Abstract
Objective
To develop a decision support system to identify patients at high risk for hyperlactatemia based upon routinely measured vital signs and laboratory studies.
Materials and methods
Electronic health records of 741 adult patients at the University of California Davis Health System who met at least two systemic inflammatory response syndrome criteria were used to associate patients’ vital signs, white blood cell count (WBC), with sepsis occurrence and mortality. Generative and discriminative classification (naïve Bayes, support vector machines, Gaussian mixture models, hidden Markov models) were used to integrate heterogeneous patient data and form a predictive tool for the inference of lactate level and mortality risk.
Results
An accuracy of 0.99 and discriminability of 1.00 area under the receiver operating characteristic curve (AUC) for lactate level prediction was obtained when the vital signs and WBC measurements were analysed in a 24 h time bin. An accuracy of 0.73 and discriminability of 0.73 AUC for mortality prediction in patients with sepsis was achieved with only three features: median of lactate levels, mean arterial pressure, and median absolute deviation of the respiratory rate.
Discussion
This study introduces a new scheme for the prediction of lactate levels and mortality risk from patient vital signs and WBC. Accurate prediction of both these variables can drive the appropriate response by clinical staff and thus may have important implications for patient health and treatment outcome.
Conclusions
Effective predictions of lactate levels and mortality risk can be provided with a few clinical variables when the temporal aspect and variability of patient data are considered.
Keywords: Sepsis, Clinical Decision Support, Mortality Prediction, Lactate Level Prediction, Machine Learning, Electronic Health Records
Background and significance
The treatment of sepsis has placed a serious burden on healthcare systems, with an estimated US$14.6 billion spent annually on hospitalizations in the USA.1 Severe sepsis and septic shock, the more serious forms of sepsis, kill one in four patients affected.2 Sepsis occurs when an infection, beginning in any tissue in the body, results in the systemic inflammatory response syndrome (SIRS).3 The SIRS criteria are heart rate >90 beats/min, respiratory rate (RR) >20 breaths/min (or partial pressure of arterial CO2 <32), temperature either >38°C or <36°C, and white blood cell count (WBC) either >12 000 or <4000 cells/mm3 (or >10% bands). For a patient to be diagnosed with sepsis, at least two of the SIRS criteria need to be present with an infection.3 Severe sepsis is defined as sepsis resulting in organ dysfunction. Septic shock occurs when there is sepsis-induced hypotension (where either the systolic blood pressure is <90 mm Hg, <40 mm Hg below baseline, or the mean arterial pressure (MAP) is <70 mm Hg) that persists despite adequate fluid resuscitation.4
Progression to severe sepsis is associated with increased mortality and morbidity, including permanent organ damage, cognitive impairment, and physical disability.1 Provision of appropriate treatment early in the development of sepsis has been associated with improved patient outcomes.4 5 The benefit of these interventions is dependent upon the underlying short-term risk for mortality of the patient. For patients at high short-term risk, aggressive treatment and broad-spectrum empiric antibiotics significantly decrease mortality risk.5 6 However, for low-risk patients, the associated risk of aggressive treatments outweighs their benefit.7 For this reason, it is imperative to rapidly and accurately stratify patients with sepsis according to risk at the onset of the syndrome.
Previous studies have demonstrated that machine learning methods can be incorporated into electronic health records (EHRs) to predict clinically relevant outcomes in patients with sepsis. Recently, Peelen et al8 developed a dynamic Bayesian network (BN), modeling the progression of organ failure based on the Sequential Organ Failure Assessment (SOFA) score in the intensive care unit (ICU).9 Additionally, neural networks have been used to predict the critical states of sepsis, where a critical state is defined as a physiologic state proximally associated with a worsening clinical condition or death.10 Progression from sepsis to severe sepsis was found to be accurately classified by using support vector machines (SVMs).11 12 Furthermore, dynamic BNs can predict mortality in sepsis and ICU patients.13 14 Previously, we employed static BNs for early prediction of sepsis,15 which was followed recently by Nachimuthu and Haug16 who used dynamic BNs for early sepsis detection in the emergency department. Taken together, these results demonstrate the potential of SVMs and dynamic BNs to model clinical states and to prognosticate the outcome in patients with sepsis.
Although the aforementioned studies demonstrate the feasibility of extracting clinically relevant information pertaining to patients with sepsis, they do not specifically deal with the early identification of sepsis. Early identification of sepsis is challenging as the infection is not always clinically evident. In addition, the signs that constitute the SIRS criteria were selected to be sensitive, but not necessarily specific for sepsis, making early diagnosis of the syndrome prone to false-positive classifications. Conversely, scoring systems such as the SOFA,9 the Acute Physiology and Chronic Health Evaluation (APACHE),17 and Simplified Acute Physiology Score (SAPS)18 have shown reasonable discrimination when classifying patients with sepsis admitted to the ICU into groups with high-risk and low-risk mortality. Although there has been some work to stratify patients with sepsis according to risk before ICU care is needed,19 further investigation is necessary for a classification system that can accurately assess risk.
Serum lactate levels are used as a biomarker for end-organ hypoperfusion, impending shock, and increased risk of death in sepsis.20 21 Adults with sepsis and hyperlactatemia appear to have a lower mortality risk if diagnosed early and enrolled in a targeted resuscitative strategy programme.6 22 For this reason, updated consensus international guidelines for the treatment of severe sepsis include new recommendations for early lactate measurement among all adults with sepsis.4 Unfortunately, testing all adults with suspected sepsis for hyperlactatemia lacks specificity (SP) (4.6–12.9% SP for hyperlactatemia in some sepsis screening studies).23–25
Compliance with the guideline for severe sepsis improves patient outcomes in disparate clinical settings,26 27 but the burden of screening large numbers of patients for hyperlactatemia has been reported as a significant barrier to compliance.24 If screening for severe sepsis could be streamlined to test only patients at risk of hyperlactatemia, and avoid unnecessary testing of low-risk patients, then a major barrier to compliance with the guideline for severe sepsis could be mitigated. If this improvement in screening efficiency led to increased guideline compliance, then patient care and outcomes could be improved while increasing healthcare efficiency and decreasing costs. We therefore sought to develop an automated decision support system to identify patients at high risk for hyperlactatemia based upon routinely measured vital signs and laboratory studies. We applied naïve Bayes (NB), Gaussian mixture model (GMM) and hidden Markov model (HMM) classification to the time series data of temperature, white blood count, RR, and MAP to predict high or low serum lactate levels in a cohort of patients meeting SIRS criteria admitted to a large tertiary care hospital.
We also used BNs to determine the underlying relationships between lactate and patient outcomes, including mortality and sepsis. Furthermore, using SVM and NB classifiers, we predicted the risk of mortality in patients with and without sepsis and compared the performance of this method with existing ICU severity of illness models.
Methods
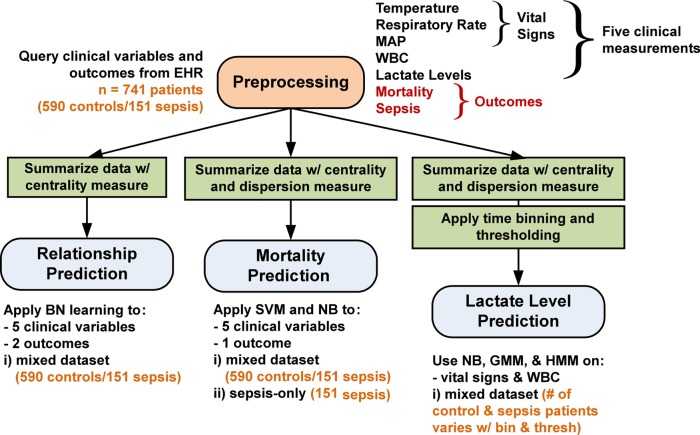
The data processing and analysis pipeline of the EHR sepsis database is summarized in figure 1. The processing pipeline has four main components, (1) database preprocessing, (2) BN structure learning, (3) mortality prediction with SVM and NB, and (4) lactate level prediction with NB, GMM, and HMM.
Figure 1.
Processing pipeline of electronic health record (EHR) sepsis database. The processing pipeline has four main components, (1) database preprocessing, (2) Bayes network structure learning, (3) mortality prediction with support vector machine (SVM) and naïve Bayes (NB), and (4) lactate level prediction with NB, Gaussian mixture model (GMM), and hidden Markov model (HMM). The variable number of patients used in lactate level prediction is given in tables 2 and 3. MAP, mean arterial pressure; WBC, white blood cell count.
