Abstract
Pax5 is an alternatively spliced transcription factor that regulates B cell development and activation. The function of specific Pax5 isoforms is unknown. Here we report the existence of seven alternatively spliced isoforms of Pax5 in the rainbow trout. We hypothesized that B cells differentially express specific Pax5 isoforms as a means of modulating Pax5 activity during cell maturation. Flow cytometric analyses using Pax5-specific antibodies recognizing the paired domain, a central (exon 6-encoding) domain, or the C-terminus, revealed the existence of distinct Pax5-expressing cell populations in trout immune tissues. Additionally, using the transcription factor EBF, we show that Pax5 isoforms lacking a paired domain are already expressed at the earliest stages of trout (B) lymphopoiesis, and unexpectedly, that minor populations of such cells reside in blood and spleen. These data support use of differentially expressed Pax5 isoforms to identify novel B cell subsets in the form of Pax5 tissue signatures, and as such, provides new biomarkers for malignancy, infectious disease, and disease resistance in trout and humans.
Keywords: Alternative splicing, Pax5, B cell development, teleost
Introduction
The expression and function of Pax5 isoforms remains an enigma in the immunological community. Thus far, multiple Pax5 isoforms have been identified, many of which are conserved across species (Adams et al., 1992; Arseneau et al., 2009; Short and Holland, 2008; Zwollo et al., 1997). Despite these gains, research has yet to demonstrate a definitive pattern of Pax5 isoforms across cell lines, immune tissues, and malignancies, and there is still no clear biological function for isoform expression. B cell differentiation, given its complex developmental pathway, provides an excellent system to investigate possible functions for alternative Pax5 isoforms.
While previous research has extensively examined mammalian B cell development, researchers have only recently made significant insights into the teleost B cell system (Barr et al., 2011; Bromage et al., 2004; Kaattari and Irwin, 1985; Kaattari et al., 1986; Zwollo et al., 2005; Zwollo et al., 2008; Zwollo et al., 2010). Teleosts possess molecular and cellular components similar to mammals while retaining a unique system of immune cell development. In contrast to mammals, teleosts lack bone marrow and alternatively utilize the anterior kidney (K1) as their primary immune organ (Zapata and Cooper, 1990; Zwollo et al., 2005). Similar to mammalian B cells, mature teleost B cells reside in the spleen (SPL) where they encounter antigen and differentiate into short-lived plasmablasts and plasma cells (Bromage et al., 2004). A small subset of these terminally differentiated, antibody-secreting cells may migrate back to the anterior kidney (K1) where they reside as long-lived plasma cells (Bromage et al., 2004). Research characterizing the similarities and differences between mammalian and teleost immune systems provides a greater understanding of both general immune system function and alternative immune system approaches.
Although the surfeit of mammalian serological reagents is unavailable for deciphering teleost stages of B cell development, recent research has employed suitable alternatives (Barr et al., 2011; Zwollo et al., 2005; Zwollo et al., 2008; Zwollo et al., 2010). Conserved transcription factor expression (see Table I) demarcates stages of B cell development in both mammals and teleosts (Zwollo, 2011). Early developing trout B cells can be characterized by the expression of the transcription factor Early B cell Factor (EBF) and the recombinase RAG1 (Zwollo, 2011). Late developing B cells lack EBF and RAG1, but express the transcription factor Pax5 and membrane immunoglobulin M (IgM), while terminally differentiated plasma cells lack Pax5 and membrane IgM, and secrete high levels of IgM (Zwollo et al., 2010). Plasmablasts continue to express Pax5 and IgM, and can be detected using proliferation marker EdU (Barr et al., 2011). Additionally, a small subset of B cells express IgT or IgD (Flajnik, 2002; Hansen et al, 2005; Ramirez-Gomez et al., 2011).
Table I. Phenotypes of developing trout B cells and putative expression patterns of developmental markers.
| PD | E6 | E10 | FSC | Proliferation | EBF | mu | ||
|---|---|---|---|---|---|---|---|---|

|
CLP/pro-B | low | ++ | low | high | + or − | + | − |
|
Large pre-B? | low | + | + | int. | + or − | + | low |

|
Late Pre-B/(im)mat B | + | + | + | low | − | low | + |

|
Act. B | ++ | ++ | ++ | int. | − | − | ++ |
|
PB | ++ | ++ | ++ | int. | + | − | ++ |

|
Unnamed | low | low | + | low | − | − | + |
Notes. CLP, common lymphocyte progenitor; Act. B, activated B cell; PB, plasmablast; FSC, forward scatter light (cell size); Proliferation, cell proliferation through EdU incorporation (see Materials and Methods). +, positive; ++, very high; -, not detected; int., intermediate. Colored arrows correspond with specific subpopulations throughout results. Shaded regions refer to Pax5-specific antibodies.
Pax5 is a highly complex gene with a large number of target genes (McManus et al., 2011), and as such, it is not surprising that it displays extensive alternative RNA splicing in order to maximize functional specificity (Zwollo, 2011; Zwollo et al., 1997). Genes encoding transcription factors commonly utilize the mechanism of alternative RNA splicing because their exons form functionally independent domains, including DNA binding-, transactivation-, regulatory, and repression-domains (Zwollo, 2011). Removal of one or more functional domains through alternative RNA splicing provides a highly efficient mechanism to modulate a transcription factor's activity.
Alternatively spliced Pax5 isoforms have been detected in multiple vertebrate species, supporting their conserved function (Arseneau et al., 2009; Short and Holland, 2008; Short et al, 2012; Zwollo et al., 1997). Most strikingly is the cross-species conservation of Pax5 isoforms lacking exon 2 (Δ2); such isoforms have been detected in all species studied so far, including mouse, amphioxus (which has Pax258), Xenopus, and human (Arseneau et al., 2009; Short and Holland, 2008; Short et al., 2012; Zwollo et al., 1997). We have shown that murine Δ2 Pax5 proteins are unable to interact with their DNA binding sites on target genes in vitro (Zwollo et al., 1997), and may function as co-repressors or -activators (Lowen et al., 2001; Zwollo et al., 1997). In addition, Pax5 isoforms that exclude exons 7, 8, and/or 9 (δ7, δ8, and/or δ9) have been detected in humans (Robichaud et al., 2004) and amphioxus (Short and Holland, 2008), reportedly altering their transactivating potential. Lastly, Pax5 isoforms that lack exons 6 through 10 have been reported in mice and humans (Robichaud et al., 2004; Zwollo et al., 1997). In mouse, deletions of exon 6 of Pax5 remove an octamer motif that interacts with Groucho proteins to inhibit gene transcription (Eberhard et al., 2000) and deletions in exon 10 result in Pax5 isoforms lacking part of an inhibitory domain (Dorfler and Busslinger, 1996).
While roles for full-length Pax5 have been described extensively, little is known about the potential functions of alternatively spliced Pax5 isoforms. Previous studies have been limited in their ability to correlate Pax5 isoforms with specific B cell stages, either at the RNA level (RT-PCR) or protein level (western blot analysis), due to the use of pooled tissue cells (Arseneau et al., 2009; Robichaud et al., 2004). As an alternative to elucidate possible functions for Pax5 isoforms, we have developed a flow cytometric approach with antibodies recognizing differentially expressed transcription factors in rainbow trout B cells (Barr et al., 2011; Zwollo et al., 2005; Zwollo et al., 2008; Zwollo et al., 2010). This has allowed us to differentiate between early developing B, late developing B, and antibody-secreting cells, as characterized through specific flow cytometric patterns or “B-cell signatures” (Zwollo et al., 2010). We use this approach here, hypothesizing that specific, alternatively spliced Pax5 isoforms are (transiently) present during B cell development and/or activation as a means of modulating Pax5 activity. Our goal was to define trout B cell subpopulations based on their combinatorial staining patterns for three functional Pax5 domains.
Using PCR and cloning techniques, we first show that at least seven alternative Pax5 splice forms are expressed in immune tissues of rainbow trout. Next, using flow cytometric analysis, we demonstrate that early developing B, late developing B, activated B cells, and plasmablasts, differentially express three Pax5 domains and that the pattern of Pax5 domain expression differs between immune tissues. We refer to these specific tissue patterns as “Pax5 signatures” (Zwollo, 2011). Lastly, we reveal that Pax5 isoforms lacking exon 2 are expressed in early B cell progenitors in trout anterior kidney, and show that a small population of such early developing B cells is also present in trout blood and spleen.
Materials and Methods
Animals and facilities
Outbred adult rainbow trout (Onchorhynchus mykiss, 20-30 cm) were from The Virginia Institute of Marine Science/The College of William and Mary Trout Facility. Fish were maintained in 100-gallon tanks with a recirculating system employing biologically-filtered water at 12° C.
