for term normalization and stemming in the distribution of MetaMap.28 Moreover, MetaMap, which was used in this study, is a software application developed to map text to corresponding biomedical concept(s) indexed with the UMLS Concept Unique Identifier (CUI) and its associated UMLS semantic type (the UMLS semantic type of each concept). Similarly to MetaMap functionality for mapping text to the UMLS concepts, NCBO also has developed a concept mapping tool, Mgrep, which has been previously compared to MetaMap.29
While the UMLS is a natural resource for mapping senses of clinical abbreviations and acronyms, the UMLS has previously been shown to have limited coverage of abbreviations and acronyms,30 although some work has shown improved coverage for a subset of acronyms. For example, Xu et al5 in 2009 found that the UMLS only covered approximately 35% of the abbreviations and acronyms that the authors examined in the clinical domain. Similarly, Liu et al31 reported coverage of 66% of examined abbreviations and acronyms with less than six characters in the clinical domain by the UMLS.
Another Database of Abbreviations in Medline
A number of rule-based and statistically generated sense inventories have been created using the assumption that the short form and the long form of an abbreviation or acronym are collocated when first introduced in biomedical literature documents (eg, SaRAD,15 ARCH,16 and ALICE17). Among them, ADAM is a representative abbreviation and acronym biomedical sense inventory resource generated from titles and abstracts via 2006 Medline.14 ADAM contains 59 403 pairs of short and long forms as a database for B-terms32 projected after filtering out insignificantly connected pairs based on length ratio rules and empiric cut-off values. B-terms represent the relevant score between two articles with title words or phrases. ADAM also provides the term frequency of different terms along with other statistical information to illustrate usage of each abbreviation or acronym within the biomedical literature. ADAM does, however, contain a significant level of redundancy between different long form expressions owing to the lack of either syntactic or semantic normalization between different expressions.
Medical dictionaries
Medical dictionaries such as Stedman's and Dorland's are currently not available as part of the UMLS and thus tend to be underutilized in the development of biomedical and clinical NLP work. These dictionaries may, however, provide an important adjunctive resource for clinical sense inventories because medical dictionaries are used commonly within the clinical domain and have a large amount of information about biomedical and clinical terms represented in texts. The definitions of terms in these resources can also be potentially used to constrain semantic information for related tasks such as WSD.33 On the other hand, potential issues with medical dictionaries include copyright restrictions, the comparative slowness of these resources to adopt new clinical terms, and the hybrid nature of these resources, which contain both clinical as well as basic science terms.
Methods
Clinical documents from four hospitals in the University of Minnesota affiliated Fairview Health Services, including the University of Minnesota Medical Center and other Fairview metropolitan hospitals in the Twin Cities, from 2004 to 2007 in our clinical document data repository were used for this study. The corpus contains primarily verbally dictated and transcribed notes stored in electronic format. These 352 267 clinical notes include admission notes, consultation notes, and discharge summaries. Table 1 describes the metadata for corpus.
Table 1.
Metadata for corpus
| Metadata | Description | Entire corpus (%) | Sampled notes* (%) |
|---|---|---|---|
| Total | 352 267 (100) | 90 907 (100) | |
| Note date (year) | 2004 | 82 433 (23.4) | 18 182 (20.0) |
| 2005 | 86 396 (24.5) | 22 400 (24.6) | |
| 2006 | 89 457 (25.4) | 23 319 (25.7) | |
| 2007 | 93 981 (26.7) | 27 006 (29.7) | |
| Note type | Admission note | 134 027 (38.1) | 31 327 (34.5) |
| Consultation note | 209 715 (59.5) | 50 008 (55.0) | |
| Discharge summary | 8525 (2.4) | 9572 (10.5) | |
| Author specialty | Adult behavioral health | 42 688 (12.1) | 560 (0.6) |
| Adult critical care | 862 (0.3) | 258 (0.3) | |
| Adult medicine (general) | 152 942 (43.4) | 46 005 (50.6) | |
| Adult medical specialty | 34 348 (9.8) | 14 133 (15.6) | |
| Surgery (general) | 22 650 (6.4) | 5214 (5.7) | |
| Surgical specialty | 32 891 (9.3) | 6468 (7.1) | |
| Obstetrics and gynecology | 27 509 (7.8) | 7764 (8.5) | |
| Pediatric behavioral health | 10 736 (3.1) | 1 (0.0) | |
| Pediatric critical care | 86 (0.0) | 49 (0.0) | |
| Pediatrics (general) | 15 637 (4.4) | 5001 (5.5) | |
| Pediatric medical specialty | 5311 (1.5) | 3279 (3.6) | |
| Missing specialty | 6607 (1.9) | 2175 (2.4) | |
| Age of patient | less than 10 | 13 918 (4.0) | 4518 (5.0) |
| 10–19 | 29 247 (8.3) | 4923 (5.4) | |
| 20–29 | 31 464 (8.9) | 7806 (8.6) | |
| 30–39 | 41 425 (11.8) | 10 098 (11.1) | |
| 40–49 | 49 142 (13.9) | 11 437 (12.6) | |
| 50–59 | 50 496 (14.3) | 13 500 (14.8) | |
| 60–69 | 43 659 (12.4) | 12 662 (13.9) | |
| 70–79 | 43 580 (12.4) | 12 423 (13.7) | |
| 80 and over | 49 336 (14.0) | 13 540 (14.9) | |
| Gender of patient | Male | 150 773 (42.8) | 38 806 (42.7) |
| Female | 201 494 (57.2) | 52 101 (57.3) |
*Sampled notes were the unique notes from which abbreviation and acronym clinical sense inventory samples were extracted.
