Abstract
Calcium-dependent protein kinases (CDPKs) are multifunctional proteins combining calcium-binding and signaling capabilities within a single gene product. This unique versatility enables multiple plant biological processes to be controlled, including developmental programs and stress responses. The genome of flowering plants typically encodes around 30 CDPK homologs that cluster in four conserved clades. In this Review, we take advantage of the recent availability of genome sequences from green algae and early land plants to examine how well the previously described CDPK family from angiosperms compares to the broader evolutionary states associated with early diverging green plant lineages. Our analysis suggests that the current architecture of the CDPK family was shaped during the colonization of the land by plants, whereas CDPKs from ancestor green algae have continued to evolve independently.
Keywords: Calcium signaling, CDPKs, Green plant lineages, Evolution
Signaling through plant calcium sensors
The calcium (Ca2+) ion has long been recognized as a versatile secondary messenger that induces signaling in all eukaryotes. Plants, for instance, use Ca2+ signaling to regulate development, as well as responses to biotic and abiotic challenges [1]. Following perception of stimuli, plant cells initiate various signaling pathways that relay information to specific primary targets, including Ca2+ channels and transporters [2]. Transport from major Ca2+ stores (i.e. apoplast and organelles) results in local Ca2+ fluxes at the plasma membrane or phospholipidic membrane of intracellular organelles. This phenomenon, conceptually referred to as the alteration of Ca2+ signatures, is constantly monitored by several sensor proteins, which convert upstream Ca2+ signals into appropriate downstream cellular responses [3,4].
Plants possess several types of Ca2+ sensors, many of which have the EF-hand motif. This helix–loop–helix structural domain coordinates a single Ca2+ ion, providing direct Ca2+-binding activity to the sensor and resulting in Ca2+-dependent conformational changes that promote signaling. The Ca2+ sensing apparatus from plants comprises somewhat complex protein families that are represented by the calmodulin (CaM) and calmodulin-like (CML) families [5,6], the family of calcineurin B-like (CBL) proteins [7–9], and the CDPKs [10–12].
Among plant Ca2+ sensors, CDPKs are unique because they unite Ca2+-binding and signaling capabilities within a single gene product. This combination probably arose following the early fusion of an upstream protein kinase (PK) gene and a downstream CaM gene [13] to enable immediate and efficient translation of input Ca2+ signals into appropriate output phosphorylation events. The aim of this Review is to trace the evolution of the CDPK family within species that represent key milestones in the evolution of green plants (Box 1; and see Table S1 in the supplementary material online). We first provide a brief description of the various structural features found in higher plant CDPKs and then assess the conservation of these features in CDPK homologs from descendants of ancestral green plant lineages.
Box 1. Evolution of green plants.
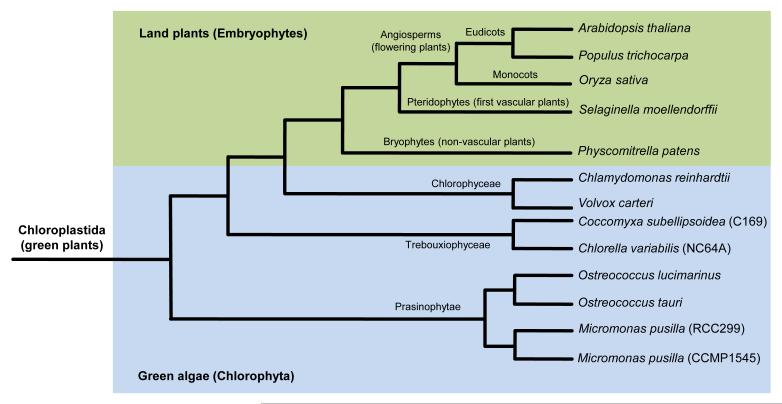
Chloroplastida (also known as green plants) is one of the three lineages derived from the single primary endosymbiosis of a cyanobacterium and a eukaryotic host cell [73,74]. Relying on their outstanding plasticity with respect to biochemistry, cell architecture, morphology and general lifestyle, green plants have gradually managed to conquer almost every ecosystem on Earth. The Chloroplastida comprises all species of green algae, as well as embryophytes (also known as land plants) [74]. Despite their diversity, these organisms form a monophyletic group divided in two divisions: the Chlorophyta and the Charophyta, which includes the Streptophyta [65,74-77]. The Chlorophyta comprise a large array of marine and freshwater green algae [65,66,74], including the Prasinophytae (e.g., Ostreococcus and Micromonas), the Trebouxiophyceae (Coccomyxa and Chlorella), and the Chlorophyceae (Chlamydomonas and Volvox) (Figure I and Table S1 in the supplementary material online). About 725 million years ago (Mya)–1200 Mya, a green alga species diverged from the Chlorophyta to become the first Streptophyta lineage [78–81]. Descendants from the first Streptophyta species nowadays constitute a small but diverse group of freshwater green algae known as the streptophyte (or charophycean) algae. The Chlorophyta–Streptophyta split was crucial because ancient streptophyte algae evolved new characters that eventually facilitated colonization of terrestrial habitats around 450 Mya–500 Mya [82–84]. The first land plants did not possess vascular tissues and descendants of these ancient lineages today consist of the liverworts, hornworts and mosses (collectively referred to as the bryophytes or non-vascular plants) (Figure I). Ancient bryophytes progressively colonized drier habitats, a trend that promoted differentiation of water-conducting tissues known as the phloem and xylem. Land plants with vascular tissues are referred to as higher plants, and the pteridophytes correspond to the most ancestral lineage within this group (Figure I). Bryophytes and pteridophytes both rely on spores for reproduction, and the differentiation of seeds by spermatophytes (seed plants) was another key breakthrough that accelerated land plant dispersal. Nowadays, spermatophytes comprise five distinct groups, including the gymnosperms and the angiosperms (Figure I). Expansion of embryophytes was one of the most important steps for the evolution of life because these organisms forever changed the atmospheric oxygen concentration and contributed significantly to the shaping of terrestrial habitats.
Figure I.
Relationships among green plant lineages used to study the evolution of CDPKs. Histones are among the most highly conserved genes and were therefore used to reflect the evolutionary path of green plant lineages used in this study. The genome assembly of each of the indicated species was searched using the predicted amino acid sequence of the Arabidopsis histone H4 gene (AT1G07660) as a query. Full-length coding sequence (CDS) of histone H4 homologs were next aligned with ClustalW, using the following alignment parameters: for pairwise alignment, gap opening, 10.0, and gap extension, 0.1; for multiple alignment, gap opening, 10.0, and gap extension, 0.20. Resulting alignments were submitted to the MEGA4 software to generate a neighbor-joining tree derived from 5000 replicates. Because the tree only depicts topology among the various taxa, branch size is not proportional to the extent of primary sequence divergence. The blue section comprises species of the Chlorophyta division, a taxonomically diverse group of green algae from which Streptophyta diverged 725 Mya–1200 Mya. Streptophyta includes a number of green algae collectively known as the streptophyte algae (not shown here), as well as embryophytes (also referred to as land plants). The green section comprises various species of land plants that represent some of the most important evolutionary milestones characterizing the evolution of terrestrial plants: colonization of the land by bryophytes; differentiation of vascular tissues in pteridophytes; seeds enclosed in fruits for protection in angiosperms.
