Abstract
Objective
The rapidly growing volume of multimodal electrophysiological signal data is playing a critical role in patient care and clinical research across multiple disease domains, such as epilepsy and sleep medicine. To facilitate secondary use of these data, there is an urgent need to develop novel algorithms and informatics approaches using new cloud computing technologies as well as ontologies for collaborative multicenter studies.
Materials and methods
We present the Cloudwave platform, which (a) defines parallelized algorithms for computing cardiac measures using the MapReduce parallel programming framework, (b) supports real-time interaction with large volumes of electrophysiological signals, and (c) features signal visualization and querying functionalities using an ontology-driven web-based interface. Cloudwave is currently used in the multicenter National Institute of Neurological Diseases and Stroke (NINDS)-funded Prevention and Risk Identification of SUDEP (sudden unexplained death in epilepsy) Mortality (PRISM) project to identify risk factors for sudden death in epilepsy.
Results
Comparative evaluations of Cloudwave with traditional desktop approaches to compute cardiac measures (eg, QRS complexes, RR intervals, and instantaneous heart rate) on epilepsy patient data show one order of magnitude improvement for single-channel ECG data and 20 times improvement for four-channel ECG data. This enables Cloudwave to support real-time user interaction with signal data, which is semantically annotated with a novel epilepsy and seizure ontology.
Discussion
Data privacy is a critical issue in using cloud infrastructure, and cloud platforms, such as Amazon Web Services, offer features to support Health Insurance Portability and Accountability Act standards.
Conclusion
The Cloudwave platform is a new approach to leverage of large-scale electrophysiological data for advancing multicenter clinical research.
Keywords: Electrophsyiological Big Data, Epilepsy and Seizure, Cloudwave, SUDEP, Ontology, MapReduce
Introduction
The unprecedented rate of multimodal data collection across scientific, business, and social networking domains is transforming research, education, and decision-making through data-driven insights and knowledge discovery tools.1–4 The large volume of these datasets is their defining characteristic, and the term ‘Big Data’ is often used to describe both the data and different aspects of their management.1 In addition to volume, the end users are often concerned with the velocity of Big Data, which describes both the high rate of data generation and the need for rapid analysis of data for critical decision-making tasks.3 The importance of business intelligence derived from timely interpretation of huge volumes of data (eg, consumer buying patterns) for actionable information is widely appreciated.5 There is a growing need to adopt a similar paradigm of ‘healthcare intelligence’ through near real-time processing of healthcare data to support preventive care, personalized medicine, and improved treatment outcome. Healthcare intelligence can be derived from the increasing amount of digital patient data exemplified by electronic health records, published literature, and especially multimodal electrophysiological data.
In many critical care and neurological monitoring applications, large volumes of physiological data, including electroencephalogram (EEG) from scalp and implantable intracranial electrodes, pulse oximetry (Spo2), and electrocardiogram (ECG), are collected. Together with sophisticated computing approaches, these large datasets are enabling the development of transformative concepts in studying various health issues, including neurological diseases.6 7 For example, continuous EEG, ECG, blood oxygen levels, and video data are now routinely collected during 5-day admission of patients in epilepsy monitoring units (EMUs). These data are characterized by both the volume (eg, terabytes (TB) of data per year) and velocity (eg, gigabytes (GB) of data per month), but existing computational approaches for processing signal data (eg, Neural Workbench8) are limited in their ability to support collaborative multicenter research studies in this domain. These tools often require the data to fit into memory of a local desktop and lack the ability to effectively leverage the growing capabilities of distributed computing approaches (eg, cloud computing and multicore processing).6 9
MapReduce is a popular programming framework introduced by Google to address computational and storage challenges for web-scale data.10 Apache Hadoop is an open-source implementation of the MapReduce framework, which can be used to store large volumes of data on the Hadoop Distributed File System (HDFS) and efficiently process the data by repeating the two steps of ‘Map’ and ‘Reduce’ on thousands of computing nodes.11 Hadoop has built-in support for automated data distribution, recovery from component failures, balancing the computational load across different nodes, and parallel computation.10 11 In this paper, we describe the Cloudwave platform, which provides a novel approach to parallelize signal-processing workflows using MapReduce. Cloudwave supports real-time user interactions over massive-scale ontology annotated electrophysiological data in collaborative multicenter research studies.
