Abstract
Background and objective
Electronic health records (EHRs) are increasingly being used to complement the FDA Adverse Event Reporting System (FAERS) and to enable active pharmacovigilance. Over 30% of all adverse drug reactions are caused by drug–drug interactions (DDIs) and result in significant morbidity every year, making their early identification vital. We present an approach for identifying DDI signals directly from the textual portion of EHRs.
Methods
We recognize mentions of drug and event concepts from over 50 million clinical notes from two sites to create a timeline of concept mentions for each patient. We then use adjusted disproportionality ratios to identify significant drug–drug–event associations among 1165 drugs and 14 adverse events. To validate our results, we evaluate our performance on a gold standard of 1698 DDIs curated from existing knowledge bases, as well as with signaling DDI associations directly from FAERS using established methods.
Results
Our method achieves good performance, as measured by our gold standard (area under the receiver operator characteristic (ROC) curve >80%), on two independent EHR datasets and the performance is comparable to that of signaling DDIs from FAERS. We demonstrate the utility of our method for early detection of DDIs and for identifying alternatives for risky drug combinations. Finally, we publish a first of its kind database of population event rates among patients on drug combinations based on an EHR corpus.
Conclusions
It is feasible to identify DDI signals and estimate the rate of adverse events among patients on drug combinations, directly from clinical text; this could have utility in prioritizing drug interaction surveillance as well as in clinical decision support.
Keywords: Electronic Health Records, Pharmacovigilance, Drug Interaction, Adverse Reactions, Data Mining, Ontology
Background and significance
Adverse drug reactions result in more than 100 000 deaths annually,1 with an associated yearly cost of over $136 billion.2 Simultaneously, there is a rise in polypharmacy, which is the use of multiple concomitant drugs to treat medical conditions; one study estimated that 29.4% of elderly patients3 are taking six or more drugs. Drug–drug interactions (DDIs) that lead to adverse reactions are potentially avoidable, if detected early, given that DDIs account for more than 30% of all adverse drug reactions.4–6
New drugs are tested for interactions with existing drugs before market approval using in vivo and in vitro methods.7 However, owing to the sheer number of ways in which drugs can interact,8 it is infeasible to test for every kind of interaction. Many DDIs manifest after a certain period of exposure and it takes several exposures for rare DDIs to occur.9 Therefore, postmarketing surveillance is necessary to detect unanticipated interactions that occur when the drug is in use in the general population. The US Food and Drug Administration (FDA) enables such surveillance via spontaneous reporting systems (SRSs) such as the FDA Adverse Event Reporting System (FAERS), and a similar role is served internationally by the World Health Organization's VigiBase. Several studies10–13 have successfully inferred DDIs from these sources, overcoming problems of reporting biases14 and duplicate reporting.15
Electronic health records (EHRs) are a source of observational data that can complement SRSs and offer the potential for active surveillance.16 Initiatives like the Observational Medical Outcomes Partnership (OMOP) in the USA and the Exploring and Understanding Adverse Drug Reactions (EU-ADR) project in Europe are focusing on building EHR based surveillance systems. Similarly, the Mini-Sentinel Pilot Program,17 which is part of the FDA's Sentinel Initiative, uses data of more than 125 million patients across the USA from over 17 data partners for active monitoring of medical products. These projects mainly utilize the structured diagnosis and prescription data from EHRs for identifying single drug adverse reactions. Most efforts aimed at finding DDIs use reported sources for signal detection and use EHRs as a means of validation. For instance, Tatonetti et al10 found 171 new DDIs from FAERS and used the EHRs at Stanford to validate them. Another study by Duke et al18 mined MEDLINE abstracts for hypotheses generation and validated the hypotheses on EHRs.
In addition to structured data, EHRs contain rich information in the unstructured notes taken by doctors, nurses, and other practitioners. By ignoring the unstructured text, we could be missing a substantial portion of adverse events.19 Many studies19 20 have shown that coded information such as the ICD-9 is inadequate to accurately build patient cohorts and there is a considerable advantage in using the unstructured text of EHRs.21 22 We argue that such an advantage would also extend to drug safety signal detection. Indeed, there has already been some work23–26 demonstrating the discovery of the adverse event profiles for single drugs using unstructured notes.
Therefore, given increasing adoption and access to medical records for research, we expect efforts to shift toward directly mining EHRs with an increased attention to the use of unstructured data for generating hypotheses about drug safety.27 In this paper, we apply data mining methods on the textual portion of EHRs to signal adverse DDIs. To our knowledge, this is the first study of its kind. We validate our methods using a gold standard built from existing knowledge bases of DDIs, by applying our methods to two independent large EHR datasets, and by comparing with signaling DDI associations directly from FAERS using established methods. We demonstrate the utility of our method for early detection of new interactions and for identifying alternatives to risky drug combinations. Finally, we publish a database of the rate of adverse events among patients on all combinations of drugs used in our study, based on the EHRs.
Materials and methods
Data sources
Electronic health records
We use the Stanford Translational Research Integrated Database Environment (STRIDE) dataset comprising 9 million unstructured clinical notes corresponding to 1 million patients as our primary source of EHR data. These textual notes span a period of 18 years (1994–2011) and include both inpatient and outpatient notes, that are a combination of radiology, pathology, and transcription reports.
For validation of our results, we use a similar dataset from the Palo Alto Medical Foundation (PAMF), comprising over 50 million outpatient notes corresponding to 1.2 million patients for encounters during 2000–2012. The dataset includes progress, problem visit, procedure, and H&P (history and physical) notes, as well as communication transcripts.
Structured data sources
To compare the accuracy of DDI signal detection from EHRs with SRSs, we use over 4 million FAERS reports covering the period from 1997 through 2012 Q1. These reports are preprocessed to remove duplicate reports, correct terminological errors, and normalize drugs. Medical Dictionary for Regulatory Activities (MedDRA) V14.1 codes are used in FAERS to code adverse events.
We use the Anatomical Therapeutic Chemical Classification (ATC) and 18 ontologies from the Unified Medical Language System (UMLS) Metathesaurus and BioPortal28 (19 ontologies in total; see online supplementary materials S1) for building a lexicon and for normalizing drugs and diseases. Additionally, we use DrugBank,29 Medi-Span Drug Therapy Monitoring System (Wolters Kluwer Health, Indianapolis, Indiana, USA), Drugs.com,30 the National Drug File–Reference Terminology (NDF-RT), and Side Effect Resource (SIDER)31 as sources of drug indications and known DDIs.