Identification of significant abbreviations and acronyms
To select meaningful and common abbreviations and acronyms, a set of heuristic rules was applied. Potential abbreviations and acronyms were identified when the word token consisted of capital letters or numbers, with or without symbols (period, comma, colon, or semicolon) using regular expressions from clinical texts. Abbreviations and acronyms expressed in lowercase letters were excluded. For the current project, we leveraged the fact that the clinical notes we used were all dictated and transcribed by professional transcriptionists. Therefore, we expected relatively consistent capitalization of abbreviations and acronyms with the exception of shortened word forms (eg, ‘cont’ for continued or ‘mdl’ for ‘middle’). Furthermore, abbreviations and acronyms that were also English words were annotated as such. We did not remove stop words with our abbreviation and acronym detection methods since many stop words are short in length and could potentially also be an acronym or abbreviation (eg, ‘AND’ in dermatology: ‘acute neutrophilic dematosis’). Combinations of symbols in front or at the back of the targeted word token were accepted as a potential abbreviation or acronym. If the token of interest was part of document formatting (eg, header, footer, or transcription formatting), it was excluded. Heuristic rules were applied to clinical notes to detect the section information for the abbreviation or acronym. Only candidate abbreviations or acronyms with a frequency of over 500 in the corpus were included, resulting in 440 abbreviations and acronyms. The surrounding text for each of the 500 instances was also extracted and included in the inventory. The instance consisted of 12 previous-word tokens and 12 post-word tokens centering the targeted abbreviation and acronym. A set of 12 word tokens was selected based on previous work in general English showing that this is more than sufficient for manual annotation.34
Identification of possible long forms from various medical areas
All 220 000 instances for the 440 abbreviations and acronyms were given to two clinical experts for manual annotation of their clinical sense. Annotated long forms were then standardized with long forms of Stedman's. We choose Stedman's among medical dictionaries because this was available electronically and had a resource specific for abbreviations and acronyms. At this stage, formatting errors were eliminated and replaced by additional samples focusing the clinical sense inventory on the overall sense distributions of our corpus. For example, ‘1. Atrial fibrillation. 2. C3. omfort cares…’; ‘C3’ is not a valid abbreviation or acronym but rather a formatting mistake. The inter-rater reliability of the annotated senses was reported with percentage agreement and with the kappa statistic with a third clinical expert who examined 11 000 random samples (25 per abbreviation or acronym; 5% of the total).
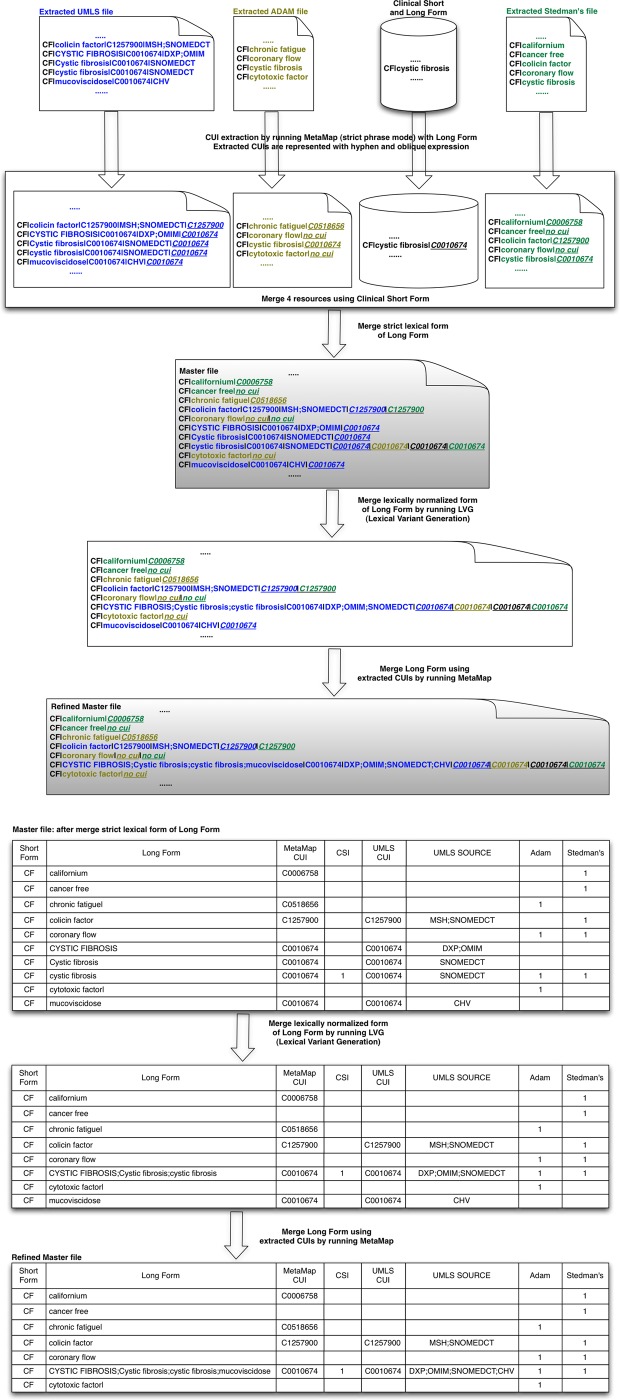
Figure 1 provides an overview of how potential long forms in the UMLS were obtained for each of the abbreviations and acronyms. As a first step, each short form of a given clinical abbreviation or acronym was mapped using the Metathesaurus MRCONSO.RRF file (UMLS 2011AB) to determine the corresponding long form(s), CUI(s) and English term type(s) (see shaded box in figure and the arrow). Second, the clinical short forms were mapped using the LRABR file to extract pairs of short forms and long forms mapped to the UMLS. These long forms from the LRABR file (the UMLS SPECIALIST Lexicon) were re-mapped to MRCONSO.RRF to get CUI(s) and English term type(s) (dotted line). Third, all identified long forms from the first and second steps were merged based on short forms (‘Merging process based on Short Form’). Fourth, collected CUIs and long forms were remapped (dotted line) to MRCONSO.RRF one more time to detect any missing variants of the long forms/information in the UMLS. The result of this process is represented as ‘Re-evaluated & Extracted UMLS file’ in figure 1.