Background and significance
Epilepsy is a chronic neurological condition that affects 65 million patients worldwide, making it the most common serious neurological disease, with more than 200 000 new cases diagnosed each year.12 Patients with epilepsy have repeated seizures that manifest as physical or behavioral changes, including changes in electrical activity of the brain, which are captured in EEG recordings, and heart rate variations, which are captured in ECG recordings.13–15 The ECG data are a vital source of clinical signs for seizure detection and for correlating effects of antiepileptic drugs on the autonomic nervous system.15 It is increasingly being studied for use in automated seizure-detection instruments.13 16 Heart rate changes in epilepsy patients are measured before seizure (pre-ictal), during seizure (ictal), between seizures (inter-ictal), and after seizure (post-ictal) to identify a variety of conditions, such as cardiac arrhythmia and conduction abnormalities.13
Identifying the QRS complex in ECG recordings, which is associated with depolarization of the right and left heart ventricles, can be used to analyze changes in heart rate —for example, increases in heart rate (also called tachycardia) or decreases in heart rate (also called bradycardia). About 75–80% of epilepsy patients with temporal lobe epilepsy have tachycardia, while bradycardia occurs more rarely in about 3% of the patients with frontal lobe epilepsy.13 17 The intervals between two consecutive heart beats, referred to as the RR interval time series, is used to derive heart rate variability (HRV) for analyzing changes in the two branches of the autonomic nervous system during seizures.18 The interval between the Q and T features in ECG (QT interval) has been found to be increased during epileptiform EEG discharges and has been studied in the context of a poorly understood phenomenon called sudden unexplained death in epilepsy (SUDEP).19
Cardiac electrophysiology in SUDEP
SUDEP is defined as sudden, unexpected, non-traumatic, non-drowning death of epilepsy patients (with or without an epileptic seizure) where no other cause of death is found.20 About 5000 epilepsy patient deaths in the USA per year are classified as SUDEP with an incidence rate of 1/200 for chronic epilepsy, with the younger population being at 24 times greater risk of death.21 22 Unlike many other disorders or diseases, the precise mechanism of death in SUDEP is unknown, and there is little understanding of the underlying risk factors that can be used for intervention or treatment.19
The effects of seizures on cardiorespiration and autonomic nervous system function are the two commonly studied aspects for identifying previously unknown risk factors and to provide greater insight into the potential mechanism(s) of SUDEP.15 19 23 For example, cardiac arrest during seizure is a known potential mechanism for SUDEP, and, similarly to some other types of epilepsy (discussed above), bradycardia and asystole during seizures have also been studied in the context of SUDEP.19 A better understanding of cardiac events and their correlation with SUDEP may enable more active intervention in terms of both seizure prevention and the management of cardiorespiratory risk factors that are implicated in the disease.
The Prevention and Risk Identification of SUDEP Mortality (PRISM) project
The PRISM project is a National Institute of Neurological Diseases and Stroke (NINDS)-funded multi-institution collaborative project that is recruiting potential SUDEP patients across multiple EMUs. It involves four EMUs located at the Case Western Reserve University-University Hospital (CWRU-UH, Cleveland), the Ronald Reagan Medical Center at the University of California Los Angeles (UCLA), the Northwestern University-Northwestern Memorial Hospital (NMH, Chicago), and the National Hospital for Neurology and Neurosurgery (NHNN, London, UK). The participating EMUs collect large-scale electrophysiological data, including ECG and EEG, over a 5-day period from the recruited patients for subsequent signal analysis.
Electrophysiological Big Data in the PRISM project
About 964 patients have been processed in the CWRU-UH EMU since January 2011 after the start of the PRISM project, and about 116 of these patients have consented to participate in the PRISM project (figure 1A). An average of 321 MB of electrophysiological data is generated from recordings of a single patient per day, and about 1.6 GB of data over a typical 5-day admission period in the EMU. This has resulted in 9.5 TB of total signal data collected in the CWRU-UH EMU and about 4 TB of data collected from patients recruited for the PRISM project since 2011. The rate of data collection in the EMU is increasing every year—for example, the volume of data at the end of 2012 was 6 TB, but 9.5 TB of data had already been collected by May 2013 (figure 1B illustrates the growth in total data collected from all patients in the EMU and patients recruited for the PRISM project). Hence, there is an acute need to define efficient algorithms and develop an effective informatics platform to manage this electrophysiological Big Data.
Figure 1.
(A) Total number of patients admitted to the Case Western University Hospital epilepsy monitoring unit (EMU) and number of patients recruited for the Prevention and Risk Identification of SUDEP Mortality (PRISM) project. (B) Cumulative growth in volume of electrophysiological signal data collected from all EMU patients and PRISM project-specific patients.
Related work
Management of Big Data is a challenging issue across the spectrum of translational medical domains, including whole-genome sequencing data24 and construction of a network view of diseases for drug development,25 especially in the context of precision medicine.26 Multiple computational solutions, such as high-performance cluster computing, cloud computing, and use of high-end graphics processing units (GPUs), have been explored in addressing Big Data challenges in biomedicine.27 The well-known BLAST framework has been implemented using a MapReduce approach,28 and recent work by White et al29 has demonstrated the effective use of large-scale web search logs for pharmacovigilance. In addition, specialized GPU-based infrastructure has been found to be significantly faster than traditional computing approaches for studying intracellular signal-transduction networks in the context of clinical outcomes and drug effectiveness.30
The GPU-based approach has also been successfully used for implementing computationally intensive signal-processing algorithms, such as ensemble empirical mode decomposition and Hilbert–Huang transformation.9 More recent work has focused on the use of the Amazon Elastic Compute Cloud (EC2) to process ECG data from wireless monitoring devices.31 To the best of our knowledge, Cloudwave is the first informatics framework to design parallel algorithms for massive-scale electrophysiological signal data management with an integrated ontology-driven web-based visualization and query interface.