Structure and activation of prototypical CDPKs
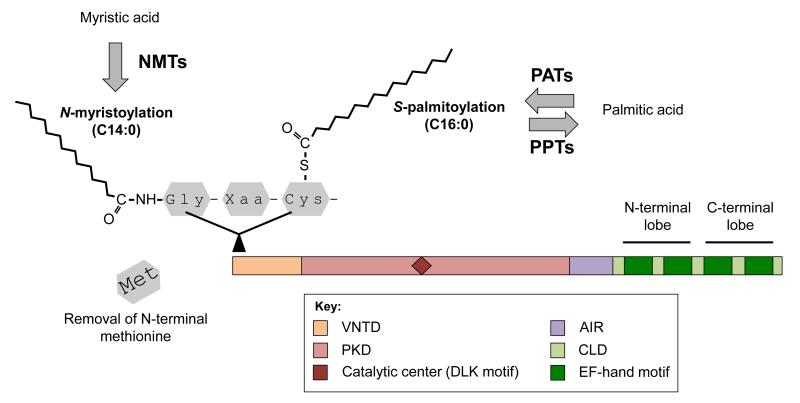
Consistent with their function as Ca2+ sensors, CDPKs harbor a C-terminal calmodulin-like domain (CLD) that typically comprises four EF-hand motifs (Figure 1). Structural analyses indicate that EF-hands function in pairs, creating two lobes characterized by differential Ca2+-binding affinities [14,15]. The C-terminal lobe has high Ca2+ affinity and binds to the ion despite the low concentration that prevails under basal conditions. Within this state, the C-terminal lobe works as a stabilizing structure by promoting intramolecular bonding between the autoinhibitory region (AIR) and the Ser/Thr protein kinase domain (PKD) (Figure 1). As its designation suggests, the AIR keeps the whole enzyme in an inactive mode by acting as a pseudosubstrate that restricts access to the kinase catalytic center [16,17]. Following the perception of a stimulus, the intracellular concentration of Ca2+ increases and EF-hand motifs from the N-terminal lobe (Figure 1) bind to the ion despite low Ca2+ affinity [18]. The whole protein then undergoes conformational changes that release auto-inhibition and promote phosphorylation of downstream substrates, including metabolic enzymes, ion channels, and transcription factors [11,19,20].
Figure 1.
Topology of a prototypical CDPK protein. Schematic view of a prototypical CDPK and organization of its conserved domains (see color-coded legend). The N-terminus of CDPKs is often targeted for N-acylation. N-myristoyl transferases (NMTs) catalyze the covalent attachment of myristic acid (a C14:0 fatty acid) to the N-terminal Gly residue (irreversible N-myristoylation). Stable anchoring to membranes also requires S-palmitoylation, which is performed by palmitoyl acyl transferases (PATs) and palmitoyl protein thioesterases (PPTs). These enzymes catalyze the reversible attachment of palmitic acid (a C16:0 fatty acid) to Cys residues. Abbreviations: AIR, autoinhibitory region; CLD, calmodulin-like domain; PKD, protein kinase domain; VNTD, variable N-terminal domain; Xaa, any amino acid.
At the N-terminus, CDPK proteins also contain the variable N-terminal domain (VNTD) (Figure 1), which varies in amino acid sequence and length [21]. Although only limited information is available about the function of the VNTD, recent studies suggest that this region is involved in specificity of substrate recognition [22,23]. The VNTD of most angiosperm CDPKs also comprises predicted N-myristoylation and S-palmitoylation sites, two features that promote protein targeting to lipid membranes [11,21,24–27] (Figure 1). Experimentally, membrane-anchoring of a CDPK was first demonstrated in rice (Oryza sativa) [28]; however, this phenomenon has since been shown for several Arabidopsis (Arabidopsis thaliana) candidates, as well as for CDPKs from other flowering plant species ([11,12] and references therein). Importantly, membrane-targeted CDPKs can also move away from membranes in response to stress signals [20]. It is likely that this feature enables CDPKs to shuttle between various subcellular compartments as a way to accomplish a broader array of cellular functions.
Evolution of green plant CDPKs
Biochemical evidence showing that CDPKs are calmodulin-independent was first provided more than 25 years ago following the purification and characterization of the first CDPK enzyme from soybean (Glycine max) [29]. Since then, CDPK homologs have been identified throughout the plant kingdom, including within species of green algae. Long thought to be plant-specific, CDPK homologs have also been found in two groups of unicellular protists: ciliates and apicomplexan parasites [13,30,31]. The emergence of the first CDPK gene therefore occurred before the basal split between green plants and alveolate protists [13].
The genome of Arabidopsis encodes 34 CDPKs that cluster in four well-resolved clades [11,21,32]. Genome sequences from other angiosperms confirm that the basal architecture of the CDPK family is conserved in other flowering plants, including monocot and eudicot species [24–27]. In a previous comparative genomics study, CDPKs from green algae were found to cluster with their flowering plant homologs, whereas CDPKs from protists grouped in a distinct protein subset [31]. The recent completion of genome sequences from early land plants [33,34] and green algae [35–41] now makes it possible to examine the whole CDPK signaling repertoires from descendants of ancient green plant lineages (Box 1 and Table S1). Our analysis provides improved phylogenetic classification of CDPK homologs from green algae. Comparison of the newly identified sequences and the rich CDPK complements from angiosperms also suggests that the basic architecture of the CDPK family is shared between all land plants, whereas homologs from green algae constitute distinct clades that had not been previously identified.
Green alga CDPKs
Numbers and clustering
Searches of publicly available databases [42,43] have identified 48 putative green alga CDPKs (Table S2 in the supplementary material online). These proteins belong to eight Chlorophyta species, a diverse group of green algae from which Streptophyta (i.e. land plants and precursor streptophyte algae) diverged before colonization of the land by plants (Box 1). Most of the identified CDPK candidates have not been characterized yet, suggesting that our survey would be a valuable resource for those who wish to investigate CDPK functions in green algae.