Figure 1.
Overview of collected long forms from the Unified Medical Language System (UMLS). CUI, Concept Unique Identifier.
Short forms of abbreviations and acronyms in the clinical domain were directly mapped to short forms of ADAM since ADAM has paired representations of short forms and long forms of abbreviations and acronyms. Additionally, we included the coverage and usage frequency of individual long forms from ADAM so as to include information about the relative usages within the biomedical literature.
Finally, for each short form, all long forms associated with a targeted clinical abbreviation or acronyms were extracted from the Stedman's. All bracketed expressions in the dictionary were reviewed to select all possible inflected forms. For example, ‘TEE’ had an original representation as ‘transesophageal echocardiograph(y) (echocardiogram)’ in Stedman's. For this, ‘echocardiogram’ and ‘echocardiography’ were kept because they have similar meanings to ‘echocardiograph’. As a result, we had three expressions for ‘TEE’: ‘transesophageal echocardiogram’, ‘transesophageal echocardiograph’, and ‘transesophageal echocardiography’.
Normalization process and analysis of the sense inventory
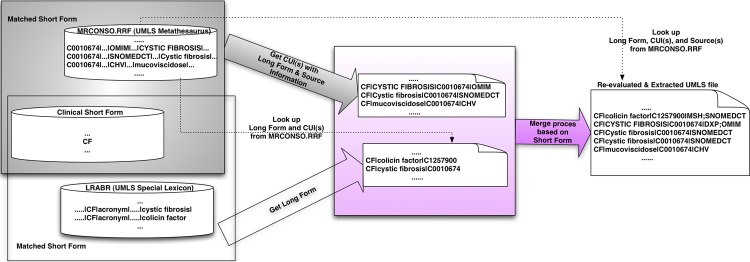
The initial sense inventory for the source clinical abbreviations and acronyms was systematically compared to each of the resources (UMLS, ADAM, Stedman's) to identify similarities and differences. Figure 2 provides an overview of the mapping processes for all acquired long forms from various medical resources. A two-step process was used to merge long forms by applying a lexical step followed by a semantic merging step.
Figure 2.
Merging process of long forms. Extracted Unified Medical Language System (UMLS) file=result from figure 1. ADAM, Another Database of Abbreviations in Medline; Stedman's, Stedman's Medical Abbreviations, Acronyms & Symbols; CUI, Concept Unique Identifier.
Before the two-step process, all previously obtained long forms were used as inputs into MetaMap. CUIs produced by MetaMap as final mappings were included only if they had a score of 1000 (highest score/confidence) to ensure exact mapping of given long forms. The MetaMap term processing option (-z) was used to obtain exact matches when MetaMap processed long forms. The ‘-z’ term processing option forces MetaMap to treat individual strings as a single phrase/unit (rather than a sentence or a full text). Therefore, with this option turned on, MetaMap processes input without splitting or rearranging it, which helps to obtain the most exact mappings for the vocabulary terms. Thus each identified long form obtained a relevant set of CUI(s) (from MRCONSO and MetaMap) that was subsequently included in the inventory.
Lexical merging of long forms was first performed to find exact matches of lexical forms of each acronym's long form in various medical resources. Only long forms with the same lexical representations were used to create the ‘Master file’ as shown in figure 2. Following this, LVG normalization with individual long forms was used to remove simple variations of lexical representations. Examples of these simple variations of lexical representations include plural expressions, word order differences, stop words (eg, ‘and’, ‘the’, ‘of’), and variation in punctuation and other symbols. Long forms with exactly the same forms after normalization through LVG were merged to represent a single concept.
Following lexical matching, semantic mapping between long forms was performed based on CUIs to enhance the quality of the sense inventory. Only perfect mappings based on CUIs from the UMLS were taken into consideration. In other words, if any set of CUIs for a given long form had an overlap of 100% to the set of CUIs for another long form, the two long forms were regarded as the same concept/meaning but had different lexical representations. These semantically equivalent long forms were mapped into a single representation in our ‘Refined Master file’ as shown in figure 2.