Methods
The Cloudwave framework aims to develop novel parallelization approaches for signal processing algorithms to achieve two primary objectives:
enable clinical researchers to have real-time interactions with massive-scale electrophysiological Big Data through (a) efficient computation of clinically relevant cardiac measurements, and (b) scalable storage using high-performance distributed file systems;
an ontology-driven web-based signal visualization and query interface that mitigates terminological heterogeneity in signal data annotation and improves data retrieval for use by clinicians and researchers.
Figure 2 illustrates the architecture of Cloudwave consisting of (1) Hadoop-based storage and computation modules with a semantic metadata access layer (figure 2A), and (2) an integrated web-based interface module that uses the epilepsy and seizure ontology (EpSO)32 for signal visualization and query (figure 2B). We describe the details of the design and implementation of the Cloudwave framework in the following sections.
Figure 2.

Architecture of the Cloudwave platform consisting of two components: (A) Hadoop-based storage and computation module with semantic metadata layer; (B) ontology-driven signal query and visualization module.
Dependency analysis of computational algorithms for cardiac measurements
The ECG data of EMU patients are usually recorded with multiple electrodes to provide both reference and redundancy. The ECG data together with electrophysiological data from other channels (eg, EEG, blood oxygen measurements) are converted into the European Data Format (EDF+), which is a widely used standard for storage and exchange of electrophysiological data.33 In the PRISM project, the four ECG channels are extracted from an EDF file and processed to compute the heart rate measurements. The two common cardiac measures used in epilepsy clinical research are (1) RR intervals and (2) instantaneous heart rate (IHR) to detect tachycardia or bradycardia. These two measures are derived from the time interval between two consecutive heartbeats requiring the accurate detection of the R-wave in one QRS complex and the accurate detection of the R-wave in the next QRS complex.34 Cloudwave uses the ‘wqrs’ open-source single-channel QRS detector algorithm and IHR algorithm developed by the PhysioNet project.34 However, these algorithms were developed for sequential execution, and several critical challenges need to be addressed to integrate them into a parallel computational workflow (designing parallelization approaches for sequential algorithms has been an active area of computer science research for the past five decades35).
Cloudwave addresses these challenges in two phases: (1) formal algorithm analysis of the cardiac-measurement workflow to characterize its degree of parallelization based on the famous Amdahl's law35; (2) defining a new parallel algorithm for the MapReduce model that can be implemented in the open-source Hadoop environment. Cloudwave uses a ‘coarse-grained parallelization’ approach for formal dependency analysis of the computational workflow36 (figure 3 illustrates the complete workflow for computing cardiac measures), where each node is considered as an atomic task (eg, QRS detection). Cloudwave uses the three conditions of ‘flow dependency’, ‘anti-dependency’, and ‘output dependency’36 to identify computational tasks that can be executed in parallel. The dependency analysis reveals that the maximum length of the critical path37 is three, spanning the tasks ‘EDF2MIT’, ‘WQRS’, and ‘IHR’, which allows ‘RDSAMP’ and ‘ANN2RR’ tasks to be executed in parallel. This dependency analysis is used to define an efficient parallel approach that conforms to the iterative two-step MapReduce framework.
Figure 3.
Cardiac measurement workflow implemented in Cloudwave to identify QRS complexes and compute RR intervals and instantaneous heart rate (IHR) values from ECG signal data. bpm, beats/min; EDF, European Data Format; HRV, heart rate variability.