Annotation of clinical text
We use sets of terms derived from 19 biomedical ontologies to define drug and event concepts (see online supplementary materials S2) as described in our previous work.26 32 For drugs, we include trade names and other forms of the drug from RxNorm, but ignore the dosage of the drug. For adverse events, we identify a SNOMED CT concept most similar to the event and systematically including synonyms and hyponyms via the is-a relationship graph. To improve the precision of recognizing drug and event concepts, based on prior work on identifying and removing non-informative terms,33 34 we remove terms that occur in common English usage from the automatically compiled term-sets.35 Finally, using frequency based sorting,36 37 we manually identify ambiguous terms that belong to more than one semantic group (drug, disease, device, procedure),36 38 and we suppress their least likely interpretation. For example, ‘clip’ is more likely to be a device than a drug in clinical text, so we suppress its interpretation as ‘corticotropin-like intermediate lobe peptide’.
We then use a fast text annotator to process clinical notes for mentions of these concepts and order the mentions by the note's timestamp, thus forming a concept timeline for every patient (figure 1). The annotator also takes into account negation and family history contextual cues to restrict concepts to positive mentions referring to the patient. For example, if the clinical note mentions ‘no evidence of active colitis’ or ‘family history of cancer’, then the note is not tagged with colitis or cancer. We follow a similar approach for the PAMF dataset. We achieve 74% sensitivity and 96% specificity in recognizing diseases on a gold standard from the 2008 i2b2 obesity challenge39 and our performance varies by condition (see online supplementary materials S3). Drug recognition is done in a similar manner using strings from RxNorm. An independent study at the University of Pittsburgh, which examined the annotations on 1960 clinical notes manually,40 estimated over 84% recall and 84% precision for recognizing drugs (personal communication, Richard Boyce).
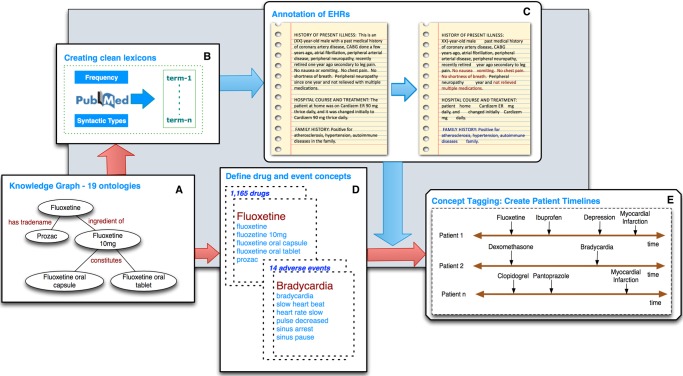
Figure 1.
The annotator workflow. (A) The annotator uses a lexicon of approximately 5.6M terms derived from the Unified Medical Language System (UMLS) and BioPortal, as well as trigger terms for NegEx and ConText. (B) It uses term frequency and syntactic type information from Medline to prune the set of strings into a clean lexicon. (C) It then uses the lexicon for exact string matching on the textual notes, followed by negation detection (red) and family history detection (blue). The output is a list of positively mentioned terms recognized in the text. (D) UMLS and BioPortal terms are used to define concepts (a set of terms), making use of the relationships in the ontologies to expand the set. (E) Each note is tagged with a concept if any one of the defining terms appears in the note as a positive mention. The concepts are ordered by the note's timestamp, creating a concept timeline for each patient.
We focus our study on a set of 14 adverse events based on existing literature,41 the availability of known DDIs causing them, their population event rate in STRIDE, and our ability to successfully detect their presence in text from EHRs. For interoperability with FAERS, which uses MedDRA codes, we manually assign to each event concept, the most closely matching MedDRA code (see online supplementary materials S4).
Computing drug–drug–event association scores
To compute the association score for a drug–drug–event tuple using EHRs, we treat the combination of two drugs as a single drug and use standard methods that measure the disproportionality of the mention of the adverse event, between exposed and comparison groups. Similarly, for FAERS based analysis, we use the Multi-Item Gamma Poisson Shrinker (MGPS)42 algorithm to measure the disproportionality of the observed number of reports for a drug–drug–event tuple compared to the expected number of reports. We signal an interaction if the association score is greater than some threshold, which is chosen based on the receiver operator characteristic (ROC) curve for the desired sensitivity and specificity.
DDI associations from EHRs
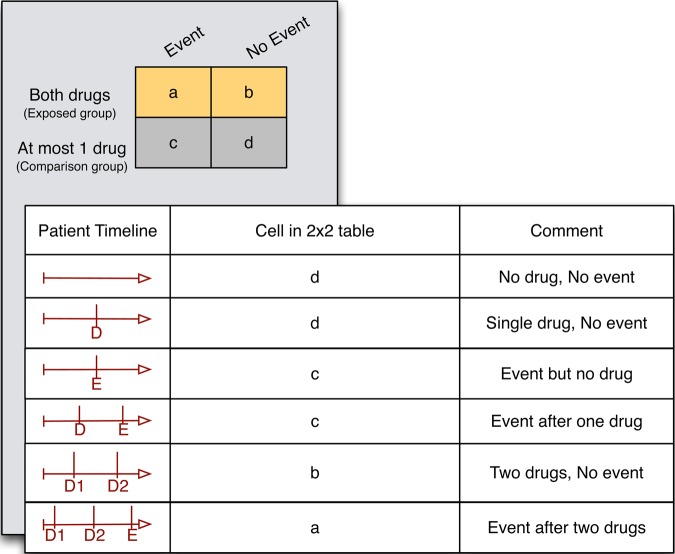
We first construct a 2×2 contingency table (figure 2) where the exposed group comprises patients who have taken both drugs and the comparison group comprises patients who have taken at most one drug (or no drug). Patients are assigned to one of the cells of the contingency table based on the ordering of the first mention of the two drugs and the event in the patient's timeline (figure 2). For example, if both drugs appear in notes whose timestamp is earlier than the note with the adverse event, the patient is counted into the ‘a’ cell. Any drugs that appear after the first occurrence of the event are ignored. The population event rate among patients on the drug combination can be calculated from this table as a/(a+b).
Figure 2.
Assignment of patients to various cells of the 2×2 contingency table. The portion of the timeline after the first occurrence of the event is ignored. D, drug; E, event.