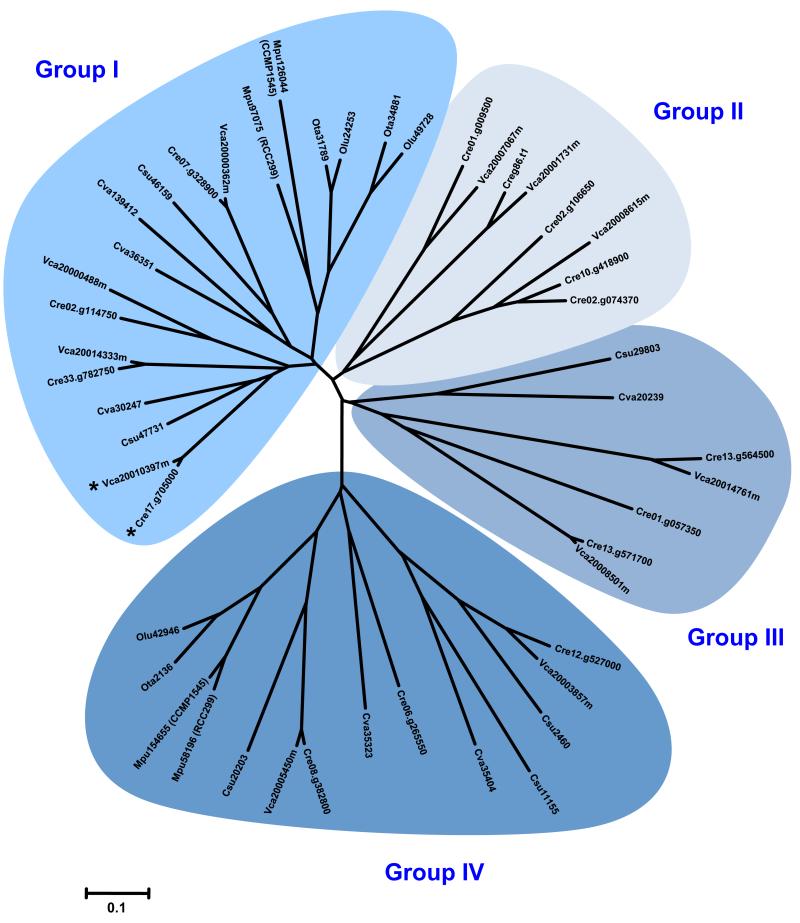
Following homology-based identification, full-length protein sequences were aligned and phylogenetic relationships were established (Figure 2). Our analysis defines four major clades of green alga CDPKs, two of which have at least one representative member from each of the eight Chlorophyta species examined (Group I and Group IV). Green algae of the Mamiellales order (e.g. the marine species Micromonas pusilla and Ostreococcus spp.) have particularly small CDPK complements (Table S2) and lack a candidate in Clades II and III (Figure 2). When compared to the freshwater algae Chlamydomonas reinhardtii and Volvox carteri, the marine species M. pusilla and Ostreococcus spp. are characterized by extensive gene loss that is associated with reduced genome size [37,39,41]. Gene loss is likely to explain the remarkably small number of CDPK candidates in these species.
Figure 2.
Phylogenetic relationships among green alga CDPKs. The genome assembly of various green algae was searched using the amino acid sequence of Arabidopsis CDPKs as queries. Retrieved gene models were accepted only if the corresponding protein had consensus sequences of the Ser/Thr PKD, including conserved Asp and Lys residues within the active site (D[L/I/V]K motif), and a CLD comprising at least one consensus EF-hand motif. Full-length CDPKs (see supplementary material online) were next aligned with ClustalW, using the following alignment parameters: for pairwise alignment, gap opening, 10.0, and gap extension, 0.1; for multiple alignment, gap opening, 10.0, and gap extension, 0.20. Resulting alignments were submitted to the Molecular Evolutionary Genetics Analysis 4 (MEGA4) software [72] to generate a neighbor-joining tree derived from 5000 replicates. To emphasize the clustering of the various CDPK clades, the unrooted distance tree visualization mode was selected. Branch size is proportional to the extent of primary sequence divergence, with the scale of ‘0.1’ representing a 10% change. The four CDPK clades are highlighted in shades of blue and a species acronym denotes the origin of each protein (Cre, Chlamydomonas reinhardtii; Csu, Coccomyxa subellipsoidea; Cva, Chlorella variabilis; Mpu, Micromonas pusilla; Olu, Ostreococcus lucimarinus; Ota, Ostreococcus tauri; Vca, Volvox carteri.). Asterisks denote unusual CDPK homologs that harbor a C2 domain at the N-terminus.
With slightly larger CDPK complements, green algae of the Trebouxiophyceae class (Coccomyxa subellipsoidea and Chlorella variabilis) retain CDPK homologs in Groups I, III, and IV, but also lack a representative member within Group II (Figure 2). In our survey, Csu29803 (C. subellipsoidea) and Cva20239 (C. variabilis) steadily clustered with Group III candidates; however, these protein kinases (PKs) diverged much earlier than their closest homologs from C. reinhardtii and V. carteri (Figure 2). Csu29803 and Cva20239 also share significant homology with candidates from Group II (data not shown), suggesting that these CDPKs may correspond to an intermediate evolutionary state that lies between Clades II and III.
Domain organization and membrane targeting
In terms of biochemical properties, CDPKs from green algae are somewhat similar to their angiosperm counterparts (see Tables S2 and S3). Shared features include comparable amino acid length, molecular weight, and isoelectric point (pI). At the structural level, three distinct prediction tools (PROSITE: http://prosite.expasy.org, SMART: http://smart.embl-heidelberg.de, and Pfam: http://pfam.sanger.ac.uk) confirm that green algae possess prototypical CDPKs with four EF-hand motifs (Figure 1), as well as atypical candidates that lack one or more Ca2+ binding site(s) at the C-terminus (Tables S2). This observation confirms that atypical CDPKs are not only found in angiosperms that hold large CDPK families, but also in green algae with markedly undersized CDPK complements (e.g. M. pusilla and Ostreococcus spp.). Recently, a survey has shown that Arabidopsis CDPKs have highly variable Ca2+-dependences for their kinase activity [44]. Interestingly, low Ca2+-sensitivity correlates with structural alterations in the predicted EF-hand motifs, although only one atypical candidate (CPK25) completely lost the ability to bind Ca2+. Conservation of atypical CDPKs throughout the green plant lineage suggests that these proteins accomplish vital functions and that better understanding of the functions associated with their unconventional CLD is a key issue in plant signaling [11,44].
Another important feature of CDPKs is the presence of N-acylation sites controlling the anchoring of proteins to membranes (Figure 1). Like their flowering plant homologs, several green alga CDPKs harbor predicted N-myristoylation and S-palmitoylation sites at the N-terminus (Table S2). However, the prevalence of these sites appears to be less common than in angiosperms where most CDPK candidates carry predicted N-acylation sites (Table S3 in the supplementary material online). Because N-acylation of cellular proteins is still poorly characterized in Chlorophyta species, we searched genomic data to identify green alga genes relevant to this process. Irreversible N-myristoylation is performed by N-myristoyl transferases (NMTs), whereas reversible S-palmitoylation is mediated by palmitoyl acyl transferases (PATs) and palmitoyl protein thioesterases (PPTs) (Figure 1). With the exception of the two Ostreococcus species for which no PPT homologs could be identified, it appears that green algae retain the minimal enzymatic machinery necessary to support N-acylation of target proteins (Table S4 in the supplementary material online). It can thus be assumed that at least some green alga CDPKs are targeted to lipid membranes.