Isolation of immune cells
Fresh trout kidney cells were obtained using methods previously described (Zwollo et al., 2005). Briefly, the trout kidney was divided into subsections of seven vertebrae beginning at the posterior end (K5) and ending in the anterior kidney (K1). K1 and K5 tissues were collected in 5 mL sterile HBSS (137 mM NaCl, 5.6 mM d-glucose, 5 mM KCl, 8.1 mM Na2HP04·2H20, and 20 mM Hepes at pH 7.05). Single cell suspensions were created by forcing cells through a 10 mL syringe followed by filtration through a 40 nm nylon filter (Falcon; BD Biosciences). Next, cells were then pelleted at 250 g for 10 minutes and resuspended in cold HBSS. Cells were then either prepared for culturing (see cell culture and mitogens) or washed in 1× PBS (1.9 mM NaH2P04·H20, 8.1 mM Na2HP04·7H20, 137 mM NaCl, and 2.6 mM KCl, pH 7.4) containing 0.02% sodium azide in preparation for fixation (see Fixation), or frozen at −80° C for RNA analysis. Blood cells were washed in cold HBSS and layered onto Histopaque 1077 cushions (Sigma Aldrich) and spun at 500 g at 4° C for 45 minutes. The peripheral blood lymphocyte (PBL) layer was removed and cells were either washed in cold HBSS for culturing or in PBS containing 0.02% azide for fixation, or pellets frozen at −80° C.
Isolation and cloning of trout Pax5 splice forms
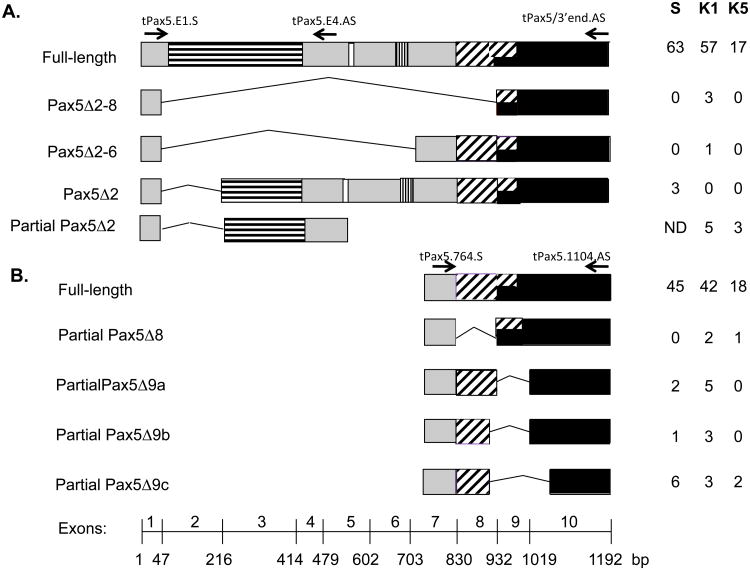
RNA was purified from SPL, K1, or K5 tissue using TRIzol Reagent (Invitrogen). cDNA was synthesized using iScript (BioRad). Alternative splice forms were PCR-amplified as described previously (Zwollo et al., 2005; Zwollo et al., 2008). Primers used for detection of full-length, Δ2, Δ2-6, and Δ2-8 were tPax5.E1.S, tPax5/3′end.AS, and tPax5.E4AS, or tPax5-E1/3.S and tPax5-E3/4.AS (Supplemental Table I). Primers for Δ8 and Δ9 isoforms were tPax5.764.S and tPax5.1104.AS (Supplemental Table I). The PCR products were cloned (without purification of individual bands) into the PCR-cloning vector pSC-A-amp/according to the manufacturer's instructions using a StrataClone PCR Cloning kit (Stratagene Inc.) Clones were collected and plasmid DNA isolated. DNA sequences were determined by the Iowa State DNA sequence facility. Sequences were analyzed using McVector software. To obtain information on the relative abundance of each isoform, the resulting sequences were then grouped by Pax5 sequence. The relative abundance of each isoform is shown in Figure 1 (right side).
Figure 1.
Alternatively spliced Pax isoforms isolated from immune cells of the raibow trout. The paired domain is indicated with horizontal stripes, the octamer sequence is in white, the homeodomain homology region is indicated with vertical stripes, the transactivation domain has diaginal stripes, and the repressor domain is in black. Numbers at the bottom indicates exons 1-10 with nucleotide position for each exon boundary indicated. Arrows indicate primers (Supplementary Table I). Primer names are listed above arrows. Number of clones for each splice form and tissue from which it was isolated, shown on the right. (A) Results using PCR primers targeting exon 2-lacking isoforms. (B) Results using primers targeting exons 7-10. ND, no data; bp, base pair. S, spleen; K1, anterior kidney; K5, posterior kidney.
Cell culture and mitogens
Freshly isolated trout PBL and SPL cells were cultured at 1.0×10̂7 cells/mL in trout culture medium (TCM) consisting of RPMI 1640 with 10 mM L-glutamine, 10% FCS, 50 μg/mL gentamicin, 50 μM 2-ME, and the nucleosides adenosine, uracil, cytosine, and guanosine (final concentration 10 μg/mL; Sigma-Aldrich). Cells were maintained at 18° C in the presence of blood gas and fed every other day with one tenth of the culture volume of a 10× tissue culture cocktail containing 500 μg/mL gentamycin, 10× essential aas, 10× non-essential aas, 70 mM l-glutamine, 70 mg/mL dextrose, 10× nucleosides, and 33% fetal bovine serum (FBS). B lymphocytes were activated using mitogen LPS (from Escherichia coli 055:B5, pasteurized for 30 min at 70° C in distilled water) at 100 μg/mL. For cell proliferation analysis, cells were incubated in the presence of 3 μM 5-ethynyl-2′-deoxyuridine (EdU; Invitrogen) for 17 hours immediately before collection.
Antibodies
The polyclonal anti-paired domain (Pax5.PD) antibody (previously called ED-1) recognizes the paired domain of vertebrate Pax5 and has been described previously (Zwollo et al., 1998). The rabbit anti-trout Pax5.E6 polyclonal antibody was raised against the peptide HGPGGRDFLRKQMRGDLF (aas 200-217 of trout Pax5). The rabbit anti-trout Pax5.E10 polyclonal antibody was raised against the peptide AASRGAGPAATATASAYDRH (aas 366-385 of trout Pax5). Pax5-E6 and Pax5-E10 antibodies were generated by GenScript Inc. The monoclonal mouse anti-trout immunoglobulin heavy chain mu (HCmu I-14) and was a generous gift from Dr. Greg Warr. The polyclonal rabbit-anti-human EBF IgG (H300; detecting aas 1–300 of EBF1, and reactive with mouse/human EBF2, 3, and 4) was purchased from Santa Cruz Biotech. Isotype control antibodies included rabbit IgG or mouse IgG (eBiosciences) conjugated to Alexa Fluor 555 or Alexa Fluor 647. For flow cytometric analyses, unlabeled antibodies were conjugated (Alexa Fluor 555 and/or Alexa Fluor 647) using protein-labeling kits according to manufacturer's instructions (Molecular Probes/Invitrogen Inc). Antibody aliquots were stored in 1% BSA at −20° C.
Fixation, permeabilization, and flow cytometry
Cell pellets were fixed in 1% ice-cold paraformaldehyde (10% stock, EM-grade; Electron Microscopy Sciences) and permeabilized in 1 mL ice-cold 80% methanol, as described previously (Zwollo et al., 2010). After overnight incubation at −20° C, cells were resuspended in permeabilizing solution (BD perm wash in PBS, BD Biosciences) and stained as described previously (Zwollo et al., 2010). For three-color flow cytometry, an indirect staining approach was used for the third antibody by staining cells with a biotinylated HCmu (Warrs) in conjunction with streptavidin APC-750 (Sigma). For cell proliferation analysis, the nucleoside analog EdU and a Click-It kit (Invitrogen) was used according to manufacturer's instructions, as described previously (Barr et al., 2011). Approximately 30,000 events were acquired per sample using a BD FACSArray (BD Biosciences). Duplicate samples were analyzed for each experiment. Experiments were repeated a minimum of three times. Contour graphs were generated using WinMDI 2-8 (J. Trotter 1993–1998) software using a 2/0.1 threshold/smooth setting. Contour graphs are shown as log algorithms with intervals of 50%. Statistics were obtained from WinMDI 2-8 software. Arithmetic means of population frequencies and standard errors were calculated for each experiment using Microsoft Excel software. Fixed and stained cell populations were sorted using a FACSCalibur. Between 5,000 and 15,000 cells were collected in PBS + SA. Cells were centrifuged after collection and cell pellets stored at −80° C until RNA isolation (below).