Results
Characteristics of clinical sense inventory
Within the overall clinical corpus of 352 267 notes, 440 common abbreviations and acronyms with 949 long forms were found occurring with a frequency of 500 or more instances in the corpus. For inter-rater reliability, the percent agreement was on average 99% and the kappa statistic was on average 0.97 between annotators. Among acronyms and abbreviations, GTT (80%, 0.25), SI (84%, 0.30), GT (84%, 0.30), US (76%, 0.35), NP (88%, 0.36), INH (88%, 0.47), ES (92%, 0.48), PCA (92%, 0.48), AP (96%, 0.49), and DP (96%, 0.49) had fair to high percent agreement and low kappa statistic (less than 0.6 in table 2).
Table 2.
Kappa statistics and sense distributions in clinical corpus
| Number of abbreviations and acronyms | |
|---|---|
| Range of value of kappa statistic | |
| 0.90–1.00 | 398 |
| 0.80–0.90 | 16 |
| 0.70–0.80 | 10 |
| 0.60–0.70 | 6 |
| Less than 0.60 | 10 |
| Total | 440 |
| Ratio of majority sense | |
| 99–100% | 323 |
| 95–99% | 42 |
| 90–95% | 14 |
| 80–90% | 21 |
| 70–80% | 11 |
| 60–70% | 8 |
| 50–60% | 14 |
| Less than 50% | 7 |
| Total | 440 |
The majority of abbreviations and acronyms in the clinical sense inventory had skewed distributions for meanings. Overall, 276 of 440 (62.7%) of abbreviations and acronyms had only one sense (long form). This majority sense prevalence was significantly different in comparison to the distributions seen in the biomedical literature. Table 2 shows the frequency distribution of the clinical senses sorted according to the baseline majority sense prevalence. Of all cases, 83% had one dominant majority sense using a conservative ratio of >95% as the definition of a dominant majority. The clinical sense inventory contained several institution-specific terms with senses that were not generalizable to the greater clinical domain. For example, in table 3, the acronym ‘FUTS’ is a short form for ‘Fairview University Transitional Services’. Another similar example, ‘FSH’ in the dataset was often used (46% of the time) to represent ‘Fairview Southdale Hospital’. In addition to these two examples, out of a total of 949 senses in the clinical sense inventory, there were 2 (0.3%) total institution-specific (Fairview-specific) abbreviation terms, 19 (0.8%) terms that were not covered by any biomedical or clinical resource, and 83 (0.1%) terms that were considered by our reviewers not to be common clinical terms.
Table 3.
Sense of FUTS and FSH
| Abbreviation | Sense | Number of instance | Coverage |
|---|---|---|---|
| FUTS | Fairview University Transitional Services | 500 | 1.00 |
| FSH | Follicle-stimulating hormone | 265 | 0.53 |
| Fairview Southdale Hospital | 231 | 0.46 | |
| Fascioscapulohumeral muscular dystrophy | 4 | 0.01 |
Overall, 335 cases of typos in acronyms were observed in the corpus used to create the clinical sense inventory. For example, the text in one instance stated: ‘…PAC pump for anesthesia…’ which should have been ‘PCA (patient-controlled analgesia)’ rather than ‘PAC’ based on the context in which the acronym occurred. In another example: ‘The patient is on Biaxin for mycobacterium AVM intracellular infection’. Here, ‘AVM’ was supposed to be ‘avium’. This error was most likely due to the transcriptionist mishearing what the physician dictated. Most frequently in our dataset, we observed mistaken use of ‘BMP’ which should have been ‘BNP’ (36 times), ‘BNP’ which should have been ‘BMP’ (18 times), ‘DT’ which should have been ‘DP’ (23 times), and PM which should have been ‘PMR’ (74 times).
An additional 306 errors were observed. An example of a mistake due to unclear meaning includes the following: ‘His factor 2 SA was 14 on admission and factor 12 SA was 62’. We represented these unsure cases as ‘unsure sense’ in our clinical sense inventory. Sometimes, the detected abbreviation or acronym was a part of word phrase that together had a particular meaning. For example, ‘Mucolytics and EC PAP device’. ‘EC PAP’ should be corrected as ‘EZ PAP’, but ‘EZ’ itself has no meaning without ‘PAP’.
Comparison among different resources
Figure 3 represents the coverage among resources. Looking only at those long forms with an exact match of lexical forms, among 24 853 total senses (long forms) of 440 abbreviations and acronyms, 224 total were matched exactly across all resources. For example, the abbreviation ABG had a single sense ‘arterial blood gas’ with the CUI ‘C0150411’. All sources (UMLS, ADAM, and Stedman's) had the long form ‘arterial blood gas’. Some long forms represented several preferred CUIs, like AVM had the sense ‘arteriovenous malformation’ with two associated CUIs: ‘C0003857’ and ‘C0334533’. Overall, these exact and completely matched long forms for all medical resources represented only 0.9% of long forms in the dataset (224 of 24 853 long forms).
Figure 3.