Domains of the unknown
Several studies have shown that domain organization is highly conserved among CDPK proteins, including more distant homologs from alveolate protists [13,31]. Nonetheless, several unusual CDPKs from apicomplexan parasites possess EF-hand motifs and a plekstrin homology domain (PHD) at the N-terminus [30]. These features are absent in CDPK homologs from green algae, an observation consistent with their closer homology with flowering plant CDPKs [31]. However, further examination of green alga protein sequences has revealed that Cre17.g705000 (C. reinhardtii) and Vca20010397m (V. carteri) each carry an unusually long VNTD that comprises a C2 domain (see asterisks in Figure 2 and Table S2). With approximately 130 residues, the C2 domain was originally identified as one of two conserved regulatory regions (C1 and C2) within protein kinase C (PKC) [45]. This domain was later found in a variety of signal transduction and membrane trafficking proteins, where it usually functions as a Ca2+-dependent module that binds to phospholipids. Given that Cre17.g705000 and Vca20010397 both lack predicted N-acylation sites (Table S2), the C2 domain is likely to offer an alternative strategy to guarantee membrane-targeting of these unusual CDPKs.
Although stand-alone C2 domain proteins are encoded by the genome of each Chlorophyta species investigated here (data not shown), CDPKs harboring a C2 domain appear to be uniquely found in green algae of the Chlamydomonadales order (Figure 2 and Table S2). This unexpected combination is likely to result from the fusion of two genes, encoding a CDPK and a C2 domain protein, respectively. It remains to be established whether this fusion is lineage-specific, or if it is a general feature that has been lost in other taxa. A search of the NCBI database (http://www.ncbi.nlm.nih.gov/) could only identify two additional CDPKs harboring a C2 domain: that is, DtCPK1 from Dunaliella tertiolecta (accession number AAF21062) [46] and CCK1 from Chlamydomonas moewusii (CAA89202). These green algae also belong to the Chlamydomonadales order, arguing in favor of a lineage-specific fusion. Overall, our search of genomic data from Chlorophyta species indicates that CDPKs from green algae cluster in four distinct clades and that these PKs have structural features that are generally similar to those observed for angiosperm homologs.
Land plant CDPKs
Numbers and clustering
Angiosperms are the most diverse subset of spermatophytes (plants that produce seeds) and represent the more advanced groups of land plants (Box 1). Although CDPKs from angiosperms have been widely studied [10–12], little is known about the CDPK homologs from more ancient embryophyte lineages, including gymnosperms, pteridophytes, and bryophytes (Box 1). The genome sequences of the bryophyte Physcomitrella patens [34] and the pteridophyte Selaginella moellendorffii [33] now offer the exciting opportunity to explore CDPK signaling within the oldest land plant lineages.
Searches of genomic data from P. patens and S. moellendorffii [42,43] have identified 35 novel CDPKs that were compared to 93 homologs from three angiosperm species, that is the monocot rice and the eudicots poplar (Populus trichocarpa) and Arabidopsis (Figure S1 and Table S3 in the supplementary material online). Analysis of the phylogenetic relationships has revealed that CDPK homologs from early land plants cluster in four distinct groups (Figure S1), which correspond to the clades formerly identified in various flowering plants [11,21,24–27,32]. Taken as a whole, these evolutionary data indicate that the basal architecture of the CDPK family is conserved among all land plants: from a primitive bryophyte species to the more sophisticated angiosperms. These results also suggest that large CDPK complements found in angiosperms evolved from the duplication of genes that are common to all embryophytes, rather than from the diversification of lineage-specific candidates that could have fashioned distinct clades of CDPKs.
Expansion through recent gene duplication
In angiosperms, the CDPK family is characterized by the presence of numerous paralogs that share high levels of homology. These closely related genes most probably emerged following recent duplication events and, therefore, have not markedly diversified yet (Figure S1). In some cases, duplication appears to predate the monocot–eudicot split (e.g. OsaCPK17/27, AthCPK1/2, and PtiCPK1/2), whereas in others, duplication appears to be specific to a particular lineage (e.g. OsaCPK12 and AthCPK29 versus PtiCPK29-1/29-2). Duplicated genes arise either by regional genomic events or genome-wide events (also called polyploidization). While polyploidization appears to be the most common mechanism of gene duplication in plants, regional events may still be responsible for a significant number of duplicated genes. Regional events include local duplication of a single gene (or tandem duplication) as well as transfer of several genes or entire chromosomal segments to more distant chromosomal locations (large segmental duplications). Examination of gene chromosomal distribution indicates that within angiosperms, few CDPK genes occur as tandem duplicates (data not shown). Notable exceptions are CPK20-1/20-3 and CPK20-2/20-4 in poplar [27], as well as a cluster of five Group II CDPKs in Arabidopsis (CPK21/22/23/27/31) [21,32]. As previously shown for mitogen-activated protein kinases (MAPKs) [47], the expansion of the CDPK family from angiosperms thus mainly relied on large-scale DNA rearrangements, namely whole-genome or large segmental duplications.
Gene duplication also appears to be a prominent feature of the CDPK family in P. patens. Although this moss is characterized by a relatively simple lifestyle, with 25 distinct members, it retains a CDPK family almost as large as the more complex angiosperms (Table S3). Within each CDPK clade, homologs from P. patens cluster tightly (Figure S1), indicating that these genes probably arose following recent duplications. It has been reported that the genome of P. patens is characterized by numerous large-scale rearrangements that contributed to the expansion of several gene families, including PKs [34]. A search of the P. patens genome could only define four pairs of tandemly duplicated CDPK genes (data not shown), suggesting that large-scale duplications are primarily responsible for the expansion of this protein family. Given that only one bryophyte has been sequenced to date, it is difficult to predict if recent large-scale duplications contributed to the expansion of the CDPK family in other non-vascular plants. This question is particularly intriguing because support for gene duplication is much less obvious for the CDPK family of S. moellendorffii (Figure S1 and Table S3). This species of pteridophyte is characterized by the absence of whole genome duplication [33], a rare feature that is likely to explain why this apparently more complex species paradoxically encodes far fewer CDPK homologs (Table S3).
Domain organization and membrane targeting
In terms of biochemical properties, CDPKs from early land plants are highly comparable to their angiosperm homologs (Table S3). For instance, CDPK proteins tend to be slightly acidic, with a pI ranging between five and six. However, a few candidates do not follow this trend and have a basic pI of eight or more. In angiosperms, basic CDPKs are mainly found within Group IV, a feature strictly conserved in P. patens and S. moellendorffii (Table S3). A basic pI may correlate with a specific subcellular localization or function of Group IV CDPKs, which accordingly correspond to the most divergent homologs within this PK family (Figure S1).