Subjects and preprocessing
An EHR database containing 1492 adult patients (≥18 years of age) meeting at least two SIRS criteria admitted to the University of California Davis Health System (UCDHS), was used for all the analyses. Of the 1492 patients, 45.0% were female, the mean length of stay was 17.0±36.7 days, and 38.0% were admitted from the emergency department. UCDHS is a tertiary care, academic medical center that did not have an active EHR alert system for the diagnosis or treatment of sepsis during the study period. All data were abstracted retrospectively from the EHR via structured query language interrogation of a de-identified relational database. Patients were included in the database if they were hospitalized and discharged between 1 January 2010 and 31 December 2010. The following seven variables were used for the modeling applications: temperature, RR, WBC, MAP, lactate levels, mortality, and sepsis occurrence (determined from the EHR diagnosis). The first five variables are measurements of a patient's condition recorded over time (all but the lactate level and WBC are considered vital signs) and the final two variables were considered outcomes for this analysis; temperature, RR, and MAP are part of the SIRS criteria. The analyses were restricted to the 741 patients for whom all seven variables were available for both the 590 non-sepsis controls and 151 patients with sepsis. All analyses were approved by the institutional review board of the University of California at Davis (IRB# 254575).
Bayesian network structure learning
The BN structure learning was performed using the R statistical software environment28 with the bnlearn package.29 To construct the BN, the network was trained using the score-based greedy search hill-climbing algorithm, wherein the four different scoring measures of Akaike information (AIC), Bayesian information, Dirichlet posterior density, and K2 criterion scores were used to guide the learning procedure. Score-based learning proceeded by evaluating the change in score after trying all possible arc permutations, including arc removals, additions, and directionality changes. The arc-specific action that maximizes the score is accepted for each edge and the procedure stops if every further arc change decreases the overall score.30
To determine the generalization error of the inference method and to avoid overfitting, 10-fold cross-validation on each network was performed to estimate the goodness of fit. The cross-validation performance is evaluated using a log-likelihood loss function, with lower values indicating better performance. Thus, the BN with the least expected loss was chosen as the ideal network.
To construct the BN, all seven variables from the mixed dataset containing 741 patients, comprising 151 with sepsis and 590 non-sepsis controls, were used as features in the model (figure 1). The five measurement variables are time series data that were summarized for each patient using the mean (BN1) or median (BN2) values. Since each variable is summarized by a centrality measure, temporal variations of the time series are attenuated, giving a static BN. The discretization of the three SIRS variables was performed using the thresholds provided by the SIRS criteria. To discretize MAP, a 70 mm Hg threshold was used and lactate level discretization was performed with the high (≥4 mmol/L) or low (<4 mmol/L) threshold used by Rivers et al,6 resulting in 125 patients with high lactate levels. Finally, the strength of the arcs within the networks was calculated using the bootstrap-based inference method31 to provide a comparison between the BN1 and BN2 structures, in which the arc strengths ranged between 1 (strongest) and 0 (weakest).
Mortality prediction
For prediction of mortality, the five measurement variables were summarized by a pair of summary statistics containing a measure of centrality and dispersion. The pairs used were the mean and SD, median and mean absolute deviation (MAD), and median and IQR. In total, 10 features were considered for the mortality prediction, which represent the five measurements by a summary statistic pair. As a consequence, although temporal information is reduced, some variation is preserved in the dispersion measure. Principal component analysis was also used to assess whether linear transformation of the feature space and dimensionality reduction can be achieved in this case.32 A filter method was applied using the area under the receiver operating characteristic curve as a ranking criterion.33
In SVM classification, the training feature vectors are mapped to a higher dimension space, in which the SVM determines a linearly separating hyperplane given by a maximal margin.34 The mapping procedure can be accomplished by kernel function such as a linear, polynomial, or radial basis function (RBF). For this study, the RBF kernel was chosen, since it can handle non-linear interactions between class labels and features. Furthermore, in certain combinations of the penalty and kernel parameters, the linear and sigmoidal kernels match the performance of the RBF kernel.35
For the SVM classification, the optimal operating point was estimated by varying the C (penalty parameter in the error term) and γ (kernel) parameters using a grid search for each combination of feature selection and dimension reduction with a 10-fold cross-validation.35 NB classification is a probabilistic classifier where, for a given a dependent output class (in this case mortality), the probability of the output class occurring can be predicted using Bayes theorem, with the assumption that the 10 input features described above are independent of each other.36
For both classification methods, two datasets containing mortality as the output target were used (figure 1), where one contained a mixture of patients with and without sepsis (741 mixed patients consisting of 590 non-sepsis controls/151 sepsis; death rate 261/741) and the other contained patients with sepsis only (151 sepsis patients; death rate 52/151). Analysis of the mixed versus the sepsis-only dataset would highlight any classification changes between the groups. In analyzing the sepsis-only patients, the sepsis classification is used as a priori information to further delineate the patients.
The mortality classification was performed with the 10-fold cross-validation MATLAB37 using the native toolboxes for the feature selection, principal component analysis, and NB, and the libsvm toolbox for SVM.35 The combinations of dimension reduction and feature selection used with the classifiers are summarized in table 1.
Table 1.
Classification using 10-fold cross-validation of mortality among patients with and without sepsis with either feature selection, dimension reduction, or both.
| Preprocessing applied | Data type | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean and SD | Median and MAD | Median and IQR | ||||||||||||||
| ACC | F | SN | SP | AUC | ACC | F | SN | SP | AUC | ACC | F | SN | SP | AUC | ||
| SVM | ||||||||||||||||
| Patients with and without sepsis (n=741) | ||||||||||||||||
| # FS | 7 | 0.715 | 0.811 | 0.944 | 0.295 | 0.713±0.047 | 0.706 | 0.805 | 0.935 | 0.284 | 0.692±0.043 | 0.706 | 0.795 | 0.883 | 0.379 | 0.689±0.039 |
| 5 | 0.704 | 0.805 | 0.940 | 0.272 | 0.685±0.044 | 0.704 | 0.804 | 0.938 | 0.276 | 0.692±0.035 | 0.711 | 0.809 | 0.944 | 0.284 | 0.709±0.033 | |
| 3 | 0.704 | 0.809 | 0.969 | 0.218 | 0.644±0.054 | 0.700 | 0.806 | 0.963 | 0.218 | 0.659±0.031 | 0.688 | 0.797 | 0.946 | 0.215 | 0.654±0.050 | |
| # PC | 7 | 0.713 | 0.809 | 0.942 | 0.291 | 0.686±0.054 | 0.709 | 0.799 | 0.892 | 0.372 | 0.693±0.044 | 0.711 | 0.803 | 0.910 | 0.345 | 0.682±0.033 |
| 5 | 0.704 | 0.808 | 0.963 | 0.230 | 0.669±0.046 | 0.711 | 0.809 | 0.946 | 0.280 | 0.693±0.035 | 0.700 | 0.803 | 0.944 | 0.253 | 0.641±0.088 | |
| 3 | 0.694 | 0.803 | 0.965 | 0.195 | 0.639±0.036 | 0.699 | 0.803 | 0.948 | 0.241 | 0.677±0.045 | 0.680 | 0.796 | 0.963 | 0.161 | 0.653±0.043 | |
| # FS, # PC | 10, 10 | 0.721 | 0.807 | 0.900 | 0.391 | 0.692±0.031 | 0.711 | 0.804 | 0.913 | 0.341 | 0.701±0.038 | 0.700 | 0.801 | 0.929 | 0.280 | 0.701±0.051 |