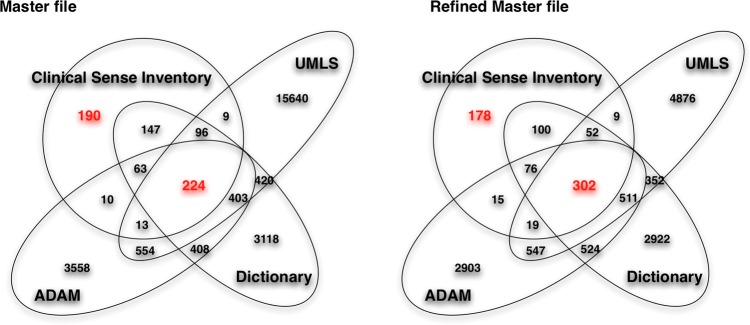
The coverage among resources. Master file and Refined Master file=result from figure 2. UMLS, Unified Medical Language System; ADAM, Another Database of Abbreviations in Medline; Stedman's, Stedman's Medical Abbreviations, Acronyms & Symbols.
The low rate of matching long forms across all resources was improved after the normalization and merging of long forms. A total of 24 853initial total long forms were merged into 17 096 long forms after performing LVG normalization (figure 2). At this stage, exact and complete matches on long forms for all resources increased to 1.7% (296 of 17 096 long forms). After we applied semantic matching for equivalent CUIs, the exact match rate increased to 2.3% (302 of 13 386 long forms).
After this three-phrase merge process, clinical long forms covered 50.9% (382 of 751 long forms) of the UMLS, 54.9% (412 of 751 long forms) of ADAM, and 70.6% (530 of 751 long forms) in Stedman's. Relative to the clinical sense inventory, the coverage of UMLS, ADAM, and Stedman's was 5.7% (382 of 6668), 8.4% (412 of 4897), and 11% (530 of 4839), respectively, of long forms in the clinical sense inventory.
We also observed that the use of abbreviations was different between the clinical and biomedical domains by comparing the clinical sense inventory with ADAM. For example, ODT is used (100%) for ‘orally disintegrating tablet’ in our clinical sense inventory but in the biomedical literature, ODT is used (100%) for ‘oculodynamic test’. Similarly, FEN means (100%) for ‘fluids, electrolytes, nutrition’ for in the clinical domain, but it is mainly used (68.1%) for ‘fenfluramine’ (C0015827) in the biomedical literature. We found different usage by domain (100% dominantly used in the clinical sense inventory but less than 50% in ADAM) with 33 abbreviations and acronyms.
We observed that some clinical senses did not correspond to long forms within any of the resources. Among 949 long forms in the clinical sense inventory, 190 had no coverage in any of the three resources using exact matches of lexical forms. This was reduced through LVG normalization and semantic matching, which reduced the number of unmatched long forms to 178. Table 4 gives some examples of long forms among the four resources.
Table 4.
Sense comparisons between the clinical sense inventory and other resources
| Short form | Long form | MetaMap CUI | CSI | Ratio in CSI | UMLS CUI | UMLS SOURCE | ADAM | Ratio in ADAM | Stedman's |
|---|---|---|---|---|---|---|---|---|---|
| AVR | Lead AVR; lead avr; aVR | C0449217 | C0449217 | CHV; LNC; SNOMEDCT | |||||
| aVR (body structure) | C0449217 | SNOMEDCT | |||||||
| Accelerated ventricular rhythm | 1 | ||||||||
| Acute vascular rejection | 1 | 0.0634 | |||||||
| Antiviral regulator | 1 | ||||||||
| Aortic valve regurgitation | C0003504 | 1 | 0.01 | ||||||
| Aortic valve replacement | C0003506 | 1 | 0.762 | C0003506 | CHV; COSTAR; NCI; SNOMEDCT | 1 | 0.8687 | 1 | |
| Aortic valve resistance | 1 | 0.008 | |||||||
| Arteriole-to-venule ratio | 1 | 0.0133 | |||||||
| Ascending vasa recta | C2952018 | C2952018 | FMA | 1 | 0.0398 | ||||
| Augmented voltage right arm | 1 | 0.206 | 1 | ||||||
| Pathogen avirulence | 1 | 0.0147 | |||||||
| BKA | Bka; CGI-35; FCF1; FCF1 gene | C1426785 | C1426785 | HUGO; MTH | |||||
| FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) | C1426785 | HUGO | |||||||
| Below-knee amputation | C0002692 | 1 | 1 | C0002692 | NCI | 1 | 0.5714 | 1 | |
| Bongkrekic acid | C0005982 | C0005982 | MSH; NDFRT | 1 | 0.4286 | ||||
| IOL | IOL; iol; Primary Intraocular Lymphoma | C0281658 | C0281658 | CHV; NCI; PDQ | |||||
| Intraocular Lymphoma; Intraocular lymphoma; intraocular lymphoma; lymphoma, intraocular; intraocular lymphoma (IOL) | C0281658; C1706527 | CHV; MTH; NCI; PDQ | |||||||
| Induction of labor | C0259787 | 1 | |||||||
| Interosseous ligament | C0447892 | 1 | 0.0968 | ||||||
| Intraocular lens; intraocular lenses | C0023311; C0023319; C0336564 | 1 | 1 | C0023319; C0336564; C1706007 | CHV; MSH; NCI; SNOMEDCT | 1 | 0.7849 | 1 | |
| Intraocular lens implantation | C0023311 | 1 | 0.1183 |
ADAM, Another Database of Abbreviations in Medline; CSI, Clinical Sense Inventory; CUI, Concept Unique Identifier; MetaMap CUI, CUI produced by running MetaMap; Stedman's, Stedman's Medical Abbreviations, Acronyms & Symbols; UMLS, Unified Medical Language System; UMLS SOURCE, source information in the UMLS.