Close examination of the protein topology has not yielded any unexpected domains that would have distinguished CDPK homologs from early land plants. It can thus be assumed that the classical organization of CDPKs is conserved among all land plants and that no striking protein innovation has marked this family since the conquest of the land by plants. The genomes of P. patens and S. moellendorffii were also found to encode prototypical CDPKs with four EF-hand motifs, as well as atypical candidates with only one to three Ca2+-binding site(s) at the C-terminus (Table S3). Interestingly, atypical homologs account for 34% of all the CDPKs found in early land plants, whereas in green algae and angiosperms, atypical homologs account for 48% and 21% of all CPDKs, respectively (these values are based on the number of EF-hand motifs as predicted by PROSITE, but similar results have been obtained with SMART or Pfam prediction tools, see Table S3). Atypical CDPKs are therefore conserved throughout the green plant lineage; however, their overall proportion compared with prototypical CDPKs tends to decrease as plants gain in complexity.
The VNTD of early land plant CDPKs has also been scrutinized and N-acylation sites were defined in several candidates (Table S3). Because genes involved in the N-acylation process are conserved in P. patens and S. moellendorffii (Table S4), it is likely that at least some early land plant CDPKs are targeted to membranes. Interestingly, CDPKs lacking N-acylation sites mainly cluster within Group I, a feature shared among all land plants (Table S3). We also noticed that 50% of green alga CDPKs are predicted to lack N-acylation sites, compared with 31% in early land plants and only 14% in angiosperms. As observed for the ratio of atypical CDPKs (see the previous section), the proportion of non-acylated CDPKs tends to decrease as plant complexity increases.
Given that CDPKs are PKs, we finally compared the predicted catalytic domain of green alga and embryophyte CDPKs. Within each domain examined, 12 characteristic PK subdomains could be defined and very few amino acid insertions interrupt the catalytic core sections (data not shown). Within PK subdomains, strictly conserved features include residues that help to anchor and orient ATP, that is the ATP binding pocket in subdomain I (GXGXXG motif), the invariant Lys in subdomain II, and the invariant Asp in subdomain VII (DFG motif). Kinase activity also requires a catalytic base for efficient phosphotransfer reaction, a function mediated by the invariant Asp in subdomain VIB (DLK motif). As above-mentioned features, we found that the DLK motif is conserved in all CDPKs examined. Overall, our analysis indicates that no particular feature is associated with the PKD of analyzed CDPKs, suggesting that this protein region is very well conserved among all green plant lineages.
Clustering and nomenclature of green plant CDPKs
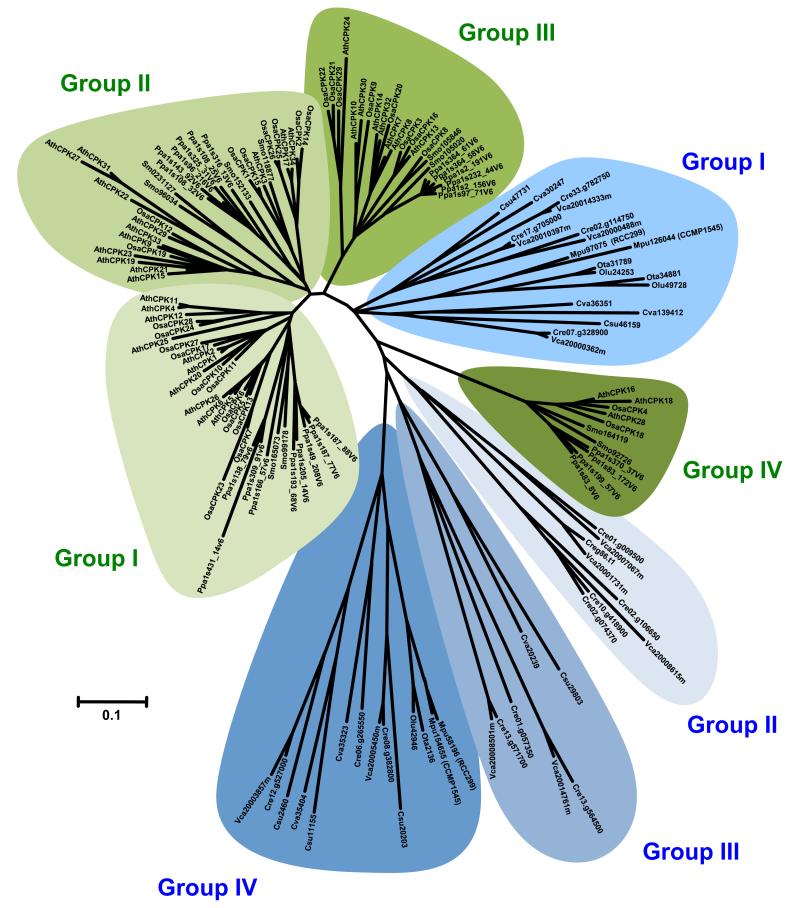
Given that green algae (Figure 2) and embryophytes (Figure S1) each comprise four distinct clades of CDPKs, it was tempting to hypothesize that when these groups were compared together they would indeed overlap because the basic architecture of the CDPK family is conserved throughout the green plant lineage. Surprisingly, phylogenetic analysis of the whole sequence dataset revealed that CDPK homologs from green algae do not cluster with counterpart proteins from land plants (Figure 3). As a result, eight CDPK clades were resolved: the first four being specific to Chlorophyta species and the second four being specific to embryophytes. Among the four groups of land plant CDPKs, Group IV appears to be more closely related to homologs from green algae. The ancestral gene that gave rise to this clade in land plants may therefore serve a general function in growth or development. When compared to CDPKs from alveolate protists, green alga CDPKs were shown to cluster with their land plant homologs [31]. However, our analysis emphasizes that fundamental differences distinguish the CDPK families from Chlorophyta and embryophytes (Figure 3).
Figure 3.