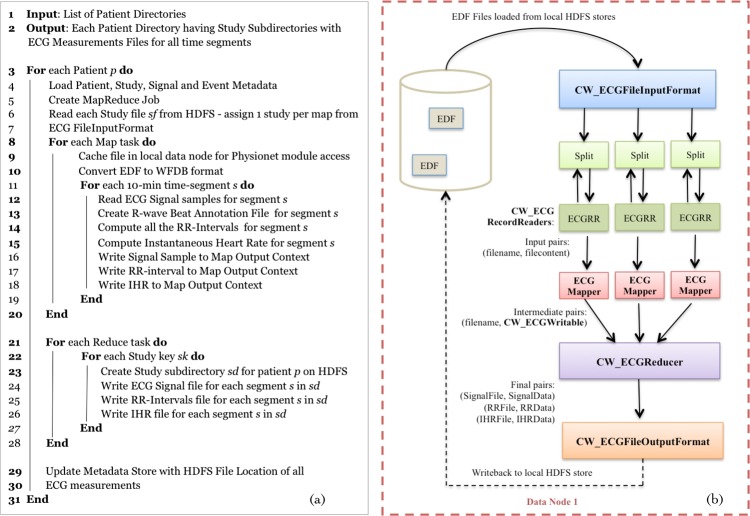
In the second phase, Cloudwave introduces a new algorithm for signal processing in the MapReduce programming model (figure 4A) that defines (1) the suitable construct for signal data partitions to achieve effective parallelization (eg, 10 min segments), (2) the set of computations that can be implemented during the Map phases, and (3) the aggregation steps that correspond to the Reduce phases. The MapReduce model operates on discrete entities of <key, value> pairs (other parallelization models such as Message Passing Interface require different data structures). The Cloudwave algorithm defines the sample identifier associated with each discrete signal measure as a ‘key’ and the signal measure as a ‘value’ for the Map phase. In the Reduce phase, the segment identifier is used as ‘key’ and the three sets of R-waves, the RR intervals, and IHR measures as ‘value’ (figure 4B). This algorithm enables the Cloudwave platform to efficiently compute cardiac measures, generate optimal-sized signal segments for visualization, and support real-time interactions for users with ontology-driven querying.
Figure 4.
(A) Cloudwave MapReduce algorithm for cardiac measurements from the ECG data. (B) Implementation of the algorithm with specialized Cloudwave classes corresponding to the Map phases and Reduce phases. EDF, European Data Format; HDFS, Hadoop Distributed File System; IHR, instantaneous heart rate.
Implementation of the Cloudwave MapReduce algorithm for electrophysiological signal processing
There is no existing support to store, access, and process ECG data in Hadoop; hence we have developed a library of specialized classes for both computations in MapReduce framework and managing electrophysiological data in HDFS. These new classes constitute an open-source Hadoop signal-processing middleware layer that can be used by other software tools for processing large-scale electrophysiological data. The Cloudwave data-storage module uses HDFS, which is a high-performance distributed file system, to address the need to store and manage TBs of signal data over multiple machines in a cluster environment.38 HDFS has built-in support to store data reliably even if some of the machines in the cluster fail and also effectively balance the distribution of data to ensure efficient retrieval.
Use of HDFS enables Hadoop MapReduce to efficiently deploy computational tasks near the location of the dataset and reduces the need to transfer large datasets across a network. Two new Cloudwave classes called CW_EDFRecordReader and CW_EDFWriteable were defined to facilitate reading and writing ECG data from the EDF files stored in HDFS.
The Cloudwave MapReduce computation module supports the use of any cardiac measurement algorithms similar to the open-source PhysioNet algorithms used in this paper, which can be easily integrated as ‘pluggable’ resources using the Cloudwave CW_ECGSignalWrapper class. Cloudwave implements the parallelization steps using four new classes:
CW_ECGSignalWrapper class, which implements three methods corresponding to algorithms used for R-wave detection and calculation of RR interval and IHR values;
CW_ECGFileInputFormat class, which uses the CW_EDFRecordReader class to access the ECG signal data from the EDF files;
CW_ECGFileOutputFormat class, which uses the CW_EDFWriteable class to transfer the results of the computations as files to be stored in HDFS;
CW_ECGProcessor class, which implements the Map and Reduce phases using the <key, value> pairs discussed above under ‘Dependency analysis of computational algorithms for cardiac measurements’.
These new classes together with user documentation for deploying Cloudwave on Hadoop installations will be made open source as part of the PRISM project. The Cloudwave computation module effectively uses the parallelized computations to perform near real-time signal-processing computations, which allows clinicians to access analysis results more rapidly than with traditional approaches (a comparative evaluation is described in the Results section ). Cloudwave also enables biomedical signal-processing researchers to implement algorithms on a large scale, which was previously not supported by desktop computing approaches. Clinical researchers can query and visualize the processed signal data using the Cloudwave ontology-driven web-based interface.
The Cloudwave signal visualization and query interface
Traditional approaches to visualizing and querying electrophysiological data use standalone software tools that are usually deployed on desktop computers—for example, Nihon Kohden.8 These tools are not suitable for multicenter collaborative studies that require researchers to simultaneously access, view, annotate, and share datasets. To address these challenges, we developed the web-based ontology-driven interface module in Cloudwave, which enables researchers to collaboratively query and visualize signal data from different EMUs.
The visual interface can selectively render one or more of the four ECG channel data and also visualize the potential differences between a ‘reference channel’ and other ECG channels (figure 5 illustrates the signal visualizer and montage builder). In addition, the interface supports querying of signal data using epilepsy-related events, such as onset of seizure, end of seizure, and EEG suppression after seizure, which are modeled as ontology classes in EpSO.32 EpSO is an epilepsy domain ontology that models epilepsy types, seizure features, the electrode placement scheme, and electrophysiological signal details using the description logic-based Web Ontology Language (OWL2).39 EpSO reuses ontology concepts from the Foundational Model of Anatomy,40 RxNorm terminological system,41 and the Neural Electromagnetic Ontologies (NEMO)42 to model anatomy, medication, and signal data metrics, respectively.