Discussion
This study provides and evaluates a sense inventory for clinical abbreviations and acronyms and compares and contrasts the long forms and short forms across three terminological resources: UMLS, ADAM, and Stedman's. The clinical sense inventory had overall highly skewed sense distributions, some local or practice-specific senses, and a number of erroneous instances. Our analysis of the 440 most common abbreviations and acronyms from clinical notes demonstrated that many long forms were not perfectly matched even after conducting lexical mappings and semantic comparisons. Despite some of the challenges and limitations encountered in the process of creating the sense inventory, we believe that the resultant resource from this study currently represents the largest sense inventory of clinical abbreviations and acronyms with accompanying examples of clinical contexts in which the acronyms occur. This resource is publically available to support the research of the greater NLP and biomedical and health informatics community. For example, NLP researchers can use this resource to validate WSD techniques, such as our work examining window size and orientation for clinical abbreviation WSD.35
We observed that vocabulary resources used in this study had uneven granularity of sense distributions as compared to each other. This created challenges in the normalization process of the inventory's long forms. For example, ADAM and the UMLS distinguished ‘total knee arthroplasty’ and ‘total knee arthroscopy’. In contrast, Stedman's collapses these two concepts in a single sense: ‘total knee arthroplasty (arthroscopy)’. Because this combined sense is not suitable for obtaining CUIs with MetaMap and has two semantic meanings, this was separated into two expressions for our study.
Another challenge encountered with the sense inventory was that of ambiguous abbreviations or acronyms within the text. For example, ‘Imdur SA 60 mg p.o. q.d.’. ‘SA’ can be either ‘slow acting’ or ‘sustained action’, which has a similar sense but different long forms. The occurrence of two meaningful senses repeatedly occurring was prominent in a few abbreviations/acronyms. These ambiguous senses were observed 373 times with ‘SA’ (‘slow acting’ or ‘sustained action’), 121 times with ‘OP’ (‘oblique presentation’ or ‘occiput posterior’), and 105 times with ‘MP’ (‘metatarsophalangeal’ or ‘metacarpophalangeal’) in the 500 samples of those particular abbreviations/acronyms. We also observed some ambiguity associated with senses related to levels. For example, abbreviation ‘C3’ has one representative sense, ‘cervical (level) 3’. Here ‘level’ can be interpreted as one of several meanings such as ‘nerve’, ‘dermatome’, ‘vertebrae’, or ‘disc’ depend on surrounding words.
Another issue with term normalization across resources was the degree of redundancy of long form terms, particularly the significant degree of redundancy in ADAM among its different long forms, where all distinct lexical forms remained separated. Additional steps are required to further reduce the redundancy of long form senses prior to mapping to ADAM long forms to other resources. While some work has been done to merge synonymous variants of the long forms,36 our sense inventory only utilized strict and exact matching processes.
The assumption used in biomedical literature and general English is generally that there is only one sense per discourse per abbreviation/acronym. This assumption stems from NLP work in general English WSD.37 We found this to be an invalid assumption for clinical documents. In some instances, even the assumption of one sense per sentence does not hold in clinical discourse, making the problem of word sense ambiguity resolution more challenging in this domain. We found several examples where two senses for an abbreviation/acronym were observed within a single sentence such as: ‘Postop MRI recently showed increase T2 signal from C2 through T2 level’. Here, the first ‘T2’ means ‘T2 (MRI phase)’ but the second ‘T2’ means ‘thoracic vertebra 2’. We did find, however, that most instances of ‘T2 (MRI phase)’ appeared in the section ‘PROCEDURE’, and the sense ‘thoracic vertebra 2’ appeared mostly in the section ‘HISTORY OF PRESENT ILLNESS’, indicating that the section may be helpful for determining the sense of an abbreviation/acronym in a clinical discourse. The section information will not be helpful in all cases, however.
One observation that has been made previously5 31 and was confirmed by our study is that the UMLS is limited as a resource for mapping short forms with long forms. The LRABR file in the UMLS contains overall 57 704 pairs of short and long forms. Of the 949 long forms, 190 in the clinical sense inventory were missing in the UMLS. This fact demonstrates challenges. With Stedman's and ADAM, there was less coverage of long forms although some other areas of coverage not afford by the UMLS, pointing to the complementary nature of these resources.
Limitations
Our study has several limitations. After performing exact lexical matching, the techniques used for normalization of senses were dependent on the automated tools we used (ie, MetaMap and LVG), which may introduce additional errors in the normalization process. Also, other aspects of the clinical notes such as specialty or the section information were not utilized to normalize senses or to narrow the scope of senses. Because our sense inventory was built based on only 500 random samples from one institution that were extracted and manually annotated, these samples may not be completely representative of the entire corpus. It is also possible that these samples exclude additional minority senses.