We calculate an unadjusted OR (UOR) from this contingency table, and use the lower bound of the 95% CI,43 which we denote as UOR025, as an unadjusted association score. To compute an adjusted OR (AOR), we use propensity score matching (PSM) using a standard caliper of 0.05 (Matching package in R44), to match 10 patients in the comparison group to each patient in the exposed group without replacement, followed by conditional logistic regression45 (Survival package in R46). Once again, we use the lower bound of the 95% CI of the AOR, denoted as AOR025 as an association score. We compute the propensity score using age, gender, race, note count, drug count, and disease count. For patients taking at least one of the drugs, we use the average age at the time of drug exposure whereas for patients not taking either drug, we use the average age of the patient in our dataset. Drug count and disease count serve as proxies for the overall health of the patient,27 47 and including the note count serves to adjust for the quantity of data for each patient.
DDI associations from FAERS
We use the MGPS, which is a commonly applied algorithm endorsed by the FDA for FAERS based disproportionality analysis,48 to generate a DDI signal based on FAERS. MGPS is based on a Bayesian framework that accounts for uncertainty of a disproportionality ratio by ‘shrinking’ it towards a baseline case of no association to an extent proportional to its variance. We make use of the EB05 measure, which is the lower 5th percentile of the posterior distribution of the disproportional reporting ratio for our drug–drug–event tuple. The MGPS algorithm performs stratification on age, gender, and year of report to adjust for confounding by these variables. Drug combinations with fewer than five reports are ignored and are given a score of 0.
Preparation of gold standard
We use three sources to define our list of drugs: RxNorm (4993 drugs, counted as unique ingredients), DrugBank (6711 drugs), and ATC (4406 drugs). We limit our study to 1165 drugs present in all three sources. We use known interactions from DrugBank and Medi-Span as positive interactions in our gold standard. Using our concept definitions to identify adverse events from the interaction monographs, we form drug–drug–event relations and manually validate them. For estimating the false discovery rate, we simulate a set of negative interactions by generating random drug–drug–event tuples, and removing any known interactions according to DrugBank, Medi-Span, or Drugs.com. We also remove tuples for which the event is an indication (from Medi-Span, DrugBank, Drugs.com, UMLS, and SIDER) for either drug individually.
Results
Characteristics of datasets
We include 1165 drugs and 14 adverse events in our study and the defining term sets are available as online supplementary materials S2. Out of 1.04 million patients, there are 565 998 patients (53% female) in the STRIDE dataset, with at least one drug or event concept from our study mentioned in their records. Out of the 1165 drugs, 858 (73.6%) appeared at least once. Similarly, out of 1.2 million patients, there are 969 511 patients (54% female) in the PAMF dataset, covering 1048 (90%) drugs. Table 1 shows the number of patients corresponding to each adverse event in STRIDE and PAMF datasets.
Table 1.
Characteristics of datasets and gold standard by adverse event
| Event | No. patients in STRIDE | No. patients in PAMF | No. positive test cases |
|---|---|---|---|
| Total | 565 998 | 969 511 | 849 |
| Cardiac arrhythmias | 88 555 | 109 601 | 65 |
| Acute renal failure | 32 197 | 28 994 | 15 |
| Bradycardia | 22 906 | 24 162 | 52 |
| Hyperglycemia | 19 189 | 41 574 | 47 |
| Neutropenia | 14 322 | 6783 | 13 |
| Hypoglycemia | 11 150 | 28 320 | 43 |
| Pancytopenia | 8718 | 2850 | 2 |
| Hypokalemia | 8405 | 10 356 | 44 |
| Hyperkalemia | 4973 | 7691 | 142 |
| Parkinsonian symptoms | 3541 | 4605 | 9 |
| Nephrotoxicity | 1460 | 639 | 83 |
| Rhabdomyolysis | 1378 | 1948 | 30 |
| QT prolongation | 1260 | 1441 | 150 |
| Serotonin syndrome | 674 | 1511 | 154 |
We consider 14 adverse events for our study; our gold standard contains a total of 849 positive test cases and 849 randomly generated negative test cases.
PAMF, Palo Alto Medical Foundation; STRIDE, Stanford Translational Research Integrated Database Environment.
Evaluation using gold standard and an independent dataset
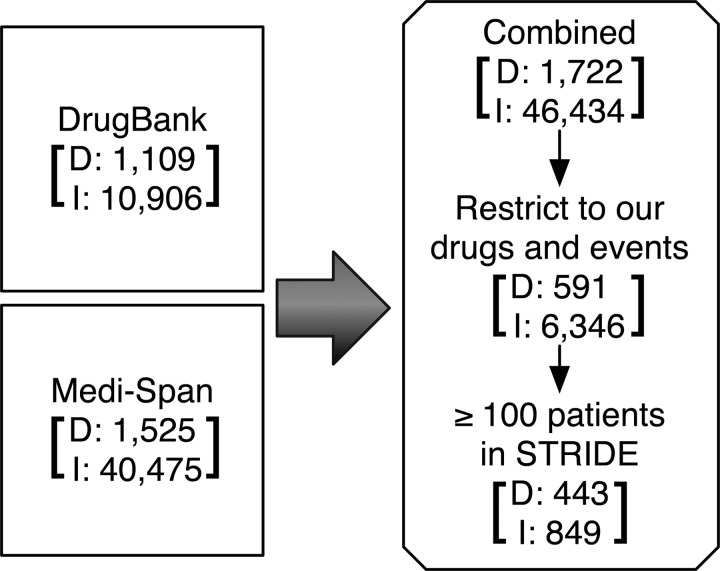
DrugBank contains 10 906 DDI monographs and Medi-Span contains 40 475 DDI monographs. Together, there were 46 434 interactions that result in 6346 drug–drug–event tuples (13.66%) corresponding to our set of drugs and events (figure 3). Of these, 849 (13.4%) interactions (comprising 443 drugs and 14 events) satisfy the support threshold of exposure of at least 100 patients in the STRIDE data and these form our set of positive test cases. Table 1 shows the number of positives test cases per event. For each event, we generate as many negative test cases as there are positives (see Methods), thus resulting in 849 randomly generated negative test cases.
Figure 3.
Preparation of Gold standard. We use known interactions from DrugBank and Medi-Span having at least 100 patients on the drug combination in the Stanford Translational Research Integrated Database Environment (STRIDE) as the true positives in our gold standard. The number of drugs (D) and interactions (I) at each stage are specified.