Isolation of RNA from paraformaldehyde-fixed cells, endpoint PCR, and DNA sequencing
RNA was isolated from paraformaldehyde-fixed, sorted cells using an RNeasy FFPE kit (Qiagen Inc.) to reverse RNA formaldehyde modification, according to the manufacturer's instructions. Approximately 50-100 ng of purified RNA was used to generate 20 μL cDNA using iScript random labeling cDNA kit (BioRad), and 2-5 μL of this cDNA was used in a 25 μL volume end-point PCR using the Platinum PCR SuperMix (Invitrogen Inc.). To amplify Pax5 Δ2 cDNA, we used a sense primer flanking exons 1 and 3 (tPax5-E1/3.S; Supplemental Table I) together with an anti-sense primer flanking exons 3 and 4 (tPax5-E3/4.AS; Supplemental Table I). Each PCR run was carried out on an ABI-Veriti 96-well Fast Thermocylcler (ABI) and consisted of 2 min. at 95° C, 35 cycles of denaturing at 95° C for 15 sec, annealing at 56° C for 15 sec, and extension at 72° C for 15 sec, and ending with 7 min. at 72° C. Lastly, 10 μL of the reaction was analyzed on a 2% agarose gel. For detection of exon 6-containing Pax5 cDNA, the following two primers were used in endpoint PCR as above: tPax5.485.S and tPax5.680AS (Supplemental Table I). To verify that the PCR products contained Pax5 sequence, PCR products were cleaned up using USB Affymetrix ExoSAP-IT PCR Product Clean-Up kit according to manufacturer's instructions. Cleaned-up sample was then used as a template in ABI BigDye v3.1 sequencing reactions. Reaction products were purified using Performa Gel-Filtration Cartridges (EdgeBio). Finally, samples were resuspended in ABI HiDi Formamide, and sequence products were resolved on an ABI3500 Genetic Analyzer and analyzed using ABI 5.4 Sequence Analysis Software.
qRT-PCR analysis
1 μg of total RNA was used to make 20 μL of cDNA using the iScript kit (BioRad). Quantitative real-time PCR was performed using a PerfeCTa SYBR Green SuperMix, ROX PCR kit (Quanta Biosciences). Each sample was measured in triplicate. To determine the amount of Pax5 δ2 cDNA, 1 mL (3 mM) of tPax5-E1/3.S (Supplemental Table I) was used together with 1 mL (3mM) tPax5-E3/4AS (Supplemental Table I), and 1.50 mL cDNA. Trout α-tubulin (Tubulin 56; Supplemental Table 1) was used as a reference. The reaction was performed on an Applied Biosystems StepOne Real-Time PCR instrument. Specificity of the Pax5 primers was confirmed by sequencing real-time PCR fragments using capillary electrophoresis on ABI 3130 Avant Genetic Analyzer. Sequences were analyzed on ABI sequencing software version 5.1. An NCBI Blast tool was used to align obtained sequences with the published Oncorhynchus sp. sequence data. Expression levels were calculated as fold-change relative to the reference samples. The fold change, or amount of target, was calculated according to the Fold Change = 2−ΔΔCT equation (Livak and Schmittgen, 2001).
Western blot analysis
Cell suspensions were centrifuged in aliquots of 1.5 × 105 cells and pellets quick-frozen at −80° C. As negative control sample, 1.5 × 105 splenic cells from 50%-Percoll-gradients were used as described elsewhere (Zwollo et al., 2005). Whole-cell protein lysates were prepared by resuspending cells in 40 μL of a sample buffer containing 5% 2-ME and proteins separated by size using denaturing 12% SDS-PAGE gels. Proteins were transferred onto a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore). Membranes were incubated in blocking solution of 5% dry milk in PBS containing 0.5% Tween (PBST) for 1 h, followed by a 2 h incubation in blocking solution in the presence of primary antibody: Pax5.PD (ED-1; 0.5 μg/mL final dilution), Pax5.E10 (1 μg/mL final dilution) or Pax5.E6 (1 μg/mL). A rabbit-anti-human α-tubulin IgG (H300; Santa Cruz Inc., which also cross-reacts with zebrafish and Xenopus α-tubulin) was used at 0.1 μg/mL as a control for protein quality and loading, by stripping the filters with 0.2N NaoH, followed by re-probing the filter. After primary staining with each antibody, four 5 min washes in PBST were then followed by a 1 h incubation with the secondary antibody goat anti-rabbit IgG-HRP conjugate (0.1 μg/mL; Zymed Laboratories) in blocking solution and membranes were washed four more times for 5 minutes each in PBST. Filters were developed using a chemiluminescence kit (ECL-Plus; GE Healthcare Life Sciences).
Results
The trout Pax5 gene is alternatively spliced
Seven different Pax5 splice forms (not including full-length Pax5) were isolated and cloned from trout anterior kidney (K1), posterior kidney (K5), and/or spleen (SPL), as shown in Figure 1. Using a sense primer (tPax5.E1.S; Supplemental Table I) in combination with an anti-sense primer targeting exon 10 (tPax5/3′end.AS), three alternative spliced isoforms were detected in addition to full-length Pax5: one that lacked exons 2-8 (Pax5Δ2-8), one that lacked exons 2-6 (Pax5Δ2-6), and one that lacked exon 2 (Pax5Δ2). With this primer set, the latter splice form was detected in SPL, but not K1 or K5. To determine if any exon 2-lacking isoforms were present in the kidney, we then used the same sense primer targeting exon 1 (tPax5.E1.S) in combination with an anti-sense primer targeting exon 4 (tPax5.E4AS). This resulted in detection of Pax5Δ2 clones in anterior and posterior kidney, suggesting that in the kidney, Pax5Δ2 isoforms lack exon 10. This exon has been reported to encode a transcriptional repressor domain in murine Pax5 (Dorfler and Busslinger, 1996). The number of clones isolated for each specific isoform, and the tissues from which they were isolated, are listed in Figure 1.
Using primers targeting the region covering exons 7-10 (tPax5.764.S and tPax5.1104.AS), four different in-frame Pax5 splice forms were detected (Figure 1B), and included isoforms that lacked exon 8 (Pax5Δ8), exon 9 (Pax5Δ9a), the 3′-end of exon 8 plus exon 9 (Pax5Δ9b), and lastly, a fairly abundant isoform (Pax5Δ9c) which lacked exon 9 as well as the 5′ part of exon 10. Both Pax5Δ9b and Pax5δ9c used alternative internal splice sites, similar to what had been observed in Amphioxus (Short and Holland, 2008). Approximate predicted protein sizes for the isoforms were as follows: Pax5Δ2, 35 kD; Pax5Δ2-6, 22 kD; Pax5Δ2-8, 12 kD; the C-terminal isoforms Pax5Δ8, Pax5Δ9a, Pax5Δ9b, and Pax5Δ9c around 42-44 kDs. Note that the isoform lacking exon 2 (Pax5Δ2; GenBank NM-001124682) is predicted (based on murine studies) to use a second, internal start-codon close to the 3′-side of exon 3, resulting in a protein that lacks all sequence encoded by exon 2 and most of the sequence encoded by exon 3 (Lowen et al., 2001; Zwollo et al., 1997). Pax5 isoforms lacking either exon 1 or exon 10 were not investigated in this study. No isoforms lacking exon 7 were detectable.
Antibodies tPax5.E6 and tPax5.E10 both recognize trout Pax5 protein
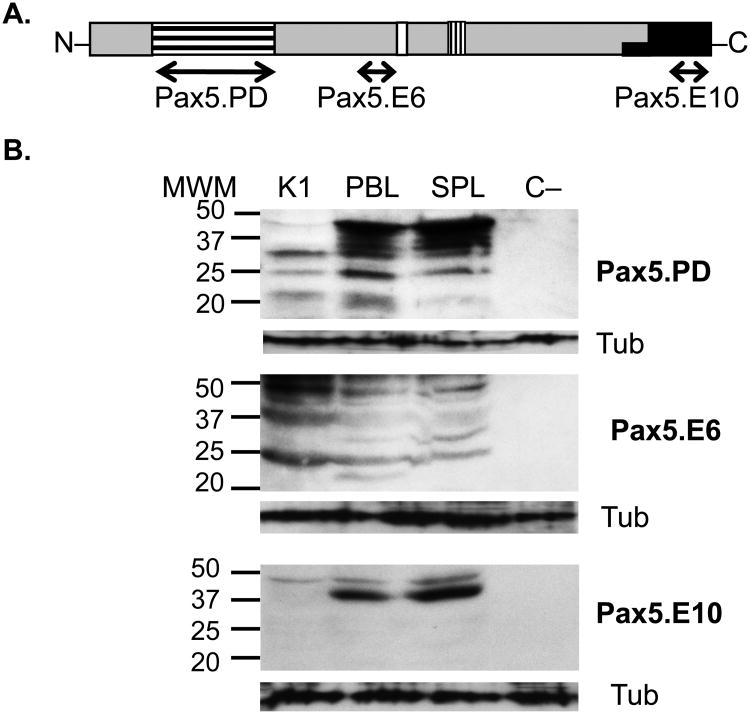
To begin investigating expression patterns of alternatively spliced Pax5 proteins in trout B cell subpopulations, three anti-Pax5 antibodies were used, each recognizing a different Pax5 domain (shown in Figure 2A). The Pax5.PD antibody recognizes the paired domain of Pax5 and had been used previously (Barr et al, 2011; Zwollo et al., 2005; Zwollo et al., 2008; Zwollo et al, 2010; Zwollo et al., 1998). A second anti-Pax5 antibody, Pax5.E6, was generated against a region encoded within exon 6 of trout Pax5 just upstream from the partial homeodomain (Figure 2A). The third antibody, Pax5.E10, was designed to recognize the C-terminus of Pax5, encoded by exon 10 (Figure 2A).