Figure 5.
Cloudwave web interface for querying signal data using epilepsy and seizure ontology (EpSO) concepts (marked as annotations such as ‘onset of clonic seizure’) and visualization (note that, during a seizure, the EKG3–EKG4 signal moves beyond the reference frame). The ‘montage builder’ allows the clinician to create different combinations of electrodes.
EpSO addresses the critical challenge of terminological heterogeneity in signal data annotation by providing a well-defined and formal reference schema for consistent description of clinical events in signal data. These semantic annotations are used in the PRISM project for both patient cohort identification in the Visual Aggregator and Explorer (VISAGE) tool43 and signal data querying in Cloudwave. Figure 5 illustrates the EpSO-driven query composer, which allows users to select a variety of signal event terms, which are subsequently used to query the signal database. The query module uses reasoning over the EpSO class structure (and ontology annotations) to (a) reconcile string mismatch (eg, the acronym PSP maps to the term ‘PolySpike’) and (b) expand the query expression to include subcategories of a term (eg, ‘abnormal EEG patterns’ includes ‘epileptiform patterns’, ‘slow activity’, and ‘special patterns’). In the next section, we present the results of a comparative evaluation of Cloudwave with traditional signal-processing approaches to illustrate the advantages of the Cloudwave platform.
Results
The PRISM project aims to enroll about 1100 patients across the four participating EMUs, and the CWRU-UH EMU has already recruited 116 patients. Cloudwave is being used to process data from 111 of these patients. Table 1 describes the characteristics of these patients. Female patients outnumbered male patients (62% vs 38%), with a range of 17–77 years and median age of 39 years. Most of the patients (75%) had focal seizures, with either both hemispheres involved (21%) or left (36%) or right (16%) hemispheres. Only 25% of the patients had generalized seizures, and in other cases the origin of the seizure could not be accurately determined. The majority of patients (85%) were taking antiepileptic medication, with two of these patients taking a neuroleptic drug (olanzapine).
Table 1.
Characteristics of patients enrolled in the PRISM project
| Characteristic | Patients | |
|---|---|---|
| Number | Percentage | |
| Sex | ||
| Female | 69 | 62 |
| Male | 42 | 38 |
| Age (years) | ||
| Median | 39 | |
| Range | 17–77 | |
| Seizure classification (patients experienced seizure during admission) | ||
| Epileptic paroxysmal episode | 81 | 73 |
| Non-epileptic psychogenic episode | 8 | 7.3 |
| Organic paroxysmal episode | 3 | 2.7 |
| Other (paroxysmal episode) | 19 | 17 |
| Medication | ||
| Antiepileptic | 94 | 85 |
| Neuroleptic | 2 | 1.8 |
| Antidepressant | 1 | 0.2 |
| Cardiac events | ||
| HRV | 16 | 14 |
| Asystole | 0 | |
| Etiology | ||
| Genetic | 7 | 6 |
| Structural | 23 | 21 |
| Unknown | 81 | 73 |
HRV, heart rate variability; PRISM, Prevention and Risk Identification of SUDEP Mortality.
During their stay in the EMU, 62 patients experienced a seizure event (55%). The patient seizure classification shows that 81 patients experienced an epileptic paroxysmal episode (73%), and the others experienced a non-epileptic psychogenic paroxysmal episode or an organic paroxysmal episode. Only a small number of patients had HRVs (14%), which were computed using a new algorithm developed as part of the PRISM project. The HRV was always associated with an epileptic paroxysmal event, which potentially highlights the correlation between epileptic seizures and autonomic functions. In addition, none of the patients had asystole.
Comparative evaluation of cardiac measure computations on Cloudwave and traditional standalone computing infrastructure
The comparative evaluation was performed on (1) a desktop computer with an Intel Core i7 2.93 GHz processor (16 GB main memory and 8 MB cache), (2) a single-node cluster implementation of Hadoop on the same desktop computer configuration, and (3) a Hadoop implementation on a multi-node cluster with six nodes. In the multi-node cluster implementation, the master node uses a dual quad-core Intel Xeon 5150 2.66 GHz processor, while the other nodes use dual quad-core Intel Xeon 5450 3.0 GHz processors with 16 GB of memory, and the nodes are connected by a 10 Gigabit Ethernet (GigE). The three primary objectives of the comparative test are to evaluate:
the time taken to identify R-waves on a single 640 MB size EDF file with subsequent computation of RR intervals and IHR values;
the time taken to identify the R-waves, and to compute the RR intervals and IHR cardiac measure on the largest dataset supported by the desktop computer with 3.2 GB of signal data from five EDF files;
the impact of optimization approaches that divide the signal data into 10 min segments for faster signal rendering on the Cloudwave visualization and query interface.