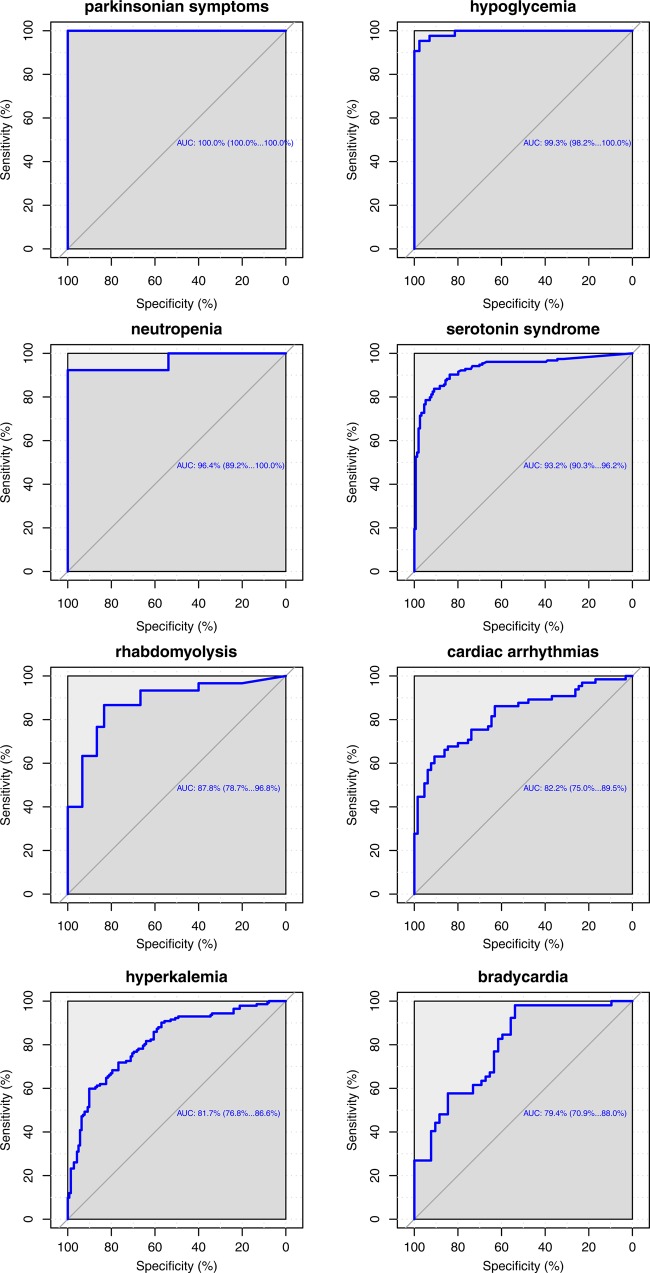
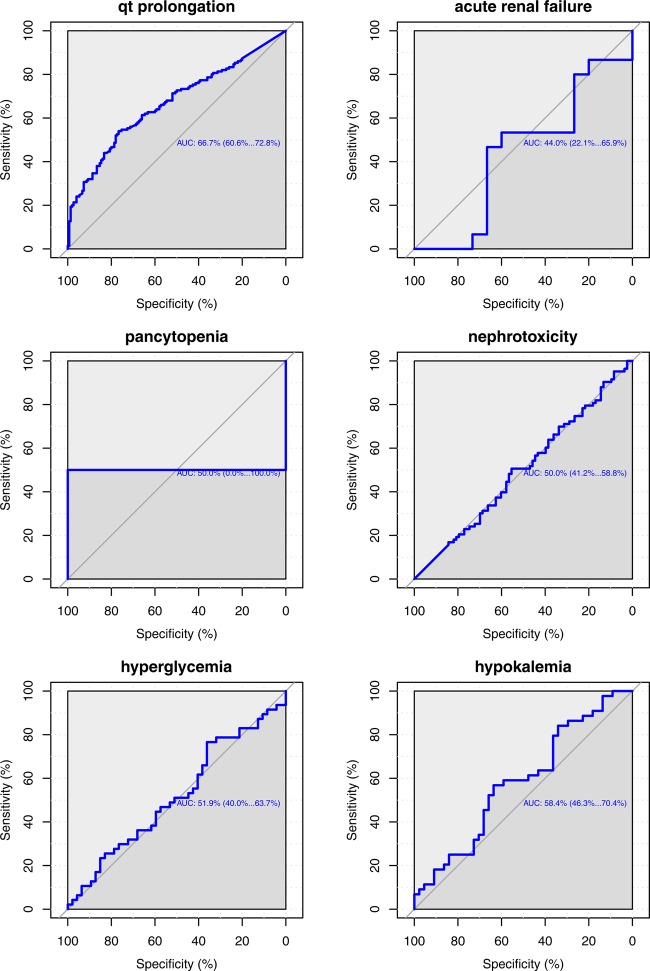
We use ROC curves that show all possible values of sensitivity and specificity that can be achieved by our association scores on our gold standard by varying the threshold parameter, as a measure of performance. Using UOR025, the area under the ROC (AUC) curve is 71.4% and we obtain a specificity of 81.27% and a positive predictive value (PPV) of 72.78% at a sensitivity of 50%. We found that acute renal failure (ARF), nephrotoxicity, hypokalemia, and hyperglycemia did not perform well (AUC ≈0.5, see Discussion). On removing these events (1320 remaining interactions), the AUC improves to 78.3% and the use of AOR025 further increases the AUC to 82.3% (figure 4). Additionally, specificity improves to 94.24%, with a PPV (precision) of 89.67% at 50% sensitivity (recall) (see online supplementary materials S5 for precision–recall curves). Overall performance suggests that the unstructured text contains relevant information for signaling DDIs. We also observe differential performance when we look at event specific ROC curves (figure 5).
Figure 4.