Figure 2.
Trout Pax5 antibody specificity. (A) Pax5.PD antibody recognizes the paired domain of Pax5 (encoded by exons 2-3); Pax5.E6 recognizes a region encoded by exon 6; Pax5.E10 recognizes the C-terminus encoded by exon 10 (repressor domain). Labeling of domains as in Figure 1. (B) Western blot analysis on trout immune cells using three Pax5-specific antibodies. Left: MWM, molecular weight markers, in kD. Cell lysate from 1.5 × 105 cells were loaded per well. Tub (α-tubulin) staining shown underneath each blot, detected by stripping and reprobing of the same filter. SPL, spleen; PBL, peripheral blood lymphocytes; K1, anterior kidney, C-, negative control cells (SPL 50%Percoll; see Methods).
To verify that the antibodies recognized trout Pax5 protein, cell lysates from trout SPL, PBL, and K1 were analyzed by western blot analysis. Figure 2B shows that all three antibodies recognize multiple proteins with the largest and most abundant protein being slightly smaller than 50 kD in all three tissues, in agreement with this being the full-length form of trout Pax5 (Zwollo et al., 2005). The smaller size bands represent either partially degraded Pax5 protein, and/or Pax5 splice forms, although western blot cannot distinguish between these two possibilities. This high sensitivity to proteolysis has previously been reported for murine Pax5 proteins (Anspach et al., 2001; Lowen et al., 2001).
Using the Pax5.E10 antibody, we observed patterns which suggest the majority of Pax5 proteins containing the C-terminal repressor domain are full length. However, the overall Pax5 signal using either Pax5.PD or Pax5.E10 antibodies stained more weakly in K1 cells compared to SPL and PBL. This was expected based on the distribution of B cells in each tissue, as determined previously by flow cytometry (Zwollo et al., 2010). In contrast, the Pax5.E6 antibody stained at least as strongly in K1 as in PBL and SPL, which would suggest it is expressed more abundantly in developing B cells, as K1 is a primary immune organ. A negative control cell sample (C-) that lacked Pax5-expressing cells as reported previously (50% Percoll-purified splenic cells; (Zwollo et al., 2005) did not stain with any of the three Pax5 antibodies, as expected.
Together, the western blot results demonstrate that all three trout-Pax5 antibodies recognized trout Pax5 protein. The Pax5.PD antibody should therefore detect isoforms Pax5Δ8, Pax5Δ9a Pax5Δ9b and Pax5Δ9c, but not (or very weakly) exon-2 lacking isoform Pax5Δ2, and not Pax5Δ2-6, and Pax5Δ2-8. The Pax5.E6 antibody should detect isoforms that lack the paired domain or the C-terminus (encoded by exon 10), but should not stain with Pax5Δ2-6 or Pax5Δ2-8. The Pax5.E10 antibody should detect all isoforms except those with a deletion in exon 10, or isoforms lacking exon 7 (which results in a frame-shift).
Patterns of Pax5-expression using three Pax5 antibodies in flow cytometry
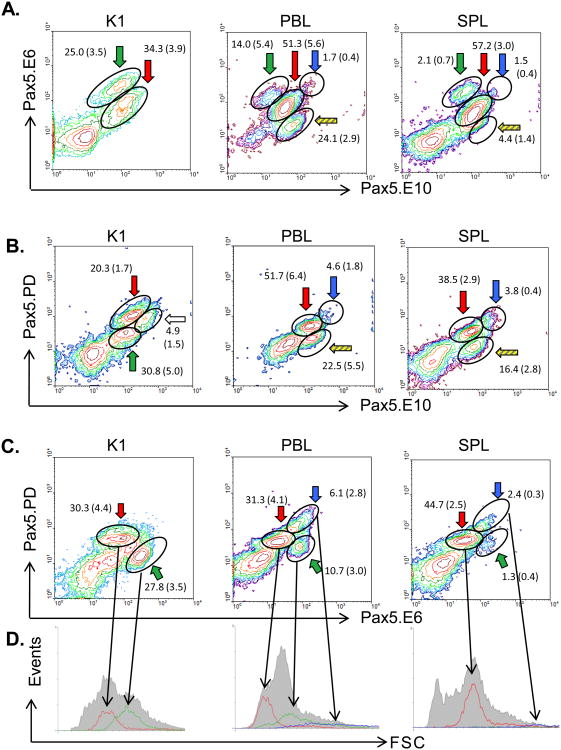
Permeabilized K1, PBL, and SPL cells were first co-stained with the Pax5 antibodies Pax5.E6 and. Pax5.E10, and analyzed by two-color flow cytometry (Figure 3A). Multiple B cell populations were detected for all three tissues. Their phenotypes and putative developmental stage are summarized in Table I. The cell population that co-stained with both antibodies (E6+/E10+) was the most abundant population in PBL and SPL (Figure 3A; red arrow), and was made up of small cells (low Forward Scatter, or FSC, data not shown). These populations most likely represent resting mature B cells, as previously described (Barr et al, 2011; Zwollo et al., 2005; Zwollo et al., 2008; Zwollo et al., 2010). For K1, the co-staining population (Figure 3A; red arrow) also consisted of small low FSC cells and based on its abundance likely represents late-pre-B or (im)mature B cells (Zwollo et al., 2010).
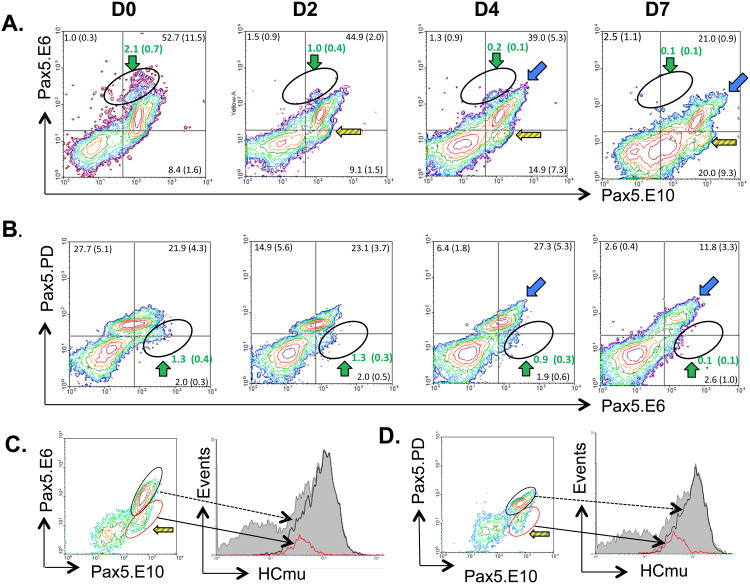
Figure 3.
Two-color flow cytometry of trout immune cells using three different Pax5 antibodies. Frequencies of each population are given as mean % gated lymphocyte population +/− S.E. N ≥ 4. Colors of arrows correspond to putative stage of the B cell (Table I). (A) Pax5.E6 and Pax5.E10 antibodies. (B) Pax5.PD and Pax5.E10 antibodies. (C) Pax5.PD and Pax5.E6 antibodies. (D) Histographs representing cell size (FSC; x-axis) of gated populations and number of cells (Events; y-axis) per subpopulation. Grey shaded region represents all gated lymphocytes. Green arrow (green line on histograph): putative early developing cells; red arrow (red line on histograph): (im)mature B cells; blue arrow (blue line on histograph): activated B cells. K1, anterior kidney; PBL, peripheral blood lymphocytes; SPL, spleen.
A population of relatively large cells with strong E6 staining, but low E10 staining (E6++/E10 low) was most abundant in K1, intermediate in PBL and scarce in SPL (Figure 3A; green arrow). Conversely, cells that expressed Pax5 proteins that stained for the C-terminus-detecting E10 but not E6 (E6−/E10+) were not detectable in K1, scarce in spleen, but fairly common in PBL (Figure 3A; yellow striped arrow for SPL and PBL). Although these cells lacked the exon 6-encoded mid-region of Pax5, they presumably expressed other, C-terminus-containing isoforms, including Pax5Δ2-6 and/or Pax5Δ2-8.