Phylogenetic relationships among green plant CDPKs. The genome assembly of various green plants was searched using the amino acid sequence of Arabidopsis CDPKs as queries. Retrieved gene models were accepted only if the corresponding protein had consensus sequences of the Ser/Thr PKD, including conserved Asp and Lys residues within the active site (D[L/I/V]K motif), and a CLD comprising at least one consensus EF-hand motif. Full-length CDPKs (see supplementary material online) were next aligned with ClustalW, using the following alignment parameters: for pairwise alignment, gap opening, 10.0, and gap extension, 0.1; for multiple alignment, gap opening, 10.0, and gap extension, 0.20. Resulting alignments were submitted to the MEGA4 software to generate a neighbor-joining tree derived from 5000 replicates. To emphasize the clustering of the various CDPK clades, the unrooted distance tree visualization mode was selected. Branch size is proportional to the extent of primary sequence divergence, with the scale of ‘0.1’ representing a 10% change. CDPK clades from green algae and land plants are highlighted in shades of blue and green, respectively. A species acronym identifies the origin of each protein (Ath, Arabidopsis thaliana; Cre, Chlamydomonas reinhardtii; Csu, Coccomyxa subellipsoidea; Cva, Chlorella variabilis; Mpu, Micromonas pusilla; Olu, Ostreococcus lucimarinus; Osa, Oryza sativa ssp. japonica; Ota, Ostreococcus tauri; Ppa, Physcomitrella patens; Smo, Selaginella moellendorffii; Vca, Volvox carteri). For clarity purposes, CDPK homologs from poplar were not included in this analysis.
In light of their differential clustering, we did not attempt to expand the nomenclature of Arabidopsis CDPKs [21] to homologs from Chlorophyta species. The authors from a recent comparative genomics survey reached a similar conclusion regarding the nomenclature of green plant MAPKs [48]. By contrast, CDPK homologs from embryophytes form conserved clades (Figure 3) that may rationalize the establishment of a uniform nomenclature. To further support this initiative, we made use of various web-based tools [43,49] to investigate orthologous relationships between the CDPK homologs from land plants.
With the exception of candidates from poplar and Arabidopsis, none of the bioinformatics tools was able to predict reliably orthologous relations between embryophyte CDPKs. As a result, we do not propose an expanded nomenclature for CDPK homologs from early land plants and, hence, we used the previously published nomenclature of rice CDPKs [24]. In the case of poplar and Arabidopsis, the prediction tools were generally able to yield clear one-to-one affiliations that matched the phylogenetic classification (Figure S1). Based on this pairing, we named poplar CDPKs in a way that reflects sequence relatedness with the closest homologs from Arabidopsis (e.g. PtiCPK1 is most similar to AthCPK1). In some cases, additional numbering was included to discriminate between clearly paralogous forms of a given kinase (e.g. PtiCPK3-1 and PtiCPK3-2 versus AtCPK3) [47,50]. We recognize that such a naming and numbering protocol is strictly driven by predicted evolutionary relationships and that conservation of biological functions within these relationships remains to be established. Nonetheless, systematic nomenclature should facilitate species-to-species comparisons and help functional characterization of CDPK homologs showing clear orthologous relationships with candidates from the model plant species Arabidopsis.
Concluding remarks and future outlook
Our analysis has revealed that the basic architecture of the CDPK family is conserved among all land plants (Figure S1). This includes a bryophyte species that is related to the first green plant lineage that successfully colonized terrestrial habitats (Box 1). By contrast, CDPK homologs from Chlorophyta form alga-specific clades that apparently kept evolving independently (Figure 3). Taken as a whole, these findings suggest that the Chloropyhta–Streptophyta split (Box 1) played a key role in the evolution of CDPKs. However, major architectural changes that affected this protein family were essentially completed after the colonization of the land by plants (Figure S1). As a result, we propose that plant adaptation to terrestrial habitats has worked as a driving force that provided a basis for modern CDPK complements of embryophytes.
Plant lineages that have progressively developed the capacity to survive in dry habitats obviously required a wide range of structural and physiological adaptations to tolerate the novel stresses associated with life on land. These include enhanced tolerance to desiccation and adaptation to direct UV radiation. Interestingly, abiotic stress responses are among the most frequently reported functions of CDPK homologs in angiosperms [10–12]. Notably, Arabidopsis CDPKs from Group I (CPK4/6/11), Group II (CPK3/21/23), and Group III (CPK10) have been shown genetically and/or biochemically to be involved in a complex regulatory network controlling stomatal movements in response to drought and abscisic acid [51–56]. Given that stomata control the essential balance between water loss and CO2 uptake, this useful structural feature is conserved in all land plants, with the notable exception of liverworts, one of the three bryophyte subgroups (Box 1). Interestingly, recent data confirm that stomata from the bryophyte P. patens and the pteridophyte Selaginella uncinata respond to environmental signals in a comparable way to those of flowering plants [57,58]. These studies also provide strong evidence that intracellular signaling pathways controlling stomatal aperture are similar in both ancestral and modern land plant lineages. Regulation of stomatal aperture by CDPKs could therefore correspond to one of the key functions acquired during the conquest of the land by plants.
CDPK homologs from various plant species have also been frequently involved in the control of biotic stress responses [11,12]. In particular, six Arabidopsis CPDKs from Group I have been shown to play a role in innate immunity. CPK1/2 control the onset of the hypersensitive response (HR) and along with CPK4/11, these PKs phosphorylate NADPH oxidases for reactive oxygen species (ROS) production [20]. Treatments with a bacterial elicitor result in transient activation of CPK4/5/6/11, which work as transcriptional regulators of the convergent signaling network downstream of diverse pattern recognition receptors [59]. The prolonged activation of CPK4/5/6/11 by various bacterial type-III effectors and their cognate NLR (nucleotide-binding domain leucine-rich repeat) intracellular immune sensors leads to the phosphorylation of specific WRKY transcription factors and enhanced plant protection against pathogenic bacteria [20]. Interestingly, some of these defense-related functions (and downstream substrates) appear to be well conserved among Group I CDPKs from various plant species. StCDPK4/5 have for instance been shown to phosphorylate RBOHB, an NADPH oxidase from potato (Solanum tuberosum) [60]. Accordingly, overexpression of an active variant of StCDPK5 results in transgenic plants that have increased ROS production and HR-like symptoms [61]. Recent studies also suggest that the Arabidopsis CPK5/NADPH oxidase activation circuit has a role in rapid defense signaling propagation [62]. In monocots, the rice OsCPK13 (also referred to as CDPK-7) induces rapid cell death, accumulation of pathogenesis-related (PR) proteins, and up-regulation of defense genes when ectopically expressed in Sorghum bicolor [63]. Given that every embryophyte lineage experiences invasions by numerous pathogens, the wide conservation of cellular functions within CDPK homologs from Group I suggests that immune signaling through CDPKs also evolved as a result of the new constraint imposed by colonization of the land by plants.
During the course of evolution, CDPK signaling repertoires of land plants have significantly expanded through gene duplications. It is likely that this phenomenon generated novel CDPK homologs that evolved to achieve highly specialized functions, helping plant species to meet a broader array of lineage-specific requirements. Experimental support for this idea also comes from the study of Arabidopsis, where 10 CDPKs genes (Group I, CPK20/25; Group II, CPK17/19/22/34; Group III, CPK14/24; and Group IV, CPK16/18) show weak or no expression in mesophyll cells [59]. Interestingly, most of these CDPKs are highly and specifically expressed in pollen (data not shown), a situation consistent with a recent report showing that CPK17/34 regulate polarized tip growth in pollen tubes [64]. Taken as a whole, these findings imply that about a third of Arabidopsis CDPKs accomplish specialized reproductive functions, instead of more general vegetative growth functions that could be shared with lower land plants and green algae. Each CDPK signaling repertoire thus probably holds a certain level of speciation, which will not necessarily be captured by the sole investigation of CDPK homologs from model plants.