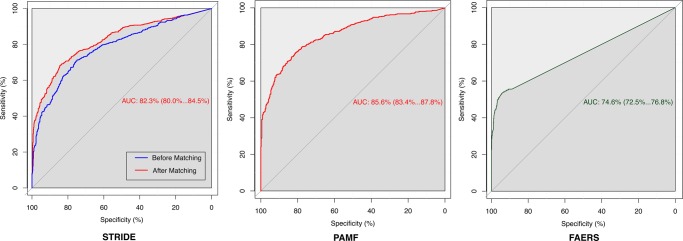
Performance on the Stanford Translational Research Integrated Database Environment (STRIDE), Palo Alto Medical Foundation (PAMF), and FDA Adverse Event Reporting System (FAERS) datasets as evaluated by the gold standard: receiver operator characteristic curves showing sensitivity and specificity levels that can be achieved by varying the threshold. Performance improves after propensity score based matching (red curve in STRIDE and PAMF). For STRIDE, we use our gold standard of 1320 interactions on 10 adverse events. 1132 out of 1320 interactions have enough support for signaling from PAMF. FAERS uses all 1320 test cases, and test cases without enough reports in FAERS were given a score of 0.
Figure 5.
Event-wise performance in the Stanford Translational Research Integrated Database Environment (STRIDE): receiver operator characteristic (ROC) curves showing sensitivity and specificity values for various thresholds on the gold standard test cases using STRIDE. Using such curves, event specific thresholds can be chosen. Hyperkalemia, acute renal failure, nephrotoxicity, and hyperglycemia did not perform well. This could be due to our inability to accurately tag notes with these concepts, or due to the gold standard itself (see Discussion). The area under the ROC curve for pancytopenia has a very large variance due to an insufficient number of tested interactions.
Of the 1320 interactions in our gold standard, 1132 interactions contain enough support (exposure of at least 100 patients) to signal an interaction in the PAMF dataset, and using AOR025, we achieve an AUC of 85.6% (figure 4), with a specificity of 95.8% and a PPV (precision) of 92.7% for a sensitivity (recall) of 50% (see online supplementary materials S5).
Evaluation using FAERS
Of the 1320 interactions, 879 did not satisfy the support threshold for FAERS (EB05=0); 293 of these 879 were known true interactions, illustrating that several interactions can be missed in FAERS. The EB05 measure on FAERS achieves an AUC of 74.6% on the gold standard (figure 4), showing that performance of our method on EHRs seems to be comparable with that of current methods on FAERS—as assessed by our gold standard. Additionally, on running our workflow on STRIDE using data up to each year between 2000 and 2011, we find that the interaction between amiodarone and haloperidol known to cause QT prolongation is signaled using STRIDE data as early as 2007. FAERS reports for this interaction started appearing in 2009, possibly correlated with a research publication at that time by Bush et al,49 demonstrating the ability of EHRs to complement FAERS for early detection.
Population rate of adverse events
Using our set of 1165 drugs and 14 events, there are 569 398 drug–drug–event tuples with at least 100 patients on both drugs in STRIDE. We publish the 2×2 contingency tables along with population event rate and UOR025 for these tuples in STRIDE as online supplementary materials S6; this population event rate information has potential utility in prioritizing DDIs in several settings. Table 2 shows the drug combinations with the highest event rates in STRIDE, for each event from our gold standard.
Table 2.
Interactions in the gold standard with the highest event rates (a/(a+b)) in the Stanford Translational Research Integrated Database Environment dataset for each event
| Adverse event | Drug1 | Drug2 | a | b | Rate (%) |
|---|---|---|---|---|---|
| Parkinsonian symptoms | Levodopa | Lorazepam | 176 | 235 | 42.82 |
| Cardiac arrhythmias | Potassium chloride | Lisinopril | 1091 | 1615 | 40.32 |
| Neutropenia | Paclitaxel | Trastuzumab | 140 | 567 | 19.8 |
| Bradycardia | Amiodarone | Metoprolol | 796 | 3671 | 17.82 |
| Hypoglycemia | Glipizide | Lisinopril | 367 | 2160 | 14.52 |
| Acute renal failure | Hydrochlorothiazide | Ibuprofen | 884 | 8375 | 9.55 |
| Hyperkalemia | Potassium chloride | Spironolactone | 349 | 3471 | 9.14 |
| Hyperglycemia | Prednisone | Salmeterol | 379 | 4612 | 7.59 |
| Nephrotoxicity | Fluconazole | Tacrolimus | 85 | 1208 | 6.57 |
| Pancytopenia | Mercaptopurine | Azathioprine | 15 | 278 | 5.12 |
| Hypokalemia | Prednisone | Salmeterol | 222 | 4982 | 4.27 |
| Serotonin syndrome | Tramadol | Duloxetine | 57 | 1301 | 4.2 |
| QT prolongation | Amiodarone | Ciprofloxacin | 46 | 2487 | 1.82 |
| Rhabdomyolysis | Ciprofloxacin | Simvastatin | 50 | 5184 | 0.96 |
Utility of association scores
We compute AOR025 for those tuples that are highly likely to represent an interaction. To choose these tuples, we use a three-step strategy. We first obtain thresholds that result in 95% specificity for UOR025 on STRIDE (threshold=4.7) and EB05 in FAERS (threshold=1.5) as assessed by our gold standard. We then compute EB05 using FAERS for all 569 398 tuples; we already have UOR025 for these 569 398 tuples. Finally, we compute AOR025 for those 9306 tuples that satisfy the UOR025 and EB05 thresholds for 95% specificity (see online supplementary materials S6).
Based on AOR025, a threshold of 1.1 gives us 95% specificity with respect to our gold standard. Of the 9306 tuples, 5983 satisfy this threshold and have a high likelihood of representing true interactions. We also find that 49 of these 5983 tuples (see online supplementary materials S7) were added to Drugs.com in the period between May 2011 and November 2012 (STRIDE data is up to March 2011). Therefore we believe that EHRs can be used for hypothesis generation to identify new DDI signals.
Table 3 shows AOR025 for the combination of statins with calcineurin inhibitors, tacrolimus, and cyclosporine A, associated with rhabdomyolysis. Using a threshold of 1.1 (95% specificity), the combination of tacrolimus and statins is relatively less associated with rhabdomyolysis. This is in agreement with a study by Lemahieu et al50 in 2005, which argues that patients exposed to statins and cyclosporine A are at an increased risk of rhabdomyolysis, whereas tacrolimus is safer. This result illustrates a potential utility of the EHR-derived association scores for clinical decision support—to choose between several viable treatment alternatives.
Table 3.
