Abstract
In the USA, the national cyberinfrastructure refers to a system of research supercomputer and other IT facilities and the high speed networks that connect them. These resources have been heavily leveraged by scientists in disciplines such as high energy physics, astronomy, and climatology, but until recently they have been little used by biomedical researchers. We suggest that many of the ‘Big Data’ challenges facing the medical informatics community can be efficiently handled using national-scale cyberinfrastructure. Resources such as the Extreme Science and Discovery Environment, the Open Science Grid, and Internet2 provide economical and proven infrastructures for Big Data challenges, but these resources can be difficult to approach. Specialized web portals, support centers, and virtual organizations can be constructed on these resources to meet defined computational challenges, specifically for genomics. We provide examples of how this has been done in basic biology as an illustration for the biomedical informatics community.
Keywords: National Cyberinfrastructure, Big Data, Genomics
Introduction
There are daily announcements of new meetings, journals, or commercial solutions to the ‘Big Data’ problems facing biomedical research, yet biomedicine is not the first discipline to face these challenges. In the USA, there is an existing system of research supercomputer centers and high speed research networks collectively known as the national cyberinfrastructure (CI).1 These resources have been used by practically all scientific disciplines, but the majority of computation has come from physical and earth scientists, and more recently biologists; the CI has been little used by biomedical researchers. Yet many of the computational challenges facing medical informatics, particularly regarding ‘discovery ‘omics’ such as genomics, can be economically solved using the national CI! Further, problems related to data movement, data management, and software optimization benefit by tapping the computational expertise of the scientists and engineers associated with the national CI.
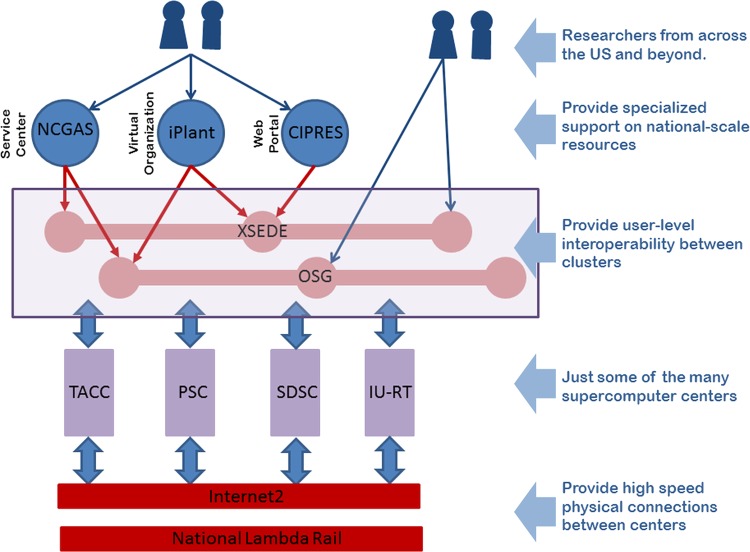
The national cyberinfrastructure in the USA is not a single entity. Rather, the term describes an assortment of large-scale computational resources available to the scientific community. Figure 1 is a stylized view showing the interrelation of some components of the CI. The core of the CI is a series of independently funded supercomputer centers. The National Science Foundation supports the Extreme Science and Engineering Discovery Environment (XSEDE), a project that brings together dozens of supercomputers and other high-performance computing (HPC) resources from 13 of these supercomputing centers, to advance science and engineering research and education across the USA.2 XSEDE provides a framework that supports the development of next generation Big Data analysis capabilities. Like XSEDE, the Open Science Grid3 (OSG) uses advanced networks like Internet2 and National Lambda Rail to provide access to CI computers and clusters. Service organizations, such as iPlant4 and the National Center for Genome Analysis Support (NCGAS),5 use the resources of XSEDE and OSG to create unified user experiences for different research communities.
Figure 1.
A Roadmap to the Research Information Superhighway: over 200 supercomputer centers are interconnected across a series of high speed physical networks. The resources in these centers are shared across organizations such as XSEDE and OSG. Specialized centers use XSEDE and OSG to support specialized user communities.
CI support for genomics
Many life sciences are already harnessing the national CI, and we feel that three examples will help demonstrate what is currently possible, and perhaps help stimulate ideas for collaborations between the biomedical and medical informatics communities and the national CI.
The iPlant Collaborative is working to enrich plant and animal sciences through the development of cyberinfrastructure—the physical computing resources, collaborative environment, virtual machine resources, and interoperable analysis software and data services—that are essential components of modern biology.6 It is a community-driven effort bringing together plant biologists, bioinformaticians, computational scientists, and HPC professionals to address grand challenges in plant and animal sciences.