In light of the exceptional diversity among land plants, our survey included members of the bryophyte, pteridophyte, and angiosperm lineages (Table S1). These plant species were rationally selected because they represent some of the most important milestones characterizing the evolution of embryophytes (Box 1). Land plants belong to the Streptophyta, early members of which diverged from green algae of the Chlorophyta division (Box 1). Streptophyta not only comprise land plants, but also several species of green algae collectively referred to as the streptophyte algae. These organisms are classified in five distinct classes [65,66] and based on different strategies, several studies have suggested that some of these classes are the sister clade of all land plants [67–71]. Although the true sister clade of land plants is still a matter of debate, it is now widely accepted that characteristics enabling successful colonization of terrestrial habitats by the first embryophytes have been inherited from ancient streptophyte algae [65]. Descendants of these lineages therefore occupy a key position with regard to our understanding of the evolution of gene families that facilitated colonization of the land by plants. Unfortunately, the study of CDPK homologs from streptophyte algae has been hindered because the genome sequences from these organisms are not yet available. Once processed, this information should help to better define the transition steps that gradually lead to a remodeling of the CDPK family from green algae, eventually leading to the contemporary CDPK signaling repertoires of embryophytes.
Supplementary Material
Highlights.
Calcium-dependent protein kinases (CDPKs) are conserved proteins
CDPKs combine Ca2+-binding and signaling capabilities
We compare CDPKs from angiosperms and early diverging green plant lineages
Current architecture of the CDPK family was shaped during land colonization by plants
Chlorophyta CDPKs constitute alga-specific phylogenetic groups that apparently kept evolving independently
We suggest that the Chloropyhta–Streptophyta split played a key role in CDPK evolution
Acknowledgements
L.-P.H. was the recipient of a postdoctoral fellowship from the Fonds Québécois de la Recherche sur la Nature et les Technologies (FQRNT). This work was also supported by a grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada to A.S., and an NIH grant (R01 GM70567) to J.S. We apologize to our colleagues whose work could not be cited here due to space limitations.
Footnotes
Supplementary data Supplementary data associated with this article can be found at doi:XXXXXXX’
Disclosure statement The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kudla J, et al. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler GL, Brownlee C. Ca2+ signalling in plants and green algae-changing channels. Trends Plant Sci. 2008;13:506–514. doi: 10.1016/j.tplants.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Batistič O, Kudla J. Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta. 2012;1820:1283–1293. doi: 10.1016/j.bbagen.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto K, Kudla J. Calcium decoding mechanisms in plants. Biochimie. 2011;93:2054–2059. doi: 10.1016/j.biochi.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 5.McCormack E, et al. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005;10:383–389. doi: 10.1016/j.tplants.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Perochon A, et al. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie. 2011;93:2048–2053. doi: 10.1016/j.biochi.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Batistič O, Kudla J. Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta. 2009;1793:985–992. doi: 10.1016/j.bbamcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Luan S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14:37–42. doi: 10.1016/j.tplants.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Weinl S, Kudla J. The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 2009;184:517–528. doi: 10.1111/j.1469-8137.2009.02938.x. [DOI] [PubMed] [Google Scholar]
- 10.Asano T, et al. CDPK-mediated abiotic stress signaling. Plant Signal. Behav. 2012;7:817–821. doi: 10.4161/psb.20351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boudsocq M, Sheen J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013;18:30–40. doi: 10.1016/j.tplants.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liese A, Romeis T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK) Biochim. Biophys. Acta. 2013;1833:1582–1589. doi: 10.1016/j.bbamcr.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 13.Zhang XS, Choi JH. Molecular evolution of calmodulin-like domain protein kinases (CDPKs) in plants and protists. J. Mol. Evol. 2001;53:214–224. doi: 10.1007/s002390010211. [DOI] [PubMed] [Google Scholar]
- 14.Christodoulou J, et al. Evidence for differing roles for each lobe of the calmodulin-like domain in a calcium-dependent protein kinase. J. Biol. Chem. 2004;279:29092–29100. doi: 10.1074/jbc.M401297200. [DOI] [PubMed] [Google Scholar]
- 15.Rutschmann F, et al. LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 2002;129:156–168. doi: 10.1104/pp.000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weljie AM, et al. Conformational changes in the Ca2+-regulatory region from soybean calcium-dependent protein kinase-alpha: fluorescence resonance energy transfer studies. J. Biol. Chem. 2003;278:43764–43769. doi: 10.1074/jbc.M306799200. [DOI] [PubMed] [Google Scholar]
- 17.Weljie AM, Vogel HJ. Unexpected structure of the Ca2+-regulatory region from soybean calcium-dependent protein kinase-alpha. J. Biol. Chem. 2004;279:35494–35502. doi: 10.1074/jbc.M311520200. [DOI] [PubMed] [Google Scholar]
- 18.Harper JF, et al. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004;55:263–288. doi: 10.1146/annurev.arplant.55.031903.141627. [DOI] [PubMed] [Google Scholar]
- 19.Curran A, et al. Calcium-dependent protein kinases from Arabidopsis show substrate specificity differences in an analysis of 103 substrates. Front. Plant Sci. 2011;2:36. doi: 10.3389/fpls.2011.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X, et al. Bifurcation of Arabidopsis NLR immune signaling via Ca2+-dependent protein kinases. PLoS Pathog. 2013;9:e1003127. doi: 10.1371/journal.ppat.1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng SH, et al. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002;129:469–485. doi: 10.1104/pp.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asai S, et al. The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J. Biol. Chem. 2013;288:14332–14340. doi: 10.1074/jbc.M112.448910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito T, et al. Alteration of substrate specificity: the variable N-terminal domain of tobacco Ca2+-dependent protein kinase is important for substrate recognition. Plant Cell. 2010;22:1592–1604. doi: 10.1105/tpc.109.073577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asano T, et al. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 2005;46:356–366. doi: 10.1093/pcp/pci035. [DOI] [PubMed] [Google Scholar]
- 25.Li AL, et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.) Plant Mol. Biol. 2008;66:429–443. doi: 10.1007/s11103-007-9281-5. [DOI] [PubMed] [Google Scholar]
- 26.Ma P, et al. Genome-wide identification of the maize calcium-dependent protein kinase gene family. Appl. Biochem. Biotechnol. 2013;169:2111–2125. doi: 10.1007/s12010-013-0125-2. [DOI] [PubMed] [Google Scholar]
- 27.Zuo R, et al. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa) Mol. Biol. Rep. 2013;40:2645–2662. doi: 10.1007/s11033-012-2351-z. [DOI] [PubMed] [Google Scholar]