| 7, 5 | 0.684 | 0.798 | 0.960 | 0.176 | 0.634±0.033 | 0.684 | 0.794 | 0.942 | 0.211 | 0.638±0.027 | 0.691 | 0.801 | 0.958 | 0.199 | 0.654±0.040 | |
| Sepsis-only patients (n=151) | ||||||||||||||||
| # FS | 7 | 0.702 | 0.809 | 0.960 | 0.212 | 0.629±0.120 | 0.715 | 0.814 | 0.949 | 0.269 | 0.639±0.083 | 0.709 | 0.812 | 0.960 | 0.231 | 0.630±0.106 |
| 5 | 0.715 | 0.814 | 0.949 | 0.269 | 0.653±0.082 | 0.702 | 0.805 | 0.939 | 0.250 | 0.683±0.121 | 0.722 | 0.821 | 0.970 | 0.250 | 0.624±0.101 | |
| 3 | 0.735 | 0.826 | 0.960 | 0.308 | 0.679±0.082 | 0.728 | 0.821 | 0.949 | 0.308 | 0.726±0.045 | 0.722 | 0.817 | 0.949 | 0.288 | 0.659±0.128 | |
| # PC | 7 | 0.715 | 0.817 | 0.970 | 0.231 | 0.641±0.104 | 0.722 | 0.819 | 0.960 | 0.269 | 0.645±0.084 | 0.715 | 0.815 | 0.960 | 0.250 | 0.594±0.119 |
| 5 | 0.702 | 0.812 | 0.980 | 0.173 | 0.578±0.081 | 0.702 | 0.809 | 0.960 | 0.212 | 0.601±0.096 | 0.715 | 0.814 | 0.949 | 0.269 | 0.641±0.104 | |
| 3 | 0.656 | 0.785 | 0.960 | 0.077 | 0.484±0.143 | 0.702 | 0.807 | 0.949 | 0.231 | 0.613±0.090 | 0.695 | 0.808 | 0.980 | 0.154 | 0.567±0.119 | |
| # FS, # PC | 10, 10 | 0.722 | 0.817 | 0.949 | 0.288 | 0.642±0.108 | 0.748 | 0.830 | 0.939 | 0.385 | 0.683±0.119 | 0.702 | 0.809 | 0.960 | 0.212 | 0.613±0.109 |
| 7, 5 | 0.695 | 0.808 | 0.980 | 0.154 | 0.621±0.111 | 0.695 | 0.808 | 0.980 | 0.154 | 0.588±0.123 | 0.68 | 0.80 | 0.96 | 0.15 | 0.620±0.119 | |
| NB | ||||||||||||||||
| Patients with and without sepsis (n=741) | ||||||||||||||||
| # FS | 7 | 0.673 | 0.787 | 0.929 | 0.203 | 0.609±0.037 | 0.671 | 0.778 | 0.892 | 0.264 | 0.629±0.048 | 0.687 | 0.785 | 0.883 | 0.326 | 0.632±0.047 |
| 5 | 0.679 | 0.790 | 0.933 | 0.211 | 0.643±0.049 | 0.688 | 0.795 | 0.931 | 0.241 | 0.647±0.053 | 0.677 | 0.784 | 0.902 | 0.264 | 0.636±0.031 | |
| 3 | 0.669 | 0.785 | 0.929 | 0.192 | 0.613±0.061 | 0.673 | 0.787 | 0.933 | 0.195 | 0.623±0.046 | 0.672 | 0.784 | 0.921 | 0.215 | 0.630±0.053 | |
| # PC | 7 | 0.691 | 0.795 | 0.927 | 0.257 | 0.653±0.046 | 0.706 | 0.802 | 0.921 | 0.310 | 0.665±0.041 | 0.692 | 0.790 | 0.892 | 0.326 | 0.653±0.032 |
| 5 | 0.676 | 0.788 | 0.931 | 0.207 | 0.644±0.041 | 0.692 | 0.798 | 0.935 | 0.245 | 0.649±0.038 | 0.673 | 0.781 | 0.900 | 0.257 | 0.639±0.052 | |
| 3 | 0.660 | 0.782 | 0.944 | 0.138 | 0.621±0.040 | 0.676 | 0.786 | 0.921 | 0.226 | 0.648±0.042 | 0.656 | 0.777 | 0.923 | 0.165 | 0.642±0.032 | |
| # FS, # PC | 10, 10 | 0.696 | 0.797 | 0.921 | 0.284 | 0.652±0.031 | 0.683 | 0.782 | 0.879 | 0.322 | 0.654±0.071 | 0.692 | 0.787 | 0.875 | 0.356 | 0.666±0.037 |
| 7, 5 | 0.663 | 0.780 | 0.921 | 0.188 | 0.602±0.044 | 0.672 | 0.781 | 0.904 | 0.245 | 0.610±0.030 | 0.667 | 0.777 | 0.898 | 0.241 | 0.595±0.034 | |
| Sepsis-only patients (n=151) | ||||||||||||||||
| # FS, # PC | 7 | 0.689 | 0.798 | 0.939 | 0.212 | 0.554±0.123 | 0.689 | 0.789 | 0.889 | 0.308 | 0.624±0.106 | 0.715 | 0.809 | 0.919 | 0.327 | 0.618±0.130 |
| 5 | 0.642 | 0.771 | 0.919 | 0.115 | 0.537±0.087 | 0.656 | 0.768 | 0.869 | 0.250 | 0.583±0.096 | 0.669 | 0.779 | 0.889 | 0.250 | 0.587±0.106 | |
| 3 | 0.649 | 0.774 | 0.919 | 0.135 | 0.560±0.074 | 0.662 | 0.773 | 0.879 | 0.250 | 0.594±0.108 | 0.642 | 0.759 | 0.859 | 0.231 | 0.578±0.084 | |
| # PC | 7 | 0.675 | 0.784 | 0.899 | 0.250 | 0.597±0.104 | 0.669 | 0.773 | 0.859 | 0.308 | 0.635±0.067 | 0.695 | 0.791 | 0.879 | 0.346 | 0.638±0.090 |
| 5 | 0.669 | 0.786 | 0.929 | 0.173 | 0.564±0.105 | 0.669 | 0.779 | 0.889 | 0.250 | 0.592±0.090 | 0.642 | 0.769 | 0.909 | 0.135 | 0.534±0.126 | |
| 3 | 0.662 | 0.788 | 0.960 | 0.096 | 0.557±0.099 | 0.636 | 0.764 | 0.899 | 0.135 | 0.577±0.065 | 0.636 | 0.764 | 0.899 | 0.135 | 0.559±0.075 | |
| # FS, # PC | 10, 10 | 0.689 | 0.795 | 0.919 | 0.250 | 0.593±0.136 | 0.715 | 0.805 | 0.899 | 0.365 | 0.635±0.086 | 0.709 | 0.798 | 0.879 | 0.385 | 0.660±0.050 |
| 7, 5 | 0.669 | 0.788 | 0.939 | 0.154 | 0.628±0.079 | 0.669 | 0.779 | 0.889 | 0.250 | 0.617±0.122 | 0.642 | 0.761 | 0.869 | 0.212 | 0.551±0.086 | |
ACC, accuracy; AUC, area under the receiver operating characteristic curve; F, harmonic mean of precision and recall; FS, features selected; MAD, median absolute deviation; NB, naïve Bayes; PC, principal components; SN, sensitivity; SP, specificity; SVM, support vector machine.
The cells highlighted in bold indicate the highest five performances achieved for each summary statistic pair.
Lactate level prediction
Prediction of the lactate level was performed with the classification algorithms of NB, clustering using GMM, and HMM. All three algorithms used the initial 741 patient dataset (590 non-sepsis controls and 151 patients with sepsis) containing the vital signs and WBC as inputs, while the target output was the lactate level for each patient, which was discretized in the same manner as in the BN structure learning section with the cut-off points of high (≥4 mmol/L) and low (<4 mmol/L) used by Rivers et al.6
The GMM performs clustering over the three vital signs and WBC through the estimation of Gaussian mixing parameters by employing the expectation-maximization algorithm.38 In our case, the GMM is assumed to be composed of two mixtures, one for high lactate level and the other low. The HMM provides the probability distribution of a set of hidden states and the observations generated over time through a Markov chain.38 In our study, the hidden states of high or low lactate levels were estimated from the observed vital signs and WBC. Both the GMM and HMM algorithms were initialized by mean and covariance estimates from the input of the vital signs and WBC.
The mean and SD were used to summarize the three vital signs and WBC, similar to the mortality prediction model, but the median/MAD and median/IQR pairs were not investigated. These two pairs were not investigated because the BN structure learnt based on the median did not show sepsis as conditionally dependent on the lactate level.
In MATLAB and with cross-validation, the GMM classification was performed using the built-in native toolboxes and software from Zhong et al,39 and the HMM classification was performed using a publicly available software toolbox.40
Time-binning and thresholding
Before applying the lactate level classification algorithms, the data were preprocessed to compare patients at similar time points for each measurement. The patient data were analyzed using three separate preprocessing schemes: (1) the patient data represented by the summary statistics only (no time-binning or thresholding), (2) the patient data thresholded by the number of points per measurement per patient, and (3) the patient data that was time-binned and then thresholded by the number of points per measurement per patient. The thresholds for the number of measurements were set at the first and fifth centiles, the 99th and 95th centiles, and 1.5 times the IQR. Thus, patients with or without an overabundance of measurements at the specific time bin for any of the vitals, WBC, or lactate were removed before the application of the classification algorithms. As a result, errors due to mismanagement of vital sign and laboratory measurements were mitigated. The time bins were set at bin widths of 6, 12, and 24 h, with two bins for each width.
Owing to time-binning and thresholding, the number of patients inputted into the algorithms varied; the number of total, high-lactate, and patients with sepsis remaining is summarized for each preprocessing scheme in tables 2 and 3. Averaged across all time-bin and thresholding combinations, the number of total patients is 347.4±248.5, high-lactate is 63.1±38.9, and sepsis is 74.9±46.6. The 10-, seven-, five-, and threefold cross-validations were performed for each classification when there was sufficient data remaining after the preprocessing. Furthermore, to concisely assess the viability and effects of time-binning and thresholding on the mortality prediction, the overall top five time-binning and thresholding combinations and the top combination in the first 24 h time bin in the lactate level prediction were used as features during SVM training.