Figure 6A shows that the multi-node Cloudwave implementation reduces the time required for computation by a factor of 3.8 (0.32 vs 1.2 min) for data from one ECG channel, and it is one order of magnitude faster (4.8 vs 0.48 min) than the desktop computer for data from four ECG channels. This dramatic improvement in performance is also seen as the size of data increases to 3.2 GB from five EDF files, where the multi-node Cloudwave implementation is 14 times faster for data from one ECG channel than single-node implementation (0.42 vs 6.08 min) and 20 times faster than the desktop computer for data from all four ECG channels (1.57 vs 32.45 min) (figure 6B). We also note that the time required for computations on the multi-node Cloudwave implementation increases only by a factor of 3.7, although the number of channels increases by a factor of 4, which corresponds to the expected impact of parallelization (figure 6B). Figure 6C illustrates the effect of a Cloudwave optimization approach that divides the signal data into 10 min segments to support efficient visualization and query of ECG data by minimizing the impact of network latency. The multi-node Cloudwave implementation is 5.3 times faster (145 vs 27 s) and 18 times faster than the desktop computer for 36 segments of data. Since the desktop computer did not support computations on the signal dataset larger than 3.2 GB, we demonstrate the scalability of Cloudwave to support larger sized data on the multi-node cluster implementation in the next section.
Figure 6.
Comparative evaluation of computing three cardiac measures of QRS complex detection, RR intervals, and instantaneous heart rate (IHR) values using a desktop computer and two implementations of Cloudwave over (A) 640 MB data from one European Data Format (EDF) file, (B) 3.2 GB data from five EDF files and (C) 10 min data segments (optimized for use by Cloudwave interface), and (D) scalability of multi-node Cloudwave implementation for one to four ECG channels.
Scalability of Cloudwave on multi-node cluster
Figure 6D illustrates the performance of Cloudwave implementation on the multi-node cluster in terms of time taken for computing cardiac measures as the size of signal data increases from 3.2 to 12 GB of data for one to four ECG channels. It is clear that Cloudwave easily scales to this dataset and takes only a maximum of 3.3 min to complete the computations. This Cloudwave implementation featured only six computing nodes with a total of 50 GB disk space, which could accommodate 12 GB of data because of the Hadoop replication factor. We are in the process of increasing the available disk space to 5 TB as we continue to load all PRISM data into Cloudwave, which can be easily supported because of the extensibility of Hadoop to hundreds or thousands of nodes.44
Discussions
Electrophysiological signal data are usually not mentioned in discussions on biomedical Big Data, but they are playing an increasingly central role in driving both patient care and clinical research in neurological diseases and sleep medicine.7 A key challenge for Big Data management is ensuring compliance with patient privacy regulations and protection from unauthorized access on cloud platforms.
Privacy on cloud platforms
Currently, Cloudwave has been deployed on a private cloud computing infrastructure that is protected by institutional firewalls with secure access control. The PRISM signal data are manually deidentified to remove all protected health information. The Health Insurance Portability and Accountability Act (HIPAA) privacy and security toolkit, as well as the HITECH privacy subsection, describes a framework for managing the security of electronic health information, especially a set of ‘safeguard principles’ to prevent unauthorized access or use. Commercial cloud infrastructures, such as Amazon Web Services (AWS) and Microsoft Windows Azure, support compliance with both HIPAA security and privacy rules with standard-based data encryption, access control, and auditing mechanisms. Specifically, AWS EC2 supports the use of 2048-bit RSA key pair generation, allowing system administrators to create user groups with distinct levels of access and restrict network traffic to EC2 instances using customized rules.45 In addition, data stored on the AWS Simple Storage Service (S3) can be encrypted using standard techniques, and fine-level access control can be maintained using an access control list for each S3 ‘object.’45
Cost–benefit analysis of the ‘pay-as-you go’ model in cloud computing
Both AWS and Windows Azure have a flexible cost structure with a ‘pay-as-you-go’ pricing mechanism, which makes it suitable for use by different categories of applications. The multi-node Cloudwave installation was deployed within a preconfigured high-performance computing (HPC) environment at CWRU. In addition, the CWRU HPC personnel estimated the Cloudwave usage cost to be comparable with the EC2 on demand ‘large’ and ‘extra large’ instances of US 24 cents and 48 cents/h, respectively.46 The storage cost for Cloudwave was also estimated to be comparable to the AWS Elastic Block Store at US 10 cents/GB for 1 month, but without the additional data-transfer cost (download bandwidth) associated with AWS that may significantly increase the total user cost.46
Conclusions
Electrophysiological signal data are increasingly characterized by both massive volume and high velocity, as they play a greater role in supporting patient care and clinical research. In this paper, we present the Cloudwave platform, which addresses the three primary requirements of electrophysiological Big Data: (a) to reliably store a large volume of signal data; (b) to efficiently perform complex ECG signal-processing computations for real-time user interactions; (c) to support ontology-driven web-based visualization and query access for collaborative research. In contrast with traditional desktop-based signal-processing approaches, Cloudwave shows a dramatic reduction in the time required to perform computations over increasing volumes of signal data. Cloudwave is a flexible and scalable platform for supporting clinical research studies using massive-scale signal data in multiple disease domains.