We also examined the coverage of 554 online medical abbreviations from Wikipedia38 on our clinical sense inventory. After the three-phrase merge process as described in the methods, the clinical long forms contained in Wikipedia covered 35.6% (267 of 751 long forms) of the clinical sense inventory. This coverage is relatively low compared to 50.9% with the UMLS, 54.9% with ADAM, and 70.6% with Stedman's. Therefore, we concluded that currently Wikipedia's coverage is not ideal for clinical abbreviation and acronym inventories. However, the fact that we found over a third of the long forms in Wikipedia is encouraging and indicates that, as Wikipedia continues to grow, it may soon become a useful resource for medical abbreviation and acronym disambiguation.
In future work, we plan to utilize semi-automated methods as previously described12 by Xu et al to enrich our sense inventory and to overcome our manual annotation with strict and exact matching processes, concentrating our effort on abbreviations/acronyms without a single dominant sense. Also, we would like to validate our sense inventory using the corpus of another institution. Nevertheless, this study and the associated resultant sense inventory represents a significant contribution and resource for others to use in the clinical NLP domain. The anonymized dataset of acronyms and abbreviations (those with dominant sense <95%) and sense inventories are publically available at http://www.bmhi.umn.edu/ihi/research/nlpie/resources/index.htm (‘Sense Inventories’ website).
Conclusion
Although abbreviations and acronyms in clinical text are used widely in clinical documentation, relatively little work has focused on building a clinical sense inventory for abbreviations and acronyms for the purposes of NLP research and dissemination to the wider scientific community. We created a clinical sense inventory with 440 common abbreviations and acronyms and compared the senses with the UMLS, ADAM, and Stedman's. From this, we were able to examine the information within and perform a gap analysis of these clinical abbreviations and acronyms among diverse resources. Moreover, this work could be used as a foundational resource with semi-automated techniques that aim to scale the disambiguation of abbreviations for real-word use.
Supplementary Material
Acknowledgments
We would like to thank Fairview Health Services for support of this research. The authors also thank Andrea Abbott for manual annotations, and Bridget McInnes for insightful comments.
Appendix
Resources generated from this study are available at http://www.bmhi.umn.edu/ihi/research/nlpie/resources/index.htm (‘Sense Inventories’ website):
The sense inventory of clinical abbreviations and acronyms (two versions)
Anonymized sentence datasets
README (two versions)
Footnotes
Contributors: SM, SP, and GM designed the methods and experiments, analyzed the data, and interpreted the results. NL and JR annotated data and SM carried out the experiments. SM drafted the manuscript. SP and GM conceived of the study, and helped to draft the manuscript. All authors read and approved the manuscript.
Funding: This work was supported by the University of Minnesota Institute for Health Informatics Research Support Grant 2011–2012, the American Surgical Association Foundation Grant 2010–2012, and the National Library of Medicine #R01 LM009623-01.
Competing interests: None.
Ethics approval: University of Minnesota Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: This work has not been published elsewhere and represents original research. Data resources deriving from this article are available at http://www.bmhi.umn.edu/ihi/research/nlpie/resources/index.htm (‘Sense Inventories’ website).
References
- 1.Pakhomov S, Pedersen T, Chute CG. Abbreviation and acronym disambiguation in clinical discourse. AMIA Annu Symp Proc 2005:589–93 [PMC free article] [PubMed] [Google Scholar]
- 2.Pakhomov S. Semi-supervised maximum entropy based approach to acronym and abbreviation normalization in medical texts. Proceedings of the 40th Annual Meeting on Association for Computational Linguistics Philadelphia, Pennsylvania: Association for Computational Linguistics; 2002:160–7 [Google Scholar]
- 3.Stetson PD, Johnson SB, Scotch M, et al. The sublanguage of cross-coverage. Proc AMIA Symp 2002:742–6 [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H, Markatou M, Dimova R, et al. Machine learning and word sense disambiguation in the biomedical domain: design and evaluation issues. BMC Bioinformatics 2006;7:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu H, Stetson PD, Friedman C. A study of abbreviations in clinical notes. AMIA Annu Symp Proc 2007:821–5 [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn IF. Abbreviations and acronyms in healthcare: when shorter isn't sweeter. Pediatr Nurs 2007;33:392–8 [PubMed] [Google Scholar]
- 7.Walsh KE, Gurwitz JH. Medical abbreviations: writing little and communicating less. Arch Dis Child 2008;93:816–17 [DOI] [PubMed] [Google Scholar]
- 8.Hunt DR, Verzier N, Abend SL, et al. Fundamentals of Medicare patient safety surveillance: intent, relevance, and transparency, Rockville, MD: Agency for Healthcare Research and Quality, 2005 [PubMed] [Google Scholar]