Table 2.
Prediction of the high and low levels of lactate from the vital signs and white blood count, using the summary statistics of mean and SD with the NB, GMM, and HMM classifiers
| Threshold centile | Classification method (10-fold CV) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients* | NB | GMM | HMM | |||||||||||||
| ACC | F | SN | SP | AUC | ACC | F | SN | SP | AUC | ACC | F | SN | SP | AUC | ||
| All data | 741/125/151 | 0.794 | 0.879 | 0.906 | 0.240 | 0.68±0.051 | 0.837 | 0.906 | 0.943 | 0.312 | 0.759±0.031 | 0.756 | 0.850 | 0.833 | 0.376 | 0.679±0.048 |
| 5th | 402/86/83 | 0.729 | 0.830 | 0.842 | 0.314 | 0.64±0.055 | 0.811 | 0.884 | 0.915 | 0.430 | 0.801±0.042 | 0.764 | 0.848 | 0.835 | 0.500 | 0.704±0.059 |
| 95th | 631/100/137 | 0.813 | 0.898 | 0.944 | 0.160 | 0.68±0.058 | 0.853 | 0.915 | 0.947 | 0.350 | 0.779±0.043 | 0.713 | 0.815 | 0.751 | 0.510 | 0.671±0.072 |
| 5th, 95th | 313/62/72 | 0.812 | 0.890 | 0.948 | 0.258 | 0.66±0.071 | 0.843 | 0.905 | 0.928 | 0.500 | 0.849±0.050 | 0.728 | 0.813 | 0.737 | 0.694 | 0.790±0.069 |
| 1st | 426/93/91 | 0.732 | 0.831 | 0.844 | 0.333 | 0.649±0.055 | 0.808 | 0.882 | 0.919 | 0.409 | 0.782±0.038 | 0.624 | 0.717 | 0.610 | 0.677 | 0.696±0.070 |
| 99th | 715/119/150 | 0.799 | 0.883 | 0.908 | 0.252 | 0.669±0.056 | 0.841 | 0.909 | 0.951 | 0.286 | 0.774±0.037 | 0.730 | 0.828 | 0.782 | 0.471 | 0.704±0.053 |
| 1st, 99th | 404/88/90 | 0.745 | 0.841 | 0.861 | 0.330 | 0.646±0.049 | 0.814 | 0.888 | 0.940 | 0.364 | 0.817±0.045 | 0.649 | 0.750 | 0.674 | 0.557 | 0.678±0.055 |
| ±1.5IQR | 581/89/121 | 0.818 | 0.897 | 0.935 | 0.169 | 0.686±0.055 | 0.857 | 0.918 | 0.943 | 0.382 | 0.793±0.051 | 0.768 | 0.857 | 0.819 | 0.483 | 0.748±0.061 |
For each classifier, the cells highlighted in bold indicate the top three performances for the lactate level prediction.
*Patients column indicates the number of total/high-lactate/sepsis patients.
ACC, accuracy; AUC, area under the receiver operating characteristic curve; CV, cross-validation; F, harmonic mean of precision and recall; GMM, Gaussian mixture model; HMM, hidden Markov model; NB, naïve Bayes; SN, sensitivity; SP, specificity.
Table 3.
Prediction of high and low levels of lactate with time-binned and thresholded vital signs and white blood count, using the summary statistics of mean and SD with the NB, GMM, and HMM classifiers
| Bin hours | Threshold centile | Classification method (10-fold CV) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients* | NB | GMM | HMM | ||||||||||||||
| ACC | F | SN | SP | AUC | ACC | F | SN | SP | AUC | ACC | F | SN | SP | AUC | |||
| 1st 24 | 5th | 280/86/80 | 0.604 | 0.715 | 0.717 | 0.349 | 0.528±0.065 | 0.800 | 0.854 | 0.845 | 0.698 | 0.833±0.071 | 0.625 | 0.673 | 0.557 | 0.779 | 0.716±0.068 |
| 95th | 580/91/111 | 0.671 | 0.794 | 0.751 | 0.242 | 0.493±0.069 | 0.826 | 0.900 | 0.924 | 0.297 | 0.793±0.038 | 0.557 | 0.668 | 0.529 | 0.703 | 0.674±0.064 | |
| 5th, 95th | 187/50/54 | 0.727 | 0.826 | 0.883 | 0.300 | 0.495±0.105 | 0.850 | 0.900 | 0.876 | 0.780 | 0.922±0.047 | 0.765 | 0.824 | 0.752 | 0.800 | 0.812±0.089 | |
| 1st | 292/90/80 | 0.627 | 0.723 | 0.703 | 0.456 | 0.565±0.061 | 0.757 | 0.827 | 0.842 | 0.567 | 0.818±0.058 | 0.640 | 0.703 | 0.614 | 0.700 | 0.723±0.085 | |
| 99th | 698/121/141 | 0.567 | 0.699 | 0.610 | 0.355 | 0.489±0.048 | 0.814 | 0.892 | 0.929 | 0.264 | 0.702±0.032 | 0.447 | 0.532 | 0.380 | 0.769 | 0.613±0.058 | |
| 1st, 99th | 264/76/74 | 0.682 | 0.779 | 0.787 | 0.421 | 0.567±0.090 | 0.750 | 0.827 | 0.840 | 0.526 | 0.830±0.051 | 0.640 | 0.704 | 0.601 | 0.737 | 0.716±0.080 | |
| ±1.5IQR | 578/87/108 | 0.562 | 0.697 | 0.593 | 0.391 | 0.486±0.071 | 0.829 | 0.900 | 0.910 | 0.368 | 0.767±0.034 | 0.493 | 0.610 | 0.466 | 0.644 | 0.652±0.061 | |
| 2nd 24 (48 h) | 5th | 123/24/29 | 0.797 | 0.879 | 0.919 | 0.292 | 0.728±0.13 | 0.951 | 0.969 | 0.950 | 0.958 | 0.991±0.014 | 0.870 | 0.914 | 0.859 | 0.917 | 0.940±0.061 |
| 95th | |||||||||||||||||
| 5th, 95th | 64/10/11† | 0.781 | 0.873 | 0.889 | 0.200 | 0.578±0.264 | 0.953 | 0.971 | 0.944 | 1.000 | 1±0 | 0.891 | 0.931 | 0.871 | 1.000 | 0.965±0.032 | |
| 1st | 124/24/29 | 0.806 | 0.885 | 0.920 | 0.333 | 0.732±0.101 | 0.944 | 0.964 | 0.940 | 0.958 | 0.990±0.020 | 0.855 | 0.905 | 0.860 | 0.833 | 0.883±0.102 | |
| 99th | 700/27/135 | 0.917 | 0.956 | 0.941 | 0.333 | 0.651±0.143 | 0.944 | 0.971 | 0.960 | 0.556 | 0.866±0.053 | 0.696 | 0.815 | 0.695 | 0.704 | 0.74±0.086 | |
| 1st, 99th | 101/20/21 | 0.802 | 0.884 | 0.938 | 0.250 | 0.694±0.135 | 0.990 | 0.994 | 0.988 | 1.000 | 1±0 | 0.950 | 0.969 | 0.963 | 0.900 | 0.906±0.126 | |
| ±1.5IQR | |||||||||||||||||
| 1st 12 | 5th | 177/61/49 | 0.646 | 0.752 | 0.773 | 0.360 | 0.543±0.065 | 0.808 | 0.852 | 0.845 | 0.738 | 0.894±0.047 | 0.718 | 0.769 | 0.716 | 0.721 | 0.800±0.053 |
| 95th | 567/87/106 | 0.647 | 0.776 | 0.726 | 0.220 | 0.492±0.046 | 0.832 | 0.904 | 0.929 | 0.299 | 0.741±0.059 | 0.580 | 0.702 | 0.583 | 0.563 | 0.634±0.067 | |
| 5th, 95th | 88/20/27‡ | 0.75 | 0.845 | 0.882 | 0.300 | 0.718±0.14 | 0.977 | 0.985 | 0.971 | 1.000 | 0.999±0.001 | 0.875 | 0.913 | 0.853 | 0.950 | 0.941±0.087 | |
| 1st | 184/64/49 | 0.598 | 0.702 | 0.725 | 0.359 | 0.595±0.111 | 0.918 | 0.938 | 0.942 | 0.875 | 0.962±0.030 | 0.832 | 0.870 | 0.867 | 0.766 | 0.865±0.051 | |
| 99th | 701/136/140 | 0.665 | 0.785 | 0.758 | 0.279 | 0.532±0.039 | 0.796 | 0.879 | 0.919 | 0.287 | 0.702±0.048 | 0.552 | 0.659 | 0.536 | 0.618 | 0.601±0.059 | |
| 1st, 99th | 159/53/42 | 0.591 | 0.683 | 0.660 | 0.453 | 0.613±0.1 | 0.855 | 0.888 | 0.859 | 0.849 | 0.954±0.024 | 0.780 | 0.821 | 0.755 | 0.830 | 0.842±0.057 | |
| ±1.5IQR | 637/107/129 | 0.732 | 0.840 | 0.845 | 0.168 | 0.515±0.057 | 0.826 | 0.898 | 0.923 | 0.346 | 0.742±0.026 | 0.611 | 0.725 | 0.617 | 0.579 | 0.648±0.050 | |
| 2nd 12 (24 h) | 5th | 116/24/37 | 0.698 | 0.817 | 0.848 | 0.125 | 0.409±0.155 | 0.957 | 0.972 | 0.946 | 1.000 | 0.997±0.0158 | 0.845 | 0.897 | 0.848 | 0.833 | 0.866±0.11 |