Potential for clinical decision support
| Event | Statin | Calcineurin inhibitor | a | b | c | d | AOR025 |
|---|---|---|---|---|---|---|---|
| Rhabdomyolysis | Nystatin | Tacrolimus | 21 | 2324 | 1357 | 562 296 | 0.67 |
| Atorvastatin | 20 | 1267 | 1358 | 563 353 | 0.96 | ||
| Nystatin | Cyclosporine | 32 | 1407 | 1346 | 563 213 | 1.78 | |
| Atorvastatin | 37 | 1331 | 1341 | 563 289 | 2.26 |
This table shows AOR025 scores for the association of rhabdomyolysis with the combination of calcineurin inhibitors like tacrolimus or cyclosporine A, and statins. It appears that tacrolimus is relatively safe and this agrees with in vivo experiments.
AOR, adjusted OR.
Discussion
The early identification of DDIs is important and testing for interactions between all drugs by experimental methods is infeasible. Significant research exists on generating models for metabolic interactions of drugs,51 mainly for interactions related to CYP enzymes. There is also work on predicting interactions based on molecular similarity with drugs already known to interact.52 Although these methods are successful at finding new interactions, their results are limited by their modeling assumptions and the limited mechanistic understanding of known interactions. A data-driven way to identify interactions is by analyzing the effect of drugs in the general population via postmarketing surveillance.
Most existing methods for such surveillance use SRS databases to identify interactions, and use coded information present in EHRs for validation and prioritization. We have shown that it is possible to identify significant (drug–drug–event) associations and compute their event rates, directly from the unstructured text of EHRs. This can serve as an active monitoring tool for signaling unknown interactions for new and existing drugs; advancing phase IV surveillance of drugs and meaningful use of EHRs simultaneously.
We perform text mining on two large corpora of clinical records to successfully detect DDIs with a simple and fast approach to text annotation,53 where we sacrifice some accuracy in tagging concepts at the note level in exchange for population level trends (see online supplementary materials S3). We test our results using a gold standard comprising known DDIs as positive examples and an equal number of randomly generated interactions as negative examples. Testing AOR025 using all unknown interactions as negative examples poses computational problems; however, the AUC for UOR025 using all negatives is almost identical to that using sampled negatives, indicating minimal biases due to sampling. Most interaction studies52 54 use similar techniques to build a gold standard and the generation of an ROC aids in comparison of results. We see differential performance per event; for example, the performance of the method for parkinsonian symptoms, hypoglycemia, and neutropenia is significantly better than the others (in terms of AUC). This finding reflects similar findings in the EU-ADR project,55 and suggests the need for event specific thresholds.56
Our approach did not perform well for four events, perhaps owing to limitations that we discuss here. The performance of our workflow heavily depends on the accuracy of concept recognition in EHRs using lexical approaches. In this study, we use frequency based methods36 37 and manual curation to remove ambiguous terms corresponding to concepts—at the expense of reducing sensitivity. Nevertheless, some concepts are hard to detect in EHRs using lexicon based methods; for example, the occurrence of hypokalemia or hyperglycemia is sometimes stated in quantitative terms as changes in potassium or sugar levels, which would be missed by our annotator. On the other hand, having too few terms reduces our sensitivity; for example, the term set for nephrotoxicity comprised only three terms. Our current method cannot signal DDIs that are dependent on drug dosage and resolving these issues requires advanced natural language processing methods.57 We focus solely on the note's timestamp to determine the time of occurrence of a concept; we acknowledge that doing so is based on the assumption that an event described as historical in the note will be mentioned in some previous note as a current event, and this can reduce performance. We do not use drug interaction eras and drug exposures spaced far away in time may cause false associations. We are currently examining the annotations for the utility of the last mention of concepts, sentence-level co-occurrences, and temporal density of mentions to address this question. We use standard disproportionality analysis adapted to longitudinal data to signal an interaction, however several other measures of interaction have been proposed in the literature58–60 and may prove to perform better. To adjust for possible confounding factors, we use PSM to choose a comparison group that is similar to the exposure group and then use matched conditional logistic regression to generate an AOR. We acknowledge that this approach is one of several known methods to use PSM61 and that other methods could be equally effective. The databases from which the known interactions are derived are not exhaustive and some interactions are based on unsubstantial evidence; for example, several monographs for ARF claim that ‘ARF was reported on rare occasions’. We use several sources of data in this work, each having its own method for coding drugs and events, and the mapping from concepts in one data source to the other is another source of errors; for example, the mapping from our event concepts to MedDRA PTs for use with FAERS. Finally, we note that our association scores do not indicate causality—which means that in some cases, the adverse event might not be caused by a drug interaction in the mechanistic sense but might be associated with the exposure to the drugs. The goal is to provide early warnings of drug combinations that require investigation.
Using standard methods to analyze FAERS, we show its performance on our gold standard as a reference to enable an evaluation of the comparative utility of EHRs. Many known interactions from our gold standard that appear in EHR data do not contain a sufficient number of reports in FAERS, possibly owing to under-reporting. EHRs may therefore have utility in the early signaling of DDIs. Furthermore, we demonstrate a potential use of EHR derived DDI association strengths to choose between drugs used in combination therapy; for example, the results on the combination of calcineurin inhibitors with statins. Such analysis is not possible with FAERS alone owing to the differential reporting rates for drugs.
EHRs have good longitudinal coverage of patient history, a larger number of measured covariates, and thus are likely to provide an accurate measure of the real world rate of adverse events among patients on a particular drug combination. The pre-computed population event rate information and association scores could find application in clinical decision support. For example, augmenting existing DDI databases with this event rate information could potentially be useful to prioritize interaction alerts in computerized physician order entry (CPOE) systems,62 where at present 49–96% of all alerts are overridden.63 Finally, calculating population event rate information could directly serve as a means to enable stage 3 of the meaningful use measures that are to be implemented by 2016.64 To this end, we publish the event rates (see online supplementary materials) for 569 398 drug–drug–event combinations in our study. We emphasize that the limitations of our method, as highlighted above, must be accounted for when using the estimated rates of events. With the intent of finding novel DDIs, we also publish the adjusted association scores for 9306 interactions that are signaled by both EHR sources and FAERS. However, the number of predictions are still far too many to be experimentally tested; it may be possible to identify the most promising interactions using a weighted score based on features such as the number of affected people and the cost of the drugs, or by searching for a mechanistic explanation as proposed by Bauer-Mehren et al.65
Conclusion
We use data from two independent sites to demonstrate the feasibility of using the textual notes from EHRs for signaling DDIs and to estimate the rate of events among patients on various drug combinations. We develop a gold standard of DDIs that may be useful for detailed characterization of future methods and our evaluations show that we can signal interactions from EHRs with as good performance as established methods on SRSs (FAERS). We find that in some cases we can signal interactions many years before the first report in FAERS, which could expedite the discovery of new interactions and demonstrates the complementarity of our approach. Additionally, we publish a first-of-its-kind resource: a database of the rate of each event for all drug–drug pairs from our EHR corpus, which may be useful to triage alerts in CPOE systems or to identify drug combinations with a lower chance of adverse events. We conclude that the text portion of EHRs can complement existing sources for postmarketing surveillance for DDIs.