Next, trout tissues were stained with the same anti-Pax5.E10 antibody but now in combination with the anti-Pax5.PD (the paired domain-specific) antibody. Data are shown in Figure 3B. In SPL and PBL, the majority of B cells co-stained with both antibodies (PD+/E10+; Figure 3B; red arrow), and this population most likely represents mature B cells. Two other, minor populations were detected in PBL and SPL, namely a PDlow/E10+ (Figure 3B, striped arrow), and a PD++/E10++ (blue arrow) population. K1 had multiple co-staining populations, including one population with low-level co-staining and high FSC (Figure 3B; PD low/E10 low; green arrow), one population of high co-staining cells and low FSC (Figure 3B; red arrow), and a third population with higher E10 staining, lower PD staining, and intermediate FSC (Figure 3B, white arrow). The mean percentages of gated lymphocytes in each population ± S.E. are shown in the figure and the phenotypes of the cell populations are summarized in Table I.
Next, two-color flow cytometry was performed using the remaining combination of anti-Pax5 antibodies, namely anti-Pax5.PD and anti-Pax5.E6 (Figure 3C). SPL and PBL both contained one major population of cells that co-stained with PD and E6 (PD+/E6+; Figure 3C, red arrow), and were small in size (Fig 3D; red histograph line), suggesting that this cell population expresses full-length Pax5 proteins and represents mature B cells. Anterior kidney (K1) also had a population displaying this PD+/E6+ pattern (Figure 3C, red arrow), but additionally had another, rather abundant population of cells that was lower in Pax5.PD staining (PDlow/E6++; Figure 3C, green arrow and 3D, green histograph line). Unexpectedly, a minor population of these cells (PDlow/E6++) was also detected in the SPL and, more abundantly, in the PBLs (Figure 3C, green arrow). Together, the tissue distribution for these cells suggested that they could be early developing B cells with the Pax5 phenotype PDlow/E6++/E10low (indicated by green arrows, also see Table I). This was further supported by their cell size: paired domain-lacking, E6++ cells were large (high FSC, Figure 3D; green lines). These findings are in agreement with a previous study in trout and mice, which demonstrated the existence of an early developing B cell population both in anterior kidney of trout, and in bone marrow of mice, that strongly expressed the transcription factor Early B cell Factor (EBF) but lacked Pax5 paired domain expression (Zwollo et al., 2010). This was further explored below.
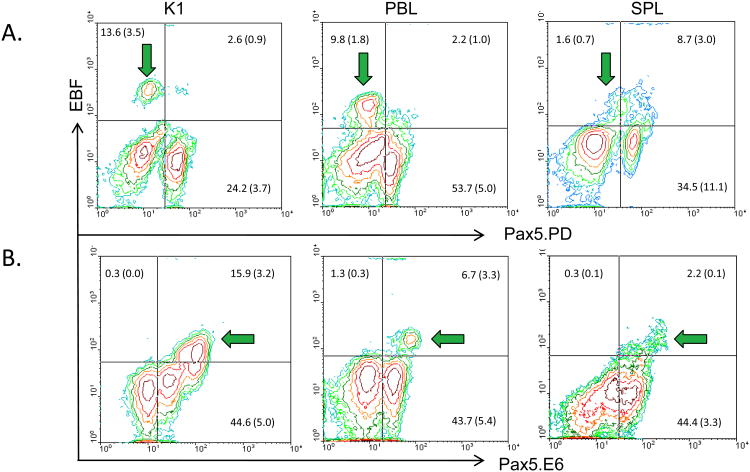
Pax5 isoforms in early developing B cells
Early developing B cells express the transcription factor EBF; in mammalian species, it is first expressed at the CLP stage and then further upregulated in pro-B and early pre-B cells (Lukin et al., 2008). EBF is also identified in early developing trout B cells (Zwollo et al., 2010), and its expression is greatly reduced in later stages of B cell maturation in both mammals and trout. Hence, EBF provides an accurate marker for early B cell development. Using an anti-EBF antibody to stain K1 cells we detected a population of large, EBF-expressing cells (high FSC; data not shown) that co-stained with E6 but not with PD, as shown in Figures 4B and 4A respectively (green arrows, left graphs). Earlier work had already shown that EBF-expressing cells in K1 do not co-stain with HCmu expressing cells, supportive of them being early developing B cells (Zwollo et al., 2010). Together, this supports that EBF-expressing, early developing B cells in K1 express Pax5 isoforms that lack a paired domain, but contain the central region of Pax5 encoded by exon 6. Next, we tested whether this putative early progenitor population could be the same population that had been detected in PBL and SPL (green arrows in Figure 3). Indeed, in PBLs, the majority of E6++ cells co-stained with EBF, while the majority of EBF-expressing PBL cells lacked PD-staining (Figure 4, middle graphs). Lastly, SPL also possessed a population of E6++/EBF+ cells, but many of these cells also co-stained with the PD antibody (Figure 4, right graphs), which is addressed in the Discussion.
Figure 4.
Detection of early progenitor cells using two-color flow cytometry on K1, PBL and SPL cells. Green arrows, see Table I. N ≥ 4. Quadrant frequencies given in figure as mean % gated population ± S.E. (A) Using EBF and Pax5.PD antibodies. (B) Using EBF and Pax5.E6 antibodies.
Differential expression of Pax5 isoforms during in vitro LPS-activation
To investigate whether any of the observed Pax5 patterns changed during B cell activation, trout SPL cells were cultured in vitro in the presence of the B cell mitogen LPS. Cells were collected on days 0, 2, 4, and 7 after activation and analyzed by two-color flow cytometry. Four different LPS-induced changes were observed. First, there was a loss of putative progenitor cells from 1.3 - 2.1% on Day 0 (introduced in Figures 3 and 4; green arrows), with very few cells remaining after 7 days of culture (Figure 5A,B; circled populations indicated by green arrows). To test if these SPL cells lacked HCmu, as expected for progenitor B cells, the cells were stained with a combination of E6 and HCmu antibodies. As shown in Figure 6A (green arrow), the cells stained weakly with HCmu. This is addressed in the Discussion.
Figure 5.
Flow cytometric analysis on LPS-activated spleen cells. D, day after activation. Colored arrows represent B cell populations, see text and Table I. Mean % and S.E. of gated lymphocytes in each quadrant. N ≥ 4. (A) and (B): two-color flow cytometry. Green text: percentage of cells within the circled populations, corresponding to CLP/pro-B cells (green arrow). (A) Pax5.E6 and Pax5.E10 antibodies. (B) Pax5.PD and Pax5.E6 antibodies. (C) and (D): Tricolor staining using Pax5 antibodies and IgM marker HCmu on SPL Day 4 post LPS. Grey shaded regions on histographs represent all gated lymphocytes. (C) Pax5.E6+/E10+, black circle on contour graph, and dotted black line pointing to black line on histograph; red circle: E6low/E10+, red circle and black arrow pointing to red line on histograph. (D) Pax5.PD+/Pax5.E10+ cells, black circle and black dotted line pointing to black line on histograph; Pax5.PDlow/Pax5.E10+, red circle and black arrow pointing to red line on histograph.
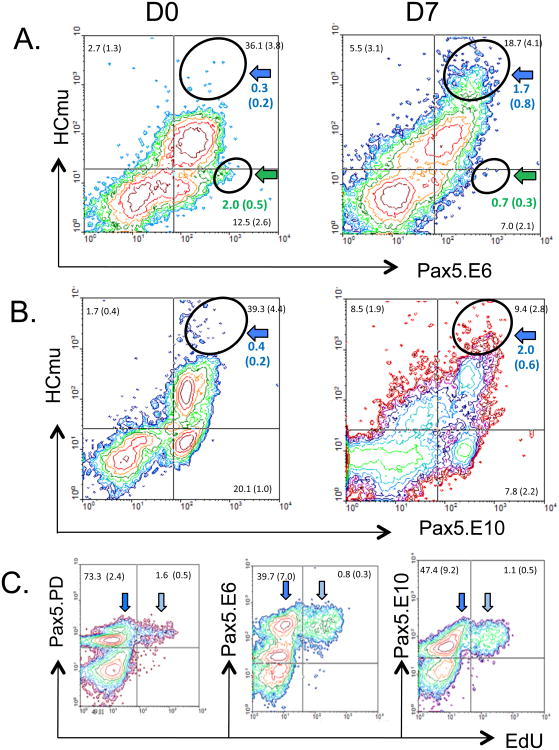
Figure 6.
Two-color flow cytometry on LPS-activated spleen cells. Colored arrows represent B cell populations, see text and Table I. D, day. Mean % and S.E. of gated lymphocytes in each quadrant. N ≥ 4. (A) HCmu and Pax5.E6 antibody staining. Black circles in upper right quadrants compare the percentage of HCmu++/E6++ cells on day 0 and 7. Similarly, black circles in lower right quadrants show the percentage of HCmu low/E6++ cell populations. (B) HCmu and Pax5.E10 antibody staining. Black circles in upper right quadrants show the percentage of HCmu++/E10++ cells on both days. (C) Day 4 spleen cells stained with each Pax5 antibody in combination with proliferation marker EdU. Light blue arrow, plasmablasts; dark blue arrow, non-proliferating B cells.