- 9.Fan JW, Friedman C. Word sense disambiguation via semantic type classification. AMIA Annu Symp Proc 2008:177–81 [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman C, Liu H, Shagina L, et al. Evaluating the UMLS as a source of lexical knowledge for medical language processing. Proc AMIA Symp 2001:189–93 [PMC free article] [PubMed] [Google Scholar]
- 11.Schuemie MJ, Kors JA, Mons B. Word sense disambiguation in the biomedical domain: an overview. J Comput Biol 2005;12:554–65 [DOI] [PubMed] [Google Scholar]
- 12.Xu H, Stetson PD, Friedman C. Methods for building sense inventories of abbreviations in clinical notes. J Am Med Inform Assoc 2009;16:103–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi M, Pakhomov S, Pedersen T, et al. A comparative study of supervised learning as applied to acronym expansion in clinical reports. AMIA Annu Symp Proc 2006:399–403 [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W, Torvik VI, Smalheiser NR. ADAM: another database of abbreviations in MEDLINE. Bioinformatics 2006;22:2813–18 [DOI] [PubMed] [Google Scholar]
- 15.Adar E. SaRAD: a Simple and Robust Abbreviation Dictionary. Bioinformatics 2004;20:527–33 [DOI] [PubMed] [Google Scholar]
- 16.Wren JD, Garner HR. Heuristics for identification of acronym-definition patterns within text: towards an automated construction of comprehensive acronym-definition dictionaries. Method Inform Med 2002;41:426–34 [PubMed] [Google Scholar]
- 17.Ao H, Takagi TI. ALICE: An algorithm to extract abbreviations from MEDLINE. J Am Med Inform Assn 2005;12:576–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang JT, Schutze H, Altman RB. Creating an online dictionary of abbreviations from MEDLINE. J Am Med Inform Assoc 2002;9:612–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu HF, Johnson SB, Friedman C. Automatic resolution of ambiguous terms based on machine learning and conceptual relations in the UMLS. J Am Med Inform Assn 2002;9:621–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz AS, Hearst MA. A simple algorithm for identifying abbreviation definitions in biomedical text. Pac Symp Biocomput 2003:451–62 [PubMed] [Google Scholar]
- 21.Savova GK, Coden AR, Sominsky IL, et al. Word sense disambiguation across two domains: biomedical literature and clinical notes. J Biomed Inform 2008;41:1088–100 [DOI] [PubMed] [Google Scholar]
- 22.Stedman's medical abbreviations, acronyms & symbols. 4th ed. Lippincott Williams & Wilkins, 2008 [Google Scholar]
- 23.NIH. 2010. Unified Medical Language System.
- 24.Leroy G, Rindflesch TC. Using symbolic knowledge in the UMLS to disambiguate words in small datasets with a naive Bayes classifier. Stud Health Technol Inform 2004;107(Pt 1):381–5 [PubMed] [Google Scholar]
- 25.Whetzel PL, Noy NF, Shah NH, et al. BioPortal: enhanced functionality via new web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res 2011;39(Web Server issue):W541–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Browne AC, McCray AT, Srinivasan S. The SPECIALIST Lexicon. NLM, 2000 [Google Scholar]
- 27.McCray AT, Aronson AR, Browne AC, et al. UMLS knowledge for biomedical language processing. Bull Med Libr Assoc 1993;81:184–94 [PMC free article] [PubMed] [Google Scholar]
- 28.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001:17–21 [PMC free article] [PubMed] [Google Scholar]
- 29.Nigam HS, Nipun B, Clement J, et al. Comparison of concept recognizers for building the Open Biomedical Annotator. BMC Bioinformatics 2009;10(Suppl 9):S14;105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McInnes BT, Pedersen T, Carlis J. Using UMLS Concept Unique Identifiers (CUIs) for word sense disambiguation in the biomedical domain. AMIA Annu Symp Proc 2007:533–7 [PMC free article] [PubMed] [Google Scholar]
- 31.Liu HF, Lussier YA, Friedman C. A study of abbreviations in the UMLS. J Am Med Inform Assn 2001:393–7 [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson DR, Smalheiser NR. An interactive system for finding complementary literatures: a stimulus to scientific discovery. Artif Intell 1997;91:183–203 [Google Scholar]
- 33.McInnes BT, Pedersen T, Pakhomov SV. UMLS-Interface and UMLS-Similarity: open source software for measuring paths and semantic similarity. AMIA Annu Symp Proc 2009;2009:431–5 [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan A. An experimental study of ambiguity and context. Mechanical Translation 1950;2:39–46 [Google Scholar]
- 35.Moon S, Pakhomov S, Melton G. Automated disambiguation of acronyms and abbreviations in clinical texts: window and training size considerations. AMIA Annu Symp Proc 2012:1310–19 [PMC free article] [PubMed] [Google Scholar]
- 36.Melton GB, Moon S, McInnes BT, et al. Automated Identification of Synonyms in Biomedical Acronym Sense Inventories. The Louhi 2010: Workshop on Text and Datamining of Health Documents. Los Angeles, CA, 2010:46–52 [Google Scholar]
- 37.Gale WA, Church KW, Yarowsky D. One sense per discourse. Proceedings of the workshop on Speech and Natural Language Harriman, New York: Association for Computational Linguistics; 1992:233–7 [Google Scholar]
- 38.Wikipedia. List of medical abbreviations, Acronyms in healthcare, List of acronyms for diseases and disorders http://en.wikipedia.org/wiki/Category:Lists_of_medical_abbreviations, http://en.wikipedia.org/wiki/Acronyms_in_healthcare, http://en.wikipedia.org/wiki/List_of_acronyms_for_diseases_and_disorders (accessed 5 Apr 2013)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.