| 95th | |||||||||||||||||
| 5th, 95th | |||||||||||||||||
| 1st | 118/24/37 | 0.686 | 0.808 | 0.830 | 0.125 | 0.351±0.133 | 0.95 | 0.967 | 0.947 | 0.958 | 0.995±0.01 | 0.856 | 0.903 | 0.840 | 0.917 | 0.900±0.096 | |
| 99th | 687/23/135 | 0.904 | 0.949 | 0.929 | 0.174 | 0.474±0.169 | 0.952 | 0.975 | 0.964 | 0.609 | 0.903±0.058 | 0.884 | 0.937 | 0.898 | 0.478 | 0.842±0.059 | |
| 1st, 99th | 98/16/30‡ | 0.541 | 0.662 | 0.537 | 0.563 | 0.546±0.16 | 0.959 | 0.975 | 0.963 | 0.938 | 0.996±0.01 | 0.857 | 0.910 | 0.866 | 0.813 | 0.855±0.13 | |
| ±1.5IQR | |||||||||||||||||
| 1st 6 | 5th | 65/25/20† | 0.523 | 0.635 | 0.675 | 0.280 | 0.482±0.11 | 0.923 | 0.937 | 0.925 | 0.920 | 0.979±0.043 | 0.831 | 0.853 | 0.800 | 0.880 | 0.892±0.060 |
| 95th | 478/76/73 | 0.638 | 0.763 | 0.692 | 0.355 | 0.513±0.083 | 0.849 | 0.912 | 0.933 | 0.408 | 0.778±0.037 | 0.521 | 0.624 | 0.473 | 0.776 | 0.655±0.076 | |
| 5th, 95th | |||||||||||||||||
| 1st | 65/25/20 | Same as 5th | Same as 5th | Same as 5th | |||||||||||||
| 99th | 658/129/130 | 0.561 | 0.690 | 0.607 | 0.372 | 0.473±0.061 | 0.781 | 0.867 | 0.887 | 0.349 | 0.73±0.033 | 0.485 | 0.579 | 0.441 | 0.667 | 0.608±0.082 | |
| 1st, 99th | 43/13/15§ | 0.442 | 0.586 | 0.567 | 0.154 | 0.436±0.193 | 0.884 | 0.912 | 0.867 | 0.923 | 0.964±0.063 | 0.837 | 0.873 | 0.800 | 0.923 | 0.931±0.144 | |
| ±1.5IQR | |||||||||||||||||
| 2nd 6 (12 h) | 5th | 75/22/19† | 0.733 | 0.825 | 0.887 | 0.364 | 0.668±0.080 | 0.947 | 0.963 | 0.981 | 0.864 | 0.976±0.015 | 0.827 | 0.869 | 0.811 | 0.864 | 0.900±0.684 |
| 95th | |||||||||||||||||
| 5th, 95th | |||||||||||||||||
| 1st | 80/23/19† | 0.775 | 0.855 | 0.930 | 0.391 | 0.658±0.175 | 0.925 | 0.947 | 0.947 | 0.870 | 0.967±0.030 | 0.800 | 0.846 | 0.772 | 0.870 | 0.824±0.094 | |
| 99th | |||||||||||||||||
| 1st, 99th | |||||||||||||||||
| ±1.5IQR | |||||||||||||||||
For each classifier the cells highlighted in bold indicate the top five performances for the lactate level prediction. Blank rows indicate that the classification was not performed owing to insufficient data.
*Patients column indicates the number of total/high-lactate/sepsis patients.
†Indicates fivefold CV; ‡indicates sevenfold CV; §indicates threefold CV.
ACC, accuracy; AUC, area under the receiver operating characteristic curve; CV, cross-validation; F, harmonic mean of precision and recall; GMM, Gaussian mixture model; HMM, hidden Markov model; NB, naïve Bayes; SN, sensitivity; SP, specificity.
Performance evaluation
The following metrics were calculated to assess performance of the classifiers in the cross-validations: accuracy (ACC, proportion of true results), F-measure (F, harmonic mean of precision and recall), sensitivity (SN), SP, and the area under the receiver operating characteristic curve (AUC) using vertical averaging.41 The AUC was calculated for its ability to evaluate the average specificity for the full range of sensitivities.42 Thus, the best performance was defined as that having the highest AUC followed by the highest accuracy (in the case of a tie). The positive/negative classes for mortality and lactate level predictions are defined as alive/deceased and low/high, respectively.
Results
The results for each analysis technique described above in the processing pipeline are summarized below.
Bayesian network structure learning
For both BN1 (mean data) and BN2 (median data) network structures, the AIC scoring criteria provided the goodness-of-fit estimate with the least loss. Consequently, the structure provided by AIC was the preferred network structure for both types of data (figure 2); the log-likelihood loss was 4.076 for BN1 and 3.942 for BN2. In a Wilcoxon signed rank test for all four scoring criteria for both BNs, there was no significant difference (p=0.125) between the network scores.
Figure 2.
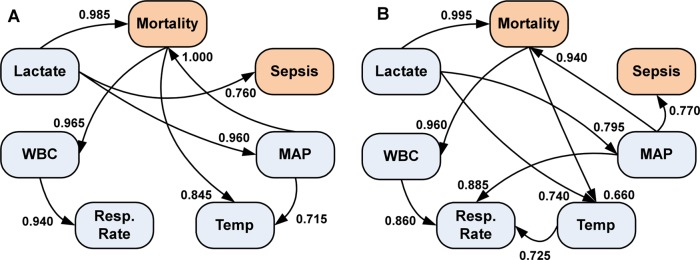
Bayesian networks from the five clinical measurements and two outcomes. (A) The Bayesian network structure determined from the mean of the five clinical measurements. (B) The Bayesian network structure from the median of five clinical measurements. On each directed arc, the strength of the arc is indicated, ranging from 0 to 1, with 1 being the strongest. MAP, mean arterial pressure; WBC, white blood cell count.
The network structure learnt for both BN1 (figure 2A) and BN2 (figure 2B) shows that both the WBC and temperature, which are both part of the SIRS criteria, are conditionally dependent on the in-hospital risk of mortality. Additionally, mortality, sepsis, and MAP levels in the BN1 structure are conditionally dependent on the lactate levels. However, for BN2 the lactate level has an indirect path to sepsis occurrence through MAP. In both BNs, lactate level is the only conditionally independent variable. The common arcs in both BNs did not show a statistically significant difference (Wilcoxon signed rank test, p=0.313). In BN1, the arc pointing from MAP to mortality was the strongest arc (strength=1.000). The strongest arc in BN2 points from the lactate levels to mortality (strength=0.995); in BN1 this was the second strongest arc (strength=0.985).
Mortality prediction
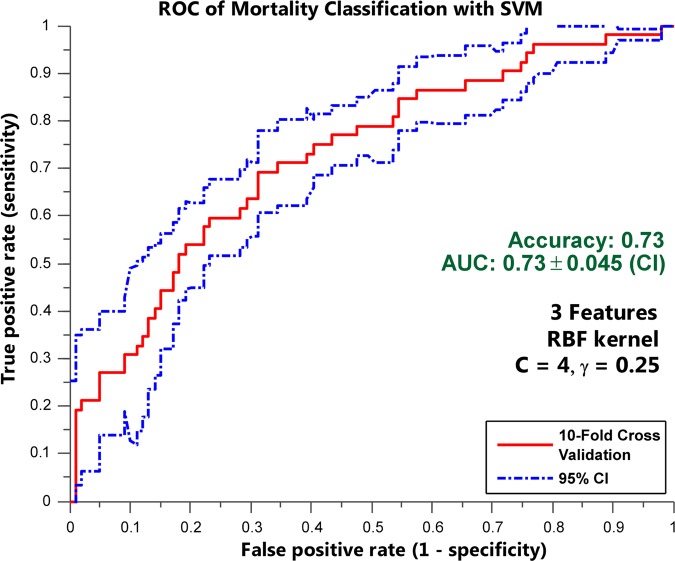
Table 1 summarizes mortality prediction performances for each summary statistic pair using the SVM and NB classifiers. The best prediction of mortality (AUC=0.713±0.047, ACC=0.715, F=0.811, SN=0.944, SP=0.295) among the mixed dataset of patients with and without sepsis was provided using SVM (C=8 and γ=0.25) with the top seven features (mean lactate, mean MAP, SD of RR, mean temperature, mean WBC, SD of lactate, and SD of MAP) summarized by the mean/SD pair. Among the sepsis-only patients, the best prediction of mortality (AUC=0.726±0.045, ACC=0.728, F=0.821, SN=0.949, SP=0.308) was again provided by SVM (C=4 and γ=0.25) using the top three features of (1) median lactate levels, (2) the MAD of RR, and (3) median MAP (figure 3).