- 28.Martín ML, Busconi L. Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 2000;24:429–435. doi: 10.1046/j.1365-313x.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- 29.Harmon AC, et al. A calcium-dependent but calmodulin-independent protein kinase from soybean. Plant Physiol. 1987;83:830–837. doi: 10.1104/pp.83.4.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Billker O, et al. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe. 2009;5:612–622. doi: 10.1016/j.chom.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harper JF, Harmon A. Plants, symbiosis and parasites: a calcium signalling connection. Nat. Rev. Mol. Cell Biol. 2005;6:555–566. doi: 10.1038/nrm1679. [DOI] [PubMed] [Google Scholar]
- 32.Hrabak EM, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132:666–680. doi: 10.1104/pp.102.011999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banks JA, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 35.Blanc G, et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012;13:R39. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanc G, et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derelle E, et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palenik B, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prochnik SE, et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 42.Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:1178–1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Bel M, et al. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 2012;158:590–600. doi: 10.1104/pp.111.189514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boudsocq M, et al. Characterization of Arabidopsis calcium-dependent protein kinases: activated or not by calcium? Biochem. J. 2012;447:291–299. doi: 10.1042/BJ20112072. [DOI] [PubMed] [Google Scholar]
- 45.Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Pinontoan R, et al. Cloning of a cDNA encoding a 66-kDa Ca2+-dependent protein kinase (CDPK) from Dunaliella tertiolecta (Chlorophyta) J. Phycol. 2000;36:545–552. doi: 10.1046/j.1529-8817.2000.99185.x. [DOI] [PubMed] [Google Scholar]
- 47.Hamel LP, et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11:192–198. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 48.Dóczi R, et al. Exploring the evolutionary path of plant MAPK networks. Trends Plant Sci. 2012;17:518–525. doi: 10.1016/j.tplants.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Östlund G, et al. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamel LP, et al. Mitogen-Activated Protein Kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant Cell. 2012;24:1327–1351. doi: 10.1105/tpc.112.096156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brandt B, et al. Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. USA. 2012;109:10593–10598. doi: 10.1073/pnas.1116590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geiger D, et al. Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 2011;4:ra32. doi: 10.1126/scisignal.2001346. [DOI] [PubMed] [Google Scholar]
- 53.Geiger D, et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA. 2010;107:8023–8028. doi: 10.1073/pnas.0912030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mori IC, et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 2006;4:e327. doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu SY, et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell. 2007;19:3019–3036. doi: 10.1105/tpc.107.050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou JJ, et al. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2+-mediated stomatal regulation in response to drought stress. Plant Physiol. 2010;154:1232–1243. doi: 10.1104/pp.110.157545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruszala EM, et al. Land plants acquired active stomatal control early in their evolutionary history. Curr. Biol. 2011;21:1030–1035. doi: 10.1016/j.cub.2011.04.044. [DOI] [PubMed] [Google Scholar]
- 58.Chater C, et al. Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr. Biol. 2011;21:1025–1029. doi: 10.1016/j.cub.2011.04.032. [DOI] [PubMed] [Google Scholar]
- 59.Boudsocq M, et al. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kobayashi M, et al. Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell. 2007;19:1065–1080. doi: 10.1105/tpc.106.048884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobayashi M, et al. StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol. 2012;196:223–237. doi: 10.1111/j.1469-8137.2012.04226.x. [DOI] [PubMed] [Google Scholar]
- 62.Dubiella U, et al. Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA. 2013;110:8744–8749. doi: 10.1073/pnas.1221294110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mall TK, et al. Expression of the rice CDPK-7 in Sorghum: molecular and phenotypic analyses. Plant Mol. Biol. 2011;75:467–479. doi: 10.1007/s11103-011-9741-9. [DOI] [PubMed] [Google Scholar]
- 64.Myers C, et al. Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 2009;59:528–539. doi: 10.1111/j.1365-313X.2009.03894.x. [DOI] [PubMed] [Google Scholar]
- 65.Becker B. Snow ball earth and the split of Streptophyta and Chlorophyta. Trends Plant Sci. 2013;18:180–183. doi: 10.1016/j.tplants.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 66.Guiry MD. How many species of algae are there? J. Phycol. 2012;48:1057–1063. doi: 10.1111/j.1529-8817.2012.01222.x. [DOI] [PubMed] [Google Scholar]
- 67.Laurin-Lemay S, et al. Origin of land plants revisited in the light of sequence contamination and missing data. Curr. Biol. 2012;22:R593–R594. doi: 10.1016/j.cub.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 68.Timme RE, et al. Broad phylogenomic sampling and the sister lineage of land plants. PLoS One. 2012;7:e29696. doi: 10.1371/journal.pone.0029696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turmel M, et al. The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol. Biol. Evol. 2006;23:1324–1338. doi: 10.1093/molbev/msk018. [DOI] [PubMed] [Google Scholar]
- 70.Turmel M, et al. The green algal ancestry of land plants as revealed by the chloroplast genome. Int. J. Plant Sci. 2007;23:679–689. [Google Scholar]
- 71.Wodniok S, et al. Origin of land plants: do conjugating green algae hold the key? BMC Evol. Biol. 2011;11:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura K, et al. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 73.Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adl SM, et al. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Becker B, Marin B. Streptophyte algae and the origin of embryophytes. Ann. Bot. 2009;103:999–1004. doi: 10.1093/aob/mcp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leliaert F, et al. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012;31:1–46. [Google Scholar]
- 77.Lewis LA, McCourt RM. Green algae and the origin of land plants. Am. J. Bot. 2004;91:1535–1556. doi: 10.3732/ajb.91.10.1535. [DOI] [PubMed] [Google Scholar]
- 78.Douzery EJ, et al. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl. Acad. Sci. USA. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hedges SB, et al. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yoon HS, et al. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 2004;21:809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 81.Zimmer A, et al. Dating the early evolution of plants: detection and molecular clock analyses of orthologs. Mol. Genet. Genomics. 2007;278:393–402. doi: 10.1007/s00438-007-0257-6. [DOI] [PubMed] [Google Scholar]
- 82.Gensel PG. The earliest land plants. Annu. Rev. Ecol. Evol. Syst. 2008;39:459–477. [Google Scholar]
- 83.Kenrick P, et al. A timeline for terrestrialization: consequences for the carbon cycle in the Palaeozoic. Philos. Trans. R. Soc. B. Biol. Sci. 2012;367:519–536. doi: 10.1098/rstb.2011.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanderson MJ, et al. Molecular evidence on plant divergence times. Am. J. Bot. 2004;91:1656–1665. doi: 10.3732/ajb.91.10.1656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.