Acknowledgments
This work used the High Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University. We also thank Hadrian Djohari and Mike Warfe at Case Western Advance Research Computing for their responsive support.
Footnotes
Contributors: SSS and CJ designed and developed Cloudwave with contributions from C-HC and G-QZ. SSS, CJ and G-QZ also wrote the paper and prepared the figures. SDL together with GG, SC and AB provided domain expertise and signal data and contributed to writing the paper. FK and KL contributed to the development of the Cloudwave interface and the description of signal processing in the paper.
Funding: This work was supported by NIH/NINDS grant number 1-P20-NS076965-01 and NIH/NCATS grant number UL1TR000439.
Competing interests: None.
Ethics approval: University Hospital Institutional Review Board (Cleveland, Ohio, USA) for PRISM project.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The Cloudwave signal-processing classes will be made open source and released after β testing.
References
- 1.Nature Editorial Community cleverness required. Nature 2008;455:1. [DOI] [PubMed] [Google Scholar]
- 2.Madden S. From databases to Big Data. IEEE Internet Comput 2012;16:4–6 [Google Scholar]
- 3.Agrawal D, Bernstein P, Bertino E, et al. Challenges and Opportunities with Big Data. CRA white paper 2012
- 4.Ferrucci D, Brown E, Chu-Carroll J, et al. Building Watson: an overview of the DeepQA project. AI Mag 2010;31:59–79 [Google Scholar]
- 5.Cohen J, Dolan B, Dunlap M, et al. MAD skills: new analysis practices for Big Data. Proc VLDB Endowment 2009;2:1481–92 [Google Scholar]
- 6.Wilson JA, Williams JC. Massively parallel signal processing using the graphics processing unit for real-time brain-computer interface feature extraction. Front Neuroeng 2009;2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg AT, Berkovic SF, Brodie MJ, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia 2010;51:676–85 [DOI] [PubMed] [Google Scholar]
- 8.Nihon Koden Neurology. http://www.nkusa.com/neurology_cardiology/ (accessed 16 Aug 2013).
- 9.Chen D, Wang L, Ouyang G, et al. Massively parallel neural signal processing on a many-core platform. Comput Science Engg 2011;13:42–51 [Google Scholar]
- 10.Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters. In: OSDI'04: Sixth Symposium on Operating System Design and Implementation: 2004; San Francisco, 2004 [Google Scholar]
- 11.Apache Hadoop. http://hadoop.apache.org/ (accessed 16 Aug 2013).
- 12.Epilepsy Foundation. http://www.epilepsyfoundation.org/aboutepilepsy/whatisepilepsy/statistics.cfm (accessed 16 Aug 2013).
- 13.Zijlmans M, Flanagan D, Gotman J. Heart rate changes and ECG abnormalities during epileptic seizures: prevalence and definition of an objective clinical sign. Epilepsia 2002;43:847–54 [DOI] [PubMed] [Google Scholar]
- 14.Nashef L, Walker F, Allen P, et al. Apnoea and bradycardia during epileptic seizures: relation to sudden death in epilepsy. J Neurol Neurosurg Psychiatry 1996;60:297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomson T, Ericson M, Ihrman C, et al. Heart rate variability in patients with epilepsy. Epilepsy Res 1998;30:77–83 [DOI] [PubMed] [Google Scholar]
- 16.Kerem DH, Geva AB. Forecasting epilepsy from the heart rate signal. Med Biol Eng Comput 2005;43:230–9 [DOI] [PubMed] [Google Scholar]
- 17.Marshall DW, Westmoreland BF, Sharbrough FW. Ictal tachycardia during temporal lobe seizures. Mayo Clin Proc 1983;58:443–6 [PubMed] [Google Scholar]
- 18.Bigger JTJ, Fleiss JL, Steinman RC, et al. Frequency domain measures of heart period variability and mortality after myocardial infarction. Circulation 1992;85:164–71 [DOI] [PubMed] [Google Scholar]
- 19.So EL. What is known about the mechanisms underlying SUDEP? Epilepsia 2008;49(Suppl 9):93–8 [DOI] [PubMed] [Google Scholar]
- 20.Surges R, Thijs RD, Tan HL, et al. Sudden unexpected death in epilepsy: risk factors and potential pathomechanisms. Nat Rev Neurol 2009;5:492–504 [DOI] [PubMed] [Google Scholar]
- 21.Ficker DM, So EL, Shen WK, et al. Population-based study of the incidence of sudden unexplained death in epilepsy. Neurology 1998;51:1270–4 [DOI] [PubMed] [Google Scholar]