Supplementary Material
Acknowledgments
We thank Oracle's Health Sciences Division for making available the FAERS data and analysis software. We acknowledge the assistance of Cliff Olson in accessing and processing the data from PAMF.
Footnotes
Contributors: SVI, RH, PL, and NHS wrote the manuscript. SVI, RH, PL, and NHS designed the research. SVI, RH, PL, AB-M, and NHS performed the research.
Funding: This work was supported by NIH grant number U54 HG004028 for the National Center for Biomedical Ontology and NIGMS R01 grant number GM101430.
Competing interests: None.
Ethics approval: IRB for Stanford University, IRB for the Palo Alto Medical Foundation.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 1998;279:1200–5 [DOI] [PubMed] [Google Scholar]
- 2.Johnson JA, Bootman JL. Drug-related morbidity and mortality. A cost-of-illness model. Arch Intern Med 1995;155:1949–56 [PubMed] [Google Scholar]
- 3.Bushardt RL, Massey EB, Simpson TW, et al. Polypharmacy: misleading, but manageable. Clin Interv Aging 2008;3:383–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strandell J, Bate A, Lindquist M, et al. Drug-drug interactions—a preventable patient safety issue? Br J Clin Pharmacol 2008;65:144–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirohamed M. Drug interactions of clinical importance. London: Chapman and Hall, 1998 [Google Scholar]
- 6.Huang SM, Temple R, Throckmorton DC, et al. Drug interaction studies: study design, data analysis, and implications for dosing and labeling. Clin Pharmacol Ther 2007;81:298–304 [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Zhang YD, Zhao P, et al. Predicting drug-drug interactions: an FDA perspective. AAPS J 2009;11:300–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroner BA. Common drug pathways and interactions. Diab Spectr 2002;15:249–55 [Google Scholar]
- 9.Triaridis S, Tsiropoulos G, Rachovitsas D, et al. Spontaneous haematoma of the pharynx due to a rare drug interaction. Hippokratia 2009;13:175–7 [PMC free article] [PubMed] [Google Scholar]
- 10.Tatonetti NP, Fernald GH, Altman RB. A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports. J Am Med Inform Assoc 2012;19:79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Puijenbroek EP, Egberts AC, Heerdink ER, et al. Detecting drug-drug interactions using a database for spontaneous adverse drug reactions: an example with diuretics and non-steroidal anti-inflammatory drugs. Eur J Clin Pharmacol 2000;56:733–8 [DOI] [PubMed] [Google Scholar]
- 12.Thakrar BT, Grundschober SB, Doessegger L. Detecting signals of drug-drug interactions in a spontaneous reports database. Br J Clin Pharmacol 2007;64:489–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harpaz R, Chase HS, Friedman C. Mining multi-item drug adverse effect associations in spontaneous reporting systems. BMC Bioinformatics. 2010;11(Suppl 9):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAdams M, Staffa J, Dal Pan G. Estimating the extent of reporting to FDA: a case study of statin-associated rhabdomyolysis. Pharmacoepidemiol Drug Saf 2008;17:229–39 [DOI] [PubMed] [Google Scholar]
- 15.Noren GN, Orre R, Bate A, et al. Duplicate detection in adverse drug reaction surveillance. Data Min Knowl Discov 2007;14:305–28 [Google Scholar]
- 16.Stang PE, Ryan PB, Racoosin JA, et al. Advancing the science for active surveillance: rationale and design for the Observational Medical Outcomes Partnership. Ann Intern Med 2010;153:600–6 [DOI] [PubMed] [Google Scholar]
- 17.Mini-Sentinel. http://mini-sentinel.org/
- 18.Duke JD, Han X, Wang Z, et al. Literature based drug interaction prediction with clinical assessment using electronic medical records: novel myopathy associated drug interactions. PLoS Comput Biol 2012;8:e1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Classen DC, Resar R, Griffin F, et al. ‘Global trigger tool’ shows that adverse events in hospitals may be ten times greater than previously measured. Health Aff (Millwood) 2011;30:581–9 [DOI] [PubMed] [Google Scholar]
- 20.Birman-Deych E, Waterman AD, Yan Y, et al. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Medical Care 2005;43:480–5 [DOI] [PubMed] [Google Scholar]
- 21.Xu H, Fu Z, Shah A, et al. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. AMIA Annu Symp Proc 2011;2011:1564–72 [PMC free article] [PubMed] [Google Scholar]
- 22.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012;19(1e):e162–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lependu P, Iyer SV, Fairon C, et al. Annotation analysis for testing drug safety signals using unstructured clinical notes. J Biomed Semantics 2012;3(Suppl 1):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haerian K, Varn D, Vaidya S, et al. Detection of pharmacovigilance-related adverse events using electronic health records and automated methods. Clin Pharmacol Ther 2012;92:228–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu M, McPeek Hinz ER, Matheny ME, et al. Comparative analysis of pharmacovigilance methods in the detection of adverse drug reactions using electronic medical records. J Am Med Inform Assoc 2013;20:420–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LePendu P, Iyer SV, Bauer-Mehren A, et al. Pharmacovigilance using clinical notes. Clin Pharmacol Ther 2013;93:547–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuemie MJ, Coloma PM, Straatman H, et al. Using electronic health care records for drug safety signal detection: a comparative evaluation of statistical methods. Med Care 2012;50:890–7 [DOI] [PubMed] [Google Scholar]
- 28.BioPortal. http://bioportal.bioontology.org/
- 29.Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 2008;36(Database issue):D901–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drugs.com. [cited 2011 May]. http://www.drugs.com/
- 31.Kuhn M, Campillos M, Letunic I, et al. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Lependu P, Iyer S, et al. Using temporal patterns in medical records to discern adverse drug events from indications. AMIA Summits Transl Sci Proc 2012;2012:47–56 [PMC free article] [PubMed] [Google Scholar]