The second LPS-induced change involved increased expression of Pax5 in B cells. Earlier, when analyzing non-stimulated PBL and SPL with the three Pax5 antibodies, a small population of large, high Pax5 co-staining cells (PD++, E6++, E10++) had been observed (see blue arrows in Figure 3A-C; blue histograph line in Figure 3D). Upon LPS-induction, the intensity of Pax5 staining in this population gradually increased over time, suggesting that LPS-activated B cells up-regulate full-length Pax5 protein levels compared to resting mature B cells (Figure 5A,B and 6A,B, blue arrows; Table I). Increased expression of full-length Pax-5 in activated B cells (PD++/E6++/E10++) was further supported by LPS-induced increased HCmu expression in activated SPL cells, using 3-color flow cytometry (Figure 5C, D, black histograph line). Lastly, the increase was supported by strongly increased co-expression of both E6 and HCmu (Figure 6A; blue arrows), HCmu and E10 co-staining cells (Figure 6B; blue arrows), and HCmu and PD co-staining cells (Barr et al., 2011) by Day 7 post-stimulation. Together, these data suggest that activated B cells have concurrent increases in expression of secreted IgM, as previously reported (Barr et al., 2011) and full-length Pax5, prior to the downregulation of Pax5 in plasma cells, which correlates with even higher HCmu levels (Barr et al., 2011). It is unlikely that B cells will have differentiated into plasma cells by Day 7, and this corresponds with continued expression of Pax5 up to that time point. Using the proliferation marker EdU, we could show that plasmablasts continued to express (full-length) Pax5, which contained all three Pax5 domains (light blue arrow, Figure 6C; Table I).
A third LPS-induced change in Pax5 patterns concerned the expansion of a minor population of B cells that had reduced intensity for E6 and increased intensity for E10 staining (Figure 5A; lower right quadrant, yellow striped arrows). This population expanded over time from 8.4% on Day 0 to 14.9 % by Day 4 and this correlates with a gradual decrease in the percentage of E6+/E10+-expressing cells during this period (Figure 5A, upper right quadrant). This suggests that LPS-induction causes some B cells to begin expressing Pax5 proteins that lack the central region encoded by exon 6 (E6−). This “unnamed” population (Table I) also had reduced levels of PD staining (e.g. lacking exon 2) and expressed only moderate levels of HCmu after LPS-induction, suggesting the cells were not activated (Figure 5 C-D, red histograph lines). These cells may express isoforms Pax5Δ2-6 or Pax5Δ2-8. It is possible that these non-activated B cells were in the early stages of apoptosis but this was not pursued further. Lastly, it should be noted that the percentage of these cells appears to be even higher (20.0%) on Day 7, but this is likely an overestimate, considering the emergence of a new (E10int/E6−) population in this quadrant (lower right quadrant percentage in Figure 5A, D7).
Lastly, a modest population was observed in all SPL samples that stained moderately with all three Pax5 antibodies, but lacked IgM-expression (Figure 6A,B, lower right quadrants). This population was also observed in K1 and PBLs (Figure 6B, Supplemental figure 1). The phenotype (HCmu−) and frequency of staining (12.5-20.1% cells) suggested that these cells could be IgT and/or IgD-expressing B cells. Previous research has shown that in addition to IgM, the isotype IgT is expressed in trout PBL and SPL (Hansen et al., 2005; Zhang et al., 2010) and that approximately 12% of B cells express IgT in the absence of IgM.
Alternative Pax5 splice forms are detected in early progenitor cells and cells C-terminus-expressing Pax5
To more rigorously test the hypothesis that alternatively spliced Pax5 isoforms were the cause of the observed Pax5 patterns, we quantified the level of Pax5 isoform transcripts in fixed, sorted cells. This was important because it could be argued that the observed differences of Pax5 antibody staining might have been solely the result of blocking of antigenic determinants on the full-length Pax5 protein (maybe as a result of protein-protein or protein-DNA interactions).
First, we wished to confirm that populations that did NOT stain with the Pax5.PD antibody did indeed express isoform Pax5Δ2. Primers were tested in a standard RT-PCR system using RNA isolated from freshly isolated trout immune cells. Two exon-flanking primers (tPax5-E1/3.S and tPax5-E3/4.AS; Supplemental Table I) were used in the region exons 1-4, which should result in a 219 bp amplicon, named Δ2. As a control, full-length Pax5 sequence (FL) was amplified in parallel, using primers targeting exons 2-4 (tPax5.E2.S and tPax5-E3/4AS), with a predicted 237 bp size fragment. This produced the expected 237 bp (FL) and 219 bp (Δ2) fragments, as illustrated in Figure 7A for SPL. Products were sequenced to confirm their identity. The ratio of FL to Δ2 products was then determined using quantitative RT-PCR with the trout α-tubulin as control gene target (Figure 7B). As expected based on our flow cytometry data, the ratio of FL to Δ2 transcripts was almost 10-fold higher in SPL compared to K1.
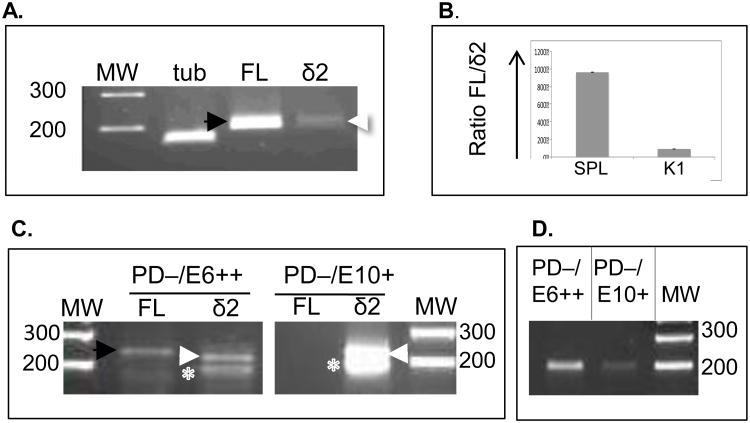
Figure 7.
Detecting alternatively-spliced Pax5 isoforms in sorted B cell populations. (A) RT-PCR using RNA from freshly isolated trout K1 or SPL cells. (B) qPCR, ratio fold-change between FL and Δ2 Pax5 isoforms in both SPL and K1. (C) RT-PCR on RNA from fixed, sorted SPL cells (either PD−/E6++ or PD−/E10+) using primers to amplify either FL Pax5 or Δ2. Expected size for FL: 237 bp (black arrow); for Δ2: 219 bp (white arrow). White asterix: non-specific 180 bp band, not Pax5. (D) End-point PCR as in C, but using primers to detect presence of exon 6-containing amplicon, 207 nts. MW, molecular weight (in bps).
Next, we determined whether exon 2-lacking Pax5 transcripts were expressed in selected cell populations, including E6++/PD−and PD−/E10+ cells. Fixed and permeabilized trout SPL or PBL cells were first stained with combinations of the three Pax5 antibodies and then cells were sorted on a FACSCalibur. Approximately 5,000-15,000 fixed cells were collected and RNA was isolated using an RNeasy FFPE kit to recover RNA from the formaldehyde-fixed cells. According to the manufacturer, this RNA will be fairly degraded, but our resulting RNA yields (between 50-200 ng) were usually enough for end-point RT-PCR. As above, primers tPax5-E1/3.S and tPax5-E3/4AS were used to detect Δ2 and FL cDNAs. Figure 7C shows PCR results from PD−/E6++ sorted splenic populations (circled population in Figure 3C; green arrows). PCR products were detected both for FL and Δ2 (237 and 219 bps respectively; Figure 7C). This may suggest that some RNA encoding exon 2 is still present in the PD−/E6++ population, or alternatively, the result of presence of PD+ cells. However, the ratio of FL to Δ2 was much reduced as compared to unsorted samples (Figure 7B). The presence of Δ2 Pax5 transcripts was also tested in a PD−/E10+ sorted splenic D4 LPS-activated population (see Figure 5D, yellow striped arrow). In these samples, the Δ2 band was strongly present, while the FL band was not detectable, as shown in Figure 7C. This suggests that PD−/E10+ sorted cells express mostly Pax5 isoforms that lack exon 2 (Δ2 cDNA). Similar results were obtained when E6++/PD− and PD−/E10+ cell populations were sorted from LPS-activated PBL (data not shown).