Figure 3.
The receiver operating characteristic (ROC) curve showing the discriminability of sepsis-only patients using the support vector machine (SVM) with only three features. The ROC curve showing the discriminability of the best prediction of mortality (area under the ROC curve (AUC)=0.726±0.045, accuracy=0.728, F=0.821, sensitivity=0.949, specificity=0.308) was provided by SVM (C=4 and γ=0.25) using the top three features of (1) median lactate levels, (2) the mean absolute deviation of respiratory rate, and (3) median mean arterial pressure. RBF, radial basis function.
An analysis of variance (ANOVA) with 2 (classifier type)×2 (patient group)×3 (summary statistic pair) between-subjects factors showed that there were no significant interactions among the factors, provided by the AUC performance data in table 1. However, there was a significant difference between the SVM and NB mortality classifications (F(1,91)=32.85, p<0.001), where the SVM had a higher mean AUC (µSVM=0.651 vs µNB=0.613). The mean AUC provided by the sepsis-only group (µsepsis=0.655) was also significantly higher (F(1,91)=49.10, p<0.001) than for the mixed group of patients (µmixed=0.609). A significant difference among the summary statistic pairs (F(2,91)=3.67, p=0.0293) on the AUC performance was found at the α=0.05 level. Further Tukey post-hoc tests on the summary statistic pair factor showed that the significant difference was between the mean/SD and median/MAD pairs, which had the higher mean AUC (p=0.0236, α=0.05, µmean/SD=0.622 vs µmedian/MAD=0.630).
Lactate level prediction
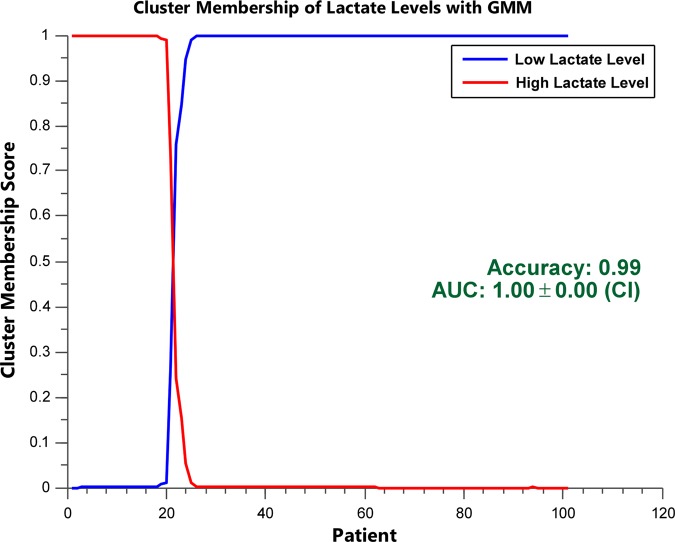
The lactate level prediction by the NB, GMM, and HMM classifiers using the thresholded and time-binned with thresholded data are summarized in tables 2 and 3, respectively, where the top five performances are highlighted for each classifier. The best overall lactate level prediction (AUC=1.000±0, ACC=0.990, F=0.994, SN=0.988, SP=1.000) was obtained with the second 24 h time bin when the vital signs and the white blood cell measurements were thresholded at the 1st and 99th centiles (table 3, figure 4). Without any thresholding or time-binning (table 2), GMM provided a reasonable lactate level prediction (AUC=0.759±0.031, ACC=0.837, F=0.906, SN=0.943, SP=0.312). For NB (AUC=0.732±0.101, ACC=0.806, F=0.885, SN=0.920, SP=0.333) and HMM (AUC=0.965±0.032, ACC=0.891, F=0.931, SN=0.871, SP=1.000), the best results were also provided with the second 24 h time bin when the measurements were thresholded at the 1st centile, and the 5th and 95th centiles, respectively (table 3).
Figure 4.
Cluster membership scores for clustering using Gaussian mixture model (GMM) in the second 24 h time bin. Cluster membership scores of patients’ lactate levels indicate the posterior probability of assigning high or low lactate levels for each patient. A sharp delineation (the presence of few 0.5 probabilities) between the high and low lactate levels indicates good separation. The cluster membership score of the second 24 h time bin with the vital signs and white blood cell count measurements thresholded at the 1st and 99th centiles is shown (area under the receiver operating characteristic curve (AUC)=1.000±0, accuracy=0.990, F=0.994, sensitivity=0.988, specificity=1.000). This is the best performance of lactate level prediction.
In an one-way ANOVA, using the AUC performance data in table 2 and the classifier type as the between-subjects factor, a significant difference was shown between the classifiers (F(2,21)=38.12, p<0.001). The Tukey post hoc test showed that the GMM classifier had the highest mean AUC (µGMM=0.794, µHMM=0.709, µNB=0.664; all comparisons p<0.05). After an ANOVA with a 3 (classifier type)×3 (time) between-subjects factors, with the AUC values in table 3, a significant difference among the three classifiers with respect to the performance was shown (F(2,82)=73.41, p<0.001). There was no significant interaction between time bins and the classifiers. Additional Tukey post-hoc tests showed that the GMM classifier had the highest mean AUC, followed by HMM (µGMM=0.889, µHMM=0.791, µNB=0.553; all comparisons p<0.01).
Time-binning and thresholding for mortality prediction
Mortality prediction for patients with and without sepsis using SVM was applied to the top five time-binned and thresholded combinations obtained from the lactate level prediction. The best mortality prediction, albeit very poor, was given by the second 24 h time bin with the vital signs and the WBC measurements thresholded at the 1st and 99th centiles (AUC=0.553±0.107, ACC=0.604, F=0.747, SN=0.937, SP=0.0526). The other four predictions, which were also poor, had a mean AUC of 0.515±0.0265. In the first 24 h time bin thresholded at the 5th and 95th centiles, the prediction (AUC=0.512+0.151, ACC=0.636, 0.778, SN=0.952, SP=0) was again much worse than the mortality prediction without time-binning.
Discussion
The BN learnt for both the median and mean summarization of the patients’ measurement variables showed that our database reinforces the key clinical finding that serum lactate levels are associated with increased mortality risk in sepsis.6 22 Also, in both BNs we confirmed that risk of mortality is conditionally dependent on MAP, which was shown to be independently associated with mortality in sepsis.43 Interestingly, the lactate levels were conditionally independent of the vital signs, WBC, and MAP, although we were able to predict it through GMMs and HMMs. Nonetheless, in both of the BNs, three of the clinical measurements were conditionally dependent on mortality, which is reflected in the success of our SVM and NB classification of mortality risk.
This study demonstrates the feasibility of using SVM classification with feature selection to predict mortality risk for patients with sepsis. With only the three features (median lactate levels, median MAP, MAD of RR) the highest accuracy and discriminability achieved were 0.73 and 0.73 AUC, respectively. This result is comparable to that of Ribas et al.44 In their study logistic regression with factor analysis was employed to achieve an accuracy of 0.78 and discriminability of 0.75 AUC with 156 ICU patients with severe sepsis. The key difference between our work and that of Ribas et al44 is the nature of the observations. In our SVM model, only a summary statistic pair of the three SIRS criteria, lactate level, and MAP was necessary to perform satisfactory classification. Ribas et al44 performed their final classification with considerably more clinical variables, including the number of dysfunctional organs, mechanical ventilation, APACHE II score, and resuscitation bundles. These variables are all derivatives of aggregate scoring systems used in hospital intensive care settings and are generally not available in non-ICU settings.44 As the majority of sepsis cases are initially identified in non-ICU settings (emergency department or hospital ward), and rapid risk stratification is essential to treatment, our approach has significantly broader potential applicability. Since aggregate scoring was not used as input data in this study, our input data may be more specific to an individual patient's dynamics. Moreover, the aggregate scoring systems combine many sources of test information, which may greatly reduce the temporal aspects of the data.