- 22.Tellez-Zenteno JF, Ronquillo LH, Wiebe S. Sudden unexpected death in epilepsy: evidence-based analysis of incidence and risk factors. Epilepsy Res 2005;65:101–15 [DOI] [PubMed] [Google Scholar]
- 23.Opherk C, Coromilas J, Hirsch LJ. Heart rate and EKG changes in 102 seizures: analysis of influencing factors. Epilepsy Res 2002;52:117–27 [DOI] [PubMed] [Google Scholar]
- 24.Langmead B, Schatz MC, Lin J, et al. Searching for SNPs with cloud computing. Genome Biol 2009;10:R134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat Rev Drug Discov 2009;8:286–95 [DOI] [PubMed] [Google Scholar]
- 26.US NRC Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. USA: National Academies Press, 2011 [PubMed] [Google Scholar]
- 27.Schadt EE, Linderman MD, Sorenson J, et al. Computational solutions to large-scale data management and analysis. Nat Rev Genet 2010;11:647–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schatz MC. BlastReduce: high performance short read mapping with MapReduce. Technical Report
- 29.White RW, Tatonetti NP, Shah NH, et al. Web-scale pharmacovigilance: listening to signals from the crowd. J Am Med Inform Assoc 2013;20:404–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linderman MD, Bruggner R, Athalye V, et al. High-throughput Bayesian network learning using heterogeneous multicore computers. In: 24th ACM International Conference on Supercomputing (ICS ‘10): 2010; Japan: ACM New York, 2010:95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandeya S, Voorsluysa W, Niua S, et al. An autonomic cloud environment for hosting ECG data analysis services. Future Gener Comp Syst 2012;28:147–54 [Google Scholar]
- 32.Sahoo SS, Lhatoo SD, Gupta DK, et al. Epilepsy and seizure ontology: towards an epilepsy informatics infrastructure for clinical research and patient care. J Am Med Assoc 2014;21:82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemp B, Olivan J. European data format ‘plus’ (EDF+), an EDF alike standard format for the exchange of physiological data. Clin Neurophysiol 2003;114:1755–61 [DOI] [PubMed] [Google Scholar]
- 34.Goldberger AL, Amaral LAN, Glass L, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a New Research Resource for Complex Physiologic Signals. Circulation 2000;101:e215–20 [DOI] [PubMed] [Google Scholar]
- 35.Amdahl GM. Validity of the single processor approach to achieving large scale computing capabilities. In: Proceeding AFIPS ‘67, Spring Joint Computer Conference; 1967:483–5 [Google Scholar]
- 36.Bernstein AJ. Analysis of programs for parallel processing. IEEE Trans Electron Comput 1966;EC-15:757–63 [Google Scholar]
- 37.Topcuoglu H, Hariri S, Wu MY. Performance-effective and low-complexity task scheduling for heterogeneous computing. IEEE Trans Parallel Distrib Syst 2002;13:260–74 [Google Scholar]
- 38.Shvachko K, Kuang H, Radia S, et al. The hadoop distributed file system. In: IEEE 26th Symposium on Mass Storage Systems and Technologies (MSST); Nevada, USA: 2010:1–10 [Google Scholar]
- 39.Hitzler P, Krötzsch M, Parsia B, et al. OWL 2 web ontology language primer. W3C recommendation. World Wide Web Consortium, 2009 [Google Scholar]
- 40.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform 2003;36:478–500 [DOI] [PubMed] [Google Scholar]
- 41.Nelson SJ, Zeng K, Kilbourne J, et al. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 2011;18:441–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dou D, Frishkoff G, Rong J, et al. Development of NeuroElectroMagnetic Ontologies (NEMO): a framework for mining brain wave ontologies. In: Thirteenth International Conference on Knowledge Discovery and Data Mining (KDD2007): 2007; San Hose, CA: ACM New York, 2007:270–9 [Google Scholar]
- 43.Zhang GQ, Siegler T, Saxman P, et al. VISAGE: a query interface for clinical research. In: AMIA Clinical Research Informatics Summit San Francisco: 2010:76–80 [PMC free article] [PubMed] [Google Scholar]
- 44.Cooper BF, Baldeschwieler E, Fonseca R, et al. Building a cloud for Yahoo!. IEEE Data Eng Bull 2009;32:36–43 [Google Scholar]
- 45.Amazon Web Services—Creating Healthcare Data Applications to Promote HIPAA and HITECH Compliance. Technical Report 2012. http://aws.amazon.com/security/security-resources/ (accessed 16 Aug 2013).
- 46.Amazon EC2 Pricing. http://aws.amazon.com/ec2/pricing/ (accessed 16 Aug 2013).