- 33.Demner-Fushman D, Mork JG, Shooshan SE, et al. UMLS content views appropriate for NLP processing of the biomedical literature vs. clinical text. J Biomed Inform 2010;43:587–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCray AT, Bodenreider O, Malley JD, et al. Evaluating UMLS strings for natural language processing. Proceedings/AMIA Annual Symposium AMIA Symposium 2001:448–52 [PMC free article] [PubMed] [Google Scholar]
- 35.MOBY Project. 2000. http://icon.shef.ac.uk/Moby/
- 36.Wu S, Liu H, Li D, et al. UMLS term occurrences in clinical notes: a large scale corpus analysis. J Am Med Inform Assoc 2012;19:e149–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu R, Musen MA, Shah NH. A comprehensive analysis of five million UMLS Metathesaurus terms using eighteen million MEDLINE citations. AMIA Annu Symp Proc 2010;2010:907–11 [PMC free article] [PubMed] [Google Scholar]
- 38.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. J Biomed Inform 2003;36:414–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uzuner O. Second i2b2 workshop on natural language processing challenges for clinical records. AMIA Annual Symposium Proceedings/AMIA Symposium 2008:1252–3 [PubMed] [Google Scholar]
- 40.Marshall MS, Boyce R, Deus HF, et al. Emerging practices for mapping and linking life sciences data using RDF—a case series. Web Semantics Sci Serv Agents World Wide Web 2012;14:2–13 [Google Scholar]
- 41.Trifiro G, Pariente A, Coloma PM, et al. Data mining on electronic health record databases for signal detection in pharmacovigilance: which events to monitor? Pharmacoepidemiol Drug Saf 2009;18:1176–84 [DOI] [PubMed] [Google Scholar]
- 42.DuMouchel W. Bayesian data mining in large frequency tables, with an application to the FDA spontaneous reporting system. Am Statistician 1999;53:177–90 [Google Scholar]
- 43.Harpaz R, DuMouchel W, LePendu P, et al. Performance of pharmacovigilance signal detection algorithms for the FDA adverse event reporting system. Clin Pharmacol Ther 2013;93:539–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekhon JS. Multivariate and propensity score matching software with automated balance optimization: the matching package for R. J Stat Software 2011;42:1–52 [Google Scholar]
- 45.Breslow NE, Day NE. Statistical methods in cancer research. The analysis of case-control studies. Lyon: IARC Scientific Publications, 1980 [PubMed] [Google Scholar]
- 46.Therneau T. Survival analysis, including penalised likelihood. 2012. http://cran.r-project.org/web/packages/survival/ [Google Scholar]
- 47.Tatonetti NP, Ye PP, Daneshjou R, et al. Data-driven prediction of drug effects and interactions. Sci Transl Med 2012;4:125ra31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szarfman A, Machado SG, ONeill RT. Use of screening algorithms and computer systems to efficiently signal higher-than-expected combinations of drugs and events in the US FDAs spontaneous reports database. Drug Saf 2002;25:381–92 [DOI] [PubMed] [Google Scholar]
- 49.Bush SE, Hatton RC, Winterstein AG, et al. Effects of concomitant amiodarone and haloperidol on Q-Tc interval prolongation. Am J Health Syst Pharm 2008;65:2232–6 [DOI] [PubMed] [Google Scholar]
- 50.Lemahieu WP, Hermann M, Asberg A, et al. Combined therapy with atorvastatin and calcineurin inhibitors: no interactions with tacrolimus. Am J Transplant 2005;5:2236–43 [DOI] [PubMed] [Google Scholar]
- 51.Dickins M, van de Waterbeemd H. Simulation models for drug disposition and drug interactions. Drug Discov Today: BIOSILICO 2004;2:38–45 [Google Scholar]
- 52.Vilar S, Harpaz R, Uriarte E, et al. Drug-drug interaction through molecular structure similarity analysis. J Am Med Inform Assoc 2012;19:1066–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Halevy A, Norvig P, Pereira F. The unreasonable effectiveness of data. Intell Syst IEEE 2009;24:8–12 [Google Scholar]
- 54.Gottlieb A, Stein GY, Oron Y, et al. INDI: a computational framework for inferring drug interactions and their associated recommendations. Mol Syst Biol 2012;8:592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coloma PM, Trifiro G, Schuemie MJ, et al. Electronic healthcare databases for active drug safety surveillance: is there enough leverage? Pharmacoepidemiol Drug Saf 2012;21:611–21 [DOI] [PubMed] [Google Scholar]
- 56.Observational Medical Outcomes Partnership. [cited 2012 Dec]. http://omop.fnih.org.
- 57.D'Avolio LW, Nguyen TM, Goryachev S, et al. Automated concept-level information extraction to reduce the need for custom software and rules development. J Am Med Inform Assoc 2011;18:607–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noren GN, Sundberg R, Bate A, et al. A statistical methodology for drug-drug interaction surveillance. Stat Med 2008;27:3057–70 [DOI] [PubMed] [Google Scholar]
- 59.Gould AL. Practical pharmacovigilance analysis strategies. Pharmacoepidemiol Drug Saf 2003;12:559–74 [DOI] [PubMed] [Google Scholar]
- 60.Noren GN, Bate A, Orre R, et al. Extending the methods used to screen the WHO drug safety database towards analysis of complex associations and improved accuracy for rare events. Stat Med 2006;25:3740–57 [DOI] [PubMed] [Google Scholar]
- 61.Kurth T, Walker AM, Glynn RJ, et al. Results of multivariable logistic regression, propensity matching, propensity adjustment, and propensity-based weighting under conditions of nonuniform effect. Am J Epidemiol 2006;163:262–70 [DOI] [PubMed] [Google Scholar]
- 62.Strasberg HR. Medi-Span drug-drug interactions study. Wolters Kluwer Health 2012 [Google Scholar]
- 63.van der Sijs H, Aarts J, Vulto A, et al. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc 2006;13:138–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.First glimpse at meaningful use stage 3 measures. 2012. [cited 2012 Nov]. http://www.govhealthit.com/news/health-it-panel-explores-draft-mu-3-measures.
- 65.Bauer-Mehren A, van Mullingen EM, Avillach P, et al. Automatic filtering and substantiation of drug safety signals. PLoS Comput Biol 2012; 8:e1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.