Langmead13 predicted the risk of mortality in patients with sepsis using dynamic BNs with an accuracy of 0.93. For that analysis, synthetic data were used, which limits applicability to clinical situations. Nonetheless, using SVM on their synthetic sepsis data, they achieved an accuracy of 0.81 (discriminability not provided). Furthermore, Kayaalp et al14 achieved a discriminability of 0.83 (accuracy not provided). Traditional logistic regression with standard scoring systems has provided the following performances: 0.86 AUC with APACHE II,45 0.88 AUC with APACHE IV,46 0.77 AUC with SAPS3 PIRO,47 and 0.70 with PIRO.48 We demonstrate that by focusing on routinely available clinical data, we can achieve a feasible level of accuracy and predictability for mortality. Through pairing of the measure of centrality and dispersion, we have a unique method to capture the salient temporal dynamics of the patients’ state.
However, a limitation of the mortality prediction presented in this study is that it is retrospective and when a prediction is made in the first 24 h time bin, thresholded at the 5th and 95th centiles, a poor discriminability of only 0.51 AUC is achieved. This may be due to the fact that the necessary temporal features in the time series are not captured. For the 151 patients with sepsis in the mortality prediction model, the average hospital length of stay was 13.5±19.2 days, which is a significantly longer time than the 24 h used for time-binning, indicating that there may be an ideal length of time that could optimize the mortality risk prediction by highlighting the significant features of the time series.
A significant finding of this study is that lactate levels can be predicted with high discriminability when the time scale and frequency of the measurements are considered. Specifically, when the time bins were divided into widths of 6, 12, and 24 h the performance of the GMM, HMM, and NB classifications of the lactate levels improved (table 3), even though the ANOVA did not show significant differences for time factor. Also, it was shown that when only the number of measurements was thresholded using centiles, the accuracies and discriminabilities increased compared with the results when no thresholding was applied (table 2). There have been recent efforts to use the temporal variability of patient treatments to increase the predictive power of decision support systems.49
For the first time, this study introduces the prediction of serum lactate levels from patient vital signs and WBC. The ability to predict lactate levels holds the promise of potentially identifying accurately septic patients at high risk of short-term mortality, while limiting needless testing of low-risk patients. Current guidelines call for lactate testing on all patients with sepsis and there is evidence that non-compliance with these guidelines increases mortality risk.50 Lactate testing is routinely performed in the ICU, but because testing all patients with suspected sepsis lacks specificity, its use has been limited in emergency departments.23 Non-compliance with the guideline may also be influenced by long processing times for laboratory studies in busy emergency departments.51
There has been recent interest in point-of-care (POC) lactate testing in the emergency department to alleviate the burden of processing time. Recently, it was shown that POC testing decreased the mean time between triage and lactate measurement from 2.5 h to 1.4 h.52 However, strong evidence about the ability of POC testing to determine high lactate levels (≥4 mmol/L) is lacking since only three of the 24 patients had raised lactate levels. Nonetheless, using our GMM in the 1st 6 h time bin, thresholded at the 5th centile, we were able to predict with a discriminability of 0.98 AUC (table 3) the 23 patients with high lactate levels from among 65 patients, who included 20 patients with sepsis, demonstrating the capabilities of our method. Furthermore, Karon et al53 demonstrated that POC measurements have a tendency to shift away from the serum lactate laboratory measurement starting at the ≥6 mmol/L level.
The main limitation of the lactate level prediction with GMM and HMM is that the algorithms require reasonable separation between features of the control and target groups to initialize the probability distributions used to parameterize the classifiers. This can become a problem when aligning patients according to time and a patient does not have data pertaining to that particular time. If sufficient patients are accumulated, a sparse dataset will form with correlated control and target groups. To circumvent this issue, we focused on patients with measurements at the times of interest. Another possible way to resolve this problem is to include the patients who have incomplete records and perform multiple imputation, which has been shown to provide unbiased estimates of missing data.54 The benefit of imputation is that we would be able to include patients who might have relevant features for analysis, but who were excluded owing to errors in data collection or recording, which is common in clinical practice. For future analyses we will extend our models to include incomplete records with imputation and we expect the models to remain predictive, given our focus on centrality and dispersion measures.
Overwhelmed clinical systems are stressed with needless testing and patients may be exposed to high-risk interventions that may not benefit them. A dynamic decision support system that accurately predicts hyperlactatemia could focus limited resources, avoid ‘alert fatigue’ that may accompany decision support systems, and decrease procedural complications for patients. The success of our lactate classification could be extended by including the predicted lactate levels with the measured vital signs to predict either mortality or sepsis.
Conclusion
Clustering with GMM can be used with patient vital signs to predict serum lactate levels with high discriminability. Additionally, SVM classification with feature selection can be used to predict mortality risk for patients with sepsis when the patient's measurements are summarized by a pair of summary statistics. The results demonstrate the importance of implementing timely and accurate recording systems in hospitals for the objective collection of patient data. Rather than collecting more data, premeditated collection of measurements at synchronous times can preserve important events and features of a patient’s state. In future studies, the age and sex variables with increased observations within the 6–12 h time scale will be included in the analysis to analyze the early dynamics of lactate levels and mortality risk among patients with sepsis. Overall, the new analyses presented in this study demonstrate the promise of predicting serum lactate levels, which may facilitate sepsis risk assessment and improve compliance with therapeutic guidelines, which significantly decreases mortality risk.
Footnotes
Contributors: EG performed all experiments and analyzed the data. HN provided the dataset. JPG, HN, JA and TA advised on the clinical approach and relevance of the study. IT advised on, and supervised, all computational aspects and data analysis of the study. All authors wrote the manuscript.
Funding: This work was supported by the Center for Information Technology Research in the Interest of Society (CITRIS) through seed grant #2469085 and by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), through grant #UL1 TR000002.
Competing interests: None.
Ethics approval: Institutional review board.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Upon publication of the manuscript, we will make public all code and analysis files from our laboratory website.
References
- 1.Weir LM, Levit K, Stranges E, et al. HCUP facts and figures: statistics on hospital-based care in the United States, 2008. [Internet]. Rockville, MD: Agency for Healthcare Research and Quality, 2010 [cited 1 February 2013]. http://www.hcup-us.ahrq.gov/reports.jsp. [Google Scholar]
- 2.Angus DC, Linde-Zwirble WT, Lidicker J, et al. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 2001;29:1303–10 [DOI] [PubMed] [Google Scholar]
- 3.Levy MM, Fink MP, Marshall JC, et al. 2001 sccm/esicm/accp/ats/sis international sepsis definitions conference. Intensive Care Med 2003;29:530–8 [DOI] [PubMed] [Google Scholar]
- 4.Dellinger RP, Levy MM, Rhodes A, et al. Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med 2013;39:165–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006;34:1589–96 [DOI] [PubMed] [Google Scholar]
- 6.Rivers E, Nguyen B, Havstad S, et al. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med 2001;345:1368–77 [DOI] [PubMed] [Google Scholar]
- 7.Kumar A, Safdar N, Kethireddy S, et al. A survival benefit of combination antibiotic therapy for serious infections associated with sepsis and septic shock is contingent only on the risk of death: a meta-analytic/meta-regression study. Crit Care Med 2010;38:1651–64 [DOI] [PubMed] [Google Scholar]