To confirm that the 219 bp fragment represented Δ2 Pax5 sequence, three independent PCR products from PD−/E6++ and PD−/E10+ sorted cells (from SPL or PBL) were sequenced. Using BLAST, all 219 bp sequences were confirmed to be trout Pax5Δ2 sequences (GenBank NM-001124682). Furthermore, the 237 bp “FL” band (lane 2) was also sequenced and represented full-length (exon 2 containing) trout Pax5 sequence. The 180 bp band shown in Figure 7C (asterix) did not contain Pax5 sequence, hence is a PCR artifact. Lastly, to determine the presence (or absence) of exon 6-containing Pax5 sequences in the sorted cells, end-point PCR was used with PCR primers targeting exon 5-6 (tPax5.485.S and tPax5.680.AS, Supplemental Table I). Results (Figure 7D) showed that RNA samples from sorted PD−/E6++ cells strongly expressed a 207 bp amplicon that included Pax5 transcripts with exon 6, while RT-PCR on PD−/E10+ cells resulted in a very weak band. As above, the sequence of the 207 bp amplicon was determined and confirmed that it contained Pax5, exon 6 sequence.
Discussion
This research queried the existence of Pax5 isoforms in rainbow trout immune tissues and the efficacy of flow cytometry to identify differential Pax5 domain expression in B cell subpopulations. Here, we describe the discovery of multiple N- and C-terminal Pax5 isoforms across immune tissues and demonstrate a differential expression of three Pax5 domains in early developing and activated B cell subpopulations. Furthermore, we establish the presence of alternatively-spliced Pax5 isoforms in B cell subpopulations, and identify tissue-specific Pax5 signatures using the flow cytometric approach.
Pax5 isoforms are conserved
Pax5 isoforms with paired domain and C-terminus deletions are conserved in rainbow trout immune cells. Paralleling the Pax5 splice forms found here in teleosts, Pax5 isoforms lacking exon 2 have also been identified in Amphioxus (Pax258 Δ2), Xenopus, murine B cells, and human PBLs (Arseneau et al., 2009; Heller and Brandli, 1997; Heller and Brändli, 1999; Short and Holland, 2008; Short et al., 2012; Zwollo et al., 1997). Four different C-terminal isoforms were discovered in trout (Pax5Δ8, Pax5Δ9a, Pax5Δ9b, and Pax5Δ9c) making them the most diverse isoforms, as they are within other species. For example, Amphioxus possess seven Pax258 isoforms with deletions in exons 6-11, murine splenic B cells contain at least two Pax5 isoforms with novel C-termini, and human PBLs have at least five Pax5 isoforms with deletions in exons 7-10, which vary in transactivation potential in transfection studies (Robichaud et al., 2004). Together, this suggests that C-terminal Pax5 isoforms may play similar roles in the modulation of expression of target genes across species, and strongly alludes to conserved function.
Alternative splicing of Pax5 and the resulting changes in Pax5 domain usage could result in a multitude of gene expression changes depending on the gene target(s). Mammalian Pax5 directly targets at least 45 genes necessary for B lymphopoiesis and directly represses at least 16 genes facilitating alternative immune cell fates (McManus et al., 2011). We speculate that trout Pax5 isoforms with paired domain deletions (lacking exon 2), as reported here, could play a role in the regulation of B lymphopoiesis (possibly acting as dominant negatives) and/or may promote trans-differentiation towards myeloid-cell development. Conversely, during B cell activation when Pax5 is first upregulated, and then necessarily downregulated, the degree of paired domain deletion could rapidly and reversibly regulate the rate of plasma cell differentiation.
Deletions in the Pax5 internal domains (including isoforms Pax5Δ2-6 and Pax5Δ2-8) could also likely affect target gene expression. Within the exon 2-6 region resides the octamer motif, which interacts with Groucho proteins to repress gene transcription (Eberhard et al, 2000). Loss of this region could be necessary to repress or de-repress gene targets enabling cell maturation and differentiation. Similarly, the internal partial homeodomain interacts with the retinoblastoma protein and further associates with TATA-binding proteins; its removal from Pax5 may affect cell cycle events during B cell proliferation (Eberhard and Busslinger, 1999). Alternatively, the overexpression of the central (exon 6-encoded) region in the absence of a paired domain, as observed in this study, may give the resulting Pax5 isoform powerful co-repressor or other regulatory functions, particularly during the CLPs and/or pro-B stages. This will need testing in future investigations.
Earliest B cell progenitors lack both the paired domain and C-terminus
An unexpected finding was the detection of Pax5 isoforms in very early progenitors (CLP/pro-B). Pax5 proteins in this population possessed an internal region (encoded by exon 6; E6), but lacked both a paired domain and C-terminus, and were found primarily in the anterior kidney (K1), the site of trout B cell development. The low ratio of the full-length to paired domain-lacking Pax5 isoforms in K1 compared to spleen supports this finding. Furthermore, flow cytometric analyses demonstrated that K1 cells lacking a paired domain were large cells, and expressed high levels of EBF, as would be expected for early developing B cells. Additionally, Pax5 isoforms with deletions in both the paired domain and the C-terminus were isolated from kidney tissue, but only if the antisense primer was targeting exon 4, not exon 10, in agreement with a PD−/E6++/E10− phenotype for early progenitors.
Importantly, the evidence shown here endorses the idea that developing B cells express Pax5 earlier in lymphopoiesis than previously postulated. Our data are consistent with a study using Pax5-Gfp BAC transgenic mice, which demonstrated that the murine Pax5 gene becomes activated during the CLP stage (Decker et al., 2009). The authors show that EBF-dependent chromatin opening at this stage contributes to Pax5 expression by facilitating interaction between promoter-enhancer regions. Clearly, this result will need further investigation, but the strong co-expression of exon-6-encoding Pax5 isoforms and EBF detected here lays the foundation for future studies on the role of alternative spliced Pax5 proteins in regulation of (B) lymphopoiesis.
Progenitor B cells are present in secondary immune tissues
The ability to measure differential Pax5 domain expression in individual cells also resulted in the striking finding that early developing cells are present in secondary immune tissues, specifically blood and spleen. RT-PCR analysis on fixed, sorted blood and spleen cells that stained for E6 but not PD confirmed that progenitor cells express Pax5 proteins that lack exon 2. Thus, the differential Pax5 domain expression identified using flow cytometry is likely a product of alternative RNA splicing.
The presence of early developing cells in secondary immune tissues is a novel finding for rainbow trout. However, hematopoietic cells have already been observed in the spleens of other species, including humans and porcine (Dor et al., 2006). Further, minor populations of B-1 and B-2 progenitors have reportedly been detected in the spleens of adult mice, while CD34+ progenitor cells have been detected in human PBL (Ghosn et al., 2011; Kato and Radbruch, 1993). The presence of early progenitor cells in trout secondary tissues begs the question of purpose. Considering that this population is more abundant in PBL compared to spleen, it is possible that such progenitor cells are stored in the blood of fish, until needed. Alternatively, it is possible that the blood simply provides the means to transport such cells from the anterior kidney into other secondary sites, including the spleen.
Of interest in this regard is the observation that two EBF-high expressing progenitor populations were detected in the spleen, one which lacked PD-staining, (as seen in K1 and PBL), and another population that did express the paired domain. We speculate that early progenitor cells, once they arrive in the spleen, will mature towards PD-containing, HCmu-expressing late-preB and immature B cells. Based on our data, we suspect that this maturation process is stimulated by LPS, as suggested by others (Ghosn et al., 2011). Alternatively, the observed LPS-induced loss of cells with this phenotype may simply be the result of apoptosis during the culturing period.
Activated B cells up-regulate full-length Pax5
Our data suggest that activated trout B cells up-regulate the amount of full-length Pax5 protein in the cell. The largest population of SPL cells co-stained all three Pax5 domains and the ratio of full-length Pax5 isoform to Δ2 isoforms was highest in this tissue. LPS activation strongly supported this finding, showing increases in the intensity of Pax5 domain co-staining throughout activation. These results complement previous research in mice, establishing an initial upregulation of Pax5 RNA in LPS-cultured splenocytes (Lin et al., 2002). This supports the idea that Pax5 is upregulated in activated B cells, perhaps to promote cell proliferation, until Blimp-1 levels are high enough to downregulate Pax5 for terminal B cell differentiation.
Conclusion
This research revealed that Pax5 protein domains are differentially expressed in individual cells of the trout B cell lineage. As demonstrated here, the pattern of Pax5 domain expression uniquely identifies each immune tissue, and led to the discovery that early B cell progenitors are not only present in anterior kidney of trout, but can also be found in trout blood and spleen. Flow cytometry provides a powerful tool to detect subtle or transient changes in B cell populations, particularly those populations that occur at low frequencies. This information can be used to identify the Pax5 signatures for immune tissues in individual fish, as well as any changes in Pax5 signatures during immune cell maturation and activation. Thus, it could prove useful in studies of fish disease, and also has implications for human diagnostics of aberrant B cell growth. Abnormalities in an immune tissue's Pax5 signature, such as the overabundance of a particular cell population, could provide novel biomarkers for detection of malignancy, infectious disease, and disease resistance.
Supplementary Material
Supplemental Figure 1. Two-color flow cytometry on K1, PBL, and SPL cells using HCmu in combination with all three Pax5 antibodies. (A) HCmu and Pax5.PD antibodies. (B) HCmu and Pax5.E10 antibodies. (C) HCmu and Pax5.E6 antibodies.