Currently in its sixth year, iPlant has over 9000 users and has supported over 8 million core hours per year, and securely houses 500 terabytes of user data. Further, iPlant has developed and deployed an extensive set of tools for scientific research, educating future scientists, and powering other web portals. The iPlant Discovery Environment is a web gateway providing a consistent, intuitive graphical user interface to over 350 community software packages across five XSEDE HPC systems.6 Atmosphere, iPlant's configurable cloud computing environment, currently supports around 100 concurrent virtual machines and serves over 2000 users.7 The DNA Subway, an online suite of tools for students and educators, is designed to demonstrate the fundamentals of genome analysis through hands-on exploration, and over 615 participants have used it within iPlant workshops. iPlant has educated over 600 users through workshops and tutorials, and actively collaborates with other organizations. Researchers interested in iPlant can visit http://www.iplantcollaborative.org/ for additional information.
NCGAS is an NSF funded collaboration between four computing facilities; Indiana University, the Texas Advanced Computing Center (TACC), the San Diego Supercomputer Center (SDSC), and the Pittsburgh Supercomputing Center (PSC).5 Founded with a $1.5 million initial award, NCGAS represents how the supercomputing centers are evolving to meet the needs of the life sciences. The center supplies bioinformatic support for genomics projects, particularly projects requiring large memory computation. NCGAS provides consulting services for biologists, assistance in running genome analyses, hardened and optimized genome analysis software, and supercomputing. NCGAS has a simple allocations process that gives researchers access to both its large-memory cluster as well as computing time from its XSEDE allocation. NCGAS's large-RAM cluster, with its 8.0 terabytes of aggregate RAM and associated 5 petabytes of high speed storage on the Data Capacitor parallel disk system, employs best practices security that is consistent with the technical requirements of regulations such as the Health Insurance Portability and Accountably Act, and is designed primarily for de novo sequence assembly.
In its first 18 months, NCGAS supported over 37 NSF-funded genomics projects from 16 states. Being housed across four supercomputing centers, NCGAS represents a model of a virtual genomic analysis facility that operates at a national scale. Each center provides specific expertise and resources, and they are all interconnected by a shared wide area file system that enables transparent workflows across all four sites. Transferring a terabyte of Next Generation Sequencing data generated in Hawaii into the shared file system in Indiana is a trivial problem for teams accustomed to moving petabytes of data over those distances. Researchers considering NCGAS resources can visit http://ncgas.org for allocation information.
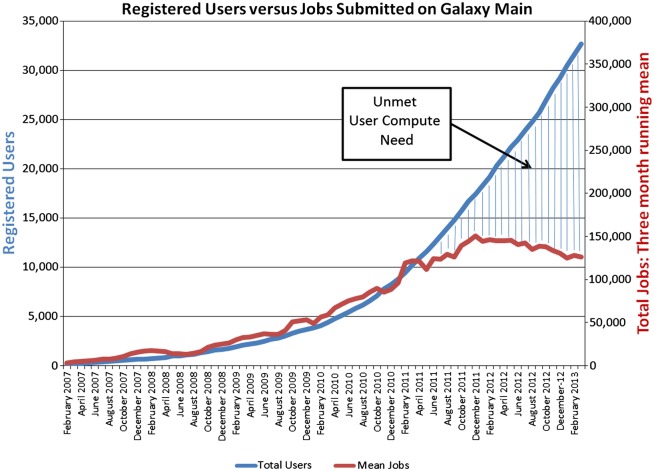
Galaxy is one of the most popular portals for genomics analysis, having seen exponential growth to approximately 35 000 users since 2005.8–10 Widely available access to high-throughput sequencing has dramatically increased the interest in and the size and complexity of genomic analyses. Thus, despite ever increasing growth in registered users, the compute capacity of Galaxy has become saturated due to compute and storage constraints, resulting in unrealized potential for scientific discovery (figure 2). Galaxy turned to PSC for support in addressing these constraints. PSC had already developed the Data Supercell11 (DSC) to help meet the challenges of large-scale, distributed data access and analysis. The DSC is a scalable, disk-based data archival system that is as cost-effective as tape archives but offers many times better bandwidth and orders of magnitude better latency than tape. The DSC uses the SLASH2 file system developed at PSC, and provides mechanisms to federate campus-level and national-level data storage systems, including inherent data replication and migration mechanisms, as well as the capability to import existing data storage systems. The DSC is currently being used to seamlessly replicate all data associated with Galaxy Main. This integration will enable analyses submitted through the Galaxy main site to run transparently on PSC's large memory systems, as well as other XSEDE systems, providing capabilities beyond what is available via commercial cloud computing providers. For example, using PSC resources, researchers recently completed the de novo assembly of over 20 primate transcriptomes to create a non-human primate reference transcriptome resource, with each transcriptome using 1.8 billion reads of RNA-sequence data. Another group completed the assembly of a soil metagenome requiring 3.5 terabytes of RAM.
Figure 2.
Users and number of compute jobs submitted through the Galaxy Main web-portal by month.
The tight integration of web platforms like Galaxy with PSC and XSEDE, together with complementary efforts and services provided by organizations like iPlant and NCGAS, brings these cutting-edge capabilities to a much larger group of researchers.
Cyberinfrastructure
XSEDE is a five-year $121 million project supported by NSF which extends the work of the previous TeraGrid program to integrate and simplify access to CI resources and services. XSEDE currently supports 15 high-performance supercomputers or computational clusters, and well over 100 petabytes of storage located at 17 partner institutions.2 More than 8000 scientists use XSEDE to complete thousands of research projects and generate more than 2000 publications annually. XSEDE works to simplify allocations and cybersecurity across supported systems, while maintaining the highest level of professional service. It also provides Extended Collaborative Support Services to help users develop projects that take advantage of XSEDE resources, and supports campus champions from over 135 different institutions who help researchers use XSEDE resources. Access to XSEDE resources is through an allocation process. Small start-up allocations can be approved within 48 h and give investigators resources to determine the feasibility of larger individual projects. Full research allocations are awarded quarterly, and large initiatives, such as NCGAS and iPlant, receive allocations sizable enough to support the computational needs of the communities they serve. To determine if your institution has an XSEDE campus champion, visit https://www.xsede.org/campus-champions; otherwise researchers can request XSEDE allocations at https://www.xsede.org.
The OSG provides an alternative for accessing the national cyberinfrastructure. Jointly funded by the NSF and the Department of Energy, the OSG enables distributed high-throughput computing for users at all scales by working with organizations to federate their computational resources. The OSG is built around the concept of virtual organizations (VO), each of which contributes capacity to other VOs, thus increasing the overall computational capacity for all projects by scavenging cycles from otherwise idle systems. This shared architecture is particularly well suited for tasks that can run on many single processors, such as BLAST. By utilizing the OSG's middleware layers the VOs can set policies to share their resources with other VOs. The 116 sites currently on the OSG submitted between 12 and 15 million jobs per month within the last year, representing 50 million CPU hours and over 30 petabytes of data transfer per month.12 Researchers interested in the OSG can visit https://www.opensciencegrid.org/.
The computational assets of the national CI are connected to each other by 10- to 100-Gbps networks. A 100-Gbps network connection allows files to move across the country faster than they can be loaded into RAM locally. For many applications file movement can be done without any awareness of the underlying network. Internet2 provides infrastructure across more than 220 institutions, 60 corporations, 70 governmental agencies, 38 regional and state networks, and 65 national research and educational networks from over 100 countries. The organization provides advanced network services that are tailored to the needs of researchers. They have network monitoring and diagnostic research networks, and they ensure high throughput data streaming not available on commodity internet connections.
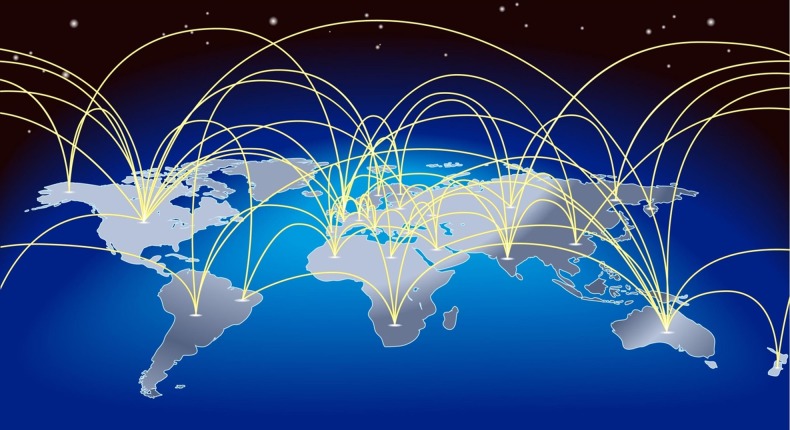
Just as domestic networks have extended their reach and speed, so have international networks. The NSF funded International Research Network Connections (IRNC) program provides high-performance network connectivity from the USA to Europe, Asia, Central and South America, and a number of other locations.13 Of course, not all international research and education (R/E) connectivity is provided by the USA. National research and educational networks in Asia and Central and South America provide additional network connections to the global R/E network fabric. The pan-European network GEANT provides connections within Europe, from Europe to the USA and from Europe to a number of other countries, ranging from Kyrgyzstan to Cambodia to China.
The network connections illustrated in figure 3 are generally 10-Gbps connections. The next generation of international networking is set to launch in mid-2013 with the implementation of 100-Gbps connectivity between the USA and Europe. This will be followed by additional 100-Gbps connections to Asia and South and Central America within the next year.
Figure 3.
Schematic representation of current international data network connectivity available for scientific research.
As an illustration of the power of this international connectivity, it is possible today for a researcher to easily transfer a large dataset from Indiana University to Tsinghua University in China using NSF and China supported connectivity. The same dataset could then be transferred from Tsinghua to University College in London using China and European supplied connectivity. And again, this dataset could be transferred from University College to Cornell University in the USA using GEANT, NSF, and Internet2 supplied connectivity, making a smooth 10-G enabled trip around the globe.
Conclusion
Consider that if there were 30 000 deployed next-generation sequencers, FT-ICR mass spectrometers, and NMR instruments across the country dedicated to biomedical research, and that each of these produced 100 GB/day of data (on average), then these instruments would produce around 1096 PB of data per year. This is a large amount of data! But, it is still less than the 2000 PB/year expected to be generated from Phase 1 of the Square Kilometer Array14 starting in 2013. What makes the biomedical informatics problem unique is the distributed nature of the data. It comes from many instruments, each used in highly unique ways, and the resulting data is analyzed with many different workflows—each workflow presenting its own computational challenge. To meet these challenges, tailored biomedical informatics solutions can and are being built on existing national CI, leveraging its scale to allow biomedical researchers to address ‘Big Data’ problems. As biomedical research increasingly pursues genomic characterization of patients and diseases, for example, resources at the scale of the national CI will be a critical component of future disease research. Further, as new paradigms emerge, for example fixed data repositories with programmable interfaces that allow workflows to move to data, the staff and scientists of the national CI have the experience to make these systems possible.
Footnotes
Contributors: RL manages the National Center for Genome Analysis Support; MV manages Life Science Computing at the Texas Advanced Computing Center, and JMF is a Research Associate there; MS is the Associate Director for Health Sciences at Internet2; JGW is the Director of International Networking for Indiana University; PDB is a Senior Scientific Specialist at the Pittsburgh Supercomputing Center; JT is the Principal Investigator for Galaxy; WKB is the Director of the National Center for Genome Analysis Support and Principal Investigator for the Open Science Grids Grid Operations Center. All authors have contributed equally to formulating the perspective presented in this manuscript.
Funding: This research is based on work supported by the National Science Foundation under grant no. ABI-1062432 (Craig Stewart, PI) and 1242759 PHY to Indiana University. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation, the National Center for Genome Analysis Support, or Indiana University. The iPlant Collaborative is funded by a grant from the National Science Foundation (#DBI-0735191).
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Stewart CA, Simms S, Plale B, et al. What is cyberinfrastructure. In Proceedings of the 38th annual ACM SIGUCCS fall conference (SIGUCCS ‘10) New York, NY, USA: ACM, 37–44 [Google Scholar]
- 2.https://www.xsede.org/web/guest/overview (accessed May 2013).
- 3.https://opensciencegrid.org/bin/view (accessed May 2013).
- 4.Goff SS, Vaughn M, McKay S, et al. The iPlant collaborative: cyberinfrastructure for plant biology. Front Plant Sci 2011;2:00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeDuc R, Wu L-S, Ganote C, et al. National Center for Genome Analysis Support Leverages XSEDE to Support Life Science Research. In Proceedings of the 2nd Conference of the Extreme Science and Engineering Discovery Environment, XSEDE'13, July 2013, in press [Google Scholar]
- 6.Lenards A, Merchant N, Stanzione D. Building an environment to facilitate discoveries for plant sciences. In Proceedings of the 2011 ACM workshop on Gateway computing environments (GCE ‘11) New York, NY, USA: ACM, 51–8 [Google Scholar]
- 7.Skidmore E, Kim S-J, Kuchimanchi S, et al. iPlant atmosphere: a gateway to cloud infrastructure for the plant sciences. In Proceedings of the 2011 ACM workshop on Gateway computing environments (GCE ‘11) New York, NY, USA: ACM, 59–64 [Google Scholar]
- 8.Goecks J, Nekrutenko A, Taylor J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 2010;11:R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blankenberg D, Kuster G, Coraor N, et al. Galaxy: a web-based genome analysis tool for experimentalists. In Curr Protoc Mol Biol Supplement 89, 2010; Chapter 19: Unit 19.10.1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giardine B, Riemer C, Hardison R, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res 2005;15:1451–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowoczynski P, Sommerfield J, Yanovich J, et al. The Data Supercell. In Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the campus and beyond (XSEDE ‘12) New York, NY, USA: ACM, Article 13,11 pages [Google Scholar]
- 12.http://display.grid.iu.edu/ (accessed May 2013).
- 13.http://irnclinks.net/ (accessed May 2013).
- 14.http://www.skatelescope.org/uploaded/21705_130_Memo_Dewdney.pdf (accessed May 2013).