- 8.Peelen L, de Keizer NF, de Jonge E, et al. Using hierarchical dynamic Bayesian networks to investigate dynamics of organ failure in patients in the intensive care unit. J Biomed Inform 2010;43:273–86 [DOI] [PubMed] [Google Scholar]
- 9.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. Intensive Care Med 1996;22:707–10 [DOI] [PubMed] [Google Scholar]
- 10.Brause R, Hamker F, Paetz J. Septic shock diagnosis by neural networks and rule based systems. In: Schmitt M, Teodorescu HN, Jain A, et al., eds. Computational intelligence techniques in medical diagnosis and prognosis. New York: Springer, 2002:323–56 [Google Scholar]
- 11.Wang SL, Wu F, Wang BH. Prediction of severe sepsis using SVM model. In: Arabnia HR. ed. Advances in computational biology. New York: Springer, 2010:75–81 [DOI] [PubMed] [Google Scholar]
- 12.Tang CHH, Middleton PM, Savkin AV, et al. Non-invasive classification of severe sepsis and systemic inflammatory response syndrome using a nonlinear support vector machine: a preliminary study. Physiol Meas 2010;31:775–93 [DOI] [PubMed] [Google Scholar]
- 13.Langmead CJ. Generalized queries and Bayesian statistical model checking in dynamic Bayesian networks: application to personalized medicine. Proceedings of the 8th International Conference on Computational Systems Bioinformatics; August 2009, 201–12 [Google Scholar]
- 14.Kayaalp M, Cooper G, Clermont G. Predicting ICU mortality: a comparison of stationary and nonstationary temporal models. In: Overhage J. ed. Converging Information, Technology, and Health Care, proceedings of AMIA Symposium; 2000, Los Angeles, USA: 2000:418–22 [PMC free article] [PubMed] [Google Scholar]
- 15.Gultepe E, Nguyen H, Albertson T, et al. A Bayesian network for early diagnosis of sepsis patients: a basis for a clinical decision support system. 2nd IEEE International Conference on Computational Advances in Bio and Medical Sciences (ICCABS); Las Vegas, NV: 23–25, 2012, 1–5 [Google Scholar]
- 16.Nachimuthu SK, Haug PJ. Early detection of sepsis in the emergency department using Dynamic Bayesian Networks. AMIA Annu Symp Proc 2012;2012:653–62 [PMC free article] [PubMed] [Google Scholar]
- 17.Knaus WA, Zimmerman JE, Wagner DP, et al. APACHE-acute physiology and chronic health evaluation: a physiologically based classification system. Crit Care Med 1981;9:591–7 [DOI] [PubMed] [Google Scholar]
- 18.Le Gall JR, Loirat P, Alperovitch A, et al. A simplified acute physiology score for ICU patients. Crit Care Med 1984;12:975–7 [DOI] [PubMed] [Google Scholar]
- 19.Shapiro NI, Howell MD, Talmor D, et al. Mortality in Emergency Department Sepsis (MEDS) score predicts 1-year mortality. Crit Care Med 2007;35:192–8 [DOI] [PubMed] [Google Scholar]
- 20.Bakker J, Gris P, Coffernils M, et al. Serial blood lactate levels can predict the development of multiple organ failure following septic shock. Am J Surg 1996;171:221–6 [DOI] [PubMed] [Google Scholar]
- 21.Nguyen HB, Rivers EP, Knoblich BP, et al. Early lactate clearance is associated with improved outcome in severe sepsis and septic shock. Crit Care Med 2004;32:1637–42 [DOI] [PubMed] [Google Scholar]
- 22.Jones AE, Brown MD, Trzeciak S, et al. The effect of a quantitative resuscitation strategy on mortality in patients with sepsis: a meta-analysis. Crit Care Med 2008;36:2734–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howell MD, Donnino M, Clardy P, et al. Occult hypoperfusion and mortality in patients with suspected infection. Intensive Care Med 2007;33:1892–9 [DOI] [PubMed] [Google Scholar]
- 24.Trzeciak S, Dellinger RP, Chansky ME, et al. Serum lactate as a predictor of mortality in patients with infection. Intensive Care Med 2007;33:970–7 [DOI] [PubMed] [Google Scholar]
- 25.Green JP, Berger T, Garg N, et al. Serum lactate is a better predictor of short-term mortality when stratified by C-reactive protein in adult emergency department patients hospitalized for a suspected infection. Ann Emerg Med 2011;57:291–5 [DOI] [PubMed] [Google Scholar]
- 26.Levy MM, Dellinger RP, Townsend SR, et al. The Surviving Sepsis Campaign: results of an international guideline-based performance improvement program targeting severe sepsis. Intensive Care Med 2010;36:222–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barochia AV, Cui X, Vitberg D, et al. Bundled care for septic shock: an analysis of clinical trials. Crit Care Med 2010;38:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team. R: a language and environment for statistical computing. [Internet]. Vienna, Austria: R Foundation for Statistical Computing, 1993 [16 May 2013; cited 24 May 2013]. http://www.R-project.org/ [Google Scholar]
- 29.Scutari M. Learning Bayesian networks with the bnlearn R package. J Stat Softw 2010;35:1–2221603108 [Google Scholar]
- 30.Hofmann R, Tresp V. Discovering structure in continuous variables using Bayesian networks. In: Touretzky DS, Mozer MC, Hasselmo ME. eds. Advances in neural information processing systems 8. MIT Press, 1996:500–6 [Google Scholar]
- 31.Friedman N, Goldszmidt M, Wyner A. Data analysis with Bayesian networks: a bootstrap approach. Proceedings of the Fifteenth conference on Uncertainty in artificial intelligence; 1999, 206–115 [Google Scholar]
- 32.Jolliffe I. Principle component analysis. New York, NY: John Wiley & Sons, 1998 [Google Scholar]
- 33.Wang R, Tang K. Feature selection for maximizing the area under the ROC curve. IEEE International Conference on Data Mining Workshops; 6 December 2009, Florida, USA: 400–5 [Google Scholar]
- 34.Vapnik V. Statistical learning theory. New York, NY: John Wiley & Sons, 1998 [Google Scholar]
- 35.Hsu CW, Chang CC, Lin CJ. A practical guide to support vector classification. [Internet]. Taipei, Taiwan: National Taiwan University, 2003 [15 April 2010; cited 15 November 2012]. http://www.csie.ntu.edu.tw/~cjlin/papers/guide/guide.pdf. [Google Scholar]
- 36.Friedman J, Hastie T, Tibshirani R. The elements of statistical learning. New York, NY: Springer, 2008 [Google Scholar]
- 37.Mathworks. 2010. MATLAB [DVD], Version r2010a, [Computer program]. Available Distributor: The MathWorks Inc. Natick, MA.
- 38.Bishop CM. Pattern recognition and machine learning. New York, NY: Springer, 2006 [Google Scholar]
- 39.Zhong Q, Busetto AG, Fededa JP, et al. Unsupervised modeling of cell morphology dynamics for time-lapse microscopy. Nat Methods 2012;9:711–13 [DOI] [PubMed] [Google Scholar]
- 40.Murphy K. Hidden Markov model (HMM) toolbox for MATLAB. [Internet]. Kevin Murphy, 1998 [8 June 2005; cited 24 May 2013]. http://www.cs.ubc.ca/~murphyk/Software/HMM/hmm.html. [Google Scholar]
- 41.Fawcett T. ROC graphs: notes and practical considerations for researchers. Mach Learn 2004;31:1–38 [Google Scholar]
- 42.Obuchowski NA. ROC analysis. Am J Roentgenol 2005;184:364–72 [DOI] [PubMed] [Google Scholar]
- 43.Varpula M, Tallgren M, Saukkonen K, et al. Hemodynamic variables related to outcome in septic shock. Intensive Care Med 2005;31:1066–71 [DOI] [PubMed] [Google Scholar]
- 44.Ribas VJ, Vellido A, Ruiz-Rodríguez JC, et al. Severe sepsis mortality prediction with logistic regression over latent factors. Expert Syst Appl 2012;39:1937–43 [Google Scholar]
- 45.Wong DT, Crofts SL, Gomez M, et al. Evaluation of predictive ability of APACHE II system and hospital outcome in Canadian intensive care unit patients. Crit Care Med 1995;23:1177–83 [DOI] [PubMed] [Google Scholar]
- 46.Zimmerman JE, Kramer AA, McNair DS, et al. Acute Physiology and Chronic Health Evaluation (APACHE) IV: hospital mortality assessment for today's critically ill patients. Crit Care Med 2006;34:1297–310 [DOI] [PubMed] [Google Scholar]
- 47.Moreno RP, Metnitz B, Adler L, et al. Sepsis mortality prediction based on predisposition, infection and response. Intensive Care Med 2008;34:496–504 [DOI] [PubMed] [Google Scholar]
- 48.Rubulotta F, Marshall JC, Ramsay G, et al. Predisposition, insult/infection, response, and organ dysfunction: a new model for staging severe sepsis. Crit Care Med 2009;37:1329–35 [DOI] [PubMed] [Google Scholar]
- 49.Buchan CA, Bravi A, Seely AJE. Variability analysis and the diagnosis, management, and treatment of sepsis. Curr Infect Dis Rep 2012;14:1–10 [DOI] [PubMed] [Google Scholar]
- 50.Gao F, Melody T, Daniels DF, et al. The impact of compliance with 6-hour and 24-hour sepsis bundles on hospital mortality in patients with severe sepsis: a prospective observational study. Crit Care 2005;9:R764–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daniels R. Surviving the first hours in sepsis: getting the basics right (an intensivist's perspective). J Antimicrob Chemother 2011;66(Suppl 2):ii11–23 [DOI] [PubMed] [Google Scholar]
- 52.Gaieski DF, Drumheller BC, Goyal M, et al. Accuracy of handheld point-of-care fingertip lactate measurement in the Emergency Department. West J Emerg Med 2013;14:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karon B, Scott R, Burritt M, et al. Comparison of lactate values between point-of-care and central laboratory analyzers. Am J Clin Pathol 2007; 128:168–71 [DOI] [PubMed] [Google Scholar]
- 54.Donders ART, van der Heijden GJMG, Stijnen T, et al. Review: a gentle introduction to imputation of missing values. J Clin Epidemiol 2006;59:1087–91 [DOI] [PubMed] [Google Scholar]