Highlights.
Pax5 is alternatively spliced in rainbow trout
Flow cytometry identifies differential Pax5 isoform domain expression in individual trout immune cells
Alternative spliced Pax5 isoforms are expressed in trout progenitor B cells
Acknowledgments
The authors thank Drs. Steve Kaattari and Jim Hagman for important feedback and discussion. We are grateful for excellent technical assistance from Alice Harman, Raaj Talauliker, Laura Stephens, and Emily Fruchterman. We also thank Tim Stevenson at the University of Alaska Anchorage, for assistance with cell sorting. This work was funded by NIH-AREA award R15 A1070249-02 to PZ, HHMI Undergraduate Summer Fellowship (AB), and graduate research funds from The College of William and Mary (LM).
Abbreviations used
- EBF
Early B cell factor
- EdU
5-ethynyl-2′-deoxyuridine
- HCmu
immunoglobulin heavy chain mu
- IgM
Immunoglobulin M
- PBL
peripheral blood lymphocyte
- RAG1
Recombinase-activated gene
- SPL
spleen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- Anspach J, Poulsen G, Kaattari I, Pollock R, Zwollo P. Reduction in DNA binding activity of the transcription factor Pax-5a in B lymphocytes of aged mice. J Immunol. 2001;166:2617–2626. doi: 10.4049/jimmunol.166.4.2617. [DOI] [PubMed] [Google Scholar]
- Arseneau JR, Laflamme M, Lewis SM, Maicas E, Ouellette RJ. Multiple isoforms of PAX5 are expressed in both lymphomas and normal B-cells. Br J Haematol. 2009;147:328–338. doi: 10.1111/j.1365-2141.2009.07859.x. [DOI] [PubMed] [Google Scholar]
- Barr M, Mott K, Zwollo P. Defining terminally differentiating B cell populations in rainbow trout immune tissues using the transcription factor XbpI. Fish Shellfish Immunol. 2011;31:727–735. doi: 10.1016/j.fsi.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromage ES, Kaattari IM, Zwollo P, Kaattari SL. Plasmablast and plasma cell production and distribution in trout immune tissues. J Immunol. 2004;173:7317–7323. doi: 10.4049/jimmunol.173.12.7317. [DOI] [PubMed] [Google Scholar]
- Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Dor FJMF, Ramirez ML, Parmar K, Altman EL, Huang CA, Down JD, Cooper DKC. Primitive hematopoietic cell populations reside in the spleen: Studies in the pig, baboon, and human. Experimental hematology. 2006;34:1573–1582. doi: 10.1016/j.exphem.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Dorfler P, Busslinger M. C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. EMBO J. 1996;15:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- Eberhard D, Busslinger M. The partial homeodomain of the transcription factor Pax-5 (BSAP) is an interaction motif for the retinoblastoma and TATA-binding proteins. Cancer Res. 1999;59:1716s–1724s. discussion 1724s-1725s. [PubMed] [Google Scholar]
- Eberhard D, Jimenez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–2303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- Ghosn EE, Sadate-Ngatchou P, Yang Y, Herzenberg LA. Distinct progenitors for B-1 and B-2 cells are present in adult mouse spleen. Proc Natl Acad Sci U S A. 2011;108:2879–2884. doi: 10.1073/pnas.1019764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JD, Landis ED, Phillips RB. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc Natl Acad Sci U S A. 2005;102:6919–6924. doi: 10.1073/pnas.0500027102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller N, Brandli AW. Xenopus Pax-2 displays multiple splice forms during embryogenesis and pronephric kidney development. Mech Dev. 1997;69:83–104. doi: 10.1016/s0925-4773(97)00158-5. [DOI] [PubMed] [Google Scholar]
- Heller N, Brändli AW. Xenopus Pax-2/5/8 orthologues: Novel insights into Pax Gene evolution and identification of Pax-8 as the earliest marker for otic and pronephric cell lineages. Developmental Genetics. 1999;24:208–219. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<208::AID-DVG4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Kaattari SL, Irwin MJ. Salmonid spleen and anterior kidney harbor populations of lymphocytes with different B cell repertoires. Dev Comp Immunol. 1985;9:433–444. doi: 10.1016/0145-305x(85)90006-0. [DOI] [PubMed] [Google Scholar]
- Kaattari SL, Irwin MJ, Yui MA, Tripp RA, Parkins JS. Primary in vitro stimulation of antibody production by rainbow trout lymphocytes. Vet Immunol Immunopathol. 1986;12:29–38. doi: 10.1016/0165-2427(86)90107-8. [DOI] [PubMed] [Google Scholar]
- Kato K, Radbruch A. Isolation and characterization of CD34+ hematopoietic stem cells from human peripheral blood by high-gradient magnetic cell sorting. Cytometry. 1993;14:384–392. doi: 10.1002/cyto.990140407. [DOI] [PubMed] [Google Scholar]
- Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–4780. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lowen M, Scott G, Zwollo P. Functional analyses of two alternative isoforms of the transcription factor Pax-5. J Biol Chem. 2001;276:42565–42574. doi: 10.1074/jbc.M106536200. [DOI] [PubMed] [Google Scholar]
- Lukin K, Fields S, Hartley J, Hagman J. Early B cell factor: Regulator of B lineage specification and commitment. Semin Immunol. 2008;20:221–227. doi: 10.1016/j.smim.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, Jaritz M, Tagoh H, Busslinger M. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30:2388–2404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Gomez F, Greene W, Rego K, Hansen JD, Costa G, Kataria P, Bromage ES. Discovery and characterization of secretory IgD in rainbow trout: secretory IgD is produced through a novel splicing mechanism. J Immunol. 2011;188:1341–1349. doi: 10.4049/jimmunol.1101938. [DOI] [PubMed] [Google Scholar]
- Robichaud GA, Nardini M, Laflamme M, Cuperlovic-Culf M, Ouellette RJ. Human Pax-5 C-terminal isoforms possess distinct transactivation properties and are differentially modulated in normal and malignant B cells. J Biol Chem. 2004;279:49956–49963. doi: 10.1074/jbc.M407171200. [DOI] [PubMed] [Google Scholar]
- Short S, Holland LZ. The evolution of alternative splicing in the Pax family: the view from the Basal chordate amphioxus. J Mol Evol. 2008;66:605–620. doi: 10.1007/s00239-008-9113-5. [DOI] [PubMed] [Google Scholar]
- Short S, Kozmik Z, Holland LZ. The Function and Developmental Expression of Alternatively Spliced Isoforms of Amphioxus and Xenopus laevis Pax2/5/8 Genes: Revealing Divergence at the Invertebrate to Vertebrate Transition. J Exp Zool B Mol Dev Evol. 2012;318:555–571. doi: 10.1002/jez.b.22460. [DOI] [PubMed] [Google Scholar]
- Zapata AG, Cooper EL. The immune system : comparative histophysiology. Wiley, Chichester; New York: 1990. [Google Scholar]
- Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, LaPatra SE, Bartholomew J, Sunyer JO. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. 2010;11:827–835. doi: 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo P. Dissecting teleost B cell differentiation using transcription factors. Dev Comp Immunol. 2011;35:898–905. doi: 10.1016/j.dci.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo P, Arrieta H, Ede K, Molinder K, Desiderio S, Pollock R. The Pax-5 gene is alternatively spliced during B-cell development. J Biol Chem. 1997;272:10160–10168. doi: 10.1074/jbc.272.15.10160. [DOI] [PubMed] [Google Scholar]
- Zwollo P, Cole S, Bromage E, Kaattari S. B cell heterogeneity in the teleost kidney: evidence for a maturation gradient from anterior to posterior kidney. J Immunol. 2005;174:6608–6616. doi: 10.4049/jimmunol.174.11.6608. [DOI] [PubMed] [Google Scholar]
- Zwollo P, Haines A, Rosato P, Gumulak-Smith J. Molecular and cellular analysis of B-cell populations in the rainbow trout using Pax5 and immunoglobulin markers. Dev Comp Immunol. 2008;32:1482–1496. doi: 10.1016/j.dci.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo P, Mott K, Barr M. Comparative analyses of B cell populations in trout kidney and mouse bone marrow: establishing “B cell signatures”. Dev Comp Immunol. 2010;34:1291–1299. doi: 10.1016/j.dci.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwollo P, Rao S, Wallin JJ, Gackstetter ER, Koshland ME. The transcription factor NF-kappaB/p50 interacts with the blk gene during B cell activation. J Biol Chem. 1998;273:18647–18655. doi: 10.1074/jbc.273.29.18647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Two-color flow cytometry on K1, PBL, and SPL cells using HCmu in combination with all three Pax5 antibodies. (A) HCmu and Pax5.PD antibodies. (B) HCmu and Pax5.E10 antibodies. (C) HCmu and Pax5.E6